Abstract
In eukaryotic cells, the unfolded protein response (UPR) pathway plays a crucial role in cellular homeostasis of the endoplasmic reticulum (ER) during exposure to diverse environmental conditions that cause ER stress. Here we report that the human fungal pathogen Cryptococcus neoformans has evolved a unique UPR pathway composed of an evolutionarily conserved Ire1 protein kinase and a novel bZIP transcription factor encoded by HXL1 (HAC1 and XBP1-Like gene 1). C. neoformans HXL1 encodes a protein lacking sequence homology to any known fungal or mammalian Hac1/Xbp1 protein yet undergoes the UPR-induced unconventional splicing in an Ire1-dependent manner upon exposure to various stresses. The structural organization of HXL1 and its unconventional splicing is widely conserved in C. neoformans strains of divergent serotypes. Notably, both C. neoformans ire1 and hxl1 mutants exhibited extreme growth defects at 37°C and hypersensitivity to ER stress and cell wall destabilization. All of the growth defects of the ire1 mutant were suppressed by the spliced active form of Hxl1, supporting that HXL1 mRNA is a downstream target of Ire1. Interestingly, however, the ire1 and hxl1 mutants showed differences in thermosensitivity, expression patterns for a subset of genes, and capsule synthesis, indicating that Ire1 has both Hxl1-dependent and -independent functions in C. neoformans. Finally, Ire1 and Hxl1 were shown to be critical for virulence of C. neoformans, suggesting UPR signaling as a novel antifungal therapeutic target.
Author Summary
The unfolded protein response (UPR) is a widely conserved signaling pathway in eukaryotic cells and protects cells from the ER stress causing accumulation of toxic unfolded or misfolded proteins. Nevertheless, the UPR pathway has been poorly exploited as a therapeutic target for treatment of eukaryotic fungal pathogens, mainly due to its evolutionarily conserved features. The present study reports unique evolution of the UPR pathway in the basidiomycetous human fungal pathogen, Cryptococcus neoformans, which causes life-threatening meningoencephalitis in both immunocompromised and immunocompetent individuals. Here, for the first time we identified and characterized the C. neoformans UPR pathway, which is composed of an evolutionarily conserved and a distinct signaling component, an ER stress sensor Ire1 and its downstream bZIP transcription factor Hxl1, respectively. Intriguingly, Cryptococcus Hxl1 is very divergent from yeast Hac1 and human Xbp1, but subject to Ire1-mediated unconventional splicing. The Ire1-Hxl1-dependent UPR pathway functions not only in the major response to ER stress, but also plays critical roles in controlling cell wall integrity, growth at host physiological temperature, antifungal drug susceptibility, and virulence of C. neoformans. Therefore we propose Hxl1 is an ideal target for antifungal drug development, based on its marked divergence from the host Xbp1 transcription factor.
Introduction
In eukaryotic cells, the endoplasmic reticulum (ER) is the site where most secreted and plasma membrane proteins are synthesized, folded, and matured. Impairment of protein folding results in accumulation of toxic unfolded or misfolded proteins, which causes ER stress and instigates intracellular signaling from the ER to the nucleus. ER stress is also induced by altered calcium homeostasis and glycosylation, nutrient starvation, pathogen infection, and oxidative stress [1]. Mounting evidence links ER stress to human diseases, as diverse as diabetes, viral infection, Alzheimer, cancer, and inflammation [2], [3]. To alleviate the ER stress, eukaryotic cells activate a conserved unfolded protein response (UPR) signaling pathway, which regulates the expression of numerous genes encoding ER chaperones and folding enzymes as well as proteins involved in diverse cellular metabolisms [1], [4].
In the budding yeast Saccharomyces cerevisiae, an unfolded protein sensor, Ire1 and its sole downstream transcription factor, Hac1, were discovered and characterized as the key signal components that mediate the UPR pathway [5]. As a type I transmembrane Ser/Thr kinase with an endoribonuclease (RNase) domain, Ire1 senses ER stress in the ER lumen and undergoes activating autophosphorylation. Subsequently activated Ire1 removes an unconventional intron from the HAC1 mRNA, which encodes a basic domain/leucine zipper (bZIP) transcription factor [6]. The unspliced HAC1 mRNA is not translated due to long-range base pairing interactions between the HAC1 5′-untranslated region and its intron. Therefore, this Ire1p-dependent unconventional splicing of the HAC1 mRNA is a critical and unique regulatory process for activation of UPR target genes [7]. Similar to yeast, upon activation of the UPR pathway, mammalian XBP1 mRNA encoding an ortholog of yeast Hac1 is spliced unconventionally by the RNase activity of Ire1α. The activated Xbp1 transcription factor is subsequently translated from the spliced XBP1 mRNA [1]. Unlike yeast Hac1, however, the unspliced XBP1 mRNA is translated, generating a negative regulator of the UPR pathway [8]. In mammalian cells, besides Xbp1, UPR is also governed by two ER-membrane-associated bZIP transcription factors, ATF6, and PERK [9]. ATF6 is a type II transmembrane protein normally retained in the ER but transported to the Golgi, where it is subjected to proteolytic process. The released N-terminal fragment of ATF6 containing bZIP DNA binding domain and transcriptional activation domains is translocated to the nucleus where it activates the transcription of ER stress response genes. Interestingly, in plants, the plant homolog for HAC1/XBP1 has not yet been reported, but two membrane-associated bZIP transcription factors, bZIP60 and bZIP28, were found to play essential roles in the ER stress response [10], [11].
Recent studies on the UPR pathway of opportunistic pathogenic fungi, including Aspergillus fumigatus and Candida albicans, reported that the HAC1 orthologs of these pathogens have an unconventional intron of 20-nt and 19-nt, respectively, which is excised in response to ER stress. Activation of the UPR pathway regulates the expression of genes which function in ER stress, protein secretion, morphological differentiation, and fungal virulence [12], [13]. In contrast to the ascomycetous fungal pathogens, here, we report the unique features of the UPR pathway in controlling growth, ER-stress response, and virulence of a basidiomycetous human fungal Cryptococcus neoformans, which causes fatal meningoencephalitis in both immunocompromised and immunocompetent individuals. A recent epidemiological study reported that cryptococcosis is responsible for more than 600,000 deaths annually in AIDS patients [14]. The present study demonstrates that the UPR pathway of Cryptococcus is composed of an evolutionarily conserved Ire1 protein kinase and a novel bZIP transcription factor, Hxl1, which is very divergent from yeast Hac1 and human Xbp1. In response to various stresses, the HXL1 mRNA is subjected to the Ire1-mediated unconventional splicing, which is essential for thermotolerance, cell wall integrity, antifungal drug susceptibility, and virulence of C. neoformans. Hence this study proposes components of the UPR pathway, particularly Hxl1, as one of ideal targets for development of novel antifungal therapies.
Results
IRE1 plays a critical role in ER-stress response, cell wall integrity, and thermotolerance in C. neoformans
To elucidate the role of the UPR pathway in C. neoformans, we first identified and characterized the function of the C. neoformans Ire1 ortholog. By BLASTp search using the protein sequence of S. cerevisiae Ire1 (ScIre1) as a query, we identified a single gene (CNAG_03670.2), which is highly homologous to ScIre1 (25% identity in amino acid sequence) in the genome database of the serotype A C. neoformans strain H99. The CNAG_03670.2 gene encodes a protein of 1,072 amino acids and contains typical Ire1-domain structures such as a luminal domain, a Ser/Thr protein kinase, and a ribonuclease domain (Figure 1A). Based on the significant sequence homology, we named it IRE1 in C. neoformans.
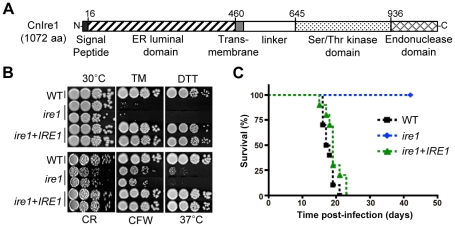
Figure 1. Functional analyses of C. neoformans ire1 strains.
(A) The conserved domain structure of the C. neoformans Ire1 protein. (B) For sensitive test to UPR- and cell wall- stresses, wild-type (H99), ire1 (YSB552 and YSB554), and ire1+IRE1 (YSB1000 and YSB1005) strains were serially diluted and spotted on YPD media with or without exposure to ER stress agents and cell wall disturbing agents (0.075 µg/ml TM, 10 mM DTT, 5 mg/ml CR, 1.5 mg/ml CFW) and incubated at 30°C for 3.5 days. (C) Virulence analysis of ire1 strains. A/Jcr mice were infected with 105 cells of MATα WT (black square, H99), ire1 (blue diamond, YSB552), ire1+IRE1 complemented (green triangle, YSB1000) strains by intranasal inhalation. Percent survival (%) was monitored for 6 weeks post-infection. P<0.0001 for WT vs. ire1 mutant and P = 0.1001 for WT vs. ire1+IRE1 complemented strain.
To functionally characterize the role of Ire1 in C. neoformans, we constructed ire1 mutants and ire1+IRE1 complemented strains in the C. neoformans H99 strain background (Table 1 and S1 and Figure S1). First we examined whether Ire1 plays a role in ER-stress response by testing sensitivity to tunicamycin (TM) and dithiothreitol (DTT), the well-known ER-stress inducing agents [5]. The ire1 mutants exhibited considerably increased sensitivity to both TM and DTT compared to the wild-type and their complemented strains (Figure 1B). Moreover, as reported in UPR-defective mutants of other yeast and fungi [12], [13], [15], [16], the ire1 mutants also exhibited hypersensitivity to both cell wall damaging agents, such as Congo red (CR) and Calcofluor white (CFW) (Figure 1B).
Table 1. Strains used in this study.
| Strain Name | Genotype | Parental strain | Reference |
| C. neoformans | |||
| H99 | Serotype A MATα | [48] | |
| KN99 | Serotype A MAT a | [49] | |
| JEC20 | Serotype D MATα | [50] | |
| JEC21 | Serotype D MAT a | [50] | |
| YSB552 | MATα ire1::NAT-STM#224* | H99 | This study |
| YSB554 | MATα ire1::NAT-STM#224 | H99 | This study |
| YSB723 | MATα hxl1::NAT-STM#229 | H99 | This study |
| YSB730 | MATα hxl1::NAT-STM#229 | H99 | This study |
| YSB1000 | MATα ire1::NAT IRE1-NEO | YSB552 | This study |
| YSB1005 | MATα ire1::NAT IRE1-NEO | YSB554 | This study |
| YSB1125 | MATα ire1::NAT HXL1-HXL1 u -NEO | YSB552 | This study |
| YSB743 | MATα ire1::NAT HXL1-HXL1 u -NEO | YSB554 | This study |
| YSB1127 | MATα ire1::NAT HXL1-HXL1 s -NEO | YSB552 | This study |
| YSB747 | MATα ire1::NAT HXL1-HXL1 s -NEO | YSB554 | This study |
| YSB762 | MATα hxl1::NAT HXL1 u -NEO | YSB723 | This study |
| KK1 | MATα cna1::NAT-STM#117 | H99 | [51] |
| KK3 | MATα mpk1:: NAT-STM#150 | H99 | [51] |
| YSB42 | MATα cac1::NAT-STM#159 | H99 | [41] |
| YSB53 | MATα ras1:: NAT-STM#150 | H99 | [41] |
| YSB64 | MATα hog1::NAT-STM#177 | H99 | [45] |
| YSB127 | MATα cpk1::NAT-STM#184 | H99 | [52] |
| YSB736 | MATα-HXL1 u -NEO | H99 | This study |
| YSB741 | MATα- HXL1 s -NEO | H99 | This study |
| YSB1320 | MATα cna1::NAT-STM#117 HXL1 u -NEO | KK1 | This study |
| YSB1325 | MATα cna1::NAT-STM#117 HXL1 s -NEO | KK1 | This study |
| C. gattii | |||
| R265 | Serotype B MATα | [53] | |
| WM276 | Serotype B MATα | [54] | |
| S. cerevisiae | |||
| BY4742 | MATα his3 leu2 lys2 ura3 | Open Biosystems | |
| 15650 | MATα his3 leu2 lys2 ura3 hac1::KanMX | Open Biosystems | |
| SCH1 | MATα his3 leu2 lys2 ura3 HAC1::CnHXL1 u | This study | |
| SCH2 | MATα his3 leu2 lys2 ura3 HAC1::CnHXL1 s | This study | |
| SCH3 | MATα his3 leu2 lys2 ura3 hac1::KanMX hac1::CnHXL1 u | This study | |
| SCH4 | MATα his3 leu2 lys2 ura3 hac1::KanMX hac1::CnHXL1 s | This study |
*Each NAT-STM# indicates the Natr marker with a unique signature tag.
