The sorting and assembly machinery (SAM) of mitochondria is essential for the sorting of β-barrel proteins. Different views have been presented on the role of polypeptide transport–associated (POTRA) domains in protein sorting. We show that the mitochondrial POTRA domain promotes the release of precursor proteins from the SAM complex.
Abstract
The mitochondrial outer membrane contains proteinaceous machineries for the translocation of precursor proteins. The sorting and assembly machinery (SAM) is required for the insertion of β‑barrel proteins into the outer membrane. Sam50 is the channel-forming core subunit of the SAM complex and belongs to the BamA/Sam50/Toc75 family of proteins that have been conserved from Gram-negative bacteria to mitochondria and chloroplasts. These proteins contain one or more N-terminal polypeptide transport-associated (POTRA) domains. POTRA domains can bind precursor proteins, however, different views exist on the role of POTRA domains in the biogenesis of β-barrel proteins. It has been suggested that the single POTRA domain of mitochondrial Sam50 plays a receptor-like function at the SAM complex. We established a system to monitor the interaction of chemical amounts of β-barrel precursor proteins with the SAM complex of wild-type and mutant yeast in organello. We report that the SAM complex lacking the POTRA domain of Sam50 efficiently binds β-barrel precursors, but is impaired in the release of the precursors. These results indicate the POTRA domain of Sam50 is not essential for recognition of β-barrel precursors but functions in a subsequent step to promote the release of precursor proteins from the SAM complex.
INTRODUCTION
Two different classes of integral membrane proteins are present in the mitochondrial outer membrane: proteins with α-helical transmembrane segments, and β-barrel proteins that are anchored in the outer membrane by multiple β-strands. β-barrel proteins are characteristic for the outer membranes of Gram-negative bacteria, mitochondria, and chloroplasts (Wimley, 2003; Gentle et al., 2005; Ruiz et al., 2006; Knowles et al., 2009; Walther and Rapaport, 2009; Schleiff and Becker, 2011). Mitochondrial β‑barrel proteins are essential for cell viability, since the central channel-forming component of the translocase of the outer membrane (TOM) is a β-barrel protein termed Tom40 (Hill et al., 1998; Suzuki et al., 2004; Becker et al., 2005). The TOM complex functions as the general entry gate for most mitochondrial proteins synthesized in the cytosol (Ryan et al., 2000; Mihara, 2003; Johnson and Jensen, 2004; Koehler, 2004; Dolezal et al., 2006; Neupert and Hermann, 2007; Chacinska et al., 2009; Endo and Yamano, 2009; Schmidt et al., 2010).
The precursors of mitochondrial β-barrel proteins are initially imported by the TOM complex, however, the TOM complex is not able to insert the β-barrel precursors into the outer membrane. Instead, the TOM complex translocates β-barrel precursors across the outer membrane to the intermembrane space side (Matouschek and Glick, 2001; Model et al., 2001; Becker et al., 2009; Walther and Rapaport, 2009; Endo and Yamano, 2010), where chaperone complexes formed by small TIM proteins bind to the β-barrel precursors and transfer them to the sorting and assembly machinery (SAM; Hoppins and Nargang, 2004; Wiedemann et al., 2004; Habib et al., 2005; Webb et al., 2006). The SAM complex is essential for the insertion of β-barrel precursors into the outer membrane and consists of four subunits termed Sam35, Sam37, Sam50, and Mdm10. Sam50 (Omp85/Tob55) is the central channel-forming subunit of the SAM complex (Kozjak et al., 2003; Paschen et al., 2003; Wiedemann et al., 2003; Gentle et al., 2004; Humphries et al., 2005; Kutik et al., 2008). Sam35 (Tob38/Tom38) cooperates with Sam50 in binding of β-barrel precursors that contain a sorting signal in the most C-terminal β‑strand (Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004; Kutik et al., 2008). Sam37 is involved in the release of β-barrel proteins from the SAM complex into the outer membrane (Chan and Lithgow, 2008; Dukanovic et al., 2009). The mitochondrial distribution and morphology protein 10 (Mdm10) associates with a fraction of SAM complexes and promotes the assembly of the TOM complex (Meisinger et al., 2004, 2006; Thornton et al., 2010; Wideman et al., 2010; Yamano et al., 2010a; Becker et al., 2011). Small subunits of the TOM complex, Tom5, Tom6 and Tom7, also function at the SAM complex to promote and modulate assembly of the precursor of Tom40 (Meisinger et al., 2006; Becker et al., 2010, 2011; Thornton et al., 2010; Yamano et al., 2010b).
Sam50 has been conserved in evolution and shows homology to the β-barrel assembly machinery (BAM) subunit A (BamA/Omp85) of Gram-negative bacteria and the translocase of the outer membrane of chloroplast (TOC) subunit 75 (Toc75; Kozjak et al., 2003; Paschen et al., 2003; Voulhoux et al., 2003; Gentle et al., 2004; Schleiff and Soll, 2005; Wu et al., 2005; Dolezal et al., 2006; Ruiz et al., 2006; Bos et al., 2007a; Kutik et al., 2009; Walther et al., 2009). Members of the BamA/Sam50/Toc75 family consist of a membrane-integral C-terminal domain that forms a β-barrel channel and a soluble N-terminal domain comprised of one or more polypeptide transport-associated (POTRA) domains (Sánchez-Pulido et al., 2003; Robert et al., 2006; Bredemeier et al., 2007; Habib et al., 2007; Kutik et al., 2008; Knowles et al., 2009; Schleiff and Becker, 2011). Different views have been presented about the function of the single POTRA domain of Sam50. Habib et al. (2007) showed that the POTRA domain binds precursor proteins and suggested a receptor-like function of the domain in the transfer of precursor proteins to the SAM complex. Kutik et al. (2008) deleted the entire POTRA domain and still observed precursor transfer to the SAM complex, suggesting that the POTRA domain of Sam50 is not essential for precursor targeting to the SAM complex.
For this study, we analyzed the role of the Sam50-POTRA domain in vivo and in organello. β-Barrel precursors could be accumulated at a POTRA-deficient SAM complex in chemical amounts; however, their release from the SAM complex was impaired. Thus, instead of functioning in the initial recognition of precursor proteins, the POTRA domain interacts with β-barrel precursors to promote their release from the SAM complex.
