Hsc70 plays a productive role during cotranslational cystic fibrosis transmembrane conductance regulator folding that is outweighed by its dominant contribution to posttranslational targeting to the ubiquitin-proteasome system. Moreover, the outcome of Hsc70 binding appears highly sensitive to the duration of its binding cycle, which is governed by regulatory cochaperones.
Abstract
The Hsp/c70 cytosolic chaperone system facilitates competing pathways of protein folding and degradation. Here we use a reconstituted cell-free system to investigate the mechanism and extent to which Hsc70 contributes to these co- and posttranslational decisions for the membrane protein cystic fibrosis transmembrane conductance regulator (CFTR). Hsc70 binding to CFTR was destabilized by the C-terminal domain of Bag-1 (CBag), which stimulates client release by accelerating ADP-ATP exchange. Addition of CBag during CFTR translation slightly increased susceptibility of the newly synthesized protein to degradation, consistent with a profolding function for Hsc70. In contrast, posttranslational destabilization of Hsc70 binding nearly completely blocked CFTR ubiquitination, dislocation from the endoplasmic reticulum, and proteasome-mediated cleavage. This effect required molar excess of CBag relative to Hsc70 and was completely reversed by the CBag-binding subdomain of Hsc70. These results demonstrate that the profolding role of Hsc70 during cotranslational CFTR folding is counterbalanced by a dominant and essential role in posttranslational targeting to the ubiquitin-proteasome system. Moreover, the degradative outcome of Hsc70 binding appears highly sensitive to the duration of its binding cycle, which is in turn governed by the integrated expression of regulatory cochaperones.
INTRODUCTION
The endoplasmic reticulum (ER) is responsible for biosynthesis, translocation, integration, folding, and trafficking of membrane and secretory proteins in eukaryotic cells (Osborne et al., 2005; Cross et al., 2009; Skach, 2009). In addition, it maintains a stringent quality control system that degrades misfolded and unassembled proteins caused by genetic mutations, viral infections, and aging (e.g., cystic fibrosis, diabetes, autoimmune disease, Parkinson's disease) (Kopito, 1997; Meusser et al., 2005). ER-associated degradation (ERAD) is a multistep process that requires substrate recognition, retrotranslocation, ubiquitination, and proteolysis (Vembar and Brodsky, 2008), in which cellular machinery within the ER lumen, ER membrane, and/or the cytosol (Carvalho et al., 2006; Denic et al., 2006) maintains the delicate balance between protein folding and degradation.
Two cytosolic heat shock proteins of 70 kDa, stress-inducible Hsp70 and constitutively expressed cognate Hsc70, play important roles in both the biosynthesis and degradation of ERAD substrates. During de novo protein folding and under conditions of proteotoxic stress, Hsp/c70 iteratively binds hydrophobic patches of partially unfolded and/or denatured proteins, thereby minimizing nonproductive aggregation in the crowded cytosolic environment (Schmid et al., 1994; Hartl, 1996; Young et al., 2004; Kampinga and Craig, 2010). Binding occurs with a hydrophobic cleft in the Hsp/c70 substrate-binding domain the access and affinity of which is controlled by nucleotide occupancy of its N-terminal ATPase domain. Weak interaction with client proteins is initiated in the ATP-bound state and stabilized by allosteric movement of a clamp-like structure upon ATP hydrolysis (Palleros et al., 1991; Hartl, 1996). Substrate recruitment and ATP hydrolysis are stimulated by a diverse family of Hsp40 (DNAJ) proteins, which result in formation of stable client–chaperone complexes (Fan et al., 2003; Summers et al., 2009; Kampinga and Craig, 2010). Similarly, nucleotide exchange factors (e.g., Bag-1, HspBP1, and Hsp110) promote ADP/ATP exchange and substrate release (Höhfeld and Jentsch, 1997; Takayama et al., 1997; Bimston et al., 1998; Takayama and Reed, 2001; Shomura et al., 2005; Andréasson et al., 2008). These interactions establish a canonical binding cycle in which protein aggregation is inhibited by transient shielding of exposed peptide loops that, when released, either become buried during productive folding or rebind Hsp/c70 (Hartl, 1996; Young et al., 2004; Kampinga and Craig, 2010). For client proteins that fold inefficiently, the Hsp/c70 C-terminal domain interacts with a different set of cochaperones via a conserved tetratricopeptide repeat (TPR) binding motif. For example, the TPR-containing protein HOP (p60) stimulates recruitment of Hsp90 chaperone complexes (Smith et al., 1993; Dittmar et al., 1996; Frydman and Höhfeld, 1997), whereas the ubiquitin E3 ligase C-terminus of Hsp/c70-interacting protein (CHIP) catalyzes covalent attachment of polyubiquitin chains to terminally misfolded substrates, and thereby targets them for degradation via the ubiquitin proteasome pathway (Connell et al., 2001; Meacham et al., 2001).
One well-studied Hsp/c70 substrate is the cystic fibrosis transmembrane conductance regulator (CFTR), a member of the ABC transporter superfamily (ABCC7). CFTR contains two transmembrane domains, each with six transmembrane segments, two cytosolic nucleotide binding domains (NBDs), and a cytosolic regulatory (R) domain (Figure 1A). In most cell types, CFTR folding is inefficient. Approximately 70% of wild-type protein and 99% of the most common inherited ΔF508 mutant are degraded via ERAD (Ward and Kopito, 1994; Ward et al. 1995; Riordan, 2008). Hsc70 is proposed to facilitate early steps of CFTR folding in conjunction with the Hsp40 cochaperone Hdj-2 (Meacham et al., 1999). Consistent with this profolding function, overexpression of stress-inducible Hsp70 and Hdj-1 stabilizes the ER form of wild-type CFTR (Farinha et al., 2002), whereas Hdj-2 knockdown blocks CFTR folding (Grove et al., 2011). Hsc70 also promotes refolding of purified NBD1 in vitro (Strickland et al., 1997). Paradoxically, the Hsc70-CHIP complex facilitates CFTR ubiquitination and increases degradation, both in cells and in vitro (Meacham et al., 2001; Younger et al., 2004). In addition, the cytosolic E3 ligase Nedd4-2, the membrane-bound E3 ligase RMA1, and the E4 ligase gp78 may also play roles in CFTR degradation through poorly understood mechanisms (Younger et al., 2006; Morito et al., 2008; Caohuy et al., 2009). A persistent question arising from these observations is the extent to which two diametrically opposed outcomes (i.e., productive folding vs. degradation) are coordinated by the Hsp/c70 system. This question has been difficult to answer unambiguously in cells, first, because protein synthesis, folding, and degradation are normally tightly coupled events and, second, because chaperone manipulation in cells is subject to robust homeostatic feedback mechanisms.
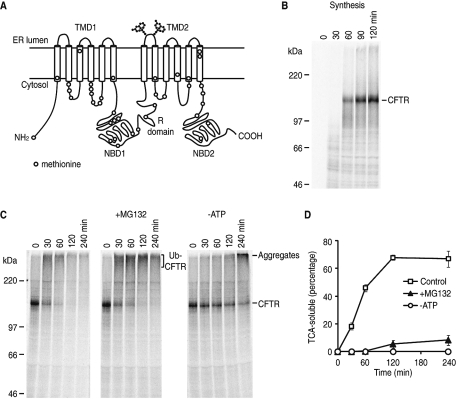
FIGURE 1:
In vitro synthesis and degradation of CFTR. (A) Schematic representation of CFTR showing membrane topology and approximate location of endogenous methionine residues. (B) Phosphorimage of CFTR translated in RRL in the presence of canine pancreas rough microsomes and [35S]Met. Aliquots were taken at indicated times and analyzed by 7% SDS–PAGE and phosphorimaging. Migration of full-length CFTR protein is indicated. (C) Microsomes containing [35S]-labeled CFTR were collected and incubated at 37°C in RRL (minus hemin) in the presence of 100 μM MG132 (+MG132) or after ATP depletion by hexokinase and 2-deoxyglucose (–ATP). Samples were analyzed by SDS–PAGE and phosphorimaging at times indicated. (D) Aliquots of degradation reaction were precipitated by TCA and analyzed by scintillation counting. The percentage of CFTR degraded into TCA-soluble fragments at each point is shown as mean ± SEM (n = 3).
To address this issue, we developed an in vitro, cell-free system that allows one to physically and temporally separate the role of Hsp/c70 on cotranslational folding from its role in ERAD (Xiong et al., 1999; Carlson et al., 2006). Synthesis is carried out in rabbit reticulocyte lysate (RRL) supplemented with ER microsomes under conditions that reconstitute membrane integration, topogenesis, and early folding events (Xiong et al., 1999; Kleizen et al., 2005), but prevent ubiquitination. ER-associated degradation is then reconstituted using microsomes containing newly synthesized, full-length CFTR. In this system, synthesis and degradation can be evaluated in native ER membranes under conditions that mimic the cytosolic environment (60–100 mg/ml protein concentration). At the same time, this system is readily amenable to biochemical manipulation of Hsp/c70 and its cochaperones both during and after protein synthesis independently of its normally tightly regulated homeostatic feedback mechanisms and other essential functions (i.e., mitochondrial import, lysosomal targeting, endocytosis) (Young et al., 2003; Chiang et al., 1989; Eisenberg and Greene, 2007). We now show that the timing of the Hsp/c70 binding cycle plays a dominant and essential role in the posttranslational decision to initiate ubiquitination and substrate degradation.