Another notable phenotype of the ire1 mutant was its extreme thermosensitivity. The ire1 mutant was unable to grow at 37°C, which is host physiological temperature (Figure 1B). This finding prompted us to investigate the virulence of the ire1 mutant. Notably, the ire1 mutant was avirulent in a murine model of systemic cryptococcosis (Figure 1C). Even after 42 days post infection, all of the mice infected with the ire1 mutant remained healthy and asymptomatic. In contrast, the mice infected with the wild-type or ire1+IRE1 complemented strains became ill by 18 to 23 days post infection (Figure 1C). In conclusion, Ire1 plays crucial roles in ER-stress response, cell wall integrity, and thermotolerance of C. neoformans.
C. neoformans HXL1, encoding a novel bZIP protein, is subject to Ire1-mediated unconventional splicing
In the conventional UPR signaling pathway, Ire1 activates its downstream transcription factor Hac1/Xbp1 upon ER-stress by removing an unconventional intron at the 3′-region of HAC1/XBP1 mRNA (Figure 2A). Therefore, we searched for the yeast HAC1 or human XBP1 ortholog. Interestingly, however, at the initial BLASTp search, no C. neoformans gene showed significant sequence homology to either full-length HAC1/XBP1, or to just the isolated bZIP domain that is relatively highly conserved among Hac1/Xbp1 orthologs. By lowering the stringency in BLASTp search (e-value = 10−2) and using only the bZIP domain sequences of Hac1/Xbp1, five putative genes were selected with a limited sequence homology to the Hac1/Xbp1 proteins (Table S2).
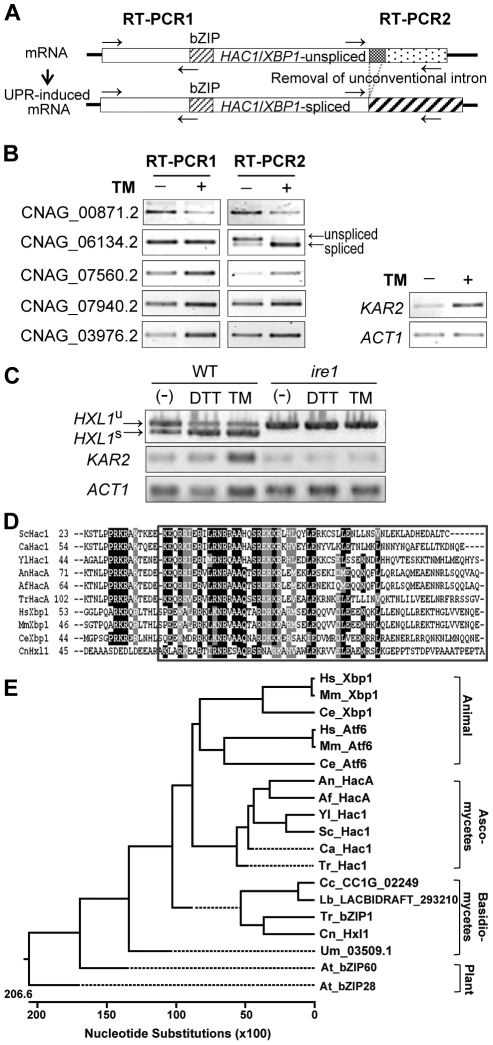
Figure 2. Identification of a novel Ire1-dependent bZIP transcription factor, Hxl1, in C. neoformans.
(A) General mRNA structures of the unspliced and spliced HAC1/XBP1. For identification of putative HAC1/XBP1 homologs, primers for RT-PCR1 and RT-PCR2 were designed to amplify the 5′-region product and the unconventionally spliced 3′-region product, respectively. (B) RT-PCR analysis for testing UPR-induced splicing. RT-PCR of CNAG_00871.2, CNAG_06134.2, CNAG_07560.2, CNAG_07940.2, CNAG_03976.2, KAR2 (CNAG_06443.2), and ACT1 (CNAG_00483.2) were performed with the cDNA samples of H99 strain cultivated in YPD medium with or without TM (5 µg/ml) treatment for 1 hr. (C) IRE1-dependent splicing of CNAG_06134.2. Strains were cultivated in YPD with or without TM (8 µg/ml) or DTT (20 mM). (D) Alignment of bZIP domains observed in Hxl1 and Hac1/Xbp1 homologs from several fungi and mammals. DNA binding domain of the bZIP domain is boxed. (E) Phylogenetic tree of the bZIP domains of transcription factors responsible for ER stress responses or Hxl1 homologs in fungi (ascomycete and basidiomycete), worm, plant, and animals. Hs: Homo sapiens Xbp1 (NP_001073007), Atf6 (NP_031374); Mm: Mus musculus Xbp1 (AAL60202), Atf6 (NP_001074773); Ce: Caenorhabditis. elegans Xbp1 (AAL60201), Atf6 (NP_510094); Sc, S. cerevisiae Hac1 (NP_116622); Ca: C. albicans Hac1 (EF655649); Yl: Yarrowia lipolytica Hac1 (XP_500811); An: Aspergillus nidulans hacA, Q8TFU8; Af: A. fumigatus HacA, ACJ61678; Tr: T. reesei hac1 (Q8TFF3); Cn: C. neoformans Hxl1 (CNAG_06134.2); Um: Ustilago maydis UM03509.1; Tf: Tremella fuciformis bZIP 1 (XP_001877531); Lb: Laccaria bicolor LACBIDRAFT_293210; Cc: Coprinopsis cinerea CC1G_02249; At: Arabidopsis thaliana bZIP28 (NP_187691), bZIP60 (NP_174998).
To identify a HAC1/XBP1-like gene in C. neoformans that would undergo a typical ER-stress induced unconventional splicing, we screened for splicing events in the 3′-regions of each mRNA by RT-PCR after TM treatment, which was shown to induce the expression of C. neoformans KAR2 (ER protein chaperone BiP) (Figure 2B), a well-known ER stress defense gene in yeasts [4]. Among the candidate genes, only CNAG_06134.2 exhibited two RT-PCR products with different sizes in the 3′-region (RT-PCR2), but a single RT-PCR product in the 5′-region (RT-PCR1) under unstressed conditions. In response to ER stress, the smaller RT-PCR2 products became more prominent (Figure 2B). Other candidate bZIP proteins did not appear to undergo unconventional splicing events although their expression levels were differentially regulated in response to ER stress. Next we examined the occurrence of unconventional splicing of CNAG_06134.2 in the C. neoformans ire1 mutant strain. CNAG_06134.2 did not show unconventional splicing event in the ire1 mutant strain in response to both TM and DTT (Figure 2C), indicating that the CNAG_06134.2 mRNA is a downstream target of Ire1 in the UPR signaling pathway of C. neoformans. Sequence analysis of the RT-PCR2 product of CNAG_06134.2 revealed that the nonconventional 56-nt intron was removed upon UPR induction. Therefore, we named CNAG_06134.2 HXL1 (HAC1 and XBP1-Like gene 1). Interestingly, about 20-30% of C. neoformans HXL1 mRNA was detected as a spliced form (HXL1 u) even under normal, unstressed condition, whereas more than 80% of HXL1 mRNA was converted to a spliced form (HXL1 s) after DTT or TM treatment (Figure 2C and S3), indicating that the UPR pathway appears to be partly activated by normal in vitro culture conditions without ER stress in C. neoformans. The increased relative ratio of HXL1 s to HXL1 u form of HXL1 mRNA upon ER stress showed a good correlation with the increased level of KAR2 transcript, suggesting that the HXL1 mRNA splicing event activated the UPR pathway. As shown in multiple sequence alignment and phylogenetic tree analysis of bZIP domains, the protein product of HXL1 (Hxl1) is only distantly related to known Hac1/Xbp1 transcription factors (Figure 2D and E), indicating that C. neoformans contains both evolutionarily conserved and distinct signaling components, Ire1 and Hxl1, respectively, for the UPR pathway.
Since the Hxl1 is structurally and phylogenetically distant from the Hac1/Xbp1, we addressed whether Hxl1 can complement the TM sensitivity of S. cerevisiae hac1 (Schac1) strain. S. cerevisiae wild type (BY4742) and Schac1 mutant strains were transformed with low copy expression vectors containing C. neoformans unspliced form of HXL1 or spliced form of HXL1. Overexpression of the spliced HXL1 s controlled by the GAL10 promoter led to severe growth inhibition even in wild type as well as in Schac1 strains (Figure 3A). Contrast to the ScHAC1 u or ScHAC1 s expression, the HXL1 s expression driven by the ScHAC1 promoter could not recover the TM sensitivity (Figure 3B) and the inositol auxotrophy of Schac1 strain (data not shown), and caused rather growth inhibition even in wild type under normal conditions (Figure 3B). In agreement with these data, a single copy integration of the CnHXL1 s gene into the ScHAC1 locus did not recover the TM sensitivity of Schac1 mutant (Figure 3C). This is in contrast to the effective compensation for the loss of HAC1 in S. cerevisiae by ascomycetes Trichoderma reesei or C. albicans Hac1 homologs [12], [17]. These results strongly suggest that the C. neoformans Hxl1 may not function properly with the S. cerevisiae transcriptional machinery due to its unique structures.
Figure 3. Heterologous expression analysis of C. neoformans HXL1 in S. cerevisiae hac1 mutants.