RESULTS
Involvement of the Sam50-POTRA domain in biogenesis of a mutant porin precursor
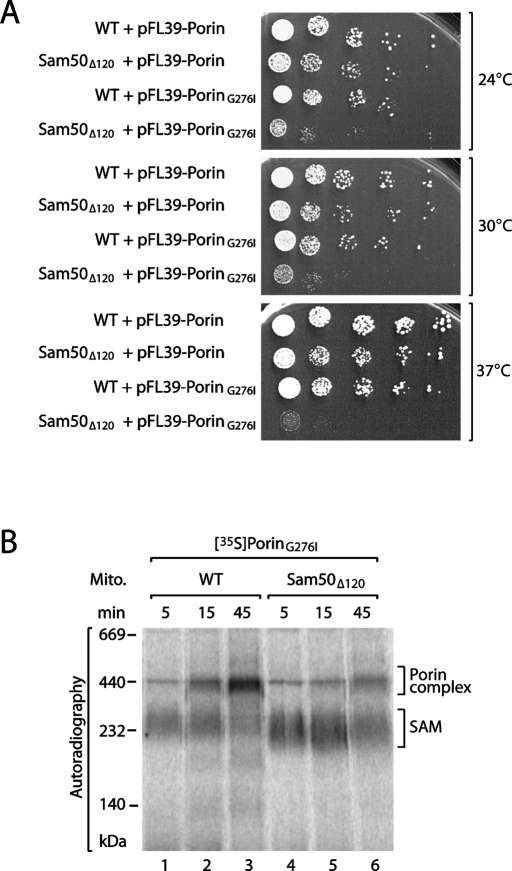
Yeast Sam50 consists of 484 amino acid residues. Residues 29–120 form the POTRA domain, whereas residues 121–484 constitute the membrane-integrated β-barrel domain (Kozjak et al., 2003; Sánchez-Pulido et al., 2003; Gentle et al., 2005; Bos et al., 2007b; Kim et al., 2007). A yeast strain with a deletion of the N-terminal 120 residues, including the entire POTRA domain, expressed growth similar to that of wild-type cells (Kutik et al., 2008). We searched for conditions where the lack of the POTRA domain is important for growth of yeast cells and coexpressed the precursor of porin, the most abundant β-barrel protein of mitochondria, from a single-copy plasmid with wild-type promoter in addition to the chromosomally expressed porin. Whereas coexpression of wild-type porin did not affect yeast growth, we observed that coexpression of a mutant form, porinG276I, impaired growth at 24–30°C and stopped growth of yeast at 37°C in Sam50Δ120, but not in wild-type yeast (Figure 1A). In porinG276I, a conserved glycine residue of the last β-strand (β-signal) has been replaced by isoleucine; this alteration does not affect recognition of the porin precursor by the SAM complex, but impairs its release from the complex (Kutik et al., 2008).
FIGURE 1:
The mitochondrial POTRA domain is required for the biogenesis of a mutant porin precursor. (A) Serial dilutions of wild-type (WT) and Sam50Δ120 yeast cells expressing an additional copy of either porin or porinG276I were plated onto minimal medium containing glucose as a carbon source and grown at the indicated temperatures. (B) Isolated mitochondria from wild-type or Sam50Δ120 yeast cells were incubated with radiolabeled [35S]porinG276I. The mitochondria were lysed with digitonin, and protein assembly was analyzed by blue native electrophoresis and digital autoradiography.
To directly study the role of the POTRA domain for the import of porinG276I into mitochondria, we used an in organello assay and analysis by native electrophoresis (Krimmer et al., 2001; Wiedemann et al., 2003; Meisinger et al., 2006; Kutik et al., 2008). PorinG276I, was synthesized and radiolabeled in a cell-free system (reticulocyte lysate) and imported into isolated yeast mitochondria. On lysis of the mitochondria with the nonionic detergent digitonin, mitochondrial protein complexes were separated by blue native electrophoresis, revealing mature porin complexes at ∼440 kDa and SAM-bound porin precursor at ∼250 kDa (Figure 1B; Kutik et al., 2008). In Sam50Δ120 mitochondria, the formation of mature porin was delayed, yet the accumulation of porinG276I at the SAM complex was enhanced (Figure 1B, lanes 4–6).
Accumulation of porin precursor at the SAM core complex in POTRA-deficient mitochondria
For in organello assays, mitochondrial precursor proteins are typically synthesized in rabbit reticulocyte lysates, leading to small (radiochemical) and variable amounts of precursor proteins. Recent studies showed that some precursor proteins could be efficiently synthesized in chemical amounts in a wheat germ–based translation system, and the mitochondrial import system challenged with saturating amounts of substrate (Becker et al., 2010, 2011; Thornton et al., 2010).
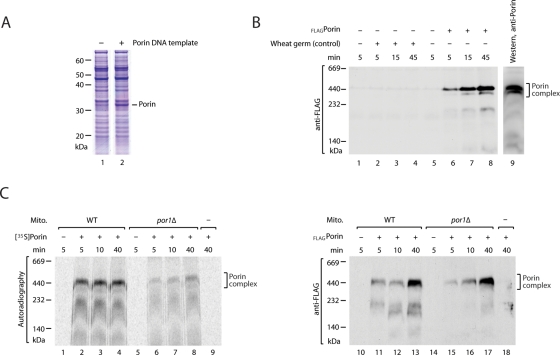
We asked if the wheat germ system could produce large amounts of import-competent porin precursor. On programming of the wheat germ system with porin DNA template, we observed a very efficient production, such that the porin precursor was directly visible in the complete wheat germ system by staining of an SDS-polyacrylamide gel with Coomassie Brilliant Blue R-250 (Figure 2A). To distinguish imported porin from endogenous mitochondrial porin, we synthesized the porin precursor with an N-terminal FLAG-tag in the wheat germ system. On incubation with isolated yeast wild-type mitochondria, imported porin formed complexes with a blue native mobility of ∼440 kDa (Figure 2B, lanes 6–8), similar to the mature endogenous porin complexes observed by decoration with porin-specific antibodies (Figure 2B, lane 9; Krimmer et al., 2001; Meisinger et al., 2001; Gentle et al., 2004; Waizenegger et al., 2004; Kozjak-Pavlovic et al., 2007; Kutik et al., 2008; Yamano et al., 2010a, 2010b). (It is likely that factors in the wheat germ system, such as molecular chaperones, are important for keeping β-barrel precursors in an import-competent state, since our previous attempts to import β-barrel precursors from urea-denatured inclusion bodies were not successful.) Previous studies using radiolabeled porin precursor showed that the precursor was efficiently assembled in wild-type mitochondria but not in por1Δ mitochondria that lacked the major mitochondrial porin1 (Figure 2C, left; Krimmer et al., 2001); the assembly of porin involves homooligomerization and the imported radiolabeled precursor molecules are apparently present in too small amounts to promote this interaction (Krimmer et al., 2001). We therefore assayed for the assembly of porin in por1Δ mitochondria to test whether the wheat germ system produced chemical amounts of import-competent porin. The porin precursor indeed efficiently assembled into mature porin complexes of ∼440 kDa (Figure 2C, right). We conclude that the wheat germ–based translation system leads to the synthesis of large amounts of assembly-competent porin precursor.
FIGURE 2:
Mitochondrial import of chemical amounts of porin. (A) Porin precursor was synthesized in vitro using a wheat germ–based cell-free expression system, followed by SDS–PAGE and staining with Coomassie Brilliant Blue R-250. (B) Wild-type (WT) mitochondria were incubated with chemical amounts of FLAGporin precursor produced in the wheat germ system (lanes 6–8). Control lanes (2–4), wheat germ system not programmed with porin DNA was added. Lanes 1 and 5, no wheat germ system added. The mitochondria were lysed by digitonin. Protein assembly was analyzed by blue native electrophoresis and Western blotting using antibodies against the FLAG epitope. For comparison, wild-type mitochondria were analyzed with antibodies against endogenous porin molecules (lane 9). (C) Left, radiochemical amounts of [35S]porin precursors were incubated with wild-type or por1Δ mitochondria and protein assembly was analyzed by blue native electrophoresis and digital autoradiography. Right, chemical amounts of FLAGporin were imported as above and analyzed by blue native gel electrophoresis, followed by Western blotting using antibodies against the FLAG epitope.
When chemical amounts of wild-type porin precursor were imported into Sam50Δ120 mitochondria, the assembly into mature porin complexes still occurred efficiently (Figure 3A). Only small amounts of porin precursor were observed in the range of the SAM-intermediate, both in wild-type mitochondria and Sam50Δ120 mitochondria (Figures 2, B and C, right, and 3A), indicating that the Sam50-POTRA domain was not crucial for the biogenesis of wild-type porin. A different result, however, was obtained when chemical amounts of the mutant precursor porinG276I were imported. In Sam50Δ120 mitochondria, porinG276I almost quantitatively accumulated in the 250-kDa region, whereas in wild-type mitochondria the precursor was found in both mature porin complexes and the SAM region (Figure 3B).