RESULTS
Reconstitution of CFTR synthesis and ERAD in vitro
To determine the relative role of Hsp/c70 in CFTR folding and degradation, we used a well-characterized, cell-free reticulocyte lysate-based (RRL) system that reconstitutes CFTR biosynthesis, core glycosylation, membrane integration, early folding, and ER-associated degradation (Xiong et al., 1999; Carlson et al., 2005; Matsumura et al., 2011). CFTR transcript was translated in RRL containing ER microsomes and [35S]methionine to generate a full-length, radiolabeled, ∼160 kDa protein (Figure 1B) that is membrane integrated (Carlson et al., 2006) and contains both nonglycosylated and core-glycosylated forms (Xiong et al., 1999). At 24°C, CFTR does not undergo ubiquitination but remains as a stable, full-length protein (Xiong et al., 1999). Moreover, because endogenous mRNA is digested, CFTR is the only major protein synthesized, and it is radiolabeled at 38 methionine residues distributed broadly throughout its sequence (Figure 1A and Carlson et al., 2006). This system therefore allows for convenient isolation of native ER microsomes that contain newly synthesized, full-length CFTR protein. When added to RRL lacking exogenous hemin and incubated at 37ºC, CFTR is rapidly converted into a high molecular weight (HMW) ubiquitinated species (Figure 1C, left panel), and subsequently degraded (Xiong et al., 1999).
Under these conditions ∼70% of radiolabeled CFTR protein is converted into trichloroacetic acid (TCA)-soluble peptide fragments within 2 h (Figure 1D), at which time the degradation activity of RRL declines (Carlson et al., 2006). Importantly, CFTR cleavage is ATP-dependent and inhibited (∼90%) by the proteasome inhibitor MG132 (Figure 1D and Oberdorf et al., 2001). Note that MG132 does not block formation of ubiquitinated CFTR, whereas ATP depletion both stabilized full-length protein (Figure 1C) and prevented degradation (Figure 1D). In the absence of ATP, CFTR aggregates are also observed upon prolonged incubation (240 min) (Xiong et al., 1999; Carlson et al., 2006). These results demonstrate that CFTR synthesis and degradation, two events that normally occur asynchronously in cells, can be temporally separated and independently analyzed.
CBag destabilizes Hsc70–CFTR interactions in vitro
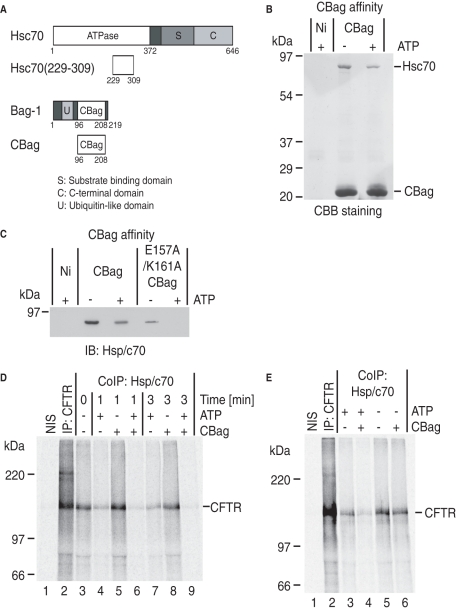
The RRL system provides a unique opportunity to evaluate the effects of Hsp/c70 manipulation in the absence of homeostatic feedback mechanisms normally present in cells. Hsc70 manipulation was accomplished using a C-terminal fragment of Bag-1 (CBag; residues 96–208 aa; Figure 2A and Supplemental Figure S1, A and B). CBag dissociates Hsp/c70, but not Hsp90, from client proteins by binding the ATPase domain of Hsp/c70 and promoting ADP–ATP exchange (Höhfeld and Jentsch, 1997; Takayama et al., 1997; Young and Hartl, 2000; Brehmer et al., 2001; Sondermann et al., 2001; Young et al., 2003). Recombinant CBag also stimulated Hsc70 ATPase activity in vitro when incubated in the presence of Hdj-2 (Supplemental Figure S1D). Immobilized His-tagged CBag also recovered a single primary 70 kDa protein from RRL (Figure 2B) that was identified to be constitutive Hsc70 by immunoblot using anti-Hsp/c70 antisera (Figure 2C). Inducible Hsp70 was undetected by immunoblot, as it is not present in substantial amounts in RRL (Brown et al., 1993). Trypsin digestion and mass spectrometry analysis of the recovered protein confirmed its identity to be Hsc70 (Supplemental Figure S2, A and B). As expected, CBag–Hsc70 interactions were destabilized by ATP, consistent with CBag dissociation after ADP–ATP exchange (Figure 2, B and C, and Takayama et al., 1997). In addition, E157A/K161A CBag, which contains a mutated Hsc70 binding site (Briknarová et al., 2001), exhibits reduced Hsc70 binding that is further decreased by ATP (Figure 2C). From these studies, we conclude that Hsc70 is the predominant CBag target in RRL, and binding to CBag is dependent on its ATPase cycle (Young et al., 2003).
FIGURE 2:
CBag destabilizes Hsc70-CFTR interactions. (A) Schematic representation of Hsc70, Hsc70(229–309), Bag-1, and CBag. The peptide Hsc70(229–309) is the minimal CBag binding subdomain. (B) CBB staining of CBag affinity pull-down of RRL in the presence and absence of ATP. The predominant interacting partner in RRL is Hsc70. (C) Hsc70 immunoblot of wild type and E157A/K161A CBag pull-down from RRL showing ATP-dependent binding to CBag and reduced binding of mutant CBag. (D) Isolated microsomes containing newly synthesized CFTR were incubated with or without CBag and ATP for 1 or 3 min and immediately solubilized in Triton X-100. Samples were immunoprecipitated with nonimmune sera (NIS, lane 1), anti-CFTR antisera (lane 2), or anti-Hsp/c70 antisera (lanes 3–9). CBag + ATP decreased Hsc70 binding (lanes 6 and 9) compared with ATP or CBag alone. (E) Following CFTR synthesis, CBag was added to the translation reaction +/– hexokinase (to deplete ATP) for 10 min, and microsomes were collected, solubilized, and immunoprecipitated with NIS, anti-CFTR antisera (lane 2), or anti-Hsp/c70 antisera. Incubation with CBag reduced Hsc70 binding in an ATP-dependent manner.
Because Hsp/c70 interacts with substrate through iterative binding cycles, an important and often overlooked aspect of function is the amount of time that it remains associated with substrate in a given cycle. To address this difficult question, microsomes containing Hsc70–CFTR complexes were isolated, and newly synthesized CFTR was coimmunoprecipitated with Hsp/c70 antisera following solubilization (Figure 2D, lane 3). Because cytosolic Hsc70 is removed during microsome pelleting, it is not available for rebinding, thus allowing us to measure the duration of Hsc70 association to membrane-bound CFTR during a single release cycle. In the absence of ATP, Hsc70–CFTR complexes were remarkably stable (Figure 2D, lanes 3, 5, and 8) and persisted for several hours after solubilization (Xiong et al., 1999). On ATP addition, some Hsc70 release from CFTR was observed within 1 min, but complexes were still present at 3 min (Figure 2D, lanes 4 and 7). Some Hsc70 release is presumably due to the low intrinsic ADP–ATP exchange activity of substrate-bound Hsc70. In contrast, addition of CBag to isolated microsomes stimulated release of nearly all Hsc70 from CFTR within 3 min (Figure 2D, lanes 3, 6, and 9). The increased rate of ADP–ATP exchange induced by CBag (Supplemental Figure S1D) therefore increases Hsc70 cycling as predicted, and markedly reduces the duration of substrate association. Importantly, CBag plus ATP also reduced CFTR interaction with Hsc70 in RRL, which contains endogenous Hsc70 and other cochaperones, indicating that stimulating substrate release also decreases the steady-state occupancy of CFTR by Hsc70 (Figure 2E, lanes 3–5). Thus CBag provides a means to investigate the physiological consequences of Hsc70–client interactions in a native-like cytosolic environment.
Stable Hsc70–CFTR interactions are required for ERAD
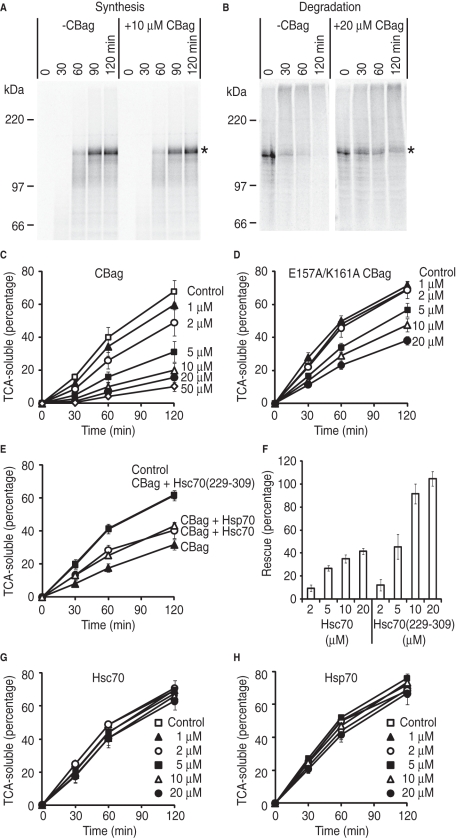
To define the relative contribution of Hsc70 to CFTR maturation, we tested how CBag affected CFTR synthesis and degradation. CBag (approximately eightfold excess over endogenous Hsc70 in the translation reaction) had no discernible effect on CFTR synthesis (Figure 3A), but caused a dose-dependent decrease in CFTR degradation as evidenced by stabilization of full-length protein (Figure 3B and Supplemental Figure S3, A and B) and inhibition of CFTR conversion into TCA-soluble peptides (Figure 3C). E157A/K161A mutant CBag, which exhibits reduced Hsc70 binding activity (Figure 2C and Briknarová et al., 2001), was also less efficient at blocking CFTR degradation (Figure 3D). Average degradation rates during the initial 60 min (expressed as % of CFTR converted into TCA-soluble fragments/min) were 0.67 ± 0.10%/min in the absence of CBag, 0.27 ± 0.06%/min in 5 μM CBag, and 0.56 ± 0.05%/min in 5 μM E157A/K161A mutant CBag. The apparent IC50 values for CFTR degradation were 3.8 μM for wild-type CBag (approximately twofold excess over Hsc70 in the degradation reaction) and >20 μM for mutant CBag (Supplemental Figure 4). Thus the ability of CBag to block CFTR degradation depends on its Hsc70-binding activity.
