The current study provides mechanistic insight into the overlapping dynamics by which glycoprotein folding and quality control use distinct intracellular compartments as part of the proteostasis network in mammalian cells.
Abstract
The Golgi complex has been implicated as a possible component of endoplasmic reticulum (ER) glycoprotein quality control, although the elucidation of its exact role is lacking. ERManI, a putative ER resident mannosidase, plays a rate-limiting role in generating a signal that targets misfolded N-linked glycoproteins for ER-associated degradation (ERAD). Herein we demonstrate that the endogenous human homologue predominantly resides in the Golgi complex, where it is subjected to O-glycosylation. To distinguish the intracellular site where the glycoprotein ERAD signal is generated, a COPI-binding motif was appended to the N terminus of the recombinant protein to facilitate its retrograde translocation back to the ER. Partial redistribution of the modified ERManI was observed along with an accelerated rate at which N-linked glycans of misfolded α1-antitrypsin variant NHK were trimmed. Despite these observations, the rate of NHK degradation was not accelerated, implicating the Golgi complex as the site for glycoprotein ERAD substrate tagging. Taken together, these data provide a potential mechanistic explanation for the spatial separation by which glycoprotein quality control components operate in mammalian cells.
INTRODUCTION
A current challenge in both cell biology and the biomedical sciences is to elucidate how the processing of encoded proteins, rather than the corresponding genomic blueprint, helps orchestrate the fidelity of expressed biological information and contributes to the pathophysiology of disease. To this end, protein biosynthetic quality control, which is part of the cellular proteostasis network (Balch et al., 2008), ensures that only properly folded and/or assembled proteins are delivered to their sites of action, which is essential for numerous normal cellular activities. Approximately one-third of newly synthesized polypeptides are translocated into the endoplasmic reticulum (ER), where they achieve native conformation, which is a prerequisite for productive transport through the secretory pathway (Ghaemmaghami et al., 2003). Those polypeptides unable to adopt native structure are eventually identified as terminally misfolded and then subjected to ER-associated degradation (ERAD) by cytoplasmic 26S proteasomes (Sommer and Wolf, 1997; Brodsky and McCracken, 1999). Although the ER is a site for the retention of many misfolded proteins (Nehls et al., 2000), evidence has suggested that numerous lumenal ERAD substrates are likely delivered to the Golgi complex and recycled back to the ER before retrograde translocation into the cytosol for degradation (Hammond and Helenius, 1994; Caldwell et al., 2001; Vashist et al., 2001). This observation implies that successful biosynthetic quality control likely involves the spatial separation of some of its components. In support of this notion, the overexpression of three traditional Golgi α1,2-mannosidases (IA, IB, and IC) was recently shown to accelerate the degradation of misfolded glycoproteins in mammalian cells (Hosokawa et al., 2007). The descriptive nature of that study, however, stopped short of exploring the dynamics of ERAD substrate generation or establishing the intracellular site where the overexpressed molecules were able to accelerate ERAD.
The capacity to distinguish between terminally misfolded proteins and properly folded wild-type intermediates as appropriate ERAD substrates is one of the earliest defining steps in protein biosynthetic quality control. Prior studies have indicated that the removal of terminal mannose units from asparagine-linked Man9GlcNAc2 generates the glycan component of an ERAD signal that operates in both yeast and mammalian cells (Lederkremer, 2009). Confusion still exists, however, in understanding how this step operates in the mammalian system because the analysis of ERManI function has exclusively involved the overexpressed recombinant protein, and both ERManI (Hosokawa et al., 2003; Avezov et al., 2008) and EDEM (ER degradation-enhancing mannosidase like protein; Hosokawa et al., 2001, 2010; Hirao et al., 2006; Olivari et al., 2006) are apparently capable of hydrolyzing the α1,2-linked mannose units.
Human ERManI consists of 699 amino acids with a calculated molecular mass of 79.5 kDa. Although ∼50% homologous to the Golgi α1,2-mannosidases IA, IB, and IC sequences (Tremblay and Herscovics, 1999), it is predicted to function as an ER resident protein based on the immunostaining of the yeast orthologue MNS1 (Burke et al., 1996) and localization of the overexpressed recombinant human orthologue in mammalian cells (Gonzalez et al., 1999). The mannosidase was first implicated as a member of the glycoprotein quality control system because deletion of the Mns1 gene in budding yeast greatly hindered the degradation of an N-glycosylated ERAD substrate (Jakob et al., 1998). The inhibition of ERAD was later observed in several mammalian systems in response to the inhibition of class I α1,2-mannosidases with kifunensine, and by small interfering RNA (siRNA)-mediated gene knockdown (Liu et al., 1999; Cabral et al., 2001, 2002; Sifers, 2003; Wu et al., 2003; Karaveg et al., 2005; Avezov et al., 2008). The mammalian orthologue appears to function in a dose-dependent manner, as its experimental overexpression accelerates the degradation of terminally misfolded glycoproteins as well as newly synthesized wild-type folding intermediates (Wu et al., 2003).
The clinical involvement of cellular proteostasis in disease pathogenesis was recently revealed by a study from our group in which homozygosity for a single nucleotide polymorphism located at the 3′-untranslated region of the human ERManI mRNA is associated with the early onset of end-stage liver disease in patients with α1-antitrypsin deficiency (Pan et al., 2009). This observation implies that the mannosidase, as part of the ER proteostasis network (Balch et al., 2008; Bouchecareilh et al., 2010), can function as a genetic modifier of disease and therefore is a potential target for the therapeutic intervention of the associated liver and lung diseases (Sifers, 2010).
Both in vitro and in vivo studies have indicated that the participation of ERManI in glycoprotein quality control stems from its weak enzymatic activity capable of cleaving one or more α1,2-mannose residues from asparagine-linked Man9GlcNAc2 (Gonzalez et al., 1999; Tremblay and Herscovics, 1999; Hosokawa et al., 2003; Karaveg et al., 2005; Avezov et al., 2008). The covalent processing event has been shown to disrupt the participation of misfolded glycoproteins in the calnexin folding cycle, allowing for their entrance into ERAD (Cabral et al., 2001). Moreover, a recent study has shown that proteasome inhibitor treatment causes experimentally overexpressed ERManI to localize to a presumed ER protein quality control center (ERQC), where misfolded glycoproteins are suspected to accumulate before their dislocation into the cytoplasm for proteasomal degradation (Avezov et al., 2008). From these observations, researchers have concluded that the enzyme might play an additional role as a lectin capable of sorting misfolded lumenal glycoproteins to a specialized subcellular compartment before ERAD. Support for this notion was recently provided by the fission yeast model (Kanehara et al., 2007). Despite these presented hypotheses, the exact functional roles played by ERManI in mammalian glycoprotein quality control are still poorly characterized.
A primary goal for the present study was to gain greater mechanistic insight into the dynamics of ERAD substrate generation. A panel of highly specific mAbs was generated against recombinant human ERManI, and was then used to elucidate the intracellular location and properties of the endogenous protein in cultured mammalian cells under basal conditions. Numerous lines of experimental evidence indicate that the endogenous mannosidase exists predominately in an O-glycosylated form that is localized exclusively to the Golgi apparatus, rather than the proposed ERQC (Kamhi-Nesher et al., 2001; Avezov et al., 2008), where it participates in the generation of a glycoprotein ERAD disposal signal. The new findings provide additional molecular insight into the dynamics by which protein folding and ERAD substrate generation operate throughout multiple compartments in mammalian cells.
RESULTS
Covalent modification of endogenous human ERManI
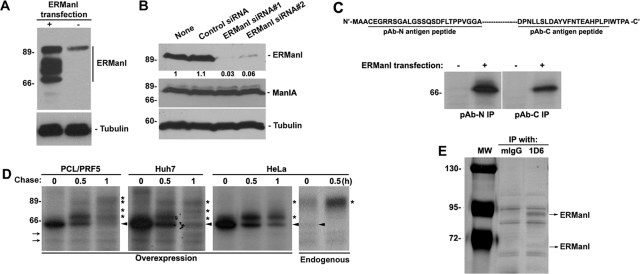
We previously generated rabbit polyclonal antisera against human ERManI to assist our mechanistic analysis of glycoprotein quality control (Wu et al., 2003). Poor detection limits, however, hindered the reproducible detection of the endogenous protein. Therefore, a panel of mAbs was generated using the GST-tagged, full-length human recombinant protein as antigen. Three hybridoma clones (1D6, 3C2, and 4H12) were first chosen on the basis of specific immunoreactivity against GST-tagged human ERManI via an enzyme-linked immunosorbent assay (unpublished data). Additional characterization involved Western blotting of detergent lysates derived from HeLa cells transfected with either empty vector or with a human ERManI cDNA expression construct. In mock-transfected HeLa cells, all three mAbs recognized a single protein band that exhibited an apparent molecular mass of ∼90 kDa, which is significantly greater than the predicted value of the newly synthesized protein (79.5 kDa). In extracts from HeLa cells transfected with recombinant human ERManI, each of the three mAbs recognized a ladder of bands migrating between ∼70 and ∼90 kDa (Figure 1A and Supplemental Figure S1). The slowest-migrating band exhibited an electrophoretic mobility in SDS–PAGE that was identical to endogenous human ERManI. The observation that the overexpression of the recombinant protein enhanced the density of the 90-kDa species implied that this species likely represents the endogenous form of the molecule. Consistent with this notion, HeLa cells separately transfected with two different siRNAs that target distinct sequences of the human ERManI mRNA diminished (by ∼95%) the intracellular concentration of the endogenous ∼90-kDa protein when normalized against untransfected control cells or when cells were transfected with a nonspecific siRNA (Figure 1B). The specificity of this manipulation was confirmed by the inability of the specific siRNAs to diminish the concentration of Golgi ManIA, which shares ∼50% sequence similarity with human ERManI. These data strongly indicated that the single ∼90-kDa protein detected by all the mAbs represents the mature form of endogenous human ERManI.