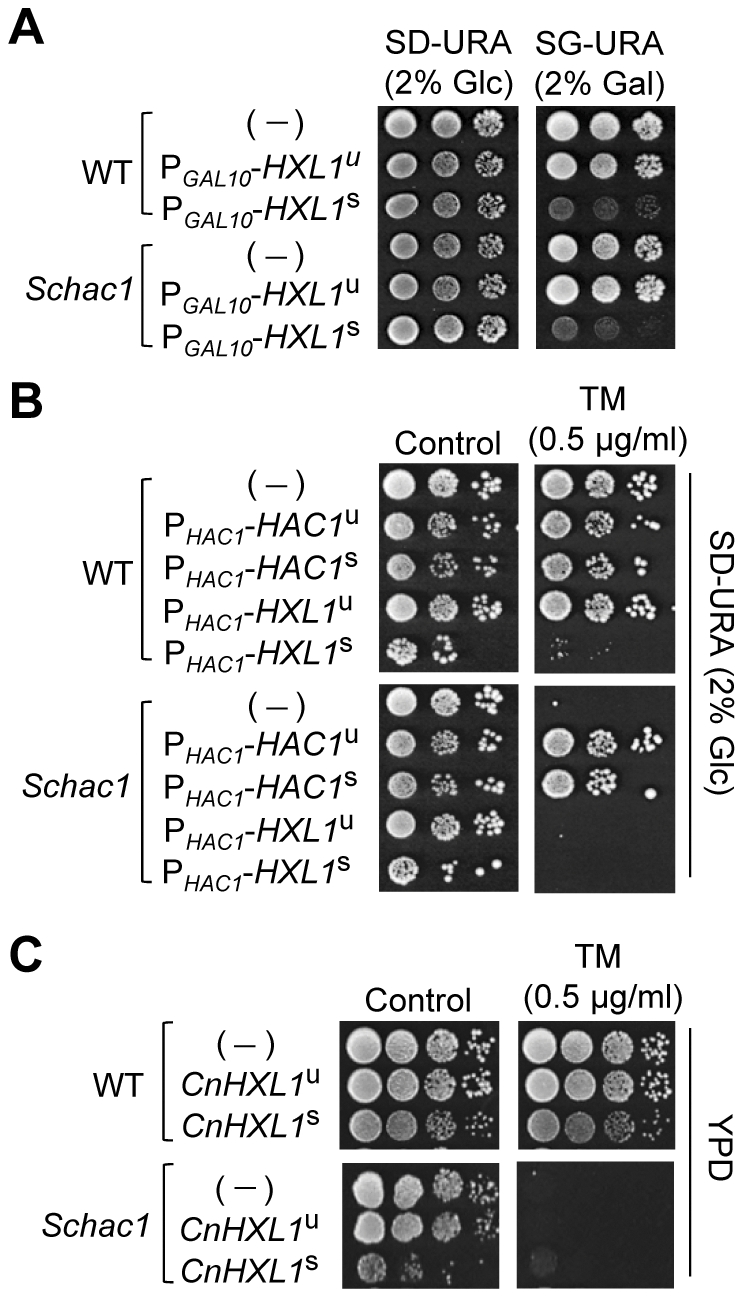
(A) Overexpression of C. neoformans HXL1 by the GAL10 promoter. S. cerevisiae wild type and hac1 strains harboring the expression vectors of unspliced or spliced forms of CnHXL under the GAL10 promoter (PGAL10-HXL1u and PGAL10-HXL1 s, respectively) were serially diluted and spotted on synthetic minimal media containing 2% glucose (repression) or 2% galactose (induction). (B) Expression of CnHXL1 by the ScHAC1 promoter. Wild- type and Schac1 strains harboring the expression vectors of unspliced or spliced forms of ScHAC1 (PHAC1-HAC1 u and PHAC1-HAC1 s, respectively) and unspliced or spliced forms of CnHXL1 under the ScHAC1 promoter (PHAC1-HXL1 u and PHAC1-HXL1 s, respectively) were serially diluted and spotted on synthetic minimal media with or without TM. Strains were incubated at 30°C for 3 days and photographed. (C) Integrative expression of CnHXL1. Wild-type (BY4742) and Schac1 strains harboring a single copy of unspliced or spliced forms of CnHXL1integrated into the ScHAC1 locus were serially diluted and spotted on complex YPD media with or without TM. Strains were incubated at 30°C for 2 days and photographed.
Structural conservation of HXL1 in different serotypes of C. neoformans
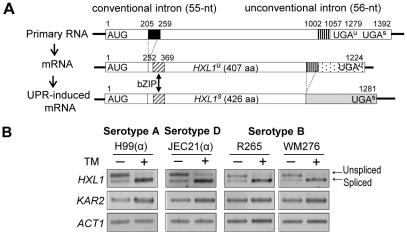
Because Hxl1 was identified in the serotype A C. neoformans strain H99 and found to be phylogenetically distinct from other Hac1/Xbp1 orthologs, we addressed whether HXL1 is present in other serotypes of C. neoformans. Serotype D strains (JEC21 and B-3501A) contain an HXL1 ortholog (CNM01380 and CNBM1240, respectively) and the serotype B strains (R265 and WM276) also contain an HXL1 ortholog (CNBG_4842.2 and CGB_M1590W, respectively). Sequence analysis of HXL1 revealed that all of the HXL1 genes of different serotypes have both a conventional intron of 55-nt and the unconventional intron of 56-nt at the upstream and downstream regions of the bZIP domain, respectively, as observed in the serotype A strain H99 (Figure 4A). All of the C. neoformans strains also exhibited UPR-induced unconventional splicing of HXL1 mRNA (Figure 4B), indicating that the structural organization of HXL1 is widely conserved in different serotypes of C. neoformans.
Figure 4. Structural conservation of the distinct HXL1 gene in different serotypes of C. neoformans.
(A) Scheme for conventional and UPR-induced non-conventional splicing of C. neoformans HXL1. The correct frame of the bZIP domain is formed after conventional splicing of the HXLI primary mRNA and shown as a diagonal box. The conventional and unconventional introns are shown as a solid black box and a vertical striped box, respectively. The C-terminal fragments of HXL1 before and after unconventional splicing are shown as the dotted box and a grayed box, respectively. (B) RT-PCR analysis of UPR-induced splicing of HXL1 in C. neoformans strains of different serotypes. The cDNA samples, synthesized from yeast cells treated with or without TM (5 µg/ml) for 1 hr, were used as templates for RT-PCR of HXL1.
Although the sequences of HXL1 were not completely identical, the position and size of unconventional intron and its border sequences were well conserved in all serotype strains of C. neoformans (Figure S2A). It is noteworthy that the HXL1 ortholog of serotype B (Cryptococcus gattii) encodes a protein of 406 (unspliced form) and 426 (spliced form) amino acids, respectively, due to loss of an alanine residue caused by a 3-bp deletion (CGC, +144 to +146) compared with those of serotype A and D strains.
Hxl1 functions as an essential downstream transcription factor of Ire1
To analyze whether Hxl1 is the target transcription factor of Ire1, we constructed hxl1 mutant strains (Table 1 and Figure S1) and compared their phenotypes with those of the ire1 mutants. Similar to the ire1 mutant, hxl1 mutants exhibited equivalent levels of sensitivity to TM, DTT, CR, or CFW (Figure 5A). Verifying these results, targeted re-integration of the HXL1 into its native locus (the hxl1+HXL1 strain) completely restored wild-type levels of resistance of the hxl1 mutant to ER stress-inducing and cell wall disturbing agents. Most importantly, the UPR-induced spliced form of HXL1 (HXL1 s), but not the unspliced form of HXL1 (HXL1 u), completely suppressed ER stress sensitivity of the ire1 mutant to the same extent observed in the wild-type and ire1+IRE1 complemented strains (Figure 5A), demonstrating that Hxl1 functions downstream of Ire1. Remarkably, the hxl1 mutant displayed a more severe growth defect than the ire1, cna1, ras1, and mpk1 strains at high temperature (Figure 5B).
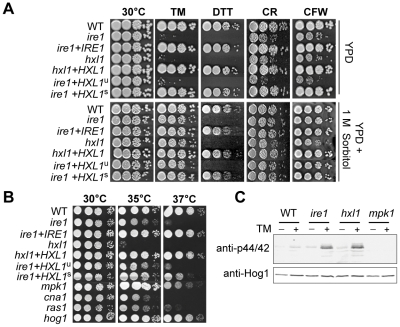
Figure 5. Functional analyses of C. neoformans HXL1 deletion and complementation.
(A) For sensitive test to UPR- and cell wall- stresses, strains were serially diluted and spotted on YPD media with or without exposure to ER stress agents and cell wall disturbing agents (0.075 µg/ml TM, 10 mM DTT, 5 mg/ml CR, 1.5 mg/ml CFW), without (top panel) or with 1 M sorbitol (bottom panel) and incubated at 30°C for 3.5 days. (B) For the thermosensitivity test, strains were spotted on YPD medium and incubated at 35°C or 37°C for 3.5 days. (C) Phosphorylation of Mpk1 induced by ER stress and loss of UPR function. Strains in early exponential phase were treated with TM (5 µg/ml) at 30°C for 2 hr. 30 µg of total protein was analyzed by Western blotting with phospho-p44/42-MAPK antibody (upper panel) for the phosphorylated Mpk1 (Mpk1-P) protein and anti-Hog1 antibody (lower panel) for the Hog1 protein as a loading control. Strains were: wild-type (H99), ire1 (YSB552), ire1+IRE1 complemented (YSB1000), hxl1 (YSB723), hxl1+HXL1 u complemented (YSB762), ire1+HXL1 u suppressed (YSB743), ire1+HXL1 s suppressed (YSB747), mpk1 (KK3), cna1 (KK1), ras1 (YSB53), and hog1 (YSB64).
TM sensitivity of the UPR mutants was fully recovered by addition of an osmotic stabilizer 1 M sorbitol, whereas DTT sensitivity was not, indicating that TM and DTT evoke ER stress in different ways (Figure 5A). TM treatment would mainly result in the defect in N-glycosylation, affecting mainly cell wall integrity, whereas DTT could act as oxidative damaging agent. Supporting this, the hypersensitivity to cell wall destabilizers of both UPR mutant strains of C. neoformans was mostly rescued by 1 M sorbitol (Figure 5A). Notably the basal level phosphorylation of the cell wall integrity-controlling Mpk1 MAPK was about threefold higher in the ire1 and hxl1 mutants compared to the wild-type even during normal growth. The levels of phosphorylated Mpk1 increased much more in the UPR mutant strains compared to the wild-type upon TM treatment (Figure 5C). Thus defects in the UPR pathway and/or exposure to ER stress generate cell wall defects, which trigger induction of the Mpk1-mediated cell wall integrity pathway in C. neoformans, as previously shown in S. cerevisiae [15], [18]. Taken together, inactivation of the Ire1-Hxl1-mediated UPR pathway may cause extreme thermosensitivity of C. neoformans not only via a cell wall integrity defect but also due to defects in several other biological processes.