FIGURE 3:
Destabilization of Hsc70 binding by CBag blocks CFTR degradation. (A) Phosphorimage of CFTR translation reaction performed in the presence or absence of 10 μM CBag. Asterisk shows full-length CFTR. (B) CFTR was synthesized in the absence of CBag and subjected to in vitro degradation in the presence (right) or absence (left) of 20 μM CBag. Asterisk shows full-length CFTR. (C) CFTR was synthesized in the absence of CBag and subjected to in vitro degradation in the presence and absence (control) of indicated CBag concentrations. The y-axis represents the percentage of initial CFTR protein that is converted into TCA-soluble fragments. Data show mean ± SEM (n = 3–7). (D) In vitro degradation assay was carried out as in (C), but with the E157A/K161A mutant CBag. (E) CFTR degradation was carried out in the presence and absence (control) of 5 μM CBag and 20 μM Hsc70, Hsp70, or Hsc70(229–309) as indicated. (F) CFTR degradation was carried out as in (E) but with indicated concentration of Hsc70 or Hsc70(229-309). The y-axis shows the percentage of rescue of CFTR degradation due to CBag inhibition that was restored by addition of Hsc70 or Hsc70(229-309) as described in Materials and Methods. Data show mean ± SEM (n = 3–6). (G) In vitro degradation assay was carried out in the presence and absence (control) of indicated Hsc70 concentrations. (H) In vitro degradation assay was carried out in the presence and absence (control) of indicated Hsp70 concentrations.
Addition of excess recombinant Hsc70 or Hsp70 (20 μM) partially restored CFTR degradation in the presence of 5 μM CBag, whereas degradation was completely restored by an Hsc70 fragment (residues 229–309 aa, Figure 2A) containing the minimal CBag binding subdomain (Brive et al., 2001) (Figure 3E). Additional experiments confirmed that the Hsc70(229–309) fragment was substantially more potent than Hsc70 at restoring CBag-inhibited CFTR degradation (Figure 3F). Importantly, addition of Hsc70, Hsp70, or Hsc70(229–309) alone had minimal effect on CFTR degradation at concentrations between 1 and 20 μM (Figure 3, G and H, and unpublished data). On the basis of these data, we conclude that CBag inhibits CFTR degradation by destabilizing Hsc70 binding to CFTR.
Hsc70 exerts a predominant and essential role in CFTR ERAD
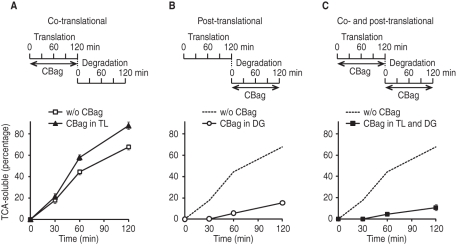
To better understand the co- and posttranslational role(s) of Hsc70, we next carried out CFTR synthesis in the presence of 10 μM CBag. Microsomes were then collected, and degradation was performed in the absence of CBag. Under these conditions, a slight but reproducible increase in CFTR degradation was observed (Figure 4A), indicating that stimulating Hsc70 release during synthesis alters cotranslational folding events such that CFTR acquires a slightly more ERAD-sensitive conformation. Similar results were obtained for ΔF508 CFTR (Supplemental Figure S5B), which is consistent with previous studies showing that wild type and ΔF508 CFTR are degraded in a similar manner in RRL (Oberdorf et al., 2005, and Supplemental Figure 5A). In contrast, regardless of whether Hsc70 binding was perturbed during synthesis, posttranslational addition of CBag strongly inhibited CFTR degradation (Figure 4, B and C). These results indicate that Hsc70 promotes CFTR folding cotranslationally, but exerts a dominant effect after synthesis is complete when it functions to target newly synthesized protein into the ubiquitin–proteasome pathway. In addition, the marked stabilization of CFTR protein by SDS–PAGE and near complete block in TCA fragment generation suggest that Hsc70 plays an essential role in CFTR degradation.
FIGURE 4:
Hsc70 binding cycle has a dominant and essential role in CFTR degradation. (A) CFTR was synthesized in the presence of 10 μM CBag and subjected to an in vitro degradation assay without CBag. The percentage of CFTR degraded into TCA-soluble fragments at each point is shown. (B) CFTR was synthesized in the absence of CBag and subjected to an in vitro degradation assay with 20 μM CBag. (C) CFTR was synthesized in the presence of 10 μM CBag and subjected to an in vitro degradation assay in the presence of 20 μM CBag. In (B) and (C), result without CBag in translation and degradation (from A) is shown as broken line for comparison. Data show mean ± SEM (n = 4–5).
FIGURE 5:
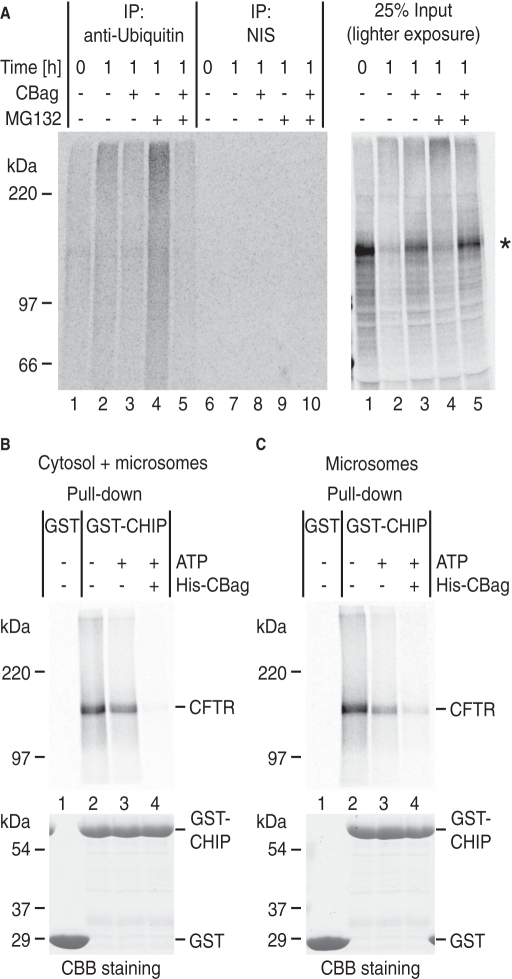
Hsc70 binding is required for CFTR ubiquitination. (A) Microsomes containing newly synthesized CFTR were subjected to an in vitro degradation assay in the presence of 20 μM CBag and 100 μM MG132 as indicated. At T = 1 h, aliquots were immunoprecipitated with anti-(mono-/poly-) ubiquitin antibody (FK2) or NIS and analyzed by SDS–PAGE and phosphorimaging. Right gel shows 25% input of sample before immunoprecipitation. (B) Following CFTR synthesis, hexokinase (lanes 1 and 2), buffer (lane 3), or His-CBag (lane 4) was incubated with translation reaction for 10 min. CFTR was then solubilized and pulled down with immobilized GST (lane 1) or GST-CHIP (lanes 2–4). (C) Isolated microsomes containing newly synthesized CFTR were incubated with or without CBag and ATP for 1 min to release Hsc70 and immediately solubilized in Triton X-100. Samples were incubated with immobilized GST(lane 1) or GST-CHIP (lanes 2–4). Bottom gels (B and C) show CBB staining of GST proteins.
The Hsc70 binding cycle is required for CFTR ubiquitination
ERAD of polytopic membrane proteins is a multistep process that requires ubiquitination, dislocation from the ER membrane, unfolding, delivery to the proteasome, and proteolytic cleavage. Because one function of Hsc70 is to recruit the E3 ligase CHIP, we reasoned that rapid release of Hsc70 from substrate might potentially block the initial step of ubiquitination. During the in vitro degradation reaction, polyubiquitinated CFTR was visualized following immunoprecipitation with anti-ubiquitin antibody, and this intermediate was further stabilized by the proteasome inhibitor MG132 (Figure 5A, lanes 1, 2, and 4). Smaller ubiquitinated CFTR fragments were also observed which result from rare cleavage events by residual proteasome activity even in the presence of MG132 (Carlson et al., 2006, and Supplemental Figure 6). Consistent with our prediction, addition of CBag strongly decreased ubiquitinated CFTR (Figure 5A, lanes 3 and 5), confirming that functional Hsc70 interactions are required for substrate ubiquitination.
Our results suggest that the timing of Hsc70 interactions is important for recruitment of one or more ubiquitin E3 ligase(s). We therefore examined whether the presence of CBag decreased the amount of CHIP associated with newly synthesized CFTR. CBag or hexokinase (to deplete ATP) was added to the translation reaction after CFTR synthesis, and microsomes were collected, solubilized, and incubated with immobilized glutathione S-transferase (GST)-tagged CHIP (Figure 5B). As expected, CFTR pull-down was increased upon ATP depletion, whereas incubation with CBag (to remove Hsc70) nearly completely blocked this interaction (Figure 5B, lanes 2–4). Similarly, when isolated microsomes containing radiolabeled CFTR were incubated with CBag and ATP for just 1 min (as in Figure 2D), CFTR pull-down by immobilized CHIP was markedly reduced (Figure 5C). In vitro binding experiments further showed that CBag did not have a direct effect on CHIP interaction with Hsc70 (Supplemental Figure 7). Thus we conclude that, by releasing Hsc70 from CFTR, CBag reduces the amount of CFTR available for CHIP. Taken together, these results indicate that the ability to recruit ubiquitination machinery is determined by the duration of Hsc70 binding cycle.