FIGURE 1:
Characterization of endogenous ERManI. (A) Cell lysates from HeLa cells transfected with ERManI (+) or empty vector (−) for 24 h were resolved by SDS–PAGE and immunoblotted with monoclonal anti-ERManI antibody clone 1D6 (top panel) and anti-tubulin antibody. (B) Cell lysates derived from untransfected HeLa cells (none) or HeLa cells transfected with control siRNA, or two distinct siRNAs against ERManI, were resolved by SDS–PAGE and blotted with antibodies against ERManI (clone 1D6), ManIA, or tubulin. Numbers beneath ERManI blot show ratios against the amount of ERManI in nontransfected cells. (C) Top panel, illustration of synthetic peptides used to generate the antibody against N terminus (pAb-N) or C terminus (pAb-C) of ERManI. Bottom panel, HeLa cells transfected with empty vector (−) or ERManI cDNA (+) for 24 h were radiolabeled with [35S] Met for 30 min, and the derived cell lysates were subjected to immunoprecipitation with pAb-N and pAb-C and observed by radiography. (D) PCL/PRF5, Huh7, and HeLa cells were transfected with ERManI cDNA. Forty-eight hours after transfection, cells were radiolabeled with [35S] Met for 30 min and chased for 0.5 and 1 h. Nontransfected HeLa cells were subjected to the same pulse-labeling and chased for 0.5 h. Cells were collected from each time point, lysed, and subjected to immunoprecipitation with a combination of pAb-N and pAb-C. *, modified forms of ERManI; arrowheads, unmodified ERManI; arrows: Nonspecific bands. (E) HeLa cell lysates were immunoprecipitated with either mouse IgG (mIgG) or anti-ERManI antibody 1D6. The immunoprecipitates were eluted and resolved by SDS–PAGE followed by silver staining. Arrows indicate the two specific bands identified by mass spectrometry as ERManI.
Next we asked whether the ladder of immunoreactive proteins detected in transfected HeLa cells represents degradation products versus covalently modified forms of the molecule. To address this issue, we took advantage of two polyclonal antibodies previously generated against synthetic peptides identical to amino acid residues located in the N terminus (pAb-N) and C terminus (pAb-C) of human ERManI (Wu et al., 2003). HeLa cells were transfected with empty vector or the recombinant human ERManI cDNA expression construct. Approximately 24 h posttransfection, cells were metabolically radiolabeled with [35S]Met for 30 min before cell lysis and immunoprecipitation with either pAb-N or pAb-C. As shown in Figure 1C, the newly synthesized ∼70-kDa band was immunoprecipitated with either antibody. In some experiments, a faint ∼70-kDa band was also detected in control cells, possibly representing the newly synthesized endogenous human ERManI (Figure 1C). Taken together, the data imply that the ∼70-kDa band represents full-length, newly synthesized human ERManI before the onset of any detectable posttranslational modifications.
To determine whether the ∼90-kDa band is derived from the newly synthesized precursor, recombinant human ERManI was experimentally overexpressed in multiple cell lines, including PCL/PRF5 and Huh7 (human hepatoma cells), as well as in HeLa cells. The de novo biosynthesis and posttranslational processing of ERManI were detected by metabolic pulse-chase radiolabeling and immunoprecipitation with a combination of the pAb-N and pAb-C polyclonal antisera. As shown in Figure 1D, human ERManI was initially synthesized as a ∼70-kDa polypeptide in all three cell lines. Most of the radiolabeled molecules were degraded, as recently reported (Wu et al., 2007), but the remainder gradually increased in apparent molecular mass, through the formation of multiple intermediates, into a stable ∼90-kDa form. The maturation of the transfected recombinant protein, which varied among the different cell lines, was least efficient in HeLa cells and most efficient in PCL/PRF5. A similar experiment was performed with untransfected HeLa cells to characterize the maturation of endogenous human ERManI. In contrast to the slow modification of the overexpressed recombinant ERManI, the endogenous molecule was quickly modified, with both the 70- and ∼90-kDa bands detectable within the 20-min pulse period. Furthermore, the 70-kDa band had disappeared at the 30-min chase, coinciding with an increase in the ∼90-kDa band. These observations demonstrate that the endogenous molecules efficiently mature to the fully modified state following biosynthesis. Considering these observations, it is entirely possible that the slower maturation of overexpressed ERManI might represent saturation of the modification system. Taken together, these results indicate that an undegraded fraction of the newly synthesized endogenous human ERManI is subjected to a series of posttranslational modifications that eventually generate a mature ∼90-kDa species.
In a final set of experiments, mass spectrometry was used as a means to validate the identity of the 70- and 90-kDa forms. HeLa cell extracts were subjected to immunoprecipitation with either mouse IgG (as negative control) or mAb1D6 (the mAb that exhibited the greatest immunoprecipitation efficiency). Although numerous proteins were detected in SDS–PAGE, the results of mass spectrometry confirmed that the 70- and 90-kDa bands represent human ERManI (Figure 1E and Supplemental Figure S2). Importantly, the 90-kDa band exhibited a greater intensity than did the 70-kDa band, confirming the notion that the former band represents the predominant intracellular species of endogenous human ERManI.
Endogenous human ERManI is O-glycosylated at multiple sites in the luminal stem domain
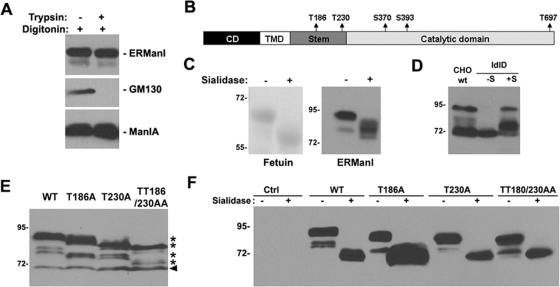
We next directed our efforts toward identifying the modification(s) responsible for altering the apparent molecular mass of newly synthesized human ERManI. First, a protease protection assay was performed with semipermeabilized HeLa cells. Our reasoning was that incubation with an exogenous protease would remove the modification, dramatically shifting the apparent molecular mass, if it occurs in the short N-terminal cytoplasmic domain. In contrast, the modification would be protected from proteolysis if present on a luminal domain. For this purpose, HeLa cells were incubated in the cold with digitonin to selectively permeabilize the plasma membrane (Liu et al., 1999), followed by incubation with trypsin. As shown in Figure 2A, the manipulation did not significantly alter the mobility of endogenous human ERManI in SDS–PAGE, consistent with proteolytic removal of only the short N-terminal cytoplasmic tail. As a positive control, GM130, which is a Golgi matrix protein localized predominantly on the cytoplasm surface of the endomembrane system (Nakamura et al., 1995), was completely susceptible to trypsin digestion. In contrast, the negative control ManIA, a type II Golgi enzyme that is primarily luminal (Bieberich and Bause, 1995), was entirely resistant to proteolysis under these conditions. The combination of these results supported the notion that the posttranslational modification of human ERManI occurs on a luminal portion of the molecule.
FIGURE 2:
ERManI is glycosylated at multiple sites in the luminal domain. (A) HeLa cells were permeabilized with digitonin followed by treatment with mock solution or trypsin. The cells were then lysed and immunoblotted with antibodies against ERManI, GM130, or ManIA. (B) Illustration of five potential O-glycosylation sites in the luminal domain of ERManI that are predicted by NetOGlyc3.1 and 2.0 software. (C) Fetuins purified from calf serum or HeLa cell extract were treated with (+) or without (−) Sialidase A. The samples were then resolved on SDS–PAGE, and the gels were either stained with Coommassie blue (for futuin) or transferred and immunoblotted with anti-ERManI antibody 1D6 (for ERManI). (D) CHO cells and ldlD cells were transfected with wild type or ERManIO-def, respectively, and were cultured in regular medium (for CHO cells) or basic medium containing no additional sugars (–S) or with additional galactose and N-acetylgalactosamine (+S) (for ldlD cells). Forty-eight hours after transfection, cells were lysed and subjected to Western blotting with anti-ERManI antibody 1D6. (E) Hepa1A cells transfected with wild type or indicated mutants of ERManI for 48 h were lysed and immunoblotted with anti-ERManI antibody 1D6. *, modified forms of ERManI; arrowhead, newly synthesized unmodified ERManI. (F) Lysates of Hepa1A cells transfected with wild type or indicated mutants of ERManI for 48 h were treated with (+) or without (−) Sialidase A, resolved by SDS–PAGE, and immunoblotted with anti-ERManI antibody 1D6.