Subsets of UPR genes are differentially regulated by Ire1 and Hxl1
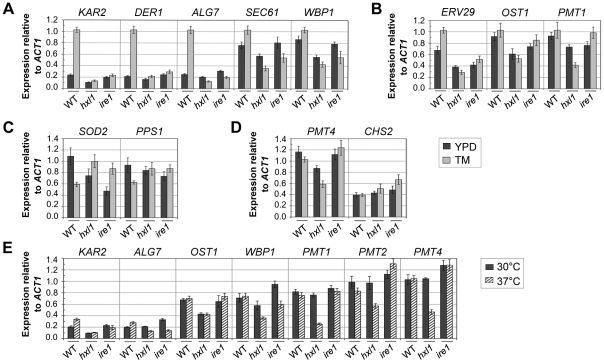
To verify further Hxl1 as a transcription factor of the UPR pathway in C. neoformans, we measured the transcript levels of putative UPR-regulated genes under ER stress conditions. A set of C. neoformans homologs of the S. cerevisiae HAC1 regulons (Figure S4) harboring the UPRE and UPRE-like sequences (CAGNGTG, TGACGTCA, CCAGC, CGTGTCGG, or TACGTG) [19], [20] in their promoters, were chosen and their transcript levels were compared between the wild-type and the UPR mutant strains after TM treatment. The C. neoformans homologs of well-established UPR target genes involved in protein secretion and glycosylation, such as KAR2, SEC61 (translocation of misfolded proteins out of ER), DER1 (ER-associated degradation), WBP1 (lipid-linked N-oligosaccharyltransferase), and ALG7 (lipid-linked N-oligosaccharide biosynthesis), were up-regulated upon TM treatment in an Ire1- and Hxl1-dependent manner (Figure 6A). However, it is notable that the induced expression of some UPR target candidates such as ERV29 (ER to Golgi vesicle-mediated transport), OST1 (lipid-linked N-oligosaccharyltransferase), and PMT1 (protein O-mannosyltransferase) homologs were dependent on Hxl1 but not on Ire1 (Figure 6B). On the other hand, the SOD2 (mitochondrial superoxide dismutase) and PPS1 (protein phosphatase S phase) homologs, which are responsive to osmotic and heat stress in C. neoformans, respectively [21], [22], were down-regulated under ER stress in an Ire1- and Hxl1-dependent manner (Figure 6C). For some C. neoformans genes such as PMT4 (protein O-mannosyltransferase) and CHS2 (chitin biosynthesis), their expression levels did not change in the wild-type or hxl1 mutants during ER stress, but significantly increased in the ire1 mutant (Figure 6D). All these results strongly indicate that although Hxl1 and Ire1 mainly work together in the UPR pathway, the loss of their function can generate differential effect on the expression of some subsets of UPR genes in response to ER stress.
Figure 6. Quantitative real-time PCR analysis of Hxl1-dependent regulation of putative UPR genes.
Strains were grown in YPD medium to early exponential phase, exposed to TM (5 µg/ml) for 1 hr (A–D) or to high temperature at 37°C for 2 hr (E), and harvested. The relative expression levels of putative UPR-regulated genes were analyzed by qRT-PCR with primers listed in Table S1 and normalized to that of ACT1. Strains were wild-type (H99), hxl1 mutant strain (YSB723), and ire1 mutant (YSB552). Error bars represent standard deviation (SD) of triplicated (A–D) or duplicated (E).
It is noteworthy that the hxl1 mutant showed a much more severe thermosensitive phenotype than the ire1 mutant and that the introduction of HXL1 s did not completely suppress the thermosensitivity conferred by ire1 mutation (Figure 5B). To understand the differences with respect to thermotolerance, we also analyzed the expression patterns of the putative UPR genes under heat stress (Figure 6E). KAR2 and ALG7 showed an induced expression pattern upon heat stress in an Ire1- and Hxl1-dependent manner. Intriguingly, however, genes involved in protein N-/O-glycosylation, such as WBP1, PMT1, PMT2, and PMT4, showed about twofold decreased expression only in the hxl1 mutant, but not in either the wild-type or the ire1 mutant (Figure 6E). These results indicate that compared to the loss of Ire1, the absence of Hxl1 might exert more negative effect on expression of some UPR genes under heat stress, partly explaining why the hxl1 mutant was more thermosensitive than the ire1 mutant.
Ire1 and Hxl1 play similar and distinct roles in controlling virulence and antifungal drug resistance of C. neoformans
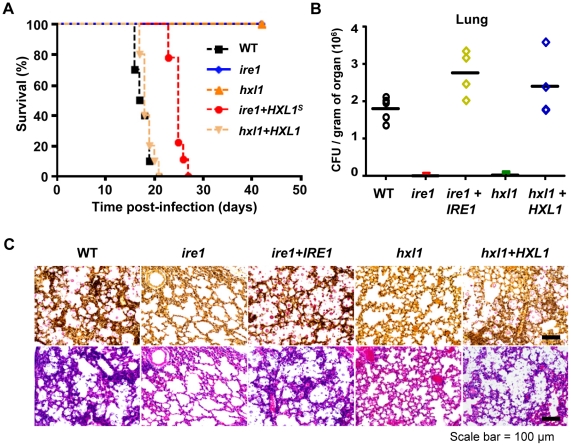
We further examined whether Hxl1p also plays a critical role in virulence of C. neoformans like Ire1p. As similar to the ire1 mutant, the hxl1 mutant was avirulent in a murine model of systemic cryptococcosis and their virulence was restored by reintroduction of the wild-type genes (Figure 7A). To ensure that Ire1 controls virulence of C. neoformans via its downstream Hxl1, we also analyzed mice infected with an ire1 strain harboring the activated, spliced HXL1 s. Interestingly, expression of the HXL1 s partially restored virulence of the ire1 mutant strain (Figure 7A). The ire1+HXL1 s strain was less virulent than the wild-type or the ire1+IRE1 strains, but still retained virulence. Taken together, it is conceivable that Ire1 controls virulence of C. neoformans both in Hxl1-dependent and -independent manners.
Figure 7. The IRE1-HXL1 UPR pathway is essential for virulence of C. neoformans.
(A) A/Jcr mice were infected with 105 cells of MATα WT (black square, H99), ire1 (blue diamond, YSB554), hxl1 (orange triangle, YSB723), ire1+HXL1 s suppressed (red circle, YSB747), and hxl1+HXL1 u complemented strains (peach inverted triangle, YSB762) by intranasal instillation. Survival (in %) was monitored for 6 weeks post-infection (P<0.0001 for WT vs. ire1, WT vs. hxl1, WT vs. hxl1+HXL1 u, and WT vs. ire1+HXL1 s strains). (B) The colony forming units (CFU) of surviving fungal cells per gram of organ was determined in the lung from sacrificed animals infected with WT (H99), ire1 (YSB552), ire1+IRE1 (YSB1000), hxl1 (YSB723), and hxl1+HXL1 (YSB762) (P<0.05 for WT vs. ire1 and WT vs. hxl1, P>0.05 WT vs. ire1+IRE1 and WT vs. hxl1+HXL1 strains). (C) Histopathological staining of infected lung tissues with mucicarmine (upper panel) and H&E (lower panel).
To elucidate whether the fully attenuated virulence of the ire1 and hxl1 mutants is caused by reduced cell survival or proliferation in the host, we performed fungal burden assay to determine the number of C. neoformans cells recovered from the sacrificed mice. The ire1 and hxl1 mutants showed significant decrease in the cell numbers compared to wild-type in the lung and brain tissues (Figure 7B and S5). Furthermore, the histopathological analysis of mucicarmine-stained tissues revealed that the wild-type (H99), ire1+IRE1, and hxl+HXL1 complemented strains proliferate extensively in lung tissues, while cells of the ire1 and hxl1 mutants were not observed (Figure 7C), suggesting that Ire1 and Hxl1 are required for proliferation in lung tissues. In the H&E staining, alveolar damage or tissue necrosis in lung tissues was observed only in mice infected with the wild type and ire1+IRE1 and hxl+HXL1 complemented strains but not with the ire1 or hxl1 mutants (Figure 7C). The result of histopathological analysis was well correlated with that of fungal burden in the lung tissues. However, in the brain tissues, we did not observe mucicarmine-stained or damaged tissues in animals infected with any strains (data not shown). Taken together, the results showed that the UPR pathway is essential for C. neoformans to survive and proliferate during infection.
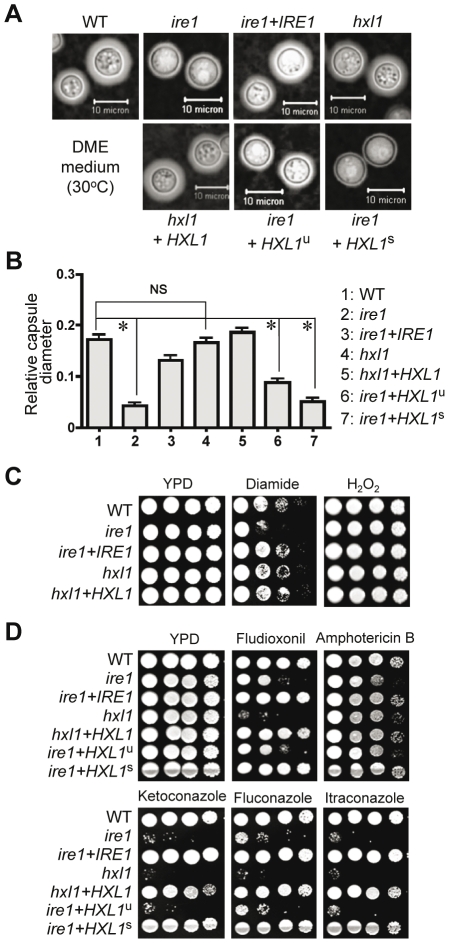
Next we investigated other virulence factors Ire1 might control in an Hxl1p-independent manner. Besides thermotolerance, antioxidant melanin and antiphagocytic capsule are two major virulence factors for C. neoformans. Intriguingly, only the ire1 mutants were defective in capsule biosynthesis whereas hxl1 mutants were as efficient in capsule production as the wild-type strain based on both qualitative and quantitative measurements (Figure 8A and B). Moreover, the ire1 mutants with a spliced HXL1 allele (ire1+HXL1 s) or an unspliced allele (ire1+HXL1 u) also exhibited a defect in capsule production. In contrast to capsule biosynthesis, the ire1 and hxl1 mutants both produced wild-type levels of melanin (data not shown). We also tested oxidative stress responses of the UPR mutants because the ability to counteract reactive oxygen species is one of key virulence factors in C. neoformans [23], [24]. The UPR mutants were as resistant to hydrogen peroxide (H2O2) as the wild-type strain (Figure 8C). Interestingly, however, the ire1 mutant, but not the hxl1 mutant, exhibited increased sensitivity to another oxidant diamide (Figure 8C). The results suggest that Ire1 controls capsule synthesis and diamide-resistance in an Hx1l-independent manner.
Figure 8. Ire1 controls capsule formation, oxidative stress responses, and antifungal drugs resistance in Hxl1 dependent- and independent manners in C. neformasn.
(A and B) Strains lacking IRE1 produce less capsule. Strains were grown at 30°C on solid DME medium for 2 days and photographed. The relative capsule diameter of cells was measured after India ink staining [*P<0.001 for WT vs. ire1 mutant, for WT vs. ire1+HXL1 u mutant, and WT vs. ire1+HXL1s mutant and NS, not significant (P>0.05)]. (C) Different sensitivity of ire1 and hxl1 mutant strains to diamide. Strains were serially diluted and spotted on YPD medium with 2 mM diamide or 3 mM hydrogen peroxide and incubated at 30°C for 3 days. (D) The UPR is involved in antifungal drug susceptibility. Strains were serially diluted and spotted on YPD medium containing the indicated concentrations of antifungal drugs including ketoconazole (0.1 µg/ml), fluconazole (14 µg/ml), itraconazole (0.5 µg/ml), fludioxonil (0.5 µg/ml), and amphotericin B (0.8 µg/ml). Cells were incubated in 30°C for 3 days. Strains were: WT (H99), ire1 (YSB552), ire1+IRE1 complemented (YSB1000), hxl1 (YSB723), hxl1+HXL1 u complemented (YSB762), ire1+HXL1 u suppressed (YSB1125), and ire1+HXL1 s suppressed (YSB1127).