Hsc70 binding is required for CFTR ER dislocation
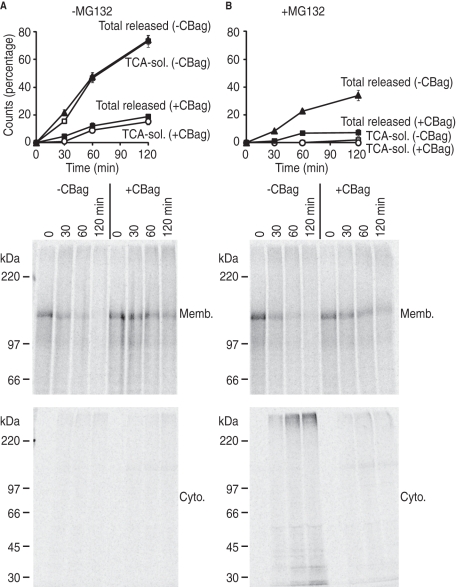
Under normal physiological conditions, CFTR dislocation and proteolysis are tightly coupled, and CFTR is released from the ER membrane as TCA-soluble fragments (Xiong et al., 1999; Oberdorf et al., 2006), because proteolytic capacity of the proteasome exceeds the ability of AAA-ATPases, p97 (VCP/CDC48) and the 19S proteasome regulatory subunit, to unfold and extract CFTR from the lipid bilayer (Ye et al., 2001; Lee et al., 2004; Carlson et al., 2006; Oberdorf et al., 2006; Matsumura and Skach, 2009). Inhibition of β-subunit activity, however, uncouples extraction from proteolysis and allows one to measure retrotranslocation independently of peptide cleavage. Because ubiquitination is generally believed to provide the signal for p97 and/or 19S recruitment, we tested whether stimulating Hsc70 release from CFTR also prevented dislocation from the membrane. Microsomes containing newly synthesized CFTR were incubated in RRL in the presence or absence of CBag, and proteolytic activity of the proteasome was inhibited by MG132. Reactions were then separated into pellet (membrane) and supernatant (cytosolic) fractions to monitor the rate at which CFTR is released from the ER. In the absence of MG132, the total amount of cytosolic (dislocated) CFTR was indistinguishable from the TCA-soluble counts (Figure 6A, graph), indicating that CFTR is released from the membranes solely as small peptide fragments (Oberdorf et al., 2006). Indeed, no protein bands were detected in the cytosolic fraction by SDS–PAGE (Figure 6A, bottom gel). Addition of CBag dramatically decreased release of CFTR into the cytosol (Figure 6A, graph) consistent with impaired degradation, although a small number of cytosolic counts were still present as TCA-soluble fragments.
FIGURE 6:
Hsc70 is required for CFTR dislocation. (A) Microsomes containing newly synthesized CFTR were subjected to in vitro degradation in the presence or absence of CBag. At indicated times, aliquots were separated into pellet (membrane) and supernatant (cytosol) fractions. Graph shows the total amount of CFTR released into the cytosol compared with percentage of TCA-soluble CFTR at each time point (mean ± SEM, n = 4). Bottom panels show phosphorimage of membrane and cytosol fractions. (B) CFTR degradation was carried out as in (A), but in the presence of both MG132 and CBag. Graph shows TCA-soluble and total CFTR released from membranes (mean ± SEM, n = 4), whereas gels show SDS–PAGE and phosphorimaging. In the presence of MG132, CFTR is released from membranes as both a HMW species and partially degraded fragments. In the presence of CBag, both of these forms were barely detected.
In the presence of MG132, nearly all CFTR released into the cytosol was in the form of TCA-insoluble fragments (Figure 6B, graph) that were visualized as both HMW (ubiquitinated) species and degradation intermediates (Figure 6B, bottom gel). Under these conditions, CBag again decreased CFTR release into the cytosol (Figure 6B, graph) and prevented the appearance of both the ubiquitinated and partially degraded fragments (Figure 6B, bottom gel). Thus stimulating release of Hsc70 from CFTR by CBag not only prevents CFTR ubiquitination, but also prevents dislocation and release from the ER membrane, even in the absence of proteolytic cleavage.
Molar excess of CBag is required to destabilize Hsp/c70 binding and inhibit CFTR degradation
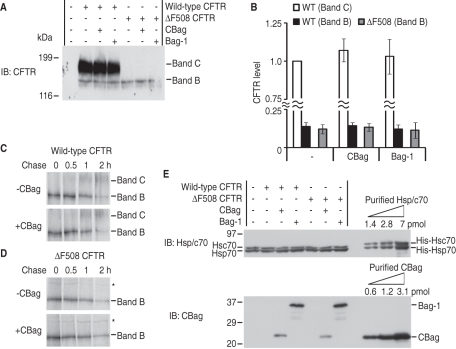
In contrast to our results, those of prior studies have shown that overexpression of full-length Bag-1 does not rescue CFTR expression in human embryonic kidney (HEK) 293 cells (Dai et al., 2005). One possibility is that the N-terminal ubiquitin-like domain of Bag-1 (Figure 1A) might have prodegradative properties (Demand et al., 2001) that counteract stabilizing properties of the C-terminal fragment. Alternatively, differences might exist between intact cells and our in vitro system. We therefore cotransfected wild type and ΔF508 CFTR with CBag or Bag-1 into HEK293 cells. For wild-type CFTR, steady-state levels of band B (ER form) and band C (post-Golgi form) CFTR were unchanged by either CBag or Bag-1 overexpression (Figure 7, A and B). For ΔF508 CFTR, band B remained unchanged and band C was not detected. Pulse-chase experiments also showed that CBag overexpression did not affect CFTR synthesis (Supplemental Figure S8) or the efficiency of CFTR processing from band B to band C in wild type (Figure 7C and Supplemental Figure S8A) or ΔF508 CFTR (Figure 7D and Supplemental Figure S8B). Apparent stability for the band B form of ΔF508 CFTR was also unchanged (T1/2 value, 50 vs. 53 min; Supplemental Figure S8). Thus the inhibitory effect of CBag on CFTR degradation is not simply due to the absence of its ubiquitin-like domain.
FIGURE 7:
Relative stoichiometry of Hsp/c70 and CBag determines CFTR stability. (A) Wild-type or ΔF508 CFTR was cotransfected with CBag or Bag-1 into HEK293 cells. Forty-eight hours after transfection, cells were harvested, and lysates were analyzed by immunoblotting with anti-CFTR antibody. Immature ER (Band B) and Golgi-processed (Band C) forms are indicated. (B) CFTR levels were quantitated by densitometry and are presented as mean ± SEM (n = 5). (C and D) Wild type (C) and ΔF508 (D) CFTR were transfected +/– CBag in HEK293 cells. Twenty-four hours after transfection, cells were pulsed with [35S]Met/Cys for 15 min, and chased for indicated times as described in Materials and Methods. Whole-cell lysate was immunoprecipitated with anti-CFTR antisera and analyzed by SDS–PAGE and phosphorimaging. Asterisk denotes a background band. (E) Immunnoblot of Hsc70, Hsp70, Bag-1, and CBag in whole-cell lysate (25 μg) from transfected cells as in (A). Purified His-tagged proteins used to quantitate results are shown in the right three lanes of each gel. Ratio of total endogenous Hsp/c70 to transfected CBag and Bag-1 are estimated to be 1:0.04 and 1:0.13, respectively (see Table 1).
Why then does CBag inhibit CFTR degradation in vitro? One advantage of cell-free systems is that the stoichiometry of CBag and Hsp/c70 can be precisely controlled, whereas this is much more difficult in cells. Quantification of endogenous Hsp/c70 in HEK293 cells (based on purified His-tagged proteins) revealed cellular expression levels of ∼1.5 pmol and ∼3 pmol/25 μg total protein, or ∼12 and ∼24 μM, respectively (see Table 1). Overexpression of CBag and Bag-1 achieved cellular concentrations of 1.6 and 4.8 μM, respectively, and a ratio of Hsp/c70 to CBag and Bag-1 of 1:0.04 and 1:0.13 (Figure 7E and Table 1). Although the actual CBag and Bag-1 levels are slightly higher when corrected for transfection efficiency (80–90%), both proteins would still be present in substoichiometric amounts relative to Hsp/c70. Importantly, CBag and Bag-1 overexpression had no detectable effect on Hsc70 and Hsp70 levels (Figure 7E) and did not appear to induce a stress response based on Hsp90β, BiP, or Hdj-2 (Supplemental Figure S9). When compared with in vitro conditions, a ratio of Hsp/c70 to CBag of 1:2.5, 1:5, and 1:10 yielded twofold, fourfold, and fivefold increases in CFTR T1/2, respectively, and no effect was observed for ratios of 1:1 or lower (Supplemental Figure 3 and Table 1). Similar results were obtained for CBag overexpression in COSm6 cells, which predominantly express Hsc70 (12 μM, Table 1). Thus enhancement of CFTR stability strongly depends on the stoichiometry, and in vitro results actually agree well with those obtained in cells in which it is exceedingly difficult, if not impossible, to achieve CBag levels that exceed endogenous Hsp/c70.