Many types of posttranslational modifications, such as phosphorylation and glycosylation, can occur in the luminal domain of a transmembrane protein, resulting in a significant shift in the apparent molecular mass. Analysis with the NetPhos2.0 algorithm predicts that human ERManI harbors several potential phosphorylation sites. Digestion of either crude HeLa cell lysates or immunoprecipitated endogenous human ERManI with a variety of commercially available phosphatases, however, did not appreciably alter the molecular mass of the endogenous ∼90-kDa species (unpublished data), implying the involvement of an alternative modification. Negative results were also obtained in response to the treatment of HeLa cell lysates or immunoprecipitated human ERManI with PNGase, an enzyme that cleaves complex type N-linked oligosaccharides, consistent with the absence of N-glycosylation sites in the primary amino acid sequence. A third possibility is that the NetOGlyc2.0 and NetOGlyc3.1 algorithms identified several potential consensus sequences for the O-glycosylation of human ERManI. As illustrated in Figure 2B, the algorithms predicted that ERManI contains multiple threonine and serine residues in the stem and catalytic domains potentially capable of undergoing O-glycosylation. To test this possibility, crude HeLa cell lysates were generated and then incubated with or without Sialidase A, an enzyme that cleaves terminal sialic acids from N- or O-glycans. Changes in the mobility of endogenous human ERManI were monitored by Western blotting following SDS–PAGE, and commercially available glycoprotein fetuin was used as a positive control. As shown in Figure 2C, treatment with Sialidase A increased the electrophoretic mobility of immunoreactive human ERManI by nearly 15 kDa, indicating that sialic acid–containing modifications were present.
To obtain additional evidence to support the observation that human ERManI is subject to O-glycosylation, we transfected the corresponding expression vector into both wild-type CHO cells and a mutant CHO cell line designated ldlD (Kozarsky et al., 1988). The ldlD cell line carries a defect in the 4-epimerase, an enzyme that is responsible for the conversion of glucose and N-acetylglucosamine into galactose and N-acetylgalactosamine, respectively. As a consequence, the ldlD cells are able to synthesize O-linked oligosaccharides only when galactose and N-acetylgalactosamine are added to the culture medium. Without these sugar additions, no mucin-type, O-linked oligosaccharide chains are synthesized (Kozarsky et al., 1988). As shown in Figure 2C, recombinant human ERManI expressed in wild-type CHO cells migrated primarily as either the 70-kDa, newly synthesized form or 90-kDa, fully modified form, with the existence of a few minor intermediate bands. In ldlD cells cultured without sugar supplementation, however, recombinant human ERManI migrated solely as the 70-kDa form. When both galactose and N-acetylgalactosamine were added to the culture medium, the electrophoretic migration of the recombinant protein shifted to a maximum of 90 kDa, although the majority of molecules were only partially modified, possibly due to saturation of the responsible system. Taken together, these observations confirmed that human ERManI is modified by mucin-type, O-linked oligosaccharides.
Our next step was to use site-directed mutagenesis to identify the amino acid residues responsible for the O-glycosylation of human ERManI. Predicted threonine and serine residues (Figure 2B) were mutated to alanine, and the constructs were then transiently expressed in mouse hepatoma cells Hepa1A because the mouse orthologue is not recognized by any of the newly generated mAbs, and therefore will not interfere with the detection of transfected recombinant human ERManI. As before, changes in electrophoretic mobility were monitored by SDS–PAGE and Western blotting. Among all five individual mutants (T186A, T230A, S370A, S393A, and T697A), only T186A and T230A exhibited detectable mobility changes in SDS–PAGE (Figure 2E and unpublished data). Moreover, a combination of these two mutations (the TT186/230AA double mutant) exhibited the most dramatic mobility shift, as would be expected if both were sites for O-linked glycosylation (Figure 2E). The double mutant, however, did not completely convert the mobility of ERManI to that of the newly synthesized 70-kDa form, implying that additional sites for O-glycosylation and/or an alternative modification(s) remained. To distinguish between these two possibilities, we treated the transfected wild-type and mutant human ERManI (TT186/230AA) with Sialidase A and monitored changes in the protein's electrophoretic mobility. As shown in Figure 2F, Sialidase A treatment converted both the wild-type and mutant ERManI to the 70-kDa form, indicating that additional O-glycosylation sites exist that were not predicted by the aforementioned algorithms.
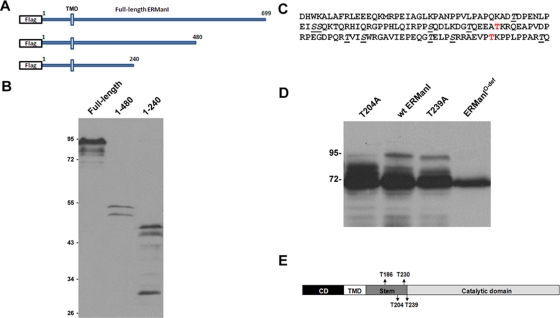
To identify additional O-glycosylation sites, we generated Flag-tagged cDNA constructs through serial truncation of the luminal domain (Figure 3A) to narrow down the region in which human ERManI undergoes O-glycosylation. Each of these constructs was transfected into Hepa1A cells, and the encoded proteins were detected by Western blotting using an anti-Flag antibody. As shown in Figure 3B, the constructs truncated at the end of the stem region (amino acids 1–240) underwent the most robust modification, whereas constructs truncated in the middle of the catalytic domain (amino acids 1–480) showed modification similar to that of the wild type, suggesting that O-glycosylation is limited to the stem region where T186 and T230 reside. The modifications within the stem region contained terminal sialic acids, as confirmed by the altered electrophoretic mobility change in response to Sialidase A digestion (unpublished data). To identify the O-glycosylation sites, each of the threonine and serine residues within the stem domain were mutated to alanine (Figure 3C) in addition to the above identified T186 and T230, and the consequences were monitored by the same approach as stated earlier in this article. Importantly, single mutations of T204 and T239 each affected the electrophoretic pattern of recombinant human ERManI (Figure 3D). Finally, a combination of these two mutated sites together with the previously identified T186A and T230A shifted the mobility of recombinant human ERManI to 70 kDa (Figure 3D), and was designated O-glycosylation–deficient ERManI (ERManIO-def). Based on these observations, we concluded that four threonine residues in the luminal stem domain function as the primary sites for O-glycosylation (Figure 3E).
FIGURE 3:
Identification of additional O-glycosylation sites of ERManI. (A) Illustration of full-length or truncated ERManI constructs that contain an N-terminal Flag tag. (B) The full-length or truncated flag-tagged ERManI constructs were each transfected into Hepa1A cells. Forty-eight hours after transfection, cells were lysed, and the total protein extracts were separated by SDS–PAGE followed by Western blotting with 1D6. (C) Sequence of the stem region of ERManI. Amino acids in red represent T186 and T230 identified in earlier experiments. Underlined amino acids represent the residues mutated to Alanine. (D) Wild-type or mutant ERManI as indicated was transfected into Hepa1A cells. Forty-eight hours after transfection, cells were lysed and subjected to Western blotting with 1D6. (E) Experimentally validated positions of the four O-linked glycans detected on human ERManI are depicted.
Endogenous human ERManI is localized to the Golgi apparatus
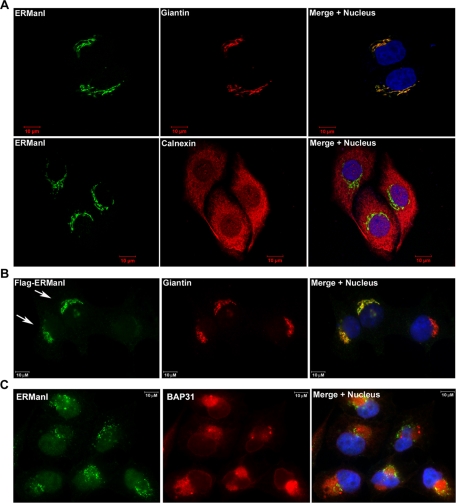
Because O-glycosylation occurs predominantly in the Golgi apparatus, we next asked whether the endogenous human ERManI actually resides in this organelle, which is inconsistent with its ER localization that has been widely assumed (Burke et al., 1996; Gonzalez et al., 1999). The intracellular localization of endogenous human ERManI in HeLa cells was determined by indirect immunofluorescence microscopy using a combination of three anti-human ERManI mAbs. Cells were costained with either the Golgi marker Giantin (Linstedt and Hauri, 1993) or the ER marker Calnexin (Bergeron et al., 1994) to examine organelle-specific colocalization. As shown in Figure 4A, the anti-ERManI mAbs stained a juxtanuclear area in the cytoplasm, which overlapped with the Golgi complex, but not the ER. The staining pattern was specific because the fluorescence signal was abolished upon siRNA-mediated knockdown of endogenous human ERManI (Supplemental Figure S3). Importantly, the human ERManI mAbs chosen for immunofluorescence staining exhibited similar patterns in Western blotting, suggesting that similar epitope(s) on the molecule are recognized (unpublished data). To eliminate the possibility that the epitope(s) were accessible only to the Golgi-localized molecules, HeLa cells were transfected with Flag-tagged human ERManI, and then detected with an antibody against that added epitope. As shown in Figure 4B, Flag-tagged ERManI colocalized with endogenous Giantin, confirming that endogenous human ERManI is actually a resident of the Golgi complex.