Finally we examined the potency of Ire1 or Hxl1 as a target for combination therapy with currently available clinical drugs for treatment of cryptococcosis. There are also reports that A. fumigatus hacA or C. albicans ire1 mutations enhance activity of antifungal drugs such as amphotericin B, caspofungin, and azoles [13], [16]. The loss of IRE1 or HXL1 caused dramatically enhanced susceptibility to azoles such as ketoconazole, fluconazole and itraconazole, and phenylpyrroles such as fludioxonil, but only slightly increased susceptibility to polyenes such as amphotericin B (Figure 8D). The ire1 mutants expressing the spliced HXL1 allele exhibited wild-type levels of antifungal drug sensitivity, indicating that the Ire1-Hxl1-mediated UPR pathway in C. neoformans regulates antifungal drug resistance.
The Ire1-Hxl1-mediated UPR is induced by various environmental cues in C. neoformans
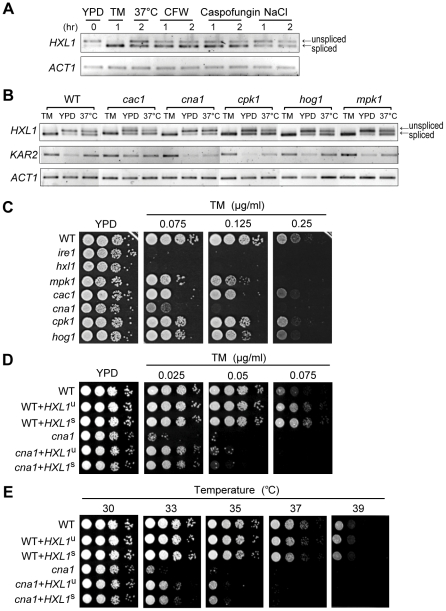
To investigate the possible induction of the Ire1-Hxl1-mediated UPR pathway by other environmental stresses and potential crosstalk with other signaling pathways, we examined the activation of HXL1 splicing under various stress conditions. Besides ER stress, other stress conditions, including treatment with cell wall-disturbing reagents such as CFW or caspofungin, heat shock, and high salt, were shown to activate unconventional splicing of the HXL1 mRNA, albeit to a lesser extent than TM, in C. neoformans (Figure 9A).
Figure 9. Induction of the UPR pathway by other stresses and potential crosstalk with other signaling pathways.
(A) Analysis of the HXL1 splicing pattern during cell wall and osmotic stresses. RT-PCR of HXL1 and ACT1 were performed with the cDNA samples of H99 strain untreated or treated with TM (5 µg/ml), CFW (20 mg/ml), caspofungin (6 µg/ml), and NaCl (1 M) at 30°C, or at 37°C, respectively, for the indicated times. (B) Analysis of the HXL1 expression pattern in signaling mutants under ER or heat stress conditions. RT-PCR of HXL1, KAR2, and ACT1 was performed with cDNA samples of strain H99 untreated or treated with 5 µg/ml TM for 1 hr at 30°C, or at 37°C for 2 hr, respectively. (C) Analysis of ER-stress sensitivity of various signaling mutants. Strains were spotted serially and incubated on YPD media with or without indicated concentrations of TM at 30°C for 2.5 days. (D and E) Phenotypic analyses of WT (H99) and cna1 mutant strains having increased expression of HXL1 against TM and thermotolerance. Each strain [WT, WT+HXL1 u (YSB736), WT+HXL1 s (YSB741), cna1, cna1+HXL1 u (YSB1320), and cna1+HXL1 s (YSB1325)] was spotted on YPD medium with indicated concentrations of TM and incubated at 30°C for 3 days for TM sensitivity or was incubated on YPD medium at indicated temperatures for 3 days for thermotolerance test.
This finding led us to examine whether multiple stress-activated signaling pathways play roles in inducing UPR via HXL1 splicing. In multiple signaling mutant strains we tested, including cac1 (adenylyl cyclase), cna1 (calcineurin), cpk1 (the pheromone responsive MAPK), hog1 (the stress-activated MAPK), and mpk1 (the cell wall integrity MAPK), most of HXL1 u mRNA was spliced unconventionally in response to TM as in the wild type strain (Figure 9B). This result indicates that cAMP/PKA, calcineurin, and any of MAPK pathways do not directly control the HXL1 splicing under the ER stress condition in C. neoformans. Interestingly, however, under high temperature condition, the ratio of HXL1 s to HXL1 u apparently decreased in the cna1 mutants compared to the wild type strain (Figure 9B and S3D). The cna1 mutant is known to exhibit significant high temperature sensitivity (Figure 5B). Thus these results imply that impairment of the calcineurin signaling pathway can affect regulation of HXL1 at high temperature directly or indirectly. Furthermore, the induction of KAR2 was also found to be significantly reduced in the cna1 mutant (Figure 9B), implicating the potential involvement of the calcineurin pathway in the UPR-mediated thermotolerance. Supporting this, the cna1 mutant exhibited hypersensitivity to TM, albeit to a lesser extent than the UPR mutant (Figure 9C). Furthermore, increased expression of HXL1 by integration of HXL1 u or HXL1 s into the native HXL1 locus clearly increased TM resistance in both wild-type and cna1 mutant strains, suggesting that the UPR pathway work parallel with the calcineurin pathway for ER stress response and adaptation (Figure 9D). In contrast, the increased expression of HXL1 u or HXL1 s partially recovered thermotolerance of the cna1 mutants, without any positive effect on wild-type strains (Figure 9E), suggesting that the defective thermotolerance caused by inhibition of the calcineurin pathway may partly result from insufficient induction of the UPR pathway. All these data strongly support the potential interaction between the calcineurin and UPR pathways in C. neoformans. Taken together, our data suggest that besides ER stress, other stress conditions could also activate the IRE1-HXL1-mediated UPR via regulation of the unconventional splicing of HXL1 mRNA in C. neoformans.
Discussion
In eukaryotic cells, protein folding and secretion in the ER is tightly regulated by the ER quality control system, including ER-associated degradation (ERAD) and N-glycan-dependent protein folding (calnexin cycle), which are closely associated with the Ire1-mediated UPR [25]. Although the pathogenic fungus C. neoformans has a evolutionally distinct dolichol-PP-linked Man9GlcNAc2 N-glycans lacking the terminal glucoses residues, this yeast still has a functional UDP-glucose:glycoprotein glucosylatransferase (UGGT), an essential sensor for misfolded glycoproteins in the calnexin cycle, which can be connected to the ERAD pathway [26]. Besides the calnexin cycle sensor UGGT, the presence of the UPR sensor Ire1 and other folding and degradation enzymes were indicated by bioinformatic analysis of C. neoformans homologs involved in ER quality control [27]. However, little is known about the UPR signal transduction pathway in C. neoformans because the presence of its key major transcription factor, Hac1/Xbp1 ortholog, has not been found. Here, for the first time we identified and characterized the C. neoformans UPR pathway, which is composed of an evolutionarily conserved and a distinct signaling component, an ER stress sensor Ire1 and its downstream bZIP transcription factor Hxl1, respectively. We demonstrated that the Ire1-mediated UPR signaling pathway plays essential roles in ER stress response, thermotolerance, cell wall integrity, and virulence via Hxl1-dependent and -independent pathways in C. neoformans.
Whereas Ire1 orthologs are evolutionarily well conserved in terms of structure and function in unfolded protein response and adaptation among most eukaryotes, Hac1/Xbp1 orthologs appear to be relatively less conserved. In this study, we showed that the C. neoformans Hxl1 is phylogenetically very divergent from the Hac1/Xbp1 transcription factors in the ascomycetous fungi and animals (Figure 2E). As the case of C. neoformans, a recent in silico analysis of the corn smut fungus Ustilago maydis reported the presence of putative homologs for UPR signaling, Um03841 and Um05045, encoding ScIRE1 and ScGCN2 homologs, respectively, but did not find any homolog for ScHAC1 [28]. Furthermore, the plant homolog for Hac1/Xbp1 has not yet been reported [10], [11]. It is notable that as shown in Figure 2D, the BLASTp analysis using the bZIP domain of C. neoformans Hxl1 as query identified several hitherto uncharacterized Hxl1 homologs in several basidiomycetous fungi, such as Tremella fuciformis bZIP 1 (e-value = 4e-07), Coprinopsis cinerea CC1G_02249 (6e-06), Laccaria bicolor LACBIDRAFT_293210 (1e-05), and U. maydis UM03509.1 (1e-04). Analysis of the roles of these Hxl1 homologs in other basidiomycetous fungi could provide more clues on the relationship of bZIP transcription factors evolved to function in the UPR pathway to cope with ER stress.
Despite that Hxl1 is phylogenetically very divergent from the Hac1/Xbp1 transcription factors in the ascomycetous fungi and animals, the borders of the unconventional intron of HXL1 mRNA are predicted to form a typical stem-loop structure of intron regions (Figure S2B), which is probably recognized by Ire1 [29]. The conserved nucleotides CAG at both of the intron borders were identified as the most likely splice sites of HXL1 mRNA (Figure S2C). However, the highly conserved bipartite element, which is important for HAC1 mRNA splicing in vivo and found in the stem-loop of 3′ untranslated region (UTR) of all HAC1 orthologs identified thus far [30], [31], was not detected in HXL1, indicating the presence of a different splicing mechanism of HXL1 by Ire1. Moreover, the complementary sequences to the HAC1 introns, involved in the translational inhibition of unspliced HAC1 mRNA in S. cerevisiae [7], was not detected in the 5′ UTR of HXL1 mRNA, indicating that the regulation of HXL1 expression at translational level might be different from that of HAC1 in S. cerevisiae. Length of the unconventional intron of CnHXL1 mRNA (56-nt) is longer than that of mammalian XBP1 mRNA or some other fungal orthologs (19∼29-nt), but much shorter than that of S. cerevisiae HAC1 mRNA (252-nt) [12], [13], [29], [31], [32], [33]. In contrast to the intron of S. cerevisiae HAC1, the shorter introns of other fungal HAC1 ortholog and mammalian XBP1 mRNAs do not contain the sequences complementary to their 5′ UTR region and thus no translational attenuation as a regulatory means of UPR activation occurs [7], [32]. Since we could not detect the protein product of the unspliced HXL1 mRNA (Figure S6), it is speculated that the HXL1 intron may evolve a functionally distinct role in regulation of UPR in C. neoformans. Interestingly, the protein product of spliced HXL1 mRNA was observed only under ER stress condition, but not under normal, unstressed condition regardless of the presence of 20–30% splicing event of HXL1 mRNA (Figure S6 and Figure 2C). Although its reason is not clear at this point, we speculate that HXL1 mRNA may partially undergo splicing event, but its protein product is not stably maintained under unstressed condition. This hypothesis is currently under investigation.