FIGURE 3:
Accumulation of mutant porin precursor at the SAM complex in Sam50Δ120 mitochondria. (A) Chemical amounts of FLAG-tagged wild-type porin precursors were incubated with wild-type (WT) or Sam50Δ120 mitochondria and protein assembly was analyzed by blue native electrophoresis and Western blotting with antibodies against the FLAG epitope. (B) FLAGPorinG276I precursors were synthesized in chemical amounts, imported into wild-type or Sam50Δ120 mitochondria, and analyzed by blue native electrophoresis. (C) SAM complex accumulation of FLAGporinG276I precursor in wild-type or Sam50Δ120 mitochondria was analyzed by blue native electrophoresis and Western blotting using antibodies against Sam35. * indicates SAM-precursor intermediate. (D) Growth at 30°C on minimal medium containing glucose as a carbon source of wild-type and Sam50Δ120 yeast cells harboring either the high-copy number plasmid YEp352 alone, or expressing Sam37, in addition to the plasmid pFL39 expressing either porin or porinG276I, as shown.
To determine whether porinG276I was indeed accumulated at the SAM complex, we analyzed the mobility of Sam35 on blue native electrophoresis by Western blotting. In wild-type mitochondria, two major blue native forms of the SAM complex can be distinguished: a SAMcore complex of ∼200 kDa consisting of Sam35, Sam37, and Sam50, and a larger SAMholo complex that additionally contains Mdm10 (Figure 3C, lane 1; Meisinger et al., 2004, 2006; Thornton et al., 2010; Yamano et al., 2010a, 2010b; Wideman et al., 2010; Becker et al., 2011). On import of chemical amounts of porinG276I, an additional Sam35-containing blue native band migrating between SAMcore and SAMholo became apparent (Figure 3C, lane 2, asterisk). In wild-type mitochondria, the intensity of this intermediate band decreased after longer import times, correlating to the dissociation of porinG276I from the SAM complex (Figure 3C, lanes 2–4, compared with Figure 3B, lanes 2–4). Thus the accumulation of chemical amounts of porinG276I at the SAM complex can be monitored by the blue native mobility of the SAM complex.
When porinG276I was imported into Sam50Δ120 mitochondria, the SAMcore complex was almost quantitatively shifted to the intermediate band (Figure 3C, lanes 6–8), in agreement with the massive accumulation of the precursor at the SAM complex (Figure 3B, lanes 6–8). Taken together, the in vivo experiments with coexpression of porin (Figure 1A) and the in organello experiments with chemical amounts of porin precursor (Figure 3, B and C) agree well, since both the growth defect and the massive accumulation of precursor at the SAM complex are observed when porinG276I and the Sam50Δ120 mutant are combined.
It has been shown that Sam37 assists release of substrates from the SAM complex (Chan and Lithgow, 2008). We therefore asked whether expression of Sam37 from a high-copy number plasmid could suppress the growth defect of Sam50Δ120 yeast expressing porinG276I; however, overexpression of Sam37 did not alter the growth behavior of the cells (Figure 3D). Thus overexpression of Sam37 cannot substitute for the POTRA domain, suggesting that Sam37 and the POTRA domain play distinct functions in the biogenesis of β‑barrel proteins.
Involvement of the POTRA domain in Tom40 release from the SAM complex
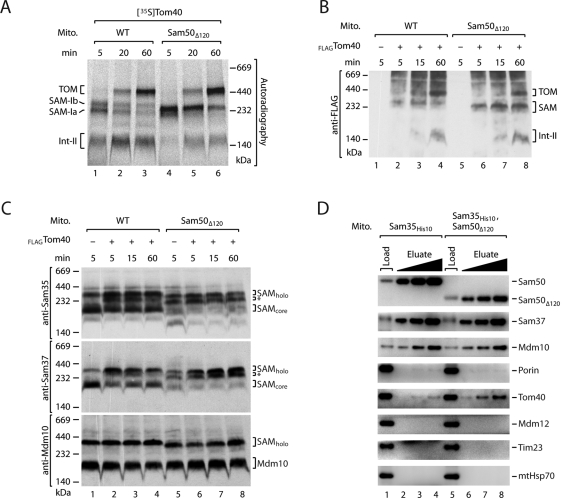
Tom40 is the second-most abundant β-barrel protein of the mitochondrial outer membrane. As the wild-type precursor of Tom40 stably interacts with the SAM complex during its biogenesis in vitro, Tom40 has become an important model substrate for analyzing the steps of β-barrel maturation at the SAM complex (Model et al., 2001; Paschen et al., 2003; Wiedemann et al., 2003; Gentle et al., 2004; Ishikawa et al., 2004; Humphries et al., 2005; Kutik et al., 2008). Recent studies resolved two stages of Tom40 interaction with the SAM complex that can be visualized with radiolabeled Tom40 precursor and high-resolution blue native electrophoresis: an initial stage of Tom40 binding to SAM, termed stage SAM-Ia; and a subsequent stage, SAM-Ib, that involves the association of small Tom proteins with the Tom40 precursor (Figure 4A, lane 1; Becker et al., 2010, 2011). The SAM-Ib stage is followed by release of the Tom40 precursor, leading to the smaller intermediate II (Int-II) and, subsequently, the assembly of Tom40 with Tom22 and other Tom subunits to form the mature TOM complex of ∼450 kDa (Figure 4A, lanes 2 and 3; Model et al., 2001; Wiedemann et al., 2003; Meisinger et al., 2004; Becker et al., 2010, 2011; Thornton et al., 2010). In Sam50Δ120 mitochondria, SAM-Ib was present in increased amounts, whereas the formation of Int-II was delayed (Figure 4A, lanes 4–6), supporting the view that Sam50Δ120 mutant mitochondria are slower in release of Tom40 precursor from the SAM complex. The overall efficiency of Tom40 assembly into the TOM complex of Sam50Δ120 mitochondria was either not or only mildly affected, compared with wild-type mitochondria, indicating the delayed release from the SAM complex was not rate-limiting with the tiny amounts of radiochemical precursor (Figure 4A; Kutik et al., 2008).
FIGURE 4:
Accumulation of Tom40 precursor at the SAM complex of Sam50Δ120 mitochondria. (A) [35S]Tom40 precursors were incubated with wild-type or Sam50Δ120 mitochondria. The mitochondria were lysed with digitonin and protein assembly was analyzed by blue native electrophoresis and digital autoradiography. (B)FLAGTom40 precursors were synthesized in chemical amounts and incubated with wild-type or Sam50Δ120 mitochondria. Protein import was analyzed by blue native gel electrophoresis and Western blotting with antibodies against the FLAG epitope. (C) SAM complex accumulation of FLAGTom40 precursor was analyzed as for (B), but with antibodies against SAM subunits. *, SAM-precursor intermediate. SAMholo can include two different forms of the SAM complex, SAM-Mdm10 and SAM-Tom5/Tom40 (Becker et al., 2010, 2011; Thornton et al., 2010). (D) Sam35His10 mitochondria or Sam35His10-Sam50Δ120 mitochondria were lysed with digitonin and subjected to metal affinity purification, SDS–PAGE, and immunoblotting. Load, 13% (3.25% for porin and Tom40); elution, 33%, 66%, and 100%.