FIGURE 4:
Endogenous ERManI is a Golgi-resident protein. (A) Confocal images of HeLa cells coimmunostained with the anti-ERManI mAbs and anti-Giantin or anti-calnexin antibodies. ERManI was visualized using Alexa 488–conjugated anti–mouse secondary antibody (green). Giantin or calnexin was visualized using Alexa 555–conjugated anti–rabbit secondary antibodies (red). Cell nuclei were counterstained with To-PRO3. (B) Flag-tagged ERManI was transfected into HeLa cells. Forty-eight hours after transfection, cells were fixed and coimmunostained with anti-Flag and anti-Giantin antibodies. Flag-ERManI was visualized using Alexa 488–conjugated anti–mouse secondary antibody (green). Giantin was visualized using Alexa 555–conjugated anti–rabbit secondary antibodies (red). Arrows indicate the cells transfected with Flag-ERManI. (C) HeLa cells treated with 10 μM MG132 for 6 h were costained for ERManI (green) and BAP31 (red). Cell nuclei were counterstained with DAPI.
Endogenous human ERManI does not localize to the ERQC in response to proteasome-inhibitor treatment
Previous studies have shown that, in response to treatment with proteasome inhibitors, overexpressed recombinant ERManI localizes to a specialized intracellular compartment designated ERQC (Avezov et al., 2008). Therefore, we asked whether the same might be true for the endogenous protein. For this, HeLa cells were incubated for 12 h with the proteasome inhibitor MG132 before fixation and staining with the mixture of human ERManI mAbs and an antibody against BAP31, the latter of which has been reported to reside in the ERQC (Wakana et al., 2008). As shown in Figure 4C, endogenous BAP31 was concentrated in a perinuclear area, representing the ERQC. In contrast, endogenous human ERManI exhibited a distinct punctate distribution adjacent to the ERQC without any demonstrable colocalization with BAP31. Importantly, an identical observation was made using another ERQC marker calnexin (Kamhi-Nesher et al., 2001) or in cells incubated with different concentrations of lactacystin (unpublished data), another proteasome inhibitor (Tomoda and Omura, 2000). This combination of experimental results indicates that the endogenous human ERManI does not colocalize with the ERQC, even under conditions of proteasomal inhibition.
O-glycosylation influences the solubility of human ERManI
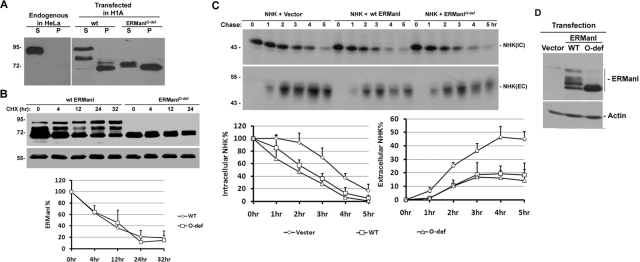
O-glycosylation is generally associated with changes in the solubility, stability, localization, and/or function of a protein (Van den Steen et al., 1998; Goto, 2007). We therefore tested the capacity of O-glycosylation to influence different aspects of human ERManI biology. To test the modification's capacity to regulate solubility of the enzyme, cells were first lysed in buffer containing selected biological detergents, followed by centrifugation. The supernatant and pellet were collected as soluble and insoluble portions, respectively, and resolved by SDS–PAGE followed by Western blotting for human ERManI. As a transmembrane protein, endogenous human ERManI is readily extracted by a variety of nonionic and ionic detergents. In response to our routine cell lysis buffer containing 0.5% NP-40, the entire population of the endogenous ERManI in HeLa cells was extracted into the supernatant (Figure 5A, left panel). In Hepa1A cells that overexpress the wild-type recombinant protein, multiple immunoreactive bands between 70 and 90 kDa are displayed in response to variable extents of O-glycosylation (Figure 5A, right panel). Whereas the fully O-glycosylated 90-kDa form of the transfected recombinant wild-type protein was completely extracted into the NP-40 supernatant, a significant portion of the partially modified molecules and the majority of the unmodified form (∼70 kDa) remained in the pellet (Figure 5A, left panel). Overall, the percentage of soluble and insoluble portions of the wild-type protein was 51 and 49%, respectively (Figure 5A, right panel). In contrast, the majority (60%) of the transfected O-glycosylation–deficient ERManIO-def remained in the pellet. Moreover, the NP-40 soluble fraction displayed a slightly slower electrophoretic mobility than did the insoluble fraction (Figure 5A, right panel), suggesting that a minor posttranslational modification still exists. This minor modification is resistant to sialidase treatment, and its identity is currently under investigation. All in all, the correlation between O-glycosylation and extraction with nonionic detergent implies that the modification stabilizes the conformation of human ERManI, possibly by preventing its intracellular aggregation.
FIGURE 5:
Functional characterization of ERManI O-glycosylation. (A) Untransfected HeLa cells or Hepa1A cells transfected with either wild type or O-glycosylation–deficient ERManIO-def were lysed in buffer containing 0.5% NP-40. Soluble proteins (S) were separated from insoluble proteins (P) by centrifugation, and both fractions were resolved on SDS–PAGE and subjected to Western blotting with 1D6. (B) Hepa1A cells transfected with either wild type or ERManIO-def for 24 h were treated with 100 μg/ml cycloheximide (CHX) for indicated times, the stability of ERManI was examined by Western blotting, and the protein bands were quantified (from three experiments) by densitometry using NIH ImageJ software. (C) NHK was cotransfected into HeLa cells together with wild type or ERManIO-def. Forty-eight hours after transfection, cells were labeled with 35S[Met] for 30 min and chased for indicated times. NHK was immunoprecipitated from cell lysates (IC) and medium (EC), collected at each time point, resolved by SDS–PAGE, and detected by autoradiography. Bar graph at bottom shows the quantification of the level of NHK by densitometry (from three experiments). (D) A representative Western blot shows the expression of wild type and ERManIO-def using whole-cell lysates derived from transfected HeLa cells as stated in (C).
To investigate the possibility that O-glycosylation might play a role in promoting ERManI's intracellular stability, Hepa1A cells were transfected with either recombinant wild type or ERManIO-def. The intracellular turnover of the expressed molecules was then monitored by Western blotting of cell lysates in cells treated with cycloheximide to block new protein synthesis. No significant differences were observed regarding the stability of wild type and ERManIO-def (Figure 5B), indicating that the absence of O-glycosylation is not sufficient to accelerate the intracellular turnover of ERManI.
Because mucin-type glycans are generated in the Golgi complex, and apparently contribute to the solubility (i.e., conformational stability) of the deployed molecule, we asked whether the modification contributes to the subcellular localization of ERManI. To address this question, Hepa1A cells were transfected with wild type or ERManIO-def, followed by detection by immunofluorescence microscopy. Both wild type and ERManIO-def localized predominantly to the Golgi apparatus (Supplemental Figure S4), implying that either the deployment or Golgi retention of the molecules is independent of O-glycosylation.
Finally, we asked whether O-glycosylation might play a role in the capacity of ERManI to tag misfolded luminal N-linked glycoproteins for ERAD in mammalian cells (Wu et al., 2003). To test this hypothesis, a mutant variant of the glycoprotein α1- antitrypsin, designated NHK (Sifers et al., 1988), that misfolds upon biosynthesis was cotransfected with either wild type or ERManIO-def into HeLa cells. Forty-eight hours posttransfection, cells were subjected to [35S]Met metabolic labeling, and NHK was immunoprecipitated from cell lysates and media. Consistent with our previous findings (Wu et al., 2003), coexpression of recombinant wild-type ERManI accelerated the rate at which NHK was subjected to ERAD, and decreased the extent of its already impaired secretion as compared with control (Figure 5C). Importantly, with an equivalent total level of expression (Figure 5D), the absence of O-glycosylation did not significantly impair the capacity of transfected ERManIO-def to accelerate the intracellular degradation of NHK over that of wild type. Rather, a moderate increase in ERAD was observed, which led to a slight decrease in NHK secretion as compared with recombinant wild-type ERManI. Taken together, these data indicate that the absence of O-glycosylation, although impairing the intracellular solubility of ERManI, is not sufficient to significantly disrupt its role played in glycoprotein quality control.