We reported that the C. neoformans strains lacking Ire1 or Hxl1 display multiple defects not only in ER function but also in cell wall integrity, thermotolerance, and virulence. However, the most notable and significant function of the UPR pathway in C. neoformans we discovered in this study is its absolute requirement for survival and proliferation at 37°C and within the mouse host. The ability to grow and survive at physiological body temperature is an essential virulence attribute of human pathogenic microorganisms. Although the precise reasons for the extreme thermosensitivity of the UPR mutants are not clear at this point, one possibility can be deduced from the recent reports of transcriptome analysis of C. neoformans during growth at high temperature [21], [34], [35]. These analyses showed that genes involved in stress response, cell wall integrity, filamentation, oxidative metabolism, protein targeting, and fatty acids metabolism are induced during growth at 37°C. We observed that in C. neoformans, the expression of ALG7 and KAR2 was up-regulated by the Ire1/Hxl1-dependent pathway under heat stress and expression of some other genes involved in N-/O-glycosylation was also significantly dependent on the presence of Hxl1 during heat stress. Particularly, defects of N-/O-glycosylation were shown to cause thermosensitivity, decreased stability and increased misfolding of glycoproteins with various cellular functions [36], [37]. Thus, down-regulation of those genes by loss of the UPR function in C. neoformans may cause severe defects in several cellular processes upon heat stress. Besides controlling thermotolerance, the UPR pathway might control other virulence factors of C. neoformans. During host infection, C. neoformans may activate the secretory pathway for secretion of host degrading enzymes, such as urease, phospholipases, proteinases, as well as transport of polysaccharide-filled vesicles. In this process, the UPR pathway could be induced to enhance the secretion and folding capacity. Therefore, we stress that the Ire1-Hxl1-mediated UPR system plays important roles in ER quality control and is essential during growth and cell survival for cryptococcal disease.
Interestingly, the ire1 strain expressing the activated HXL1 s was not fully recovered for thermotolerance, capsule formation, or virulence compared with the ire1+IRE1 complemented strain (Figure 5, 7, and 8). Furthermore, we suggest that Ire1 and Hxl1 govern the UPR of C. neoformans by similar and distinct mechanisms through expression analysis of a few UPR target genes in UPR mutant strains (Figure 6). Similarly, in the nematode Caenorhabditis elegans, many canonical UPR-induced genes are regulated by both ire-1 and xbp-1 [38]. However, some subsets of constitutive UPR genes, whose expression are not induced by TM but dependent on ER stress transducers, showed a different requirement for either ire-1 or xbp-1 [38]. In Drosophila and mammalian cells, Ire1 also has an Xbp1-independent role, as part of the regulated Ire1-dependent decay pathway, where it can selectively degrade groups of mRNAs encoding secreted or integral membrane proteins to relieve the burden on the ER [39], [40]. Collectively, we suggest that Ire1 has a broad range of physiological roles in C. neoformans in both Hxl1-dependent and -independent manners.
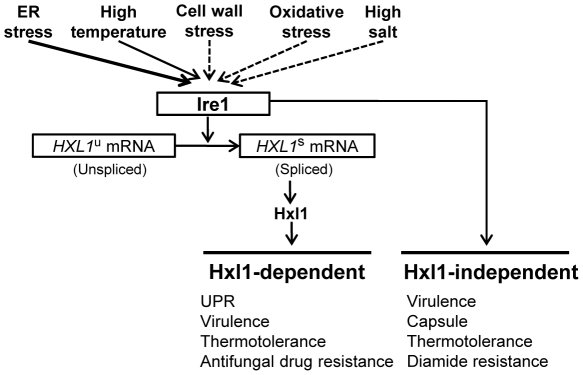
In conclusion, we demonstrated that a unique UPR pathway composed of an evolutionarily conserved Ire1 protein kinase and a novel Hxl1 transcription factor functions not only in ER stress response, but also in controlling cell wall integrity, growth at host physiological temperature, and virulence of the human pathogenic yeast C. neoformans. Moreover, the Mpk1 MAPK pathway regulating cell wall integrity was shown to be activated by TM treatment or defects in UPR, supporting the essential role of UPR as a common defense mechanism for both ER and cell wall stress in C. neoformans. Based on our data, we propose that the Ire1-Hxl1-mediated UPR serve as a core stress response pathway to various environmental cues in C. neoformans, cross-talking with other signaling pathways directly or indirectly (Figure 10). Furthermore, our findings suggest a novel antifungal therapeutic target for specific treatment of C. neoformans. Intriguingly, Hxl1 is widely conserved in different serotypes of C. neoformans and C. gattii. We propose Hxl1 is an ideal target for antifungal drug development, based on its marked divergence from the host Xbp1 transcription factor. In the future, genome-wide comparative expression analysis of ire1 and hxl1 mutant strains under several stress conditions, including ER stress and thermal stress, will shed light to further understand and identify UPR target genes and differential regulation of the Hxl1-dependent and -independent Ire1 signaling pathways.
Figure 10. The proposed role of the UPR pathway as core stress response to various stresses in C. neoformans.
The Ire1-Hxl1-mediated UPR system plays important roles in ER quality control and is essential during growth and cell survival for cryptococcal disease. Notably, Ire1 has a broad range of physiological roles in C. neoformans not only in Hxl1-dependent but also Hxl1-independent manners. Solid arrow indicates direct involvement of the UPR pathway (e.g., ER stress) and dotted arrow indicates the direct or indirect activation of the UPR pathway via potential cross-talk with other signaling pathways.
Materials and Methods
Ethics statement
The animal studies at Duke University Medical Center were in full compliance with all of the guidelines of the Duke University Medical Center Institutional Animal Care and Use Committee (IACUC) and the United States Animal Welfare Act (Public Law 98–198). The Duke University Medical Center IACUC approved all of the vertebrate studies. The studies were conducted in Division of Laboratory Animal Resources (DLAR) facilities that are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Strains, media, plasmids, and primers
The C. neoformans strains used in this study are listed in Table 1. Yeast strains were maintained and cultured in YPD (1% yeast extract, 2% peptone, 2% glucose) medium. Niger seed medium for melanin production, and agar-based Dulbecco's modified Eagle's medium for capsule production were prepared as described previously [41]. The plasmids and primers used in this study listed in the Table S1. Information on genomic DNA sequences for each gene was obtained from the C. neoformans serotype A genome database (Duke university/Broad Institute of Harvard), and the C. gattii serotype B genome database (Broad Institute of Harvard/MIT).
Gene disruption and complementation of IRE1 and HXL1
Gene disruption of IRE and HXL1 was carried out in the C. neoformans serotype A H99 (MATα) strain background by using overlap PCR or double joint (DJ)-PCR strategies combined with biolistic transformation as described previously [42], [43] and schemes of their gene disruption were illustrated in Figure S1. For the IRE1 disruption, the first 5′ and 3′ UTR regions of the IRE1 gene were PCR-amplified with the primer sets B1644/B1645, B1646/B1647, respectively, by using H99 genomic DNA as a template, and the dominant selectable marker NAT (nourseothricin acetyltransferase) was amplified with the M13Fe/M13Re primers from pNATSTM#224. A 3.6 kb IRE1 disruption cassette was generated by overlap PCR with the primers B1644/B1647 from the combined templates of the first PCR products. The IRE1 disruption cassettes were precipitated onto 600 µg of gold microcarrier beads (0.6 µm, Bio-Rad) and strain H99 was biolistically transformed. For the HXL1 gene disruption, 5′ and 3′ UTR regions were amplified with primer sets B1881/B1882 and B1883/B1884 from H99 genomic DNA as template, and the NAT markers were amplified with primer sets M13Fe/B1455 and M13Re/B1454 from the vector pNATSTM#229. A HXL1 gene disruption cassette generated by DJ-PCR was precipitated onto 600 µg of gold microcarrier beads and biolistically transformed into the strain H99 in serotype A. Stable transformants selected on YPD medium containing nourseothricin were initially screened by diagnostic PCR and the correct gene disruptions were further confirmed by Southern blot analysis (Figure S1).
To generate ire1+IRE1 complemented strains, an IRE1 gene fragment containing its 0.78 kb promoter, 3.52 kb ORF, and 0.53 kb terminator was PCR-amplified using the primers B1969/B1970 including NotI restriction sites from the genomic DNA of strain H99. The 4.8 kb PCR product of the IRE1 gene was subcloned into pTOP-V2 (Enzynomics), generating a plasmid pTOP-IRE1. Then the IRE1 gene insert was subcloned into plasmid pJAF12 [41] containing the NEO marker (neomycin/G418-resistant marker) to produce pJAF12-IRE1. MluI-digested linearized pJAF12-IRE1 was re-integrated into the native IRE1 locus of the ire1 mutants (YSB552 and YSB554) by biolistic transformation, generating the ire1+IRE1 (YSB1000 and YSB1005) strains. To generate hxl1+HXL1 complemented strains, the 1 kb promoter and the 0.6 kb terminator fragments of the HXL1 gene were PCR-amplified with two primer sets C35/C36 and C37/C38, respectively, from the genomic DNA of strain H99and fused by the second round PCR using primers C35/C38. Then the fusion-PCR product was subcloned into pJAFS1 which is derived from pJAF12 by deleting a SacI site, resulting in a vector pJAFS1-HXL1PT. An unspliced version of the HXL1 gene (HXL1 u) and a spliced version of the HXL1 gene (HXL1 s) were PCR-amplified using the primers C39/C40 from the cDNAs generated from total RNAs isolated from cells treated without or with TM, respectively. The HXL1 u and HXL1 s PCR products were then cloned into pJAFS1-HXL1PT, generating pJAFS1-HXL1u and pJAFS1-HXL1s, respectively. The vectors were digested at a unique SacI site of the HXL1 promoter and reintegrated into the native HXL1 locus of the ire1 (YSB552 and YSB554) strain. To generate an hxl1+HXL1 complemented strain, pJAFS1-HXL1u digested with SacI was reintegrated into the native HXL1 locus of the hxl1 (YSB723) strain.
Total RNA preparation, RT-PCR, and qRT-PCR
For UPR induction, cells at OD600 (optical density at 600 nm) = 0.15 were grown to mid-logarithmic phase (OD600 = 0.5) in YPD at 30°C and then treated with 5 or 8 µg/ml TM, or 20 mM DTT, for 1 or 2 hr. The cell pellets were washed with diethylpyrocarbonate-treated water twice, immediately frozen in liquid nitrogen, and disrupted thoroughly using a mortar and pestle. Total RNAs were extracted using the RNeasy Mini Kit (Qiagen) and subsequently single stranded cDNA was synthesized using a MMLV reverse transcriptase (Invitrogen). Reverse transcriptase-polymerase chain reaction (RT-PCR) was conducted using serially diluted cDNA with Maxime PCR PreMix (i-Taq) (iNtRON Biotechnology). Quantitative real-time PCR (qRT-PCR) was performed using a CFX96 Real-Time PCR detection system (Bio-Rad). Normalized fold expression (2−ΔΔCT) was calculated with the CFX manager software using ACT1 as a reference gene and graphed with Microsoft Excel. All PCR reactions were analyzed in duplicate and all primers used in the study are listed in Table S1.