TABLE 1:
Relationship between stoichiometry of Hsp/c70 to CBag and stability of full-length CFTR.
| Hsc70 (μM) | Hsp70 (μM) | Total Hsp/c70 (μM) | Exogenous CBag or Bag-1 (μM) | Total Hsp/c70 : Exogenous CBag or Bag-1 | T1/2 of full-length CFTR (min) | |
|---|---|---|---|---|---|---|
| RRL | 2 | n.d. | 2 | – | – | 25 |
| RRL + 2 μM CBag | 2 | n.d. | 2 | 2 | 1 : 1 | 34 |
| RRL + 5 μM CBag | 2 | n.d. | 2 | 5 | 1 : 2.5 | 55 |
| RRL + 10 μM CBag | 2 | n.d. | 2 | 10 | 1 : 5 | 94 |
| RRL + 20 μM CBag | 2 | n.d. | 2 | 20 | 1 : 10 | 116 |
| HEK293 | 12 | 24 | 36 | - | – | 50 |
| HEK293 + CBag | 12 | 24 | 36 | 1.6 | 1 : 0.04 | 53 |
| HEK293 + Bag-1 | 12 | 24 | 36 | 4.8 | 1 : 0.13 | n.e. |
| COSm6 | 12 | n.d. | 12 | - | n.e. | |
| COSm6 + CBag | 12 | n.d. | 12 | 1.6 | 1 : 0.13 | n.e. |
T1/2 values in RRL are from stability of full-length, wild-type CFTR in in vitro degradation assays (Supplemental Figure 3), whereas those in HEK293 cells are from stability of band B form of ΔF508 CFTR in pulse-chase assays (Supplemental Figure 8). n.d., not detected; n.e., not examined.
DISCUSSION
In this study, we used CBag, a nucleotide exchange factor, to selectively destabilize co- and posttranslational interactions between Hsc70 and a known client substrate CFTR. This strategy enabled us to distinguish the role of Hsc70 in CFTR maturation during and immediately following synthesis, from its role in posttranslational processing and ERAD. We show that destabilizing Hsc70–client binding during synthesis had no effect on CFTR translation but resulted in a protein product that was more susceptible to degradation. This finding supports those of previous studies that Hsc70 assists de novo folding of CFTR in the cellular environment (Meacham et al., 1999; Younger et al., 2004; Grove et al., 2011). The profolding effect we observed, however, was modest, possibly because the major population of CFTR is already subject to degradation (Ward and Kopito, 1994), or alternatively because Hsc70-mediated folding is relatively insensitive to the duration of the Hsc70–CFTR binding cycle. In contrast, selective destabilization of posttranslational Hsc70 binding almost completely inhibited CFTR degradation. This inhibition was manifest by a marked decrease in the rate and extent of CFTR ubiquitination, dislocation from the ER membrane, and proteasome-mediated cleavage. Taken together, these results reveal that Hsc70 plays a dominant role in targeting CFTR to the ERAD pathway.
Studies in vitro, in mammalian cells, and in yeast have shown that Hsp/c70 promotes CFTR (re)folding and degradation (Strickland et al., 1997; Meacham et al., 1999 , 2001; Zhang et al., 2001; Younger et al., 2004; Grove et al., 2011). It has been difficult, however, to determine the extent of profolding and prodegradative functions of Hsp/c70 because both processes occur simultaneously in cells, and because manipulating Hsp/c70 function induces homeostatic feedback regulation of chaperone and cochaperone pools (Okiyoneda et al., 2010). The cell-free system used here overcomes these limitations. RRL lacks endogenous transcriptional mechanisms but maintains a native-like proteostatic environment with a complete complement of biosynthetic machinery (Nimmesgern and Hartl, 1993; Frydman et al., 1994) and a robust ubiquitin–proteasome system that reflects the terminally differentiated state of reticulocytes (Xiong et al., 1999; Oberdorf and Skach, 2002; Matsumura and Skach, 2009). Thus it provides the unique ability to 1) manipulate (co)chaperones without inducing compensatory feedback mechanisms; 2) analyze stoichiometry of (co)chaperones; 3) monitor specific steps of membrane integration, ubiquitination, dislocation, and proteolysis; and 4) directly evaluate substrate fate by both SDS–PAGE and conversion to TCA-soluble peptides (Frydman et al., 1994; Terada et al., 1997; Carlson et al., 2006; Matsumura and Skach, 2009). Most importantly, our conditions allow the effects of chaperone function during synthesis to be analyzed independently of degradation processes. Hsc70 is the major Hsp70 family member present in RRL and the predominant CBag binding partner. The striking inhibitory effect of CBag on CFTR degradation, the loss of inhibition by a mutant with impaired Hsc70 binding, and restoration of degradation by Hsc70 or the Hsc70(229-309) fragment confirm that CBag effects on CFTR stability are mediated via its action on Hsc70.
Although Hsc70 and Hsp70 are generally considered to have similar functions in cells, a recent study showed that Hsc70 overexpression increased turnover of ENaC channels at the plasma membrane, whereas Hsp70 overexpression increased surface expression, possibly by improving trafficking (Goldfarb et al., 2006). RRL contains ∼2 μM of endogenous Hsc70 but undetectable Hsp70. Interestingly, addition of recombinant Hsc70 or Hsp70 had minimal effect on CFTR degradation at concentrations between 1 and 20 μM (Figure 3, G and H). Furthermore, addition of recombinant Hsp70 partially restored CBag inhibition of CFTR degradation to a similar extent as Hsc70 (Figure 3E). Therefore, differential effects of Hsc70 and Hsp70 were not observed here for CFTR, suggesting that they may be either substrate specific or dependent on levels of other (co)chaperones.
Based on stimulation of Hsc70 ATPase activity and rapid release of Hsc70 from CFTR (<1–3 min), it is likely that CBag acts by increasing the off-rate for Hsc70 binding (Sondermann et al., 2001). Such an effect should increase Hsc70 cycling and thereby decrease the time that a client remains bound during a given ATP binding cycle. When this occurs during synthesis, the effect on cotranslational folding is minor, and CFTR is rendered only slightly more susceptible to degradation. Inhibition of posttranslational Hsc70 binding, however, dramatically stabilizes newly synthesized substrate. This finding suggests that the timing of Hsc70 association, rather than binding per se, may be the most important determinant of degradation. Given that ubiquitination is directly inhibited by CBag, our results indicate that the ability to recruit ubiquitination machinery is determined by the duration of Hsp/c70 binding to unfolded substrates. When the Hsp/c70–CFTR complex cycles slowly, ubiquitination machineries have adequate time to carry out ubiquitination. It is notable that the Hsc70 cochaperone CHIP also slows ATP hydrolysis by Hsp/c70 and stabilizes Hsp/c70–client interaction, which may further favor ubiquitination (Ballinger et al., 1999; Meacham et al., 2001; Rosser et al., 2007). In contrast, rapid dissociation of the Hsp/c70–CFTR complex prevents this process. In our hands, CHIP failed to recognize CFTR after the Hsc70–CFTR complex was dissociated in the presence of CBag (Figure 5, B and C). Although it is exceedingly difficult to determine precisely how long Hsc70 remains bound to client, we observed that newly formed Hsc70–CFTR complexes in isolated microsomes remained detectable for at least 3 min upon addition of physiological ATP concentration (coimmunoprecipitation, Figure 2D) but were nearly completely abolished within 1–3 min in the presence of CBag plus ATP. A crude approximation of time needed for Hsc70 to initiate CFTR ubiquitination would therefore be on the order of a few minutes, although it is likely that many factors could influence this process.
The finding that Hsc70 is essential for CFTR ubiquitination has important mechanistic implications for ERAD. At least three ubiquitin E3 ligases – CHIP, RMA1, and Nedd4-2 – and one E4 ligase (gp78) have been shown to mediate CFTR ubiquitination (Meacham et al., 2001; Younger et al., 2006; Morito et al., 2008; Caohuy et al., 2009). CHIP is a cytosolic U-box protein that forms a ternary complex with Hsc70–CFTR through its TPR motif, and is proposed to act in concert with the E2 enzyme, UbcH5 (Meacham et al., 2001; Younger et al., 2004). In contrast, RMA1 is a RING domain ER protein proposed to recognize misfolded CFTR transmembrane domains (Younger et al., 2006). Interestingly, RMA1 was recently found to be associated with Hsc70–CFTR (Grove et al., 2011). Hsp70 also recruits substrate to a different transmembrane E3 ligase (Doa10p) in yeast (Nakatsukasa et al., 2008). The precise mechanisms by which Hsp/c70 functions to bridge clients and diverse ubiquitination machineries at the mammalian ER therefore remains an important question.
In eukaryotic cells, homeostasis of newly synthesized proteins is maintained by the balance between chaperone/cochaperone folding machinery and the ubiquitin proteasome system. This proteostatic environment, and hence the fate of unstable client proteins, is tightly regulated by cell type, differentiation state, signaling pathways, stress, aging, and so forth (Balch et al., 2008; Mu et al., 2008; Hutt et al., 2009). To date, exploitation of the Hsp/c70 axis (including Hsp40/DnaJ proteins and nucleotide exchange factors) as a means to overcome defective CFTR folding has met with limited success (Alberti et al., 2004; Youker et al., 2004; Arndt et al., 2005; Ahner et al., 2007). For example, overexpression of cysteine string protein (Hsp40 homolog) (Zhang et al., 2006) or Bag-2 (Dai et al., 2005) slightly increased steady-state levels of ER forms of wild type or ΔF508 CFTR but had relatively little effect on the efficiency of CFTR trafficking. Similarly, inhibition of Hsc70 with the small molecule, deoxyspergualin, increased CFTR chloride channel activity but failed to achieve a biochemically detectable correction (Jiang et al., 1998). High concentrations of sodium 4-phenylbutyrate, which inhibits histone deacetylases (Chen et al., 2003), down-regulates Hsc70 (Rubenstein and Zeitlin, 2000; Rubenstein and Lyons, 2001) and possibly up-regulates Hsp70 (Choo-Kang and Zeitlin, 2001) but minimally restores CFTR function in cells and cystic fibrosis patients (Rubenstein et al., 1997; Rubenstein and Zeitlin, 1998). One difficulty encountered by these approaches is that Hsp/c70 functions are influenced by cochaperone levels, which may explain, in part, the more robust CFTR correction observed for the histone deacetylase inhibitor, suberoylanilide hydroxamic acid (Hutt et al., 2010). Because knockdown of CHIP ubiquitin ligase in cells increases both immature and mature CFTR (Grove et al., 2009), an attractive strategy might be to target Hsp/c70 interactions that specifically couple to ubiquitination machinery. Such compounds could specifically inhibit the prodegradative role of Hsp/c70 without interfering with its profolding role and could be used to restore CFTR function in patients.