ER localization is not required for ERManI's function in ERAD
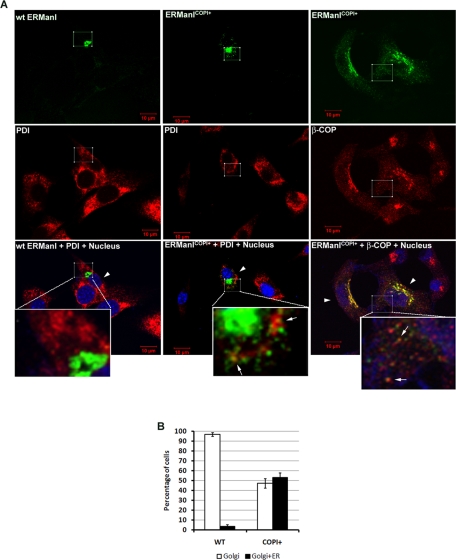
Our finding that endogenous human ERManI is actually localized to the Golgi complex raises the question of where the molecule resides when tagging glycoprotein substrates for ERAD. Presumably, either the Golgi-localized molecules play this role, or a fraction might actually recycle back to the ER. If the latter is true, by facilitating Golgi-to-ER retrotranslocation of ERManI, an increase in degradation of ERAD substrates should be observed. The cytoplasmic domain of ERManI contains several di-basic residues. None, however, are within the range (the first five amino acids at the N terminus) necessary for efficient COPI binding, which is required for Golgi-to-ER retrotranslocation (Schutze et al., 1994; Lowe and Kreis, 1998). We therefore generated an N-terminal COPI binding site by mutating MAACEGRRS into MSRRRS (ERManICOPI+; Letourneur et al., 1994; Schutze et al., 1994). By doing so, a facilitated ER recycling of ERManI and an accelerated intracellular loss of transfected NHK are expected if ERManI participates in ERAD in the ER. To determine whether the incorporated COPI binding motif facilitates Golgi-to-ER retrotranslation of ERManI, both wild type and the ERManICOPI+ were transfected into the immortalized mouse fibroblast cell line NIH/3T3 because of its superior intracellular organelle morphology. The intracellular localization of both molecules was then examined by indirect immunofluorescence using the confocal microscope. The signal associated with the recombinant wild-type protein completely colocalized with the endogenous Golgi marker Giantin. In contrast, ERManICOPI+ exhibited only a partial overlap. The remainder exhibited a punctate staining pattern throughout the cytoplasm where it partially overlapped with endogenous β-COP, a component of COPI (Waters et al., 1991), and the ER marker PDI (Figure 6A). Subsequent quantification indicated that, although nearly 100% of cells transfected with wild-type ERManI showed Golgi localization of the molecule, ∼50% of the cells transfected with ERManICOPI+ showed a combination of both Golgi and ER localization (Figure 6B). These results confirmed that the manipulation was sufficient to alter the intracellular distribution of recombinant ERManI.
FIGURE 6:
COPI binding motif alters the intracellular distribution of ERManI. (A) Wild type or the ERManICOPI+ was transfected into NIH/3T3 cells. After culture on coverslips for 48 h, the cells were fixed and coimmunostained with anti-ERManI mAbs and anti-PDI or anti-β-COP antibodies. ERManI was visualized using Alexa 488–conjugated anti–mouse secondary antibody (green). PDI or β-COP was visualized using Alexa 555–conjugated anti–rabbit secondary antibodies (red). Cell nuclei were counterstained with To-PRO3. The inset on the bottom of each merged image shows a highly magnified view of the squared area. Arrowheads point to the cells transfected with wild type or mutant ERManI. Arrows show the colocalization of ERManICOPI+ with PDI or β-COP. (B) NIH/3T3 cells transfected and stained as in (A) were counted double-blinded in three repeated experiments. The bar graph shows the percentage of cells showing ERManI localization solely in the Golgi (Golgi) or in both Golgi and ER (Golgi+ER). Error bars represent SD from three repeated experiments.
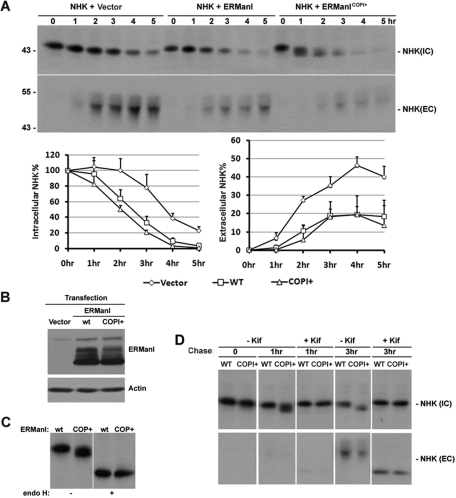
Presumably, if a fraction of Golgi-situated ERManI recycles back to the ER to tag substrates for ERAD, then the forced ER recycling of recombinant ERManI is predicted to accelerate the intracellular loss of transfected NHK. To test this hypothesis, we transfected either wild type or ERManICOPI+ into HeLa cells and examined changes in the rate at which cotransfected NHK was subjected to ERAD. Seventy-two hours posttransfection, cells were subjected to [35S]Met metabolic labeling followed by the immunoprecipitation of NHK from cell lysates. As expected, transfection with wild-type ERManI increased the rate of NHK intracellular degradation, with a concomitant decrease in the extent of its secretion (Figure 7A). Consistent with the facilitated recycling of ERManICOPI+ into the ER, the electrophoretic mobility of newly synthesized NHK in SDS–PAGE was greatly accelerated at 1 h of chase. The capacity of endoglycosidase H treatment to remove the mobility difference (Figure 7C) validated that the anomaly represented an accelerated rate at which N-glycans were trimmed. Furthermore, the increased electrophoretic mobility was completely abrogated in cells treated with kifunensine, indicating that the modification observed in untreated cells represents the removal of terminal α1,2-linked mannose units (Figure 7D), as predicted. With equivalent amounts of wild-type ERManI and ERManICOPI+ expressed (Figure 7B), no significant differences were detected in the rates of either intracellular NHK degradation or NHK secretion (Figure 7A). The combination of these experimental results argues against the current notion that the ERManI-mediated removal of mannose units to initiate ERAD occurs in the ER. Rather, the data favor the idea that the glycan-based tagging of substrates for ERManI takes place in the Golgi complex.
FIGURE 7:
COPI binding motif does not promote ERManI's function in ERAD. (A) NHK was cotransfected with empty vector, wild-type ERManI, or ERManICOPI+ into HeLa cells. Forty-eight hours after transfection, cells were labeled with 35S[Met] for 30 min and chased for indicated times. NHK was immunoprecipitated from cell lysates (IC) and medium (EC) collected at each time point and resolved on SDS–PAGE followed by autoradiography. Line graphs show the level of NHK quantified by densitometry, and the error bars represent SD values from three experiments. (B) A representative Western blot shows the equivalent expression of wild type and ERManICOPI+ using whole-cell lysates derived from transfected HeLa cells as in (A). (C) HeLa cells transfected with wild-type ERManI (wt) or ERManICOPI+ (COPI+) for 48 h were pulse-labeled with 35S[Met] for 30 min and chased for 1 h. NHK was immunoprecipitated, and the immunoprecipitates were treated with or without endoglycosidase H. The proteins were then eluted and resolved on SDS–PAGE followed by autoradiography. (D) HeLa cells transfected with wild-type ERManI (wt) or ERManICOPI+ (COPI+) for 48 h were pulse-labeled with 35S[Met] for 30 min and chased for indicated times in the presence or absence of kifunensine (Kif). NHK was immunoprecipitated from cell lysates, and culture medium was collected from each time point. Immunoprecipitates were resolved on SDS–PAGE followed by autoradiography.
DISCUSSION
The cotranslational addition of asparagine-linked glycans provides a scaffold by which the cellular proteostasis network is allowed to engage the productive folding and selective elimination of nascent misfolded proteins translocated into the secretory pathway. In the present study a panel of highly specific mAbs was used to demonstrate that endogenous human ERManI, which plays a pivotal rate-limiting role in ERAD substrate generation, is actually localized to the Golgi apparatus and therefore physically separated from the ER. Evaluating the consequences of appending a COPI binding motif to the N terminus of the recombinant protein, and identifying four sites for O-linked glycosylation, allowed us to establish that the enzyme functions in this organelle, rather than in the ER, to participate in the generation of a covalent tag that promotes the eventual intracellular disposal of specific N-glycosylated ERAD substrates.