We therefore synthesized the Tom40 precursor in the wheat germ system in order to challenge the import system with chemical amounts of wild-type precursor. The assembly steps of FLAG-tagged Tom40 were monitored by Western blot analysis of blue native gels (Figure 4B). The formation of mature TOM complex was delayed in Sam50Δ120 mitochondria, and substantial amounts of precursor accumulated at the SAM complex (Figure 4B, lanes 6–8). The accumulation of wild-type precursor at the SAM complex could be directly shown by a Western blot analysis for SAM subunits. The SAMcore complex was efficiently shifted to the SAM-precursor intermediate form in Sam50Δ120 mitochondria (Figure 4C, top and middle, lanes 6–8). In wild-type mitochondria, the formation of the SAM-precursor intermediate was observed after short import periods (Figure 4C, top and middle, lanes 2 and 3), whereas the intensity of the intermediate band decreased after longer import times concomitant with the formation of mature TOM complex (Figure 4, B and C, lane 4). The Mdm10-containing SAMholo complex was not affected by the chemical amounts of Tom40 in wild-type mitochondria or in Sam50Δ120 mitochondria (Figure 4C, bottom) in agreement with the role of SAMholo in the assembly of Tom22 (Meisinger et al., 2004; Thornton et al., 2010; Becker et al., 2011). (Only the faster mobility of SAMholo from Sam50Δ120 mitochondria compared with wild-type mitochondria was observed, due to the truncation of Sam50; Figure 4C, lower panel.) Taken together, lack of the Sam50-POTRA domain leads to an accumulation of wild-type Tom40 precursor at the SAM complex and delays the formation of mature TOM complex.
Remarkably, the SAM-precursor intermediate band could also be observed with Sam50Δ120 mitochondria in the absence of imported precursor (Figures 3C, lane 5, and 4C, top and middle, lane 5); upon import of chemical amounts of porinG276I or Tom40, the intensity of the band increased (Figures 3C and 4C). In contrast, in wild-type mitochondria without imported precursor, the SAM-precursor intermediate was not detected (Figures 3C, lane 1, and 4C, top and middle, lane 1). This finding raised the possibility that POTRA-deficient mitochondria may accumulate an endogenous β-barrel protein at the SAM complex. To study this possibility, we generated a Sam50Δ120 yeast strain carrying a His-tag fused to Sam35. The coding region for the His-tag was chromosomally inserted into the SAM35 gene, and the tagged Sam35 was expressed from its endogenous promoter (Milenkovic et al., 2004). Mitochondria were lysed by digitonin, and the SAM complex was purified via nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. All subunits of the SAM complex, Sam50, Sam37, and Mdm10, were efficiently copurified with tagged Sam35 independently of the presence or absence of the POTRA domain (Figure 4D). A considerable fraction of endogenous Tom40 molecules were copurified with the SAM complex of Sam50Δ120 mitochondria, whereas only small amounts were copurified from wild-type mitochondria (Figure 4D; Thornton et al., 2010). In contrast, porin was only copurified in very small amounts, and this occurred independent of the presence or absence of the POTRA domain. Further control proteins were not copurified with tagged Sam35, including outer membrane Mdm12, inner membrane Tim23, and matrix heat shock protein 70 (mtHsp70; Figure 4D).
We conclude that the POTRA-deficient mitochondria efficiently bind β-barrel precursors to the SAM complex, however, the release from the complex is impaired. The accumulation of wild-type Tom40 at the SAM complex is observed not only with imported precursor but also with endogenous Tom40.
Alteration of the SAM-Mim1 machinery upon partial deletion of the POTRA domain
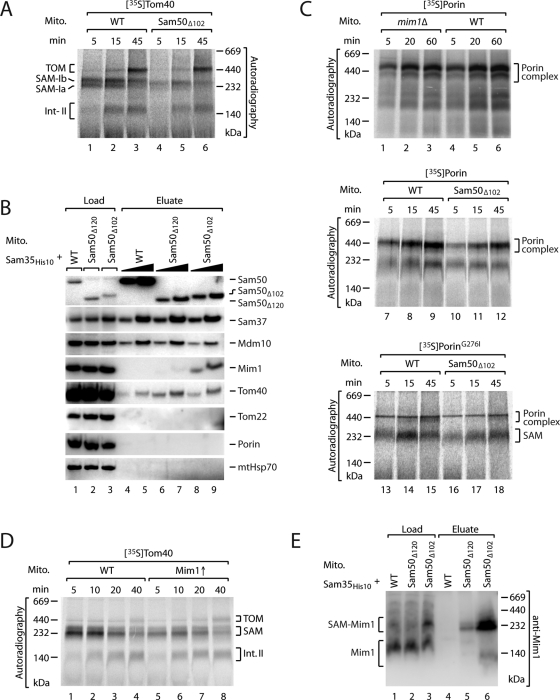
Finally, we asked why different phenotypes were reported for yeast deletion mutants of the Sam50-POTRA domain. Habib et al. (2007) deleted the N-terminal 102 amino acid residues of Sam50 that include the major part of the POTRA domain; they observed a growth defect of the yeast mutant cells and an impaired binding of the Tom40 precursor to the SAM complex. We deleted the N-terminal 120 residues of Sam50, including the entire POTRA domain, and did not observe a growth defect (Kutik et al., 2008) unless a mutant precursor was coexpressed (Figure 1A). The opposite result was obtained for the Tom40-SAM interaction: the binding of precursor to SAM was not inhibited, but increased amounts of precursor accumulated at the SAM complex (Figure 4). To exclude the possibility that different yeast strains were responsible for the observed effect, we also generated a yeast strain with the Δ102 partial POTRA deletion and confirmed the growth defect of the mutant yeast strain (Kutik et al., 2008). Mitochondria of Sam50Δ102 yeast were impaired in binding of Tom40 to the SAM complex (Figure 5A, lanes 4 and 5) in agreement with Habib et al. (2007). Thus the additional 18 amino acid residues (103–120) at the SAM complex exert a negative effect on β-barrel biogenesis and yeast growth.
FIGURE 5:
Partial deletion of the POTRA domain leads to accumulation of Mim1 at the SAM complex. (A) [35S]Tom40 precursors were incubated with wild-type (WT) or Sam50Δ102 mitochondria, and protein assembly was analyzed by blue native electrophoresis. (B) Mitochondria isolated from wild-type, Sam50Δ120, or Sam50Δ102 yeast cells containing Sam35His10 were subjected to affinity purification and analyzed by SDS–PAGE and Western blotting. Load: 12.5% for Sam50, Sam37, Mdm10, and Mim1; 6.25% for porin and mtHsp70; 3% for Tom40 and Tom22; elution, 50% and 100%. (C) Top, [35S]porin precursors were incubated with wild-type or mim1Δ mitochondria, and protein assembly was analyzed by blue native electrophoresis. [35S]porin (middle) and [35S]porinG276I (bottom) precursors were incubated with wild-type or Sam50Δ102 mitochondria and analyzed as above. (D) [35S]Tom40 precursors were incubated with either wild-type mitochondria or Mim1↑ mitochondria that were isolated from a strain overexpressing an additional copy of MIM1 from a high-copy number plasmid under galactose induction. Protein assembly was analyzed by blue native electrophoresis. (E) Affinity purification was performed as for (B), but analyzed by blue native electrophoresis and Western blotting with antibodies against Mim1. Load, 10%; elution, 100%.