MATERIALS AND METHODS
Plasmids
Mammalian expression plasmids pc-CFTR and pc-CFTRΔF508 were generated using pSPCFTR (Xiong et al., 1997; Carlson et al., 2006) by two-step ligation of a ∼2.2 kb HindIII–EcoRI CFTR 5′-cDNA fragment and a ∼4.1 kb XbaI–XbaI 3′-cDNA fragment into pcDNA3. Rat Hsc70 cDNA (provided by Jason C. Young; Tzankov et al., 2008) was subcloned into the NdeI and BamHI sites of pET15b to obtain pET15b–Hsc70. The Hsc70 cDNA fragment encoding amino acids 229–309 was amplified by PCR and ligated into the NcoI and XhoI sites of pET30 to obtain pET30–Hsc70(229–309). E157A/K161A mutant CBag plasmid was generated from pProEx-CBag (encoding residues 96–208 of human Bag-1, provided by Jason C. Young; Bhangoo et al., 2007) by PCR overlap extension using complementary sense and antisense oligonucleotides. Mouse Bag-1 cDNA from plasmid pcI-FLAG–Bag-1 (provided by Shinichi Takayama; Takayama et al., 1997) was amplified by PCR and ligated into the XhoI site of pET15b to obtain pET15b–Bag-1. A HindIII fragment of His6-tagged CBag cDNA was generated by PCR and ligated into the HindIII site of pcDNA3 to obtain pc-CBag. Human CHIP cDNA, provided by D.M. Cyr (Younger et al., 2004), and Hsc70 cDNA were subcloned into the BamHI and XhoI site of pGEX4T.1 to obtain pGEX4T.1–CHIP and pGEX4T.1–Hsc70, respectively. All PCR-amplified sequences were confirmed by DNA sequencing. Detailed cloning procedures are available upon request.
In vitro transcription and translation
Noncapped CFTR transcript was synthesized in 80 mM HEPES-NaOH (pH 7.5); 16 mM MgCl2; 2 mM spermidine; 3 mM each ATP, CTP, UTP, and GTP; 40 mM dithiothreitol (DTT); 0.2 U/μl RNase inhibitor; 5 μg/ml His-tagged SP6 RNA polymerase; and 50 ng/μl nonlinearized pSP-CFTR plasmid at 40°C for 2 h. After transcription, a 60% volume of 7.5 M LiCl and 50 mM EDTA was added, incubated for 1 h at –20°C, and centrifuged at 16,000 × g at 4°C for 20 min. The RNA pellet was rinsed three times with 70% ethanol and once with 95% ethanol followed by centrifugation at 16,000× g at 4°C for 1 min and dissolved into ddH2O (Matsumura et al., 2011).
CFTR protein was translated at 24°C for 2 h in a reaction containing 50 ng/μl CFTR transcript, 40% (vol/vol) nuclease-treated RRL, canine pancreas microsomes (3–4 A280), 1 mM ATP, 1 mM GTP, 12 mM creatine phosphate, 40 μM each 19 essential amino acids (except methionine), 1 μCi/ml Trans[35S]-label (MP Biomedicals, Solon, OH), 40 μg/ml creatine kinase, 0.1 mg/ml bovine calf tRNA, 0.1 U/ml RNase inhibitor, 10 mM Tris-acetate (pH 7.5), 100 mM potassium acetate, 2 mM magnesium acetate, 0.15 mM spermidine, and 2 mM DTT as described (Matsumura et al., 2011). Microsomes containing newly synthesized CFTR were pelleted by centrifugation at 180,000× g for 10 min, through 0.5 M sucrose in buffer A (50 mM HEPES-NaOH, pH 7.5, 100 mM KCl, 5 mM MgCl2, and 1 mM DTT). The membrane pellet was washed once with 0.1 M sucrose in buffer A without resuspension, and resuspended in the same buffer at half volume of original translation reaction.
In vitro degradation and dislocation assay
Microsomal membranes containing newly synthesized radiolabeled CFTR were added to RRL (15–18% and 60–70% [vol/vol] in final concentration, respectively) supplemented with 1 mM ATP, 12 mM creatine phosphate, 5 mM MgCl2, 3 mM DTT, 10 mM Tris-HCl (pH 7.5), and 80 μg/ml creatine kinase, and incubated at 37°C as described previously (Carlson et al., 2005). Purified recombinant proteins were added at the concentration indicated. At indicated times, aliquots were precipitated in 20% (wt/vol) TCA and centrifuged at 16,000 × g for 10 min. Supernatants (TCA soluble) were added to ScintiSafe scintillation fluid (Fisher Scientific, Pittsburgh, PA) and counted in a Beckmann LS6500 scintillation counter. Total incorporated [35S] in each sample was determined by directly counting an aliquot of the degradation reaction. Mock reactions were used as a control to correct for nonspecific association of [35S] and translation of any remnant mRNAs that survived nuclease treatment during lysate preparation. The percentage of protein degraded into TCA-soluble peptide fragments at each time point was determined using the following formula:
where Tn and T0 were TCA-soluble counts at T = n and T = 0 min, respectively.
Percent rescue was defined as the restoration of degradation (at T = 60 min) observed upon addition of Hsc70 or Hsc70(229–339) to degradation reactions containing CBag protein using the following formula:
where %TCAsol is control reaction without CBag, %TCAsolCBag is reaction with CBag, and %TCAsolCBag/Hsc70 is reaction with CBag and Hsc70 or Hsc70(229–339). Values are presented as mean ± SEM of three or more experiments.
For the dislocation assay, microsomes were pelleted at each time point by centrifugation at 180,000 × g for 10 min through 0.5 M sucrose in buffer A. Supernatant was then either counted directly (total CFTR released from membrane), counted after precipitation in 20% TCA (TCA-soluble counts released), or analyzed by SDS–PAGE and phosphorimaging. The percentage of total CFTR protein released from the membrane and the percentage of TCA-soluble counts released from the membrane at each time point were calculated as described earlier in text. Gels were placed onto phosphor screens and analyzed with a Personal FX PhosphorImager and QuantityOne software (Bio-Rad, Hercules, CA).
Cell culture experiments
HEK293 and COSm6 cells were grown at 37°C under 5% CO2 in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum and penicillin/streptomycin as described previously (Matsumura et al., 2008; Pratt et al., 2009). For transient transfection, HEK293 cells (3 × 105) were seeded into 35-mm dishes or six-well plates and, after 24 h, were cotransfected with 1.25 μg of pcDNA3, pc-CBag, or pcI-FLAG-Bag-1 and 1.25 μg of pc-CFTR or pc-CFTR-ΔF508 by calcium-phosphate precipitation (Kingston et al., 2003). For transfection into COSm6 cells, TransIT-LT transfection reagent (Mirus, Madison, WI) was used as described (Buck and Skach, 2005). Transfected cells were cultured for 48 h, harvested, flash frozen, and stored at −80°C until immunoblot analysis. For pulse-chase experiments, transfected cells were cultured for 24 h in six-well plates, placed in cysteine and methionine-free medium for 30 min, pulsed with 100 μCi/well of Trans35S-label for 15 min, and chased for 0–2 h in fresh medium with cysteine and methionine, harvested, flash frozen, and stored at −80°C until analysis.
Immunoblot, immunoprecipitation, and His and GST protein pull-down assay
Cells were lysed in 500 μl of buffer B (20 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% [vol/vol] Triton X-100, 0.1% [wt/vol] SDS, 0.5% [wt/vol] sodium deoxycholate) containing complete protease inhibitor (PI) mixture (Roche Applied Science, Indianapolis, IN) (Matsumura et al., 2006). After clarification of lysate at 16,000 × g at 4°C for 20 min, aliquots (25 or 50 μg of protein) were separated on an SDS–PAGE gel and transferred to a polyvinylidene fluoride membrane (Bio-Rad Laboratories). Membranes were blocked with 5% (wt/vol) skim milk in TTBS (20 mM Tris-HCl, pH 7.5, 137 mM NaCl, and 0.05% [vol/vol] Tween 20) for 1 h and incubated with the following antibodies/sera overnight: 1) rat monoclonal anti-CFTR antibody 3G11 (provided by CFTR Folding Consortium; 2) mouse anti-Hsp/c70 antibody (N-27; gift of William J. Welch; Brown et al., 1993), which recognizes both Hsp70 and Hsc70, 1:5000; 3) rabbit anti-CBag antisera (gift of F. Ulrich Hartl; Gamerdinger et al., 2009), 1:5000; 4) rabbit anti-Hsp90β antibody (gift of Ineke Braakman After being washed five times with TTBS, membranes were incubated for 1 h with the following secondary antibodies: 1) goat anti–rat immunogloblin G (IgG) horseradish peroxidase conjugate (HRP; Sigma), 1:40,000; 2) goat anti–mouse IgG-HRP (BioRad), 1:5000; or 3) goat anti–rabbit IgG-HRP (BioRad), 1:10,000. After five washes with TTBS, signals were visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Waltham, MA) and Fuji Film (Light Labs, Dallas, TX). For CFTR detection, SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) was used.