The Saccharomyces cerevisiae orthologue, designated MNS1, was originally demonstrated to function as an ER-resident protein. The conclusion was based on its major enzymatic product (asparagine-linked Man8GlcNAc2), which is predominantly associated with glycoproteins that accumulate in budding yeast bearing the sec18 mutation, which disrupts the vesicular transport of protein cargo between the ER and Golgi (Esmon et al., 1984). Subsequently, immunofluorescence and immunoelectron microscopy localized the yeast orthologue (MNS1p) to the ER (Burke et al., 1996). The human orthologue, ERManI, was identified and cloned on the basis of its close sequence homology and was therefore predicted to localize and function in the ER (Gonzalez et al., 1999; Tremblay and Herscovics, 1999), irrespective of its more than 50% sequence homology with Golgi α1,2-mannosidases IA, IB, and IC (Tremblay and Herscovics, 1999). In previous studies, subcellular localization of the human orthologue to the ER was limited to the overexpressed or tagged recombinant molecule (Gonzalez et al., 1999), whereas others reported a juxtanuclear localization reminiscent of the Golgi complex (Avezov et al., 2008). To our knowledge, the present study provides the first unambiguous evidence demonstrating that endogenous ERManI is predominantly localized to the Golgi apparatus in mammalian cells. In support of this notion, a recent study reported that the plant Arabidopsis thaliana orthologue is localized to the Golgi complex (Liebminger et al., 2009). The Golgi localization of ERManI is apparently necessary for its function in ERAD, because overexpression of the COPI signal-modified recombinant molecule, which exhibited a partial intracellular redistribution and promoted the removal of α1,2-mannose units, was not capable of further accelerating misfolded glycoprotein degradation. These observations, together with the previous finding that Golgi α1,2-mannosidase IA, IB, and IC are all involved in glycoprotein quality control (Hosokawa et al., 2007), support a functional explanation as to why some N-glycosylated ERAD substrates must apparently cycle through the Golgi complex as a prerequisite for proteasomal destruction. Considering this arrangement, one might speculate that the evolutionary placement of ERManI in the Golgi complex (where all of the α1,2-mannosidases reside in higher eukaryotes), rather than in the ER, functions to prevent any direct competition between the folding and degradation systems. Nevertheless, we cannot rule out the possibility that ERManI might play a more complex role in ERAD than merely to remove α1,2-linked mannose units. Importantly, our present analysis of endogenous ERManI indicates that Lederkremer's (Avezov et al., 2008) detection of the recombinant protein in the ERQC likely represents an unfortunate artifact associated with its overexpression and treatment with proteasome inhibitors. In fact, our data support the original model proposed by Lederkremer and Glickman (Lederkremer and Glickman, 2005) in which vesicle recycling rather than strict ER retention contributes to the “timed” degradation of secretion-incompetent glycoproteins.
In budding yeast, two distinct substrate sorting mechanisms for ERAD have been reported. Whereas some substrates are constantly retained by a static ER retention mechanism before dislocation into the cytoplasm for disposal (retention pathway), others are packed into COPII vesicles and transported to the Golgi apparatus before their retrieval back to the ER for subsequent degradation (retrieval pathway) (Vashist et al., 2001). Functional heterogeneity also has been observed in other ways. For example, yeast MNS1p is required for the efficient degradation of CPY* and CPL* (Jakob et al., 1998; Hosomi et al., 2010), whereas it is dispensable for degradation of RTA and RTL (Hosomi et al., 2010). Therefore, any requirement for the mannosidase to function in ERAD is apparently substrate-dependent, and must be taken into consideration when studying glycoprotein quality control in mammalian cell lines. Cooper's group has proposed that involvement of the Golgi complex in ERAD requires that substrates contain ER exit signals (Kincaid and Cooper, 2007). No such signal has ever been identified, however, for the α1-antitrypsin molecule. Nevertheless, it is certainly possible that any role played by the mannosidase requires transient transport of the substrate to the Golgi apparatus. In support of this hypothesis, NHK was reported to partially localize to the Golgi complex under steady-state conditions (Hosokawa et al., 2007), indicating that it is capable of exiting the ER. In this system, the cycling of specific ERAD substrates through the Golgi complex apparently provides a method to spatially separate the sequential stages of glycoprotein biosynthetic quality control.
Multiple lines of evidence indicate that those molecules of ERManI that are not immediately degraded following biosynthesis and translocation in the ER lumen (Wu et al., 2007) are instead subjected to O-glycosylation following delivery to the Golgi complex. The slow rate at which sialic acid units are added to the overexpressed recombinant molecules, as compared with the endogenous wild-type counterpart, however, has precluded an accurate assessment of the real glycosylation dynamics. Nevertheless, the molecules' altered electrophoretic mobility in response to Sialidase A treatment indicates that the appendages are of the mucin type, which contains terminal sialic acid residues. O-glycosylation occurs at the stem region (amino acids 103–240), which contains a high frequency of serine, threonine, and proline residues, similar to that of the O-glycosylated ectodomain of several mucins (Jensen et al., 2010). The exact O-glycan structure is unknown. It seems to be cell type-dependent, however, as Sialidase A treatment nearly abolished all the altered electrophoretic migration detected in Huh7, but only partially removed the modification in HeLa and PLC/PRF5 cells (unpublished data). In addition, although the modification of ERManI is sensitive to Sialidase A, it is resistant to Sialidase V (Corfield et al., 1983; unpublished data). The properties and biological functions acquired by ERManI in response to O-glycosylation are currently under investigation. Based on our results, the modification is not required for localization in the Golgi compartment or its function in ERAD. However, the glycans might play an important role in preventing or promoting specific protein–protein interactions by possibly hindering the flexibility of the lumenal stem domain (Van den Steen et al., 1998; Jensen et al., 2010). In conclusion, identifying the Golgi complex as the primary location where endogenous ERManI operates has allowed us to propose a potential functional explanation as to why a class of misfolded glycoprotein ERAD substrates, such as NHK, is transported to the Golgi complex in mammalian cells.
The reported capacity of overexpressed ERManI to result in the inappropriate degradation of wild-type α1-antitrypsin and transferrin (Wu et al., 2003) implies that the wild-type folding intermediates likely recycle through the Golgi complex before reengagement with the molecular chaperone calnexin for subsequent conformational maturation. Taken together, the new findings shed additional light on the dynamics by which glycoprotein folding and ERAD operate as part of the cellular proteostasis network.
MATERIALS AND METHODS
cDNA constructs
ERManII cDNA clone MGC-1215 (IMAGE:3533651) was purchased from the American Type Culture Collection (ATCC; Manassas, VA). The gene's coding region as well as its 3′ untranslated region were amplified by PCR using forward primer 5′-ataagcttgcctgggtggcgaattc-3′ containing HindIII and EcoRI restriction endonuclease sites, and reverse primer 5′-agcggccgcatagatgcctcgag-3′containing NotI and XhoI restriction endonuclease sites. The cDNA fragment was subsequently digested with HindIII and NotI and cloned into vector pMH (Roche Applied Science, Indianapolis, IN). To generate the GST-ERManI construct, ERManII cDNA was released from the original MGC-1251 cDNA clone by EcoRI and XhoI digestion and subcloned into pEGX4T1 (GE Bio-Sciences, Piscataway, NJ). To generate the Flag-tagged ERManI construct, the ERManI cDNA was amplified using forward primer AAGACAAGCTT ATGGCTGCCTGCGAGGGCA containing HindIII site and reverse primer AATGCGGCCGCCTAGGCAGGGGTCCAGATAGGCA containing NotI site. The cDNA fragment was then digested with HindIII and NotI and cloned into pFlag-CMV vector. The O-glycosylation site mutations, as well as the N-tm mutant were generated using the QuikChange site-directed mutagenesis kit purchased from Stratagene (LaJolla, CA) following the manufacturer's instructions. The mutagenic primers are designed using the QuikChange primer design program from Stratagene, and the sequences are available upon request. All cDNA constructs were verified by nucleotide sequencing. cDNA constructs of α1-antitrypsin NHK were described previously (Wu et al., 2003).