To find a molecular explanation for the surprising differences between Sam50Δ102 mitochondria and Sam50Δ120 mitochondria, we analyzed the composition of the SAM complex. The Sam50Δ102 yeast mutant was chromosomally modified to express Sam35 with a His-tag as for the Sam50Δ120 yeast mutant. On lysis of mitochondria with digitonin, the subunits of the SAM complex were copurified with Sam35 from both Sam50Δ102 mitochondria and Sam50Δ120 mitochondria (Figure 5B). However, one striking difference was found: the mitochondrial import protein Mim1 copurified in large amounts with the SAM complex of Sam50Δ102 mitochondria (Figure 5B). The yield of copurification of Mim1 with Sam35 of Sam50Δ102 mitochondria was close to the yield observed for genuine SAM subunits, indicating that substantial amounts of Mim1 were bound to the SAM complex. In contrast, only small, substoichiometric amounts of Mim1 copurified with the SAM complex of Sam50Δ120 mitochondria (Figure 5B).
The outer membrane protein Mim1 is involved in the import of several outer membrane proteins that carry α-helical transmembrane segments and is transiently associated with the SAM complex in wild-type mitochondria (Becker et al., 2008, 2010; Hulett et al., 2008; Popov-Celeketic et al., 2008; Lueder and Lithgow, 2009). Two conceivable explanations account for the effect of the large amounts of Mim1 accumulated at the SAM complex of Sam50Δ102 mitochondria on the biogenesis of β-barrel proteins. 1) Mim1 plays a specific role in β-barrel biogenesis and the arrest of Mim1 at the SAM complex in the Sam50Δ102 mutant disturbs Mim1-specific functions; or 2) the accumulation of Mim1 in nearly stoichiometric amounts at the SAM complex impairs the access of β-barrel precursors to the SAM complex, and delays binding of the precursors to SAM.
We compared different assays to distinguish between both possibilities. 1) It has been shown that the deletion of MIM1 affects the biogenesis of Tom40 at the SAM complex (Ishikawa et al., 2004; Waizenegger et al., 2005; Becker et al., 2008; Hulett et al., 2008; Lueder and Lithgow, 2009). Mitochondria lacking Mim1 accumulate the precursor of Tom40 at the SAM-Ia stage (Becker et al., 2010). POTRA mutant mitochondria, however, affected the Tom40 biogenesis at different stages: Sam50Δ120 mitochondria accumulated Tom40 at the SAM-Ib stage, that is, after the Mim1-dependent step (Figure 4A), whereas Sam50Δ102 mitochondria were already impaired in formation of the SAM-Ia stage, that is, before the Mim1-dependent step (Figure 5A). 2) Lack of Mim1 does not inhibit the biogenesis of porin (Ishikawa et al., 2004; Waizenegger et al., 2005; Hulett et al., 2008), shown here with our mim1Δ mutant (Figure 5C, lanes 1–3). In contrast, Sam50Δ102 mitochondria show defects in the biogenesis of both Tom40 and porin (Habib et al., 2007). Our findings fully agree with those of Habib et al. (2007), in that Sam50Δ102 mitochondria are impaired in the import of both Tom40 and porin (Figures 5A, lanes 4–6, and 5C, lanes 10–12). Additionally, accumulation of the mutant precursor porinG276I at the SAM complex was delayed in Sam50Δ102 mitochondria (Figure 5C, lanes 16–18) in contrast to the increased accumulation of porinG276I at the SAM complex in Sam50Δ120 mitochondria (Figures 1B and 3, B and C). The results described in 1) and 2) show that the defects of Sam50Δ102 mitochondria cannot be attributed to Mim1-specific functions. 3) To study whether large amounts of Mim1 impair binding of β-barrel precursors to SAM, we generated a yeast strain that expressed Mim1 from a high-copy number plasmid and indeed observed a reduced binding of the Tom40 precursor to the SAM complex (Figure 5D, lanes 5–7 compared with lanes 1–3). 4) Finally, we analyzed where Mim1 accumulated in Sam50Δ102 mitochondria by combining affinity purification and blue native electrophoresis. Mim1 of wild-type mitochondria mainly migrates at ∼140 kDa on blue native gels and shows a similar behavior in Sam50Δ120 mitochondria (Figure 5E, lanes 1 and 2). With Sam50Δ102 mitochondria, however, a considerable fraction of Mim1 was found in a larger complex of ∼240 kDa (Figure 5E, lane 3). On purification of the SAM complex via His-tagged Sam35 and analysis by blue native electrophoresis, a large amount of Mim1 was selectively copurified in the high-molecular-weight form (Figure 5E, lane 6), demonstrating that this represents the SAM-bound form of Mim1, which is highly enriched in Sam50Δ102 mitochondria. Taken together, about one-half of the Mim1 molecules are arrested at the SAM complex in Sam50Δ102 mitochondria, impairing the binding of β‑barrel precursors to SAM.
DISCUSSION
We report that the single mitochondrial POTRA domain plays a role in the release of β-barrel precursors from the SAM complex. Different functions have been discussed for POTRA domains located at the N-termini of the conserved members of the BamA/Sam50/Toc75 family of translocase proteins, ranging from precursor binding to the interaction with other subunits of the translocases (Ertel et al., 2005; Bos et al., 2007a, 2007b; Bredemeier et al., 2007; Habib et al., 2007; Kim et al., 2007; Gatzeva-Topalova et al., 2008; Knowles et al., 2008, 2009; Schleiff and Becker, 2011). We observed that upon complete deletion of the mitochondrial POTRA domain, β-barrel precursors still efficiently bind to the SAM complex and are actually accumulated at SAM, excluding a scenario where the POTRA domain is crucial for the initial recognition of precursor proteins by the SAM complex.
We used two major mitochondrial β-barrel precursors, porin and Tom40, that show a different type of interaction with the SAM complex. Tom40 interacts with the SAM complex in several steps, and stable Tom40-SAM intermediates can be purified, whereas the precursor of porin only transiently interacts with the SAM complex, and large amounts of stable SAM-porin intermediates are therefore not observed with wild-type porin (Kutik et al., 2008). Deletion of the POTRA domain affected the Tom40 precursor but not the wild-type porin precursor. However, when a mutant porin precursor that interacts with the SAM complex in a prolonged manner due to an amino acid alteration in a transmembrane β-strand was used, deletion of the POTRA domain impaired the release of this precursor from SAM. Yeast cells tolerated the loss of the mitochondrial POTRA domain as long as wild-type β-barrel precursors were expressed, since their assembly pathway was delayed but not blocked. However, when the mutant form of porin was expressed in yeast, a lack of the POTRA domain led to a strong growth defect, since this β-barrel precursor was nearly quantitatively arrested at the SAM complex. We conclude that the mitochondrial POTRA domain plays a role in the biogenesis of β-barrel proteins under conditions where their interaction with the SAM complex is prolonged; the POTRA domain supports the release of these precursor proteins from the SAM complex.
Habib et al. (2007) showed that the mitochondrial POTRA domain binds β-barrel precursors. An interaction of precursor proteins/peptides with POTRA domains has also been shown in the bacterial and chloroplast systems (Ertel et al., 2005; Bos et al., 2007b; Kim et al., 2007; Gatzeva-Topalova et al., 2008; Knowles et al., 2008). Importantly, studies with bacterial POTRA domains revealed a broad specificity of binding of various β-strands by POTRA domains (Bos et al., 2007b; Kim et al., 2007; Knowles et al., 2008, 2009), supporting the view that POTRA domains do not function as selective receptors for β-barrel signals (Kutik et al., 2008). Taken together, these and our findings suggest a chaperone-like function of POTRA domains in guiding the maturation of β-barrel precursors in an efficient manner.