For immunoprecipitation, cells and microsomes were lysed with 500 or 250 μl of buffer B containing complete PI as described (Matsumura et al., 2008). For coimmunoprecipitation, SDS and sodium deoxycholate were omitted from buffer B. For the ubiquitination assay, 10 μl of degradation reaction was directly lysed in 250 μl of buffer B containing complete PI. After clarification of lysate, the following antibodies/sera (1 μl of each) were added: 1) rabbit anti-CFTR antisera directed against amino acids 45–65 (Xiong et al., 1999): 2) rabbit anti-Hsp/c70 antisera (gift of William J. Welch; Brown et al., 1993); or 3) mouse anti-(mono-/poly-)ubiquitin antibody (FK2; Biomol International, Plymouth Meeting, PA). Samples were rotated for 1 h at 4°C, and 5 μl of Affi-Gel Protein A (BioRad) was added and rotated overnight. For mouse anti-ubiquitin antibody, ImmunoPure-immobilized Protein G (Pierce Biotechnology, Rockford, IL) was used. Samples were washed five times with buffer B and three times with Tris-buffered saline (TBS; 20 mM Tris-HCl, pH 7.5, and 137 mM NaCl) and eluted with SDS sample buffer. Eluates were separated on an SDS–PAGE gel and analyzed by phosphorimaging as described earlier in text.
For His affinity copurification, 50 μg of His-CBag was loaded on 10 μl of Ni-NTA beads (Qiagen, Valencia, CA) in TNB buffer (25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM β-mercaptoethanol) and rotated at 4°C for 1 h (Carlson et al., 2005). After washing beads six times with 100 μl of TNB buffer, 50 μl of desalted RRL using PD-10 Desalting Column (GE Healthcare Biosciences, Pittsburgh, PA; pre- equilibrated in 10 mM HEPES-NaOH, pH 7.5) was added to the beads in TNB buffer containing 5 mM MgCl2 and mixed in the presence or absence of 3 mM ATP at 4°C for 4 h. Beads were washed six times with TNB buffer containing 20 mM imidazole and eluted with TNB buffer containing 500 mM imidazole. Eluates were separated on an SDS–PAGE gel and subjected to Coomassie Brilliant blue (CBB) staining or immunoblotting.
For GST pull-down, 50 μg of GST or GST-CHIP was loaded on 10 μl of glutathione resin (BD Biosciences, San Jose, CA) in 0.1% (vol/vol) Triton X-100 in TBS and rotated at 4°C for 1 h. After four washings with 0.1% (vol/vol) Triton X-100 in TBS, solubilized microsomes were added to the beads and rotated at 4°C for 1 h. Beads were washed five times with buffer B (without SDS and sodium deoxycholate) and three times with TBS and eluted with SDS sample buffer. Eluates were separated on an SDS–PAGE gel and analyzed by phosphorimaging and CBB staining.
Supplementary Material
Acknowledgments
We thank Jason C. Young for CBag, Hdj-2, and Hsc70 plasmids; Shinichi Takayama for Bag-1 plasmids; Douglas M. Cyr for CHIP plasmid; F. Ulrich Hartl for anti-CBag antisera; William J. Welch for anti-Hsp/c70 antibody/sera; Ineke Braakman for anti-Hsp90β antibody; the CFTR Folding Consortium for 3G11 anti-CFTR antibody; Zhongying Yang for technical assistance; and other members of the Skach laboratory for their helpful discussions. This work was supported by National Institutes of Health grants DK51818 and GM53457 (to W.R.S.), the American Cystic Fibrosis Foundation (to W.R.S.), the American Heart Association Grant in Aid #0755832Z (to W.R.S.), and the Manpei Suzuki Diabetes Foundation (to Y.M.).
Abbreviations used:
- CBag
C-terminal domain of Bag-1
- CBB
Coomassie Brilliant blue
- CFTR
cystic fibrosis transmembrane conductance regulator
- CHIP
C-termimus of Hsp/c70-interacting protein
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- HEK
human embryonic kidney
- HMW
high molecular weight
- HRP
horseradish peroxidase conjugate
- NBD
nucleotide binding domain
- PI
protease inhibitor
- RRL
rabbit reticulocyte lysate
- TBS
Tris-buffered saline
- TCA
trichloroacetic acid
- TPR
tetratricopeptide repeat
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0137) on June 22, 2011.
REFERENCES
- Ahner A, Nakatsukasa K, Zhang H, Frizzell RA, Brodsky JL. Small heat-shock proteins selectΔF508-CFTR for endoplasmic reticulum-associated degradation. Mol Biol Cell. 2007;18:806–814. doi: 10.1091/mbc.E06-05-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Bohse K, Arndt V, Schmitz A, Hohfeld J. The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol Biol Cell. 2004;15:4003–4010. doi: 10.1091/mbc.E04-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson C, Fiaux J, Rampelt H, Druffel-Augustin S, Bukau B. Insights into the structural dynamics of the Hsp110-Hsp70 interaction reveal the mechanism for nucleotide exchange activity. Proc Natl Acad Sci USA. 2008;105:16519–16524. doi: 10.1073/pnas.0804187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V, Daniel C, Nastainczyk W, Alberti S, Hohfeld J. BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol Biol Cell. 2005;16:5891–5900. doi: 10.1091/mbc.E05-07-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangoo MK, Tzankov S, Fan AC, Dejgaard K, Thomas DY, Young JC. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol Biol Cell. 2007;18:3414–3428. doi: 10.1091/mbc.E07-01-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimston D, Song J, Winchester D, Takayama S, Reed JC, Morimoto RI. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 1998;17:6871–6878. doi: 10.1093/emboj/17.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehmer D, Rudiger S, Gassler CS, Klostermeier D, Packschies L, Reinstein J, Mayer MP, Bukau B. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol. 2001;8:427–432. doi: 10.1038/87588. [DOI] [PubMed] [Google Scholar]
- Briknarova K, et al. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat Struct Biol. 2001;8:349–352. doi: 10.1038/86236. [DOI] [PubMed] [Google Scholar]
- Brive L, Takayama S, Briknarova K, Homma S, Ishida SK, Reed JC, Ely KR. The carboxyl-terminal lobe of Hsc70 ATPase domain is sufficient for binding to BAG1. Biochem Biophys Res Commun. 2001;289:1099–1105. doi: 10.1006/bbrc.2001.6087. [DOI] [PubMed] [Google Scholar]
- Brown CR, Martin RL, Hansen WJ, Beckmann RP, Welch WJ. The constitutive and stress inducible forms of hsp 70 exhibit functional similarities and interact with one another in an ATP-dependent fashion. J Cell Biol. 1993;120:1101–1112. doi: 10.1083/jcb.120.5.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck TM, Skach WR. Differential stability of biogenesis intermediates reveals a common pathway for aquaporin-1 topological maturation. J Biol Chem. 2005;280:261–269. doi: 10.1074/jbc.M409920200. [DOI] [PubMed] [Google Scholar]
- Caohuy H, Jozwik C, Pollard HB. Rescue ofΔF508-CFTR by the SGK1/Nedd4–2 signaling pathway. J Biol Chem. 2009;284:25241–25253. doi: 10.1074/jbc.M109.035345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson E, Bays N, David L, Skach WR. Reticulocyte lysate as a model system to study endoplasmic reticulum membrane protein degradation. Methods Mol Biol. 2005;301:185–205. doi: 10.1385/1-59259-895-1:185. [DOI] [PubMed] [Google Scholar]
- Carlson EJ, Pitonzo D, Skach WR. p97 functions as an auxiliary factor to facilitate TM domain extraction during CFTR ER-associated degradation. EMBO J. 2006;25:4557–4566. doi: 10.1038/sj.emboj.7601307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chen JS, Faller DV, Spanjaard RA. Short-chain fatty acid inhibitors of histone deacetylases: promising anticancer therapeutics? Curr Cancer Drug Targets. 2003;3:219–236. doi: 10.2174/1568009033481994. [DOI] [PubMed] [Google Scholar]
- Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- Choo-Kang LR, Zeitlin PL. Induction of HSP70 promotes ΔF508 CFTR trafficking. Am J Physiol Lung Cell Mol Physiol. 2001;281:L58–L68. doi: 10.1152/ajplung.2001.281.1.L58. [DOI] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- Dai Q, et al. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J Biol Chem. 2005;280:38673–38681. doi: 10.1074/jbc.M507986200. [DOI] [PubMed] [Google Scholar]
- Demand J, Alberti S, Patterson C, Hohfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Dittmar KD, Hutchison KA, Owens-Grillo JK, Pratt WB. Reconstitution of the steroid receptor.hsp90 heterocomplex assembly system of rabbit reticulocyte lysate. J Biol Chem. 1996;271:12833–12839. doi: 10.1074/jbc.271.22.12833. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- Fan CY, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones. 2003;8:309–316. doi: 10.1379/1466-1268(2003)008<0309:mfrohf>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha CM, Nogueira P, Mendes F, Penque D, Amaral MD. The human DnaJ homologue (Hdj)-1/heat-shock protein (Hsp) 40 co-chaperone is required for the in vivo stabilization of the cystic fibrosis transmembrane conductance regulator by Hsp70. Biochem J. 2002;366:797–806. doi: 10.1042/BJ20011717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J, Hohfeld J. Chaperones get in touch: the Hip-Hop connection. Trends Biochem Sci. 1997;22:87–92. doi: 10.1016/s0968-0004(97)01005-0. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb SB, Kashlan OB, Watkins JN, Suaud L, Yan W, Kleyman TR, Rubenstein RC. Differential effects of Hsc70 and Hsp70 on the intracellular trafficking and functional expression of epithelial sodium channels. Proc Natl Acad Sci USA. 2006;103:5817–5822. doi: 10.1073/pnas.0507903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove DE, Fan CY, Ren HY, Cyr DM. The ER-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3 dependent degradation of nascent CFTRΔF508. Mol Biol Cell. 2011;22:301–314. doi: 10.1091/mbc.E10-09-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove DE, Rosser MF, Ren HY, Naren AP, Cyr DM. Mechanisms for rescue of correctable folding defects in CFTRΔF508. Mol Biol Cell. 2009;20:4059–4069. doi: 10.1091/mbc.E08-09-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hohfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt DM, Powers ET, Balch WE. The proteostasis boundary in misfolding diseases of membrane traffic. FEBS Lett. 2009;583:2639–2646. doi: 10.1016/j.febslet.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutt DM, et al. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat Chem Biol. 2010;6:25–33. doi: 10.1038/nchembio.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Fang SL, Xiao YF, O'Connor SP, Nadler SG, Lee DW, Jefferson DM, Kaplan JM, Smith AE, Cheng SH. Partial restoration of cAMP-stimulated CFTR chloride channel activity in ΔF508 cells by deoxyspergualin. Am J Physiol. 1998;275:C171–C178. doi: 10.1152/ajpcell.1998.275.1.C171. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Chen CA, Okayama H. Calcium phosphate transfection. In: Bonifacino JS, Dasso M, Lippincott-Schwartz J, Harford JB, Yamada KM, editors. Current Protocols in Cell Biology. New York: John Wiley & Sons; 2003. pp. 20.3.1–20.3.8. [Google Scholar]
- Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. Folding of CFTR is predominantly cotranslational. Mol Cell. 2005;20:277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Kopito RR. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- Lee RJ, Liu CW, Harty C, McCracken AA, Latterich M, Romisch K, DeMartino GN, Thomas PJ, Brodsky JL. Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. EMBO J. 2004;23:2206–2215. doi: 10.1038/sj.emboj.7600232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Ban N, Inagaki N. Aberrant catalytic cycle and impaired lipid transport into intracellular vesicles in ABCA3 mutants associated with nonfatal pediatric interstitial lung disease. Am J Physiol Lung Cell Mol Physiol. 2008;295:L698–L707. doi: 10.1152/ajplung.90352.2008. [DOI] [PubMed] [Google Scholar]
- Matsumura Y, Ban N, Ueda K, Inagaki N. Characterization and classification of ATP-binding cassette transporter ABCA3 mutants in fatal surfactant deficiency. J Biol Chem. 2006;281:34503–34514. doi: 10.1074/jbc.M600071200. [DOI] [PubMed] [Google Scholar]
- Matsumura Y, Rooney LA, Skach WR. In vitro methods for CFTR biogenesis. Methods Mol Biol. 741:233–253. doi: 10.1007/978-1-61779-117-8_16. [DOI] [PubMed] [Google Scholar]
- Matsumura Y, Skach WR. Protein export from endoplasmic reticulum to the cytosol: In vitro methods. In: Finazzi-Agrò A, editor. Encyclopedia of Life Sciences. New York: John Wiley & Sons; 2009. [Google Scholar]
- Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, Tanaka K, Iwai K, Nagata K. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRΔF508. Mol Biol Cell. 2008;19:1328–1336. doi: 10.1091/mbc.E07-06-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu TW, Ong DS, Wang YJ, Balch WE, Yates JR, 3rd, Segatori L, Kelly JW. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmesgern E, Hartl FU. ATP-dependent protein refolding activity in reticulocyte lysate. Evidence for the participation of different chaperone components. FEBS Lett. 1993;331:25–30. doi: 10.1016/0014-5793(93)80290-b. [DOI] [PubMed] [Google Scholar]
- Oberdorf J, Carlson EJ, Skach WR. Redundancy of mammalian proteasome beta subunit function during endoplasmic reticulum associated degradation. Biochemistry. 2001;60:13397–13405. doi: 10.1021/bi011322y. [DOI] [PubMed] [Google Scholar]
- Oberdorf J, Carlson EJ, Skach WR. Uncoupling proteasome peptidase and ATPase activities results in cytosolic release of an ER polytopic protein. J Cell Sci. 2006;119:303–313. doi: 10.1242/jcs.02732. [DOI] [PubMed] [Google Scholar]
- Oberdorf J, Pitonzo D, Skach WR. An energy-dependent maturation step is required for release of the cystic fibrosis transmembrane conductance regulator from early endoplasmic reticulum biosynthetic machinery. J Biol Chem. 2005;280:38193–38202. doi: 10.1074/jbc.M504200200. [DOI] [PubMed] [Google Scholar]
- Oberdorf J, Skach WR. In vitro reconstitution of CFTR biogenesis and degradation. Methods Mol Med. 2002;70:295–310. doi: 10.1385/1-59259-187-6:295. [DOI] [PubMed] [Google Scholar]
- Okiyoneda T, Barrière H, Bagdány M, Rabeh WM, Du K, Höhfeld J, Young JC, Lukacs GL. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329:805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Palleros DR, Welch WJ, Fink AL. Interaction of hsp70 with unfolded proteins: effects of temperature and nucleotides on the kinetics of binding. Proc Natl Acad Sci USA. 1991;88:5719–5723. doi: 10.1073/pnas.88.13.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt EB, Yan FF, Gay JW, Stanley CA, Shyng SL. Sulfonylurea receptor 1 mutations that cause opposite insulin secretion defects with chemical chaperone exposure. J Biol Chem. 2009;284:7951–7959. doi: 10.1074/jbc.M807012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JR. CFTR function and prospects for therapy. Annu Rev Biochem. 2008;77:701–726. doi: 10.1146/annurev.biochem.75.103004.142532. [DOI] [PubMed] [Google Scholar]
- Rosser MF, Washburn E, Muchowski PJ, Patterson C, Cyr DM. Chaperone functions of the E3 ubiquitin ligase CHIP. J Biol Chem. 2007;282:22267–22277. doi: 10.1074/jbc.M700513200. [DOI] [PubMed] [Google Scholar]
- Rubenstein RC, Egan ME, Zeitlin PL. In vitro pharmacologic restoration of CFTR-mediated chloride transport with sodium 4-phenylbutyrate in cystic fibrosis epithelial cells containing ΔF508-CFTR. J Clin Invest. 1997;100:2457–2465. doi: 10.1172/JCI119788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein RC, Lyons BM. Sodium 4-phenylbutyrate downregulates HSC70 expression by facilitating mRNA degradation. Am J Physiol Lung Cell Mol Physiol. 2001;281:L43–L51. doi: 10.1152/ajplung.2001.281.1.L43. [DOI] [PubMed] [Google Scholar]
- Rubenstein RC, Zeitlin PL. A pilot clinical trial of oral sodium 4-phenylbutyrate (Buphenyl) in ΔF508-homozygous cystic fibrosis patients: partial restoration of nasal epithelial CFTR function. Am J Respir Crit Care Med. 1998;157:484–490. doi: 10.1164/ajrccm.157.2.9706088. [DOI] [PubMed] [Google Scholar]
- Rubenstein RC, Zeitlin PL. Sodium 4-phenylbutyrate downregulates Hsc70: implications for intracellular trafficking of ΔF508-CFTR. Am J Physiol Cell Physiol. 2000;278:C259–C267. doi: 10.1152/ajpcell.2000.278.2.C259. [DOI] [PubMed] [Google Scholar]
- Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296. [DOI] [PubMed] [Google Scholar]
- Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Skach WR. Cellular mechanisms of membrane protein folding. Nat Struct Mol Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DF, Sullivan WP, Marion TN, Zaitsu K, Madden B, McCormick DJ, Toft DO. Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol Cell Biol. 1993;13:869–876. doi: 10.1128/mcb.13.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268. [DOI] [PubMed] [Google Scholar]
- Strickland E, Qu BH, Millen L, Thomas PJ. The molecular chaperone Hsc70 assists the in vitro folding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1997;272:25421–25424. doi: 10.1074/jbc.272.41.25421. [DOI] [PubMed] [Google Scholar]
- Summers DW, Douglas PM, Ramos CH, Cyr DM. Polypeptide transfer from Hsp40 to Hsp70 molecular chaperones. Trends Biochem Sci. 2009;34:230–233. doi: 10.1016/j.tibs.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Bimston DN, Matsuzawa S, Freeman BC, Aime-Sempe C, Xie Z, Morimoto RI, Reed JC. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J. 1997;16:4887–4896. doi: 10.1093/emboj/16.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol. 2001;3:E237–E241. doi: 10.1038/ncb1001-e237. [DOI] [PubMed] [Google Scholar]
- Terada K, Kanazawa M, Bukau B, Mori M. The human DnaJ homologue dj2 facilitates mitochondrial protein import and luciferase refolding. J Cell Biol. 1997;139:1089–1095. doi: 10.1083/jcb.139.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzankov S, Wong MJ, Shi K, Nassif C, Young JC. Functional divergence between co-chaperones of Hsc70. J Biol Chem. 2008;283:27100–27109. doi: 10.1074/jbc.M803923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Xiong X, Bragin A, Widdicombe JH, Cohn J, Skach WR. Structural cues involved in endoplasmic reticulum degradation of G85E and G91R mutant cystic fibrosis transmembrane conductance regulator. J Clin Invest. 1997;100:1079–1088. doi: 10.1172/JCI119618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Chong E, Skach WR. Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J Biol Chem. 1999;274:2616–2624. doi: 10.1074/jbc.274.5.2616. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Young JC, Hartl FU. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Younger JM, Ren HY, Chen L, Fan CY, Fields A, Patterson C, Cyr DM. A foldable CFTRΔF508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J Cell Biol. 2004;167:1075–1085. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Schmidt BZ, Sun F, Condliffe SB, Butterworth MB, Youker RT, Brodsky JL, Aridor M, Frizzell RA. Cysteine string protein monitors late steps in cystic fibrosis transmembrane conductance regulator biogenesis. J Biol Chem. 2006;281:11312–11321. doi: 10.1074/jbc.M512013200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, Brodsky JL. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.