Construction of HXL1 expression vectors for complementation analysis in S. cerevisiae hac1
For construction of an expression vector of HXL1 under the control of the S. cerevisiae GAL10 promoter, the DNA fragments of unspliced and spliced HXL1 were amplified with two pairs of primers, C43/C44 and C43/C45, respectively, and subcloned into pRE316 harboring the GAL1/10 promoter, CEN, and URA3, resulting plasmids pRE316-CnHXL1u and pRE316-CnHXL1s, respectively. For an expression vector of HXL1 using the S. cerevisiae HAC1 promoter, the ScHAC1 promoter and the ScHAC1 u fragment were amplified with the primer pairs C46/C47 and C48/C49, and then subcloned into pRE316, generating pRE318-ScHAC1u. The ScHAC1 u fragment of pRE318-ScHAC1u was then replaced with the ScHAC1 s fragment, which was amplified with the primers C48/C50, or the HXL1 u, and the HXL1 s fragments, generating plasmids pRE318-ScHAC1s, pRE318-CnHXL1u, and pRE318-HXL1s, respectively. S. cerevisiae transformants harboring the S. cerevisiae HAC1 or C. neoformans HXL1 expression vectors were selected on media lacking uracil and assayed for the sensitivity to TM and DTT. For construction of an HXL1 integration vector under the control of the S. cerevisiae HAC1 promoter, the 2.3 kb ScHAC1 promoter-HXL1 u or -HXL1 s fragments were obtained from pRE318-CnHXL1u or CnHXL1s, respectively, via treatment of NheI/PvuII and Klenow fragment, and ligated to pTcUR3 [44] treated with BamHI and Klenow fragment, generating pTcU-CnHXL1u and pTcU-CnHXL1s, respectively. These vectors were transformed into wild-type BY4742 and Schac1 strains after AvrII digestion.
Drug sensitivity test
Strains grown in YPD medium overnight were spotted onto solid YPD medium containing the indicated concentrations of tunicamycin (TM), dithiothreitol (DTT), Congo red (CR), Calcofluor white (CFW), diamide, and antifungal drugs [azoles (ketoconazole, fluconazole, itraconazole), polyene (amphotericin B), and phenylpyrrole (fludioxonil)], incubated at 30°C, and observed as previously described [24].
Western blotting analysis of Mpk1 phosphorylation
Total proteins were extracted as described previously [45] and analyzed by Western blotting with phospho-p44/42-MAPK antibody (#4370, Cell Signaling Technology) for Mpk1-P protein and anti-Hog1 antibody (sc-9079, Santa Cruz biotechnology) for the Hog1 protein as a loading control.
Capsule and melanin assay
Levels of capsule and melanin production were qualitatively and quantitatively measured as described previously [41]. Statistical analysis of relative capsule diameter was performed by using the Bonferroni multiple comparison test.
Animal studies
Four- to six-week-old female A/Jcr mice (Jackson Laboratory, 18–22 g) were utilized in this study. For infection, strains were cultured in YPD medium overnight at 24°C, washed twice with sterile phosphate buffered saline (PBS), and resuspended in sterile PBS at 2×106 cells per ml. Serially diluted cells were plated onto YPD medium and incubated at 24°C for 72 hr to determine viability. Ten mice per strain (except strains YSB723 and YSB747 for which nine mice each were studied) were anesthetized with pentobarbital (Lundbeck Inc.) and infected via intranasal instillation with 105 cells (in 50 µl). Survival was monitored twice daily, and moribund mice were CO2-euthanized. The Kaplan-Meier survival curves were generated by Prism 5.02 (GraphPad Software), and P values were evaluated from a Log-rank (Mantel-Cox) test.
To determine fungal burden, the lungs and brains of C. neoformans-infected mice (n = 5 for each strain, except n = 4 for strains YSB1000 and YSB762 due to death after pentobarbital treatment) were dissected at day 7. Half organ portions were weighed, transferred to a 15-ml Falcon tube filled with 5 ml PBS, and homogenized for 10 s at 13,600 rpm/min (Power Gen 500, Fisher Scientific). Tissue homogenates were serially diluted, and 200 µl was plated onto a YPD plate. The plates were incubated at 24°C for 48 h to determine CFU per gram of lung or brain. The significance of differences in fungal burden was determined using one-way analysis of variance (ANOVA) and Bonferroni multiple comparison test. For histopathological analysis, half organ samples of lung and brain were fixed in 10% phosphate-buffered formalin (Fisher), and Mucicarmine and hematoxylin and eosin (H&E) staining were performed by the Department of Pathology at Duke University. After slide preparation, each sample was examined thoroughly by microscopy for analysis of Cryptococcus colonization (Mucicarmine) and tissue necrosis (H&E). Images were captured using an Olympus Vanox microscope (PhotoPath; Duke University Medical Center).
General techniques
Protein domain analysis was performed with SMART [46]. Multiple sequence alignments were constructed with CLUTAL W (http://www.genome.jp/tools/clustalw/) and shaded using Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html). Percent identity of proteins was calculated by comparing sequence pairs in relation to the phylogeny reconstructed by the CLUSTAL W method of the Lasergene MegAlign program (DNASTAR Inc.). Searches for the presence of the UPRE and UPRE-like sequences without any mismatches in the promoter region (−1000∼−1) of the putative C. neoformans UPR regulated genes were performed with RSA-tools-dna-pattern (http://rsat.ulb.ac.be/rsat/) [47].
Supporting Information
Disruption of C. neoformans IRE1 and HXL1. (A) Diagram for disruption of the IRE1 gene in serotype A strain H99 (left panel) and Southern blot analysis (right panel). (B) Diagram for disruption of the HXL1 gene in strain H99 and Southern blot analysis. Primers used for gene disruption are indicated as bent arrows. The IRE1 and HXL1 genes were specifically deleted with the NAT-dominant selectable marker in strain H99. The correct gene disruptions were confirmed by Southern blot analysis using genomic DNAs digested with the indicated restriction enzyme.
(TIF)
Structural characteristics of the HXL1 genes in C. neoformans strains of different serotypes. (A) Non-conventional spliced intron sequences of serotype A, D, and B strains. cDNA nucleotide sequences of the HXL1 genes were aligned from +901 to +1392 of serotype A and D, and +998 to +1334 of serotype B. The unconventional intron sequences are displayed in lowercase with a dotted line, and asterisks represent the stop codon for each of the unspliced and spliced HXL1 mRNAs, respectively. (B) Secondary structure of the unconventional introns of C. neoformans HXL1, C. elegans (AAL60200), and H. sapiens (NP_005071) XBP1 mRNAs. RNA secondary structure prediction was performed with the CLC RNA benchwork 4.0 Demo program (CLC bio). Conserved sequences in splicing junctions are indicated by dotted boxes. (C) Comparison of the unconventional intron region sequences of HAC1/XBP1. The mRNA sequences surrounding the unconventional introns in C. neoformans HXL1, Y. lipolytica HAC1, C. albicans HAC1, T. reesei hac1, A. nidulans hacA, C. elegans and H. sapiens XBP1 are aligned. Flanking sequences are indicated by uppercase, and intron sequences are in lowercase. Asterisks represent the conserved nucleotide sequences among the intron junctions.
(TIF)
Quantification of relative ratio of HXL1 u and HXL1 s and KAR2 mRNA levels under normal and UPR-induced conditions. (A) For specific amplification of HXL1 u mRNA by qRT-PCR, C79 and C80 primers were designed to bind only to the unconventional (UC) intron region and to the C-terminal exon region, respectively. For amplification of HXL1 s mRNA by qRT-PCR, C81 and C82 primers were designed to bind to the exon junctions and to the C-terminal exon region, respectively. (B) The qRT-PCR analysis of HXL1 u, HXL1 s, and KAR2 mRNAs. C. neoformans strains were cultivated in YPD with or without TM (8 μg/ml) or DTT (20 mM) as described in Figure 2C. The relative expression levels of HXL1 u and HXL1 s mRNAs (left panel), and KAR2 mRNA (right panel) were analyzed by qRT-PCR with primers represented in (A) and listed in Table S1 in duplicate, and normalized to that of ACT1. (C) Relative quantification of RT-PCR products of HXL1 u, HXL1 s, and KAR2 mRNAs in Figure 2C. (D) Relative quantification of RT-PCR products of HXL1 u, HXL1 s, and KAR2 mRNAs in Figure 9B. The intensities of HXL1 u, HXL1 s, KAR2, and ACT1 were quantified by analyzing the band intensity of corresponding RT-PCR product in the gels with a Quantity One 4.6.2 software (Bio-Rad) in triplicate and calculated as the relative % ratio of HXL1 u and HXL1 s mRNAs and as the relative fold intensity of KAR2 to ACT1, respectively.
(TIF)
RSA-tools-dna-pattern analysis of putative UPR genes of C. neoformans harboring the UPRE and UPRE-like sequences. A set of promoter sequences (-1000∼-1) of putative UPR regulated genes of C. neoformans were obtained from the C. neoformans genome database and analyzed for the presence of the UPRE (CAGNGTG) and UPRE-like sequences (ATGGTATCAT, TGACGTCA, CCAGC, CGTGTCGG, ACGTGTCG, CGTGTCC, TACGTG, AGTAGGAC, AGGACAAC) by RSA-tools-dna-pattern. Solid bars indicate the exactly matched sequence regions.
(TIF)
Fungal burden assay in the brain. The scatter plot represents CFU (per gram of organ) recovered in the brain from sacrificed animals infected with WT (H99), ire1 (YSB552), ire1+IRE1 (YSB1000), hxl1 (YSB723), and hxl1+HXL1 (YSB762) strains.
(TIF)
Translation of C. neoformans HXL1. An unspliced HXL1 expression vector with an N-terminal HA tag under by its own promoter (upper panel) was integrated into WT (H99) and ire1 strains. Proteins were extracted from WT (H99, lanes 1-4) and ire1 (lane 5-8) strains incubated in YPD medium treated with (lanes 3-4, lanes 7-8) or without (lanes 1-2, lanes 5-6) TM (5 μg/ml) at 30°C for 2 hr and detected by Western blotting with anti-HA antibody (lower panel).
(TIF)
Plasmids and primers used in this study.
(DOC)
Identity of overall and the bZIP domain sequences of five C. neoformans ORFs to other Hac1/Xbp1 proteins.
(DOC)
Acknowledgments
We are grateful to Dr. Tae Kwang Oh and Dr. Yin-Won Lee for their sincere comments and encouragement.
Footnotes
The authors have declared that no competing interests exist.