Our experimental results fully agree with those of Habib et al. (2007). The different conclusions of a receptor-like function of the mitochondrial POTRA domain versus a chaperone-like role in precursor release from the SAM complex can now be explained by the use of different Sam50-deletion mutants. When the entire POTRA domain is deleted (Sam50Δ120), β-barrel precursors efficiently bind to the SAM complex, but can be affected in release from SAM. However, when the POTRA domain is partially deleted, such that its last β-strand remains present in front of the β-barrel membrane domain of Sam50 (Sam50Δ102; Figure 6), the binding of β-barrel precursors to SAM is inhibited. Thus the remaining 18 amino acid residues (103–120) of the POTRA domain are not required for recognition of β-barrel precursors, but exert a negative effect. A detailed analysis revealed that the mitochondrial import protein Mim1, which only transiently interacts with the SAM complex in wild-type mitochondria (Becker et al., 2008; Lueder and Lithgow, 2009) and Sam50Δ120 mitochondria, accumulated in large amounts at the SAM complex of Sam50Δ102 mitochondria. Mim1 promotes the import of several α-helical proteins into the outer mitochondrial membrane (Becker et al., 2008; Hulett et al., 2008; Popov-Celeketic et al., 2008; Lueder and Lithgow, 2009), but only indirectly affects the biogenesis of Tom40 via the import of small Tom proteins (Ishikawa et al., 2004; Waizenegger et al., 2004; Becker et al., 2010). The defects of Sam50Δ102 mitochondria cannot be specific for Mim1 functions, since binding of the porin precursor to SAM is impaired in Sam50Δ102 mitochondria and Mim1 is not involved in the biogenesis of porin (Ishikawa et al., 2004; Waizenegger et al., 2005; Hulett et al., 2008). Additionally, with the precursor of Tom40 different import stages are affected by POTRA mutants and mim1 mutants (Becker et al., 2010; this paper). Rather, the accumulation of nearly stoichiometric amounts of Mim1 at the SAM complex leads to a nonphysiological situation that impairs the access of β-barrel precursors to the SAM complex and inhibits cell growth.
FIGURE 6:

Cartoon representation of the Sam50 POTRA model structure obtained by homology modeling using POTRA domain-containing protein structures as templates. Sequence identity among the used POTRA domains is low (<15%), yet the final model shows high structural similarity to them with root mean-square deviations between Cα-atoms ranging from 1.5 to 2.3 Å. The two α-helices and the 3-stranded mixed β-sheet, canonical elements of POTRA domains, are clearly visible in the model. The region colored in blue represents the residues H103 to P120 that remain present in front of the Sam50 β-barrel membrane domain in Sam50Δ102 mitochondria.
Bacterial BamA proteins contain five POTRA domains and chloroplast Toc75 proteins contain three POTRA domains; in addition to binding of precursor proteins, some POTRA domains in bacteria and chloroplasts are also involved in binding of further subunits of the translocase complexes (Ertel et al., 2005; Bos et al., 2007a; Bredemeier et al., 2007; Kim et al., 2007; Gatzeva-Topalova et al., 2008; Knowles et al., 2008, 2009; Hsu and Inoue, 2009; Schleiff and Becker, 2011). The single mitochondrial POTRA domain is not essential for the interaction of Sam50 with the other genuine subunits of the SAM complex, since all SAM subunits remain stably associated and can be copurified from the POTRA deletion mutants. However, as for the Sam50Δ102 mutant, the presence of a partial segment of the POTRA domain leads to a massive accumulation of a transient interaction partner, Mim1, at the SAM complex, indicating that this segment interferes with the SAM-Mim1 reaction cycle.
In summary, we conclude that the main function of the mitochondrial POTRA domain is to support the release of β-barrel precursors from the SAM complex, suggesting a chaperone-like role of the POTRA domain in β-barrel maturation. The hypothesis of a chaperone-like function also fits to the observed weak affinity and broad specificity of bacterial POTRA domains for different β-strands (Bos et al., 2007b; Clantin et al., 2007; Kim et al., 2007; Knowles et al., 2008, 2009).
MATERIALS AND METHODS
Yeast strains
Saccharomyces cerevisiae strains containing mutant or tagged SAM50 alleles were generated by plasmid shuffling in the background of the genomic deletion of SAM50 (Kozjak et al., 2003). Sam50Δ120 and Sam50Δ102 were generated as previously described (Kutik et al., 2008). For generation of the Sam35His10 strains, a 10-histidine residue coding tag (pFA-His10-HIS3MX6) was integrated 3′ to the SAM35 open reading frame (ORF) by homologous recombination (Meisinger et al., 2001; Milenkovic et al., 2004), yielding MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, sam50::ADE2, sam35::SAM35-His10-HIS3MX6 containing either pFL39-SAM50, pFL39-sam50Δ120, or pFL39-sam50Δ102. To coexpress porin from a plasmid, strains were transformed with the plasmid pFL39 encoding either porin or porinG276I (Kutik et al., 2008) under control of the endogenous POR1 promoter and terminator. Growth was assessed on SM‑TRP plates (0.67% [wt/vol] yeast nitrogen base without amino acids [BD Biosciences, San Jose, CA], 2% [wt/vol] glucose and CSM-TRP [Bio 101, MP Biomedicals, Solon, OH]). For concomitant overproduction of Sam37, strains were first transformed with either the high-copy number plasmid YEp352 (2μ) alone or encoding SAM37 under control of the endogenous SAM37 promoter and terminator. Following selection on SM-URA plates (CSM-URA [Bio 101]), strains were transformed with the pFL39 porin plasmids as described, and growth was assessed on SM‑URA‑TRP plates (CSM-URA-TRP [Bio 101]). The strain Mim1↑ and its corresponding wild-type strain were generated by transformation of the wild-type YPH499 strain (Sikorski and Hieter, 1989) with the plasmid pYES2 (Invitrogen, Carlsbad, CA), or pYES2-Mim1 harboring the MIM1 ORF cloned between HindIII and XbaI restriction sites. Precultures were grown on SM-URA media. Overexpression of Mim1 was induced by transfer of the cells to SM-URA media containing 2% [wt/vol] galactose. The por1Δ and mim1Δ strains have been described (Blachly-Dyson et al., 1990, 1997; Becker et al., 2008).
Isolation of mitochondria
The yeast strains were grown on YPG medium (1% [wt/vol] yeast extract, 2% [wt/vol] bactopeptone, and 3% [wt/vol] glycerol) at 24–30°C. Mitochondria were isolated by differential centrifugation according to standard procedures (Stojanovski et al., 2007). Protein concentrations were adjusted to 10 mg/ml in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM 3-(N-morpholino)-propanesulfonic acid [MOPS]-KOH, pH 7.2), and mitochondria were frozen in liquid nitrogen and stored at −80°C.
Mitochondrial protein import
For radiochemical in vitro import assays, mitochondrial precursor proteins were synthesized using pGEM4Z-based constructs (Kutik et al., 2008) as templates for coupled in vitro transcription and translation (Promega, Madison, WI) in the presence of [35S]methionine. For chemical amounts of mitochondrial precursor proteins, precursors were synthesized using an in vitro cell-free wheat germ–based expression system (5 Prime, Hamburg, Germany). Templates were generated by PCR from yeast DNA according to instructions by 5 Prime. To generate FLAG-tagged precursors, the coding sequence was incorporated 5′ to the ORF by primer overhang.