Antibodies
Anti-ERManI monoclonal antibodies were generated using purified GST-ERManI recombinant protein as antigen. Production and purification of GST-ERManI recombinant proteins was performed following the procedure described previously (Pan et al., 2006). Injection of the purified GST-ERManI into BALB/c mice and screening of individual hybridoma clones were performed by A&G Precision Antibody (Columbia, MD). Polyclonal antibodies against N-terminal or C-terminal synthetic peptides of ERManI were generated by Alpha Diagnostic (San Antonio, TX). Polyclonal anti-BAP31 antibodies were generous gifts from Mitsuo Tagaya (Tokyo University of Pharmacy and Life Sciences, Tokyo, Japan). Anti-actin, anti-ManIA, and anti-Calnexin polyclonal antibodies, as well as anti–Flag tagged mAb, were purchased from Sigma Aldrich (St. Louis, MO). Anti-Giantin polyclonal antibodies were purchased from Abcam (Cambridge, MA). Anti–human α1-antitrypsin antibodies were purchased from MP Biomedicals (Solon, OH). Anti-PDI polyclonal antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell lines
HeLa cells were cultured in DMEM (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Gemini Bio-Products, West Sacramento, CA) and 1% ampicillin/streptomycin (Invitrogen, Carlsbad, CA). Huh7 and PCL/PRF5 cells (provided by Gretchen J. Darlington, Baylor College of Medicine, Houston, TX) were cultured in MEM (Mediatech) supplemented with 10% FBS and 1% ampicillin/streptomycin. Wild-type CHO cells and ldlD cell lines were provided by Monty Krieger (Massachusetts Institute of Technology, Boston, MA). The wild-type CHO cells were cultured in α-MEM (Invitrogen) supplemented with10% fetal calf serum (Gemini). The ldlD cells were grown and maintained in Ham's F-12 growth medium supplemented 5% dialyzed FBS (Invitrogen). When necessary, 20 μM galactose and 200 μM N-acetylgalactosamine (Sigma Aldrich) were added to restore the O-glycosylation process.
Transient transfection and Western blotting
The day before transfection, cells were plated into six-well dishes and allowed to reach 80% confluence by the time of transfection. cDNA plasmids (4 μg) or 4 μl of 20-nM siRNA were transfected into each well with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Posttransfection (24 or 48 h), cells were lysed and immunoblotted for ERManI following the protocol described previously (Wu et al., 2003). All siRNAs used in this study were purchased from Ambion (Austin, TX). The two ERManI-specific siRNA target sequences were GCTTTGGCGAGAGCTATGA (siRNA#1) and GTTACACTTTGAAAAGGAC (siRNA #2).
Protein identification by mass spectrometry
HeLa cell pellets (∼2 ml) were lysed in buffer containing 50 mM TrisHCl, 150 mM NaCl, 0.5% NP-40, 2 mM phenylmethylsulfonyl fluoride (PMSF), and additional protease inhibitors (Sigma) on ice for 30 min. After spinning down at 10,000 × g for 30 min, the supernatant was collected and incubated with 5 mg of 1D6 antibody immobilized onto 40 μl of protein G-agarose beads at 4°C overnight. After being washed six times with the lysis buffer, the immunoprecipitates were eluted with 100 μl of Laemmli sample buffer and resolved by 1% SDS–PAGE. The gel was silver-stained following protocols described previously (Pan et al., 2006), and the protein bands specifically precipitated by 1D6 were excised and identified at the Mass Spectrometry Core facility at Baylor College of Medicine.
Immunofluorescence staining
HeLa cells were cultured on 18-mm glass coverslips placed in a 35-mm culture dish before staining. Fixation, permeabilization, and staining of the cells were performed as previously described (Pan et al., 2006). Alexa Fluor 488–conjugated goat anti–mouse antibodies, Alexa Fluor 555–conjugated goat anti–rabbit antibodies, DAPI, TO-PRO3, and Prolong Gold mounting solution were purchased from Invitrogen. Images were captured using either an inverted Carl Zeiss fluorescence microscope or a Carl Zeiss LSM 510 Meta confocal microscope with 63× or 100× immersion oil objective (Carl Zeiss, Thornwood, NY). For the confocal microscope, the pinhole was set using the optimum condition button, and pixel time was set at 3.20 μs for an average of eight scans per track on each slice. Z-stack slices were set to 0.2 μm. Collected images by fluorescence microscope were processed using AxioVision Rel. 4.6 software, and the confocal images were processed using LSM 510 Image ver 3.2 SP2 (Carl Zeiss). The confocal images were taken in the Integrated Microscopy Core Laboratory, Baylor College of Medicine (Houston, TX).
Metabolic radiolabeling and immunoprecipitation
Cells cultured for at least 24 h were starved in methionine- and cysteine-free medium for 1 h and then were subjected to metabolic pulse-radiolabeling with [35S] Met for 20 min and chased for different time points following methods described previously (Graham et al., 1990). For kifunensine treatment, cells were preincubated for 1 h in 0.1 mM kifunensine (Toronto Research Chemicals, Ontario, Canada) before starvation, and the drug was present through the entire pulse-chase period. At each time point, cells were incubated on ice in lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40, 2 mM PMSF, and additional protease inhibitors (Sigma). Following centrifugation at 4°C for 30 min, the supernatant of each sample was collected and mixed with primary antibodies and protein G agarose beads (Calbiochem, Gibbstown, NJ). The mixtures were rotated at 4°C overnight. After stringent washes with lysis buffer, the immunoprecipitates were eluted with SDS sample buffer and separated by SDS–PAGE. In some experiments, immunoprecipitated protein was subjected to digestion with endoglycosidase H (New England Biolabs, Beverly, MA) as described previously (Sifers et al., 1988). Radiolabeled proteins were detected by autoradiography and quantified by NIH ImageJ software.
Protease protection assay
Subconfluent HeLa cells were washed in CSK buffer (0.3 M sucrose, 0.1 M KCL, 2.5 mM MgCl2, 1 mM EDTA, and 10 mM PIPES (pH 6.8) before incubating with 0.05 mg/ml Digitonin at room temperature for 5 min to selectively permeabilize the plasma membrane. After washing with CSK buffer, the permeabilized cells were incubated with 10 μg/ml trypsin on ice for 15 min. The reaction was stopped by the addition of 20 μg/ml trypsin inhibitor followed by washing with phosphate-buffered saline. Cells were then lysed with SVC buffer (50 mM Tris-HCl, pH 7.5, and 150 mM NaCl) containing 0.5% NP-40 and separated by SDS–PAGE, followed by Western blotting using ERManI, GM130, and ManIA antibodies.
Sialidase A treatment
Sialidase A was purchased from ProZyme (Hayward, CA). HeLa cells were lysed in SVC buffer containing 0.5% NP-40 on ice for 30 min, and the cell extracts were obtained by centrifugation at 10,000 × g for 30 min. The cell extracts were then used for Sialidase A treatment following the manufacturer's instructions. Briefly, the cell extracts were mixed with reaction buffer supplemented with 1% SDS and 0.5% β-mercaptoethanol, followed by heat denaturing at 95ºC for 5 min. After cooling down to room temperature, the sample was mixed with 1 μl of mock solution or Sialidase A and subsequently incubated at room temperature overnight. The samples were then mixed with SDS sample buffer and subjected to SDS–PAGE, followed by Western blotting using ERManI mAb.Fetuin (30 μg) derived from FBS (Sigma Aldrich) was used as a positive control.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants RO1 DK064232 (to R.N.S.), RO1 AI080656 (to M.K.E.), and R01 DK075322 (to K.W.M.), plus grant #R06-06 from the Alpha1-Foundation (to R.N.S.), a postdoctoral research grant (to S.P.) from the Alpha-1 Foundation, and a Pilot/Feasibility Grant as part of Grant P30 DK56338 from the National Institute of Diabetes and Digestive and Kidney Diseases. We thank the Baylor College of Medicine Mass Spectrometry Core for protein identification analysis and Sandra McGill for scientific editing.
Abbreviations used:
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERQC
ER protein quality control center
- PMSF
phenylmethylsulfonyl fluoride
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0118) on June 22, 2011.