This work was in part supported by the Microbial Genomics and Applications R&D Program grant (No. 11-2008-09-003-00) from the Korean Ministry of Education, Science, and Technology (MEST) and by the National Research Foundation of Korea grants (No. 2008-0061959, No. 2008-0061963, No. 2010-0029117, No. 2010-0024569) from MEST. This work was also supported in part by RO1 Grant AI50438 from the NIH/NIAID (to J.H.) and the Duke University Center for AIDS Research (CFAR grant, 2P30 AI064518-06) (to Y.L.C.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Lin JH, Walter P, Yen TSB. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Path: Mech Dis. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 5.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 6.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 7.Ruegsegger U, Leber JH, Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- 8.Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J Biol Chem. 2006;281:5852–5860. doi: 10.1074/jbc.M509061200. [DOI] [PubMed] [Google Scholar]
- 9.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 10.Liu J-X, Howell SH. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell. 2010;22:2930–2942. doi: 10.1105/tpc.110.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urade R. The endoplasmic reticulum stress signaling pathways in plants. Biofactors. 2009;35:326–331. doi: 10.1002/biof.45. [DOI] [PubMed] [Google Scholar]
- 12.Wimalasena TT, Enjalbert B, Guillemette T, Plumridge A, Budge S, et al. Impact of the unfolded protein response upon genome-wide expression patterns, and the role of Hac1 in the polarized growth, of Candida albicans. Fungal Genet Biol. 2008;45:1235–1247. doi: 10.1016/j.fgb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, et al. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 2009;5:e1000258. doi: 10.1371/journal.ppat.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 15.Scrimale T, Didone L, de Mesy Bentley KL, Krysan DJ. The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:164–175. doi: 10.1091/mbc.E08-08-0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010;6:e1000752. doi: 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saloheimo M, Valkonen M, Penttila M. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol Microbiol. 2003;47:1149–1161. doi: 10.1046/j.1365-2958.2003.03363.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Feldman DE, Deng C, Brown JA, De Giacomo AF, et al. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol Cancer Res. 2005;3:669–677. doi: 10.1158/1541-7786.MCR-05-0181. [DOI] [PubMed] [Google Scholar]
- 19.Foti DM, Welihinda A, Kaufman RJ, Lee AS. Conservation and divergence of the yeast and mammalian unfolded protein response. Activation of specific mammalian endoplasmic reticulum stress element of the grp78/BiP promoter by yeast Hac1. J Biol Chem. 1999;274:30402–30409. doi: 10.1074/jbc.274.43.30402. [DOI] [PubMed] [Google Scholar]
- 20.Spode I, Maiwald D, Hollenberg CP, Suckow M. ATF/CREB sites present in sub-telomeric regions of Saccharomyces cerevisiae chromosomes are part of promoters and act as UAS/URS of highly conserved COS genes. J Mol Biol. 2002;319:407–420. doi: 10.1016/S0022-2836(02)00322-4. [DOI] [PubMed] [Google Scholar]
- 21.Kraus PR, Boily M-J, Giles SS, Stajich JE, Allen A, et al. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic-DNA microarray. Eukaryot Cell. 2004;3:1249–1260. doi: 10.1128/EC.3.5.1249-1260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko YJ, Yu YM, Kim GB, Lee GW, Maeng PJ, et al. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell. 2009;8:1197–1217. doi: 10.1128/EC.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Missall TA, Lodge JK, McEwen JE. Mechanisms of resistance to oxidative and nitrosative stress: Implications for fungal survival in mammalian hosts. Eukaryot Cell. 2004;3:835–846. doi: 10.1128/EC.3.4.835-846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerik KJ, Bhimireddy SR, Ryerse JS, Specht CA, Lodge JK. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell. 2008;7:1685–1698. doi: 10.1128/EC.00146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Römisch K. Endoplasmic reticulum–associated degradation. Annu Rev Cell Dev Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- 26.Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, et al. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci USA. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerjee S, Vishwanath P, Cui J, Kelleher DJ, Gilmore R, et al. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc Natl Acad Sci USA. 2007;104:11676–11681. doi: 10.1073/pnas.0704862104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernández-Álvarez A, Elías-Villalobos A, Ibeas JI. Protein glycosylation in the phytopathogen Ustilago maydis: From core oligosaccharide synthesis to the ER glycoprotein quality control system, a genomic analysis. Fungal Genet Biol. 2010;47:727–735. doi: 10.1016/j.fgb.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 30.Aragon T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, et al. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh MH, Cheon SA, Kang HA, Kim JY. Functional characterization of the unconventional splicing of Yarrowia lipolytica HAC1 mRNA induced by unfolded protein response. Yeast. 2010;27:443–452. doi: 10.1002/yea.1762. [DOI] [PubMed] [Google Scholar]
- 32.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 33.Saloheimo M, Valkonen M, Penttila M. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol Microbiol. 2003;47:1149–1161. doi: 10.1046/j.1365-2958.2003.03363.x. [DOI] [PubMed] [Google Scholar]
- 34.Perfect JR. Cryptococcus neoformans: the yeast that likes it hot. FEMS Yeast Research. 2006;6:463–468. doi: 10.1111/j.1567-1364.2006.00051.x. [DOI] [PubMed] [Google Scholar]
- 35.Rosa e Silva LK, Staats CC, Goulart LS, Morello LG, Pelegrinelli Fungaro MH, et al. Identification of novel temperature-regulated genes in the human pathogen Cryptococcus neoformans using representational difference analysis. Res Microbiol. 2008;159:221–229. doi: 10.1016/j.resmic.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 37.Nakatsukasa K, Okada S, Umebayashi K, Fukuda R, Nishikawa S-i, et al. Roles of O-mannosylation of aberrant proteins in reduction of the load for endoplasmic reticulum chaperones in yeast. J Biol Chem. 2004;279:49762–49772. doi: 10.1074/jbc.M403234200. [DOI] [PubMed] [Google Scholar]
- 38.Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollien J, Lin JH, Li H, Stevens N, Walter P, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 41.Bahn YS, Hicks JK, Giles SS, Cox GM, Heitman J. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot Cell. 2004;3:1476–1491. doi: 10.1128/EC.3.6.1476-1491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, et al. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- 43.Kim MS, Kim SY, Yoon JK, Lee YW, Bahn YS. An efficient gene-disruption method in Cryptococcus neoformans by double-joint PCR with NAT-split markers. Biochem Biophys Res Commun. 2009;390:983–988. doi: 10.1016/j.bbrc.2009.10.089. [DOI] [PubMed] [Google Scholar]
- 44.Cho EY, Cheon SA, Kim H, Choo J, Lee D-J, et al. Multiple-yapsin-deficient mutant strains for high-level production of intact recombinant proteins in Saccharomyces cerevisiae. J Biotechnol. 2010;149:1–7. doi: 10.1016/j.jbiotec.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell. 2005;16:2285–2300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, et al. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perfect JR, Ketabchi N, Cox GM, Ingram CW, Beiser CL. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol. 1993;31:3305–3309. doi: 10.1128/jcm.31.12.3305-3309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, et al. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kojima K, Bahn YS, Heitman J. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology. 2006;152:591–604. doi: 10.1099/mic.0.28571-0. [DOI] [PubMed] [Google Scholar]
- 52.Bahn YS, Geunes-Boyer S, Heitman J. Ssk2 mitogen-activated protein kinase kinase kinase governs divergent patterns of the stress-activated Hog1 signaling pathway in Cryptococcus neoformans. Eukaryot Cell. 2007;6:2278–2289. doi: 10.1128/EC.00349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 54.Kidd SE, Guo H, Bartlett KH, Xu J, Kronstad JW. Comparative gene genealogiesindicate that two clonal lineages of Cryptococcus gattii in British Columbia resemble strains from other geographical areas. Eukaryot Cell. 2005;4:1629–1638. doi: 10.1128/EC.4.10.1629-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disruption of C. neoformans IRE1 and HXL1. (A) Diagram for disruption of the IRE1 gene in serotype A strain H99 (left panel) and Southern blot analysis (right panel). (B) Diagram for disruption of the HXL1 gene in strain H99 and Southern blot analysis. Primers used for gene disruption are indicated as bent arrows. The IRE1 and HXL1 genes were specifically deleted with the NAT-dominant selectable marker in strain H99. The correct gene disruptions were confirmed by Southern blot analysis using genomic DNAs digested with the indicated restriction enzyme.
(TIF)
Structural characteristics of the HXL1 genes in C. neoformans strains of different serotypes. (A) Non-conventional spliced intron sequences of serotype A, D, and B strains. cDNA nucleotide sequences of the HXL1 genes were aligned from +901 to +1392 of serotype A and D, and +998 to +1334 of serotype B. The unconventional intron sequences are displayed in lowercase with a dotted line, and asterisks represent the stop codon for each of the unspliced and spliced HXL1 mRNAs, respectively. (B) Secondary structure of the unconventional introns of C. neoformans HXL1, C. elegans (AAL60200), and H. sapiens (NP_005071) XBP1 mRNAs. RNA secondary structure prediction was performed with the CLC RNA benchwork 4.0 Demo program (CLC bio). Conserved sequences in splicing junctions are indicated by dotted boxes. (C) Comparison of the unconventional intron region sequences of HAC1/XBP1. The mRNA sequences surrounding the unconventional introns in C. neoformans HXL1, Y. lipolytica HAC1, C. albicans HAC1, T. reesei hac1, A. nidulans hacA, C. elegans and H. sapiens XBP1 are aligned. Flanking sequences are indicated by uppercase, and intron sequences are in lowercase. Asterisks represent the conserved nucleotide sequences among the intron junctions.
(TIF)
Quantification of relative ratio of HXL1 u and HXL1 s and KAR2 mRNA levels under normal and UPR-induced conditions. (A) For specific amplification of HXL1 u mRNA by qRT-PCR, C79 and C80 primers were designed to bind only to the unconventional (UC) intron region and to the C-terminal exon region, respectively. For amplification of HXL1 s mRNA by qRT-PCR, C81 and C82 primers were designed to bind to the exon junctions and to the C-terminal exon region, respectively. (B) The qRT-PCR analysis of HXL1 u, HXL1 s, and KAR2 mRNAs. C. neoformans strains were cultivated in YPD with or without TM (8 μg/ml) or DTT (20 mM) as described in Figure 2C. The relative expression levels of HXL1 u and HXL1 s mRNAs (left panel), and KAR2 mRNA (right panel) were analyzed by qRT-PCR with primers represented in (A) and listed in Table S1 in duplicate, and normalized to that of ACT1. (C) Relative quantification of RT-PCR products of HXL1 u, HXL1 s, and KAR2 mRNAs in Figure 2C. (D) Relative quantification of RT-PCR products of HXL1 u, HXL1 s, and KAR2 mRNAs in Figure 9B. The intensities of HXL1 u, HXL1 s, KAR2, and ACT1 were quantified by analyzing the band intensity of corresponding RT-PCR product in the gels with a Quantity One 4.6.2 software (Bio-Rad) in triplicate and calculated as the relative % ratio of HXL1 u and HXL1 s mRNAs and as the relative fold intensity of KAR2 to ACT1, respectively.
(TIF)
RSA-tools-dna-pattern analysis of putative UPR genes of C. neoformans harboring the UPRE and UPRE-like sequences. A set of promoter sequences (-1000∼-1) of putative UPR regulated genes of C. neoformans were obtained from the C. neoformans genome database and analyzed for the presence of the UPRE (CAGNGTG) and UPRE-like sequences (ATGGTATCAT, TGACGTCA, CCAGC, CGTGTCGG, ACGTGTCG, CGTGTCC, TACGTG, AGTAGGAC, AGGACAAC) by RSA-tools-dna-pattern. Solid bars indicate the exactly matched sequence regions.
(TIF)
Fungal burden assay in the brain. The scatter plot represents CFU (per gram of organ) recovered in the brain from sacrificed animals infected with WT (H99), ire1 (YSB552), ire1+IRE1 (YSB1000), hxl1 (YSB723), and hxl1+HXL1 (YSB762) strains.
(TIF)
Translation of C. neoformans HXL1. An unspliced HXL1 expression vector with an N-terminal HA tag under by its own promoter (upper panel) was integrated into WT (H99) and ire1 strains. Proteins were extracted from WT (H99, lanes 1-4) and ire1 (lane 5-8) strains incubated in YPD medium treated with (lanes 3-4, lanes 7-8) or without (lanes 1-2, lanes 5-6) TM (5 μg/ml) at 30°C for 2 hr and detected by Western blotting with anti-HA antibody (lower panel).
(TIF)
Plasmids and primers used in this study.
(DOC)
Identity of overall and the bZIP domain sequences of five C. neoformans ORFs to other Hac1/Xbp1 proteins.
(DOC)