In vitro import assays were performed based on previously published procedures (Stojanovski et al., 2007), with modifications. Briefly, 50 μg mitochondria (protein amount) were incubated at 25°C with the precursor proteins in import buffer (3% [wt/vol] bovine serum albumin [BSA], 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM l-methionine, 2 mM KH2PO4, 10 mM MOPS-KOH, pH 7.2, 4 mM NADH, 4 mM ATP, 10 mM creatine phosphate, and 100 μg/ml creatine kinase). After indicated times, the import reaction was immediately cooled on ice. Mitochondria were reisolated, washed with SEM buffer, and lysed with either 1% (wt/vol) digitonin (as standard) or 0.35% (wt/vol) digitonin (to monitor porin assembly) in lysis buffer (20 mM Tris-HCl, pH 7.2, 0.1 mM EDTA, 50 mM NaCl, 10% [wt/vol] glycerol) at 1 mg protein/ml for 15 min on ice. After being clarifed via centrifugation, the solubilized material was separated by blue native gel electrophoresis as described previously (Dekker et al., 1998, Ryan et al., 2001, Stojanovski et al., 2007). Protein complexes generated by import of radiochemical precursor proteins were detected by digital autoradiography (Storm Imaging System, GE Healthcare, Waukesha, WI). Import of chemical amounts of precursors was visualized by Western blotting onto polyvinylidene fluoride membranes and immunodecoration with enhanced chemiluminescence (GE Healthcare) according to standard procedures.
Affinity purification of Sam35His10
For affinity purification of Sam35His10, mitochondria were lysed with 1% [wt/vol] digitonin in lysis buffer containing 30 mM imidazole, and subjected to Ni-NTA-agarose affinity chromatography (GE Healthcare). Unbound material was removed by washing with excess 1% digitonin in lysis buffer containing 50 mM imidazole. Proteins were eluted with Laemmli or digitonin lysis buffer containing 500 mM imidazole. Following SDS or blue native gel electrophoresis, eluted proteins were visualized by Western blotting.
Modeling of the Sam50 POTRA domain
The Sam50 POTRA sequence was aligned with homologous POTRA sequences from the Escherichia coli BamA (YaeT) protein (Kim et al., 2007), and the alignment was submitted to the HHpred server (http://toolkit.tuebingen.mpg.de/hhpred; Söding et al., 2005). Among the templates identified by HHPRED, only POTRA domains containing structures sharing the highest homology with the Sam50 POTRA domain and with a small E-value were considered for modeling. The final model was generated using MODELLER (http://salilab.org/modeller; Šali and Blundell, 1993) with POTRA domain structures of BamA (YaeT) (Kim et al., 2007; Gatzeva-Topalova et al., 2008, 2010), FhaC (Clantin et al., 2007), and Omp85 (Arnold et al., 2010; Koenig et al., 2010) as templates.
Acknowledgments
We thank Michael Forte for yeast strains and Oliver Schmidt, Natalia Gebert, and Michael Gebert for discussions. We are grateful to Agnes Schulze-Specking for expert technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746, Trinationales Graduiertenkolleg GRK 1478, Excellence Initiative of the German Federal & State Governments (EXC 294 BIOSS; GSC-4 Spemann Graduate School), Baden-Württemberg Stiftung, Bundesministerium für Bildung und Forschung, Landesforschungspreis Baden-Württemberg, and Gottfried Wilhelm Leibniz Program.
Abbreviations used:
- BAM
β-barrel assembly machinery of Gram-negative bacteria
- BSA
bovine serum albumin
- Mdm
mitochondrial distribution and morphology
- Mim1
mitochondrial import protein 1
- mtHsp70
matrix heat shock protein 70
- Ni-NTA
nickel-nitrilotriacetic acid
- ORF
open reading frame
- POTRA
polypeptide transport-associated domain
- SAM
sorting and assembly machinery of mitochondria
- TOC
translocase of the outer membrane of chloroplasts
- TOM
translocase of the outer membrane of mitochondria
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0148) on June 16, 2011.
REFERENCES
- Arnold T, Zeth K, Linke D. Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongates differs from proteobacterial Omp85 in structure and domain composition. J Biol Chem. 2010;285:18003–18015. doi: 10.1074/jbc.M110.112516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L, Bannwarth M, Meisinger C, Hill K, Krimmer T, Casadio R, Truscott KN, Schulz GE, Pfanner N, Wagner R. Preprotein translocase of the outer mitochondrial membrane: reconstituted Tom40 forms a characteristic TOM pore. J Mol Biol. 2005;353:1011–1020. doi: 10.1016/j.jmb.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Becker T, Gebert M, Pfanner N, van der Laan M. Biogenesis of mitochondrial membrane proteins. Curr Opin Cell Biol. 2009;21:484–493. doi: 10.1016/j.ceb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Becker T, Guiard B, Thornton N, Zufall N, Stroud DA, Wiedemann N, Pfanner N. Assembly of the mitochondrial protein import channel: role of Tom5 in two-stage interaction of Tom40 with the SAM complex. Mol Biol Cell. 2010;21:3106–3113. doi: 10.1091/mbc.E10-06-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Pfannschmidt S, Guiard B, Stojanovski D, Milenkovic D, Kutik S, Pfanner N, Meisinger C, Wiedemann N. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J Biol Chem. 2008;283:120–127. doi: 10.1074/jbc.M706997200. [DOI] [PubMed] [Google Scholar]
- Becker T, Wenz LS, Thornton N, Stroud DA, Meisinger C, Wiedemann N, Pfanner N. Biogenesis of mitochondria: dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J Mol Biol. 2011;405:113–124. doi: 10.1016/j.jmb.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Blachly-Dyson E, Peng S, Colombini M, Forte M. Selectivity changes in site-directed mutants of the VDAC ion channel: structural implications. Science. 1990;247:1233–1236. doi: 10.1126/science.1690454. [DOI] [PubMed] [Google Scholar]
- Blachly-Dyson E, Song J, Wolfgang WJ, Colombini M, Forte M. Multicopy suppressors of phenotypes resulting from the absence of yeast VDAC encode a VDAC-like protein. Mol Cell Biol. 1997;17:5727–5738. doi: 10.1128/mcb.17.10.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007a;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 2007b;8:1149–1154. doi: 10.1038/sj.embor.7401092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeier R, Schlegel T, Ertel F, Vojta A, Borissenko L, Bohnsack MT, Groll M, von Haeseler A, Schleiff E. Functional and phylogenetic properties of the pore-forming β-barrel transporters of the Omp85 family. J Biol Chem. 2007;282:1882–1890. doi: 10.1074/jbc.M609598200. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Lithgow T. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol Biol Cell. 2008;19:126–136. doi: 10.1091/mbc.E07-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clantin B, Delattre AS, Rucktooa P, Saint N, Méli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- Dekker PJT, Ryan MT, Brix J, Müller H, Hönlinger A, Pfanner N. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol Cell Biol. 1998;18:6515–6524. doi: 10.1128/mcb.18.11.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Dukanovic J, Dimmer KS, Bonnefoy N, Krumpe K, Rapaport D. Genetic and functional interactions between the mitochondrial outer membrane proteins Tom6 and Sam37. Mol Cell Biol. 2009;29:5975–5988. doi: 10.1128/MCB.00069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Yamano K. Multiple pathways for mitochondrial protein traffic. Biol Chem. 2009;390:723–730. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K. Transport of proteins across or into the mitochondrial outer membrane. Biochim Biophys Acta. 2010;1803:706–714. doi: 10.1016/j.bbamcr.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Ertel F, Mirus O, Bredemeier R, Moslavac S, Becker T, Schleiff E. The evolutionarily related β-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J Biol Chem. 2005;280:28281–28289. doi: 10.1074/jbc.M503035200. [DOI] [PubMed] [Google Scholar]
- Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure. 2010;18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- Habib SJ, Waizenegger T, Lech M, Neupert W, Rapaport D. Assembly of the TOB complex of mitochondria. J Biol Chem. 2005;280:6434–6440. doi: 10.1074/jbc.M411510200. [DOI] [PubMed] [Google Scholar]