REFERENCES
- Avezov E, Frenkel Z, Ehrlich M, Herscovics A, Lederkremer GZ. Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation. Mol Biol Cell. 2008;19:216–225. doi: 10.1091/mbc.E07-05-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Bergeron JJ, Brenner MB, Thomas DY, Williams DB. Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem Sci. 1994;19:124–128. doi: 10.1016/0968-0004(94)90205-4. [DOI] [PubMed] [Google Scholar]
- Bieberich E, Bause E. Man9-mannosidase from human kidney is expressed in COS cells as a Golgi-resident type II transmembrane N-glycoprotein. Eur J Biochem. 1995;233:644–649. doi: 10.1111/j.1432-1033.1995.644_2.x. [DOI] [PubMed] [Google Scholar]
- Bouchecareilh M, Conkright JJ, Balch WE. Proteostasis strategies for restoring alpha1-antitrypsin deficiency. Proc Am Thorac Soc. 2010;7:415–422. doi: 10.1513/pats.201001-016AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, McCracken AA. ER protein quality control and proteasome-mediated protein degradation. Semin Cell Dev Biol. 1999;10:507–513. doi: 10.1006/scdb.1999.0321. [DOI] [PubMed] [Google Scholar]
- Burke J, Lipari F, Igdoura S, Herscovics A. The Saccharomyces cerevisiae processing alpha 1,2-mannosidase is localized in the endoplasmic reticulum, independently of known retrieval motifs. Eur J Cell Biol. 1996;70:298–305. [PubMed] [Google Scholar]
- Cabral CM, Liu Y, Moremen KW, Sifers RN. Organizational diversity among distinct glycoprotein endoplasmic reticulum-associated degradation programs. Mol Biol Cell. 2002;13:2639–2650. doi: 10.1091/mbc.E02-02-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral CM, Liu Y, Sifers RN. Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem Sci. 2001;26:619–624. doi: 10.1016/s0968-0004(01)01942-9. [DOI] [PubMed] [Google Scholar]
- Caldwell SR, Hill KJ, Cooper AA. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J Biol Chem. 2001;276:23296–23303. doi: 10.1074/jbc.M102962200. [DOI] [PubMed] [Google Scholar]
- Corfield AP, Higa H, Paulson JC, Schauer R. The specificity of viral and bacterial sialidases for alpha(2-3)- and alpha(2-6)-linked sialic acids in glycoproteins. Biochim Biophys Acta. 1983;744:121–126. doi: 10.1016/0167-4838(83)90080-8. [DOI] [PubMed] [Google Scholar]
- Esmon B, Esmon PC, Schekman R. Early steps in processing of yeast glycoproteins. J Biol Chem. 1984;259:10322–10327. [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gonzalez DS, Karaveg K, Vandersall-Nairn AS, Lal A, Moremen KW. Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. J Biol Chem. 1999;274:21375–21386. doi: 10.1074/jbc.274.30.21375. [DOI] [PubMed] [Google Scholar]
- Goto M. Protein O-glycosylation in fungi: diverse structures and multiple functions. Biosci Biotechnol Biochem. 2007;71:1415–1427. doi: 10.1271/bbb.70080. [DOI] [PubMed] [Google Scholar]
- Graham KS, Le A, Sifers RN. Accumulation of the insoluble PiZ variant of human alpha 1-antitrypsin within the hepatic endoplasmic reticulum does not elevate the steady-state level of grp78/BiP. J Biol Chem. 1990;265:20463–20468. [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao K, et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J Biol Chem. 2006;281:9650–9658. doi: 10.1074/jbc.M512191200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, Sleno B, Kamiya Y, Wada I, Nagata K, Kato K, Herscovics A. EDEM1 accelerates the trimming of alpha1,2-linked mannose on the C branch of N-glycans. Glycobiology. 2010;20:567–575. doi: 10.1093/glycob/cwq001. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J Biol Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K. A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, You Z, Tremblay LO, Nagata K, Herscovics A. Stimulation of ERAD of misfolded null Hong Kong alpha1-antitrypsin by Golgi alpha1,2-mannosidases. Biochem Biophys Res Commun. 2007;362:626–632. doi: 10.1016/j.bbrc.2007.08.057. [DOI] [PubMed] [Google Scholar]
- Hosomi A, Tanabe K, Hirayama H, Kim I, Rao H, Suzuki T. Identification of an Htm1 (EDEM)-dependent, Mns1-independent Endoplasmic Reticulum-associated Degradation (ERAD) pathway in Saccharomyces cerevisiae: application of a novel assay for glycoprotein ERAD. J Biol Chem. 2010;285:24324–24334. doi: 10.1074/jbc.M109.095919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob CA, Burda P, Roth J, Aebi M. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PH, Kolarich D, Packer NH. Mucin-type O-glycosylation–putting the pieces together. FEBS J. 2010;277:81–94. doi: 10.1111/j.1742-4658.2009.07429.x. [DOI] [PubMed] [Google Scholar]
- Kamhi-Nesher S, Shenkman M, Tolchinsky S, Fromm SV, Ehrlich R, Lederkremer GZ. A novel quality control compartment derived from the endoplasmic reticulum. Mol Biol Cell. 2001;12:1711–1723. doi: 10.1091/mbc.12.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Kawaguchi S, Ng DT. The EDEM and Yos9p families of lectin-like ERAD factors. Semin Cell Dev Biol. 2007;18:743–750. doi: 10.1016/j.semcdb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Karaveg K, Siriwardena A, Tempel W, Liu ZJ, Glushka J, Wang BC, Moremen KW. Mechanism of class 1 (glycosylhydrolase family 47) {alpha}-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control. J Biol Chem. 2005;280:16197–16207. doi: 10.1074/jbc.M500119200. [DOI] [PubMed] [Google Scholar]
- Kincaid MM, Cooper AA. Misfolded proteins traffic from the endoplasmic reticulum (ER) due to ER export signals. Mol Biol Cell. 2007;18:455–463. doi: 10.1091/mbc.E06-08-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarsky K, Kingsley D, Krieger M. Use of a mutant cell line to study the kinetics and function of O-linked glycosylation of low density lipoprotein receptors. Proc Natl Acad Sci USA. 1988;85:4335–4339. doi: 10.1073/pnas.85.12.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19:515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Lederkremer GZ, Glickman MH. A window of opportunity: timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem Sci. 2005;30:297–303. doi: 10.1016/j.tibs.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, Demolliere C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Liebminger E, et al. Class I alpha-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell. 2009;21:3850–3867. doi: 10.1105/tpc.109.072363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xiao N, DeFranco DB. Use of digitonin-permeabilized cells in studies of steroid receptor subnuclear trafficking. Methods. 1999;19:403–409. doi: 10.1006/meth.1999.0876. [DOI] [PubMed] [Google Scholar]
- Liu Y, Choudhury P, Cabral CM, Sifers RN. Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J Biol Chem. 1999;274:5861–5867. doi: 10.1074/jbc.274.9.5861. [DOI] [PubMed] [Google Scholar]
- Lowe M, Kreis TE. Regulation of membrane traffic in animal cells by COPI. Biochim Biophys Acta. 1998;1404:53–66. doi: 10.1016/s0167-4889(98)00046-9. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls S, Snapp EL, Cole NB, Zaal KJ, Kenworthy AK, Roberts TH, Ellenberg J, Presley JF, Siggia E, Lippincott-Schwartz J. Dynamics and retention of misfolded proteins in native ER membranes. Nat Cell Biol. 2000;2:288–295. doi: 10.1038/35010558. [DOI] [PubMed] [Google Scholar]
- Olivari S, Cali T, Salo KE, Paganetti P, Ruddock LW, Molinari M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem Biophys Res Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- Pan S, Huang L, McPherson J, Muzny D, Rouhani F, Brantly M, Gibbs R, Sifers RN. Single nucleotide polymorphism-mediated translational suppression of endoplasmic reticulum mannosidase I modifies the onset of end-stage liver disease in alpha1-antitrypsin deficiency. Hepatology. 2009;50:275–281. doi: 10.1002/hep.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Wang R, Zhou X, He G, Koomen J, Kobayashi R, Sun L, Corvera J, Gallick GE, Kuang J. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J Biol Chem. 2006;281:34640–34650. doi: 10.1074/jbc.M602263200. [DOI] [PubMed] [Google Scholar]
- Schutze MP, Peterson PA, Jackson MR. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 1994;13:1696–1705. doi: 10.1002/j.1460-2075.1994.tb06434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifers RN. Cell biology. Protein degradation unlocked. Science. 2003;299:1330–1331. doi: 10.1126/science.1082718. [DOI] [PubMed] [Google Scholar]
- Sifers RN. Intracellular processing of alpha1-antitrypsin. Proc Am Thorac Soc. 2010;7:376–380. doi: 10.1513/pats.201001-011AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SL. A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem. 1988;263:7330–7335. [PubMed] [Google Scholar]
- Sommer T, Wolf DH. Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J. 1997;11:1227–1233. doi: 10.1096/fasebj.11.14.9409541. [DOI] [PubMed] [Google Scholar]
- Tomoda H, Omura S. Lactacystin, a proteasome inhibitor: discovery and its application in cell biology. Yakugaku Zasshi. 2000;120:935–949. doi: 10.1248/yakushi1947.120.10_935. [DOI] [PubMed] [Google Scholar]
- Tremblay LO, Herscovics A. Cloning and expression of a specific human alpha 1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2 isomer B during N-glycan biosynthesis. Glycobiology. 1999;9:1073–1078. doi: 10.1093/glycob/9.10.1073. [DOI] [PubMed] [Google Scholar]
- Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- Vashist S, Kim W, Belden WJ, Spear ED, Barlowe C, Ng DT. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana Y, Takai S, Nakajima K, Tani K, Yamamoto A, Watson P, Stephens DJ, Hauri HP, Tagaya M. Bap31 is an itinerant protein that moves between the peripheral endoplasmic reticulum (ER) and a juxtanuclear compartment related to ER-associated degradation. Mol Biol Cell. 2008;19:1825–1836. doi: 10.1091/mbc.E07-08-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MG, Serafini T, Rothman JE. “Coatomer”: a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature. 1991;349:248–251. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
- Wu Y, Swulius MT, Moremen KW, Sifers RN. Elucidation of the molecular logic by which misfolded alpha 1-antitrypsin is preferentially selected for degradation. Proc Natl Acad Sci USA. 2003;100:8229–8234. doi: 10.1073/pnas.1430537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Termine DJ, Swulius MT, Moremen KW, Sifers RN. Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J Biol Chem. 2007;282:4841–4849. doi: 10.1074/jbc.M607156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.