- Habib SJ, Waizenegger T, Niewienda A, Paschen SA, Neupert W, Rapaport D. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial β-barrel proteins. J Cell Biol. 2007;176:77–88. doi: 10.1083/jcb.200602050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hoppins SC, Nargang FE. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J Biol Chem. 2004;279:12396–12405. doi: 10.1074/jbc.M313037200. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Inoue K. Two evolutionarily conserved essential β-barrel proteins in the chloroplast outer envelope membrane. Biosci Trends. 2009;3:168–178. [PubMed] [Google Scholar]
- Hulett JM, Lueder F, Chan NC, Perry AJ, Wolynec P, Likic' VA, Gooley PR, Lithgow T. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J Mol Biol. 2008;376:694–704. doi: 10.1016/j.jmb.2007.12.021. [DOI] [PubMed] [Google Scholar]
- Humphries AD, Streimann IC, Stojanovski D, Johnston AJ, Yano M, Hoogenraad NJ, Ryan MT. Dissection of the mitochondrial import and assembly pathway for human Tom40. J Biol Chem. 2005;280:11535–11543. doi: 10.1074/jbc.M413816200. [DOI] [PubMed] [Google Scholar]
- Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AE, Jensen RE. Barreling through the membrane. Nat Struct Mol Biol. 2004;11:113–114. doi: 10.1038/nsmb0204-113. [DOI] [PubMed] [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, Overduin M, Henderson IR. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol. 2009;7:206–214. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]
- Koehler CM. New developments in mitochondrial assembly. Annu Rev Cell Dev Biol. 2004;20:309–335. doi: 10.1146/annurev.cellbio.20.010403.105057. [DOI] [PubMed] [Google Scholar]
- Koenig P, Mirus O, Haarmann R, Sommer MS, Sinning I, Schleiff E, Tews I. Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85. J Biol Chem. 2010;285:18016–18024. doi: 10.1074/jbc.M110.112649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- Kozjak-Pavlovic V, Ross K, Benlasfer N, Kimmig S, Karlas A, Rudel T. Conserved roles of Sam50 and metaxins in VDAC biogenesis. EMBO Rep. 2007;8:576–582. doi: 10.1038/sj.embor.7400982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimmer T, et al. Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J Cell Biol. 2001;152:289–300. doi: 10.1083/jcb.152.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Kutik S, Stroud DA, Wiedemann N, Pfanner N. Evolution of mitochondrial protein biogenesis. Biochim Biophys Acta. 2009;1790:409–415. doi: 10.1016/j.bbagen.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Lueder F, Lithgow T. The three domains of the mitochondrial outer membrane protein Mim1 have discrete functions in assembly of the TOM complex. FEBS Lett. 2009;583:1475–1480. doi: 10.1016/j.febslet.2009.03.064. [DOI] [PubMed] [Google Scholar]
- Matouschek A, Glick BS. Barreling through the outer membrane. Nat Struct Biol. 2001;8:284–286. doi: 10.1038/86140. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Ryan MT, Hill K, Model K, Lim JH, Sickmann A, Müller H, Meyer HE, Wagner R, Pfanner N. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol Cell Biol. 2001;21:2337–2348. doi: 10.1128/MCB.21.7.2337-2348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Wiedemann N, Rissler M, Strub A, Milenkovic D, Schönfisch B, Müller H, Kozjak V, Pfanner N. Mitochondrial protein sorting: differentiation of β-barrel assembly by Tom7-mediated segregation of Mdm10. J Biol Chem. 2006;281:22819–22826. doi: 10.1074/jbc.M602679200. [DOI] [PubMed] [Google Scholar]
- Mihara K. Cell biology: moving inside membranes. Nature. 2003;424:505–506. doi: 10.1038/424505a. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Kozjak V, Wiedemann N, Lohaus C, Meyer HE, Guiard B, Pfanner N, Meisinger C. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J Biol Chem. 2004;279:22781–22785. doi: 10.1074/jbc.C400120200. [DOI] [PubMed] [Google Scholar]
- Model K, Meisinger C, Prinz T, Wiedemann N, Truscott KN, Pfanner N, Ryan MT. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat Struct Biol. 2001;8:361–370. doi: 10.1038/86253. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Popov-Čeleketić J, Waizenegger T, Rapaport D. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J Mol Biol. 2008;376:671–680. doi: 10.1016/j.jmb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Robert V, Volokhina EB, Senf F, Bos MP, van Gelder P, Tommassen J. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy T. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- Ryan MT, Voos W, Pfanner N. Assaying protein import into mitochondria. Meth Cell Biol. 2001;65:189–215. doi: 10.1016/s0091-679x(01)65012-x. [DOI] [PubMed] [Google Scholar]
- Ryan MT, Wagner R, Pfanner N. The transport machinery for the import of preproteins across the outer mitochondrial membrane. Int J Biochem Cell Biol. 2000;32:13–21. doi: 10.1016/s1357-2725(99)00114-4. [DOI] [PubMed] [Google Scholar]
- Šali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sánchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. POTRA: a conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem Sci. 2003;28:523–526. doi: 10.1016/j.tibs.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Becker T. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat Rev Mol Cell Biol. 2011;12:48–59. doi: 10.1038/nrm3027. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Soll J. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 2005;6:1023–1027. doi: 10.1038/sj.embor.7400563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Meth Cell Biol. 2007;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kadowaki T, Maeda M, Sasaki H, Nabekura J, Sakaguchi M, Mihara K. Membrane-embedded C-terminal segment of rat mitochondrial TOM40 constitutes protein-conducting pore with enriched β-structure. J Biol Chem. 2004;279:50619–50629. doi: 10.1074/jbc.M408604200. [DOI] [PubMed] [Google Scholar]
- Thornton N, Stroud DA, Milenkovic D, Guiard B, Pfanner N, Becker T. Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of α-helical outer membrane proteins. J Mol Biol. 2010;396:540–549. doi: 10.1016/j.jmb.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Waizenegger T, Habib SJ, Lech M, Mokranjac D, Paschen SA, Hell K, Neupert W, Rapaport D. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep. 2004;5:704–709. doi: 10.1038/sj.embor.7400183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger T, Schmitt S, Zivkovic J, Neupert W, Rapaport D. Mim1, a protein required for the assembly of the TOM complex of mitochondria. EMBO Rep. 2005;6:57–62. doi: 10.1038/sj.embor.7400318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Rapaport D. Biogenesis of mitochondrial outer membrane proteins. Biochim Biophys Acta. 2009;1793:42–51. doi: 10.1016/j.bbamcr.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM. Crystal structure of the mitochondrial chaperone TIM9-10 reveals a six-bladed α-propeller. Mol Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Wideman JG, Go NE, Klein A, Redmond E, Lackey SWK, Tao T, Kalbacher H, Rapaport D, Neupert W, Nargang FE. Roles of the Mdm10, Tom7, Mdm12 and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol Biol Cell. 2010;21:1725–1736. doi: 10.1091/mbc.E09-10-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Truscott KN, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem. 2004;279:18188–18194. doi: 10.1074/jbc.M400050200. [DOI] [PubMed] [Google Scholar]
- Wimley WC. The versatile β-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Yamano K, Tanaka-Yamano S, Endo T. Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep. 2010a;11:187–193. doi: 10.1038/embor.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Tanaka-Yamano S, Endo T. Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J Biol Chem. 2010b;285:41222–41231. doi: 10.1074/jbc.M110.163238. [DOI] [PMC free article] [PubMed] [Google Scholar]