Claspin mediates the activation of checkpoint kinase 1 (Chk1) by ATM- and Rad3-related kinase (ATR) in response to genomic stress. This process depends upon phosphorylation of Claspin on two critical residues. These phosphorylations allow docking of Claspin with Chk1. In this study, we identified CK1γ1 as a kinase that carries out these key phosphorylations of Claspin.
Abstract
The mediator protein Claspin is critical for the activation of the checkpoint kinase Chk1 during checkpoint responses to stalled replication forks. This function involves the Chk1-activating domain (CKAD) of Claspin, which undergoes phosphorylation on multiple conserved sites. These phosphorylations promote binding of Chk1 to Claspin and ensuing activation of Chk1 by ATR. However, despite the importance of this regulatory process, the kinase responsible for these phosphorylations has remained unknown. By using a multifaceted approach, we have found that casein kinase 1 gamma 1 (CK1γ1) carries out this function. CK1γ1 phosphorylates the CKAD of Claspin efficiently in vitro, and depletion of CK1γ1 from human cells by small interfering RNA (siRNA) results in dramatically diminished phosphorylation of Claspin. Consequently, the siRNA-treated cells display impaired activation of Chk1 and resultant checkpoint defects. These results indicate that CK1γ1 is a novel component of checkpoint responses that controls the interaction of a key checkpoint effector kinase with its cognate mediator protein.
INTRODUCTION
In eukaryotic cells, duplication of the genome and cell division must be coordinated faithfully. Otherwise, genomic defects might be propagated to progeny cells. In vertebrates, the checkpoint kinase Chk1 plays a critical role in preventing the continuation of the cell cycle if the genome has undergone any perturbation (Cimprich and Cortez, 2008). Genomic disruptions trigger the prompt activation of Chk1, which in turn phosphorylates key cell cycle control enzymes such as Cdc25 and Wee1. These reactions dampen the activation of cyclin-dependent kinases (Cdks) until the cell can rectify lesions in the DNA. The activation of Chk1 involves phosphorylation of its C-terminal regulatory domain on multiple sites by a master checkpoint regulatory kinase called ATM- and Rad3-related kinase (ATR). ATR exists in a tight complex with a partner called ATR-interacting protein (ATRIP; Cortez et al., 2001; Zou and Elledge, 2003). Upon occurrence of replication blockages in the nucleus, for example, the ATR–ATRIP complex undergoes activation in a manner that depends upon an intricate upstream regulatory pathway. In vertebrates, the culmination of this pathway results in the physical association of ATR–ATRIP with a specific activating protein known as topoisomerase IIβ-binding protein 1 (TopBP1; Kumagai et al., 2006; Mordes et al., 2008).
Significantly, the activated ATR–ATRIP complex also requires the assistance of a mediator or adaptor protein to effectively phosphorylate Chk1. In vertebrates, this protein is known as Claspin (Kumagai and Dunphy, 2000; Chini and Chen, 2003; Lee et al., 2003; Lin et al., 2004). ATR has a higher propensity to recognize the Claspin–Chk1 complex than the isolated Chk1 protein (Kumagai et al., 2004; Lindsey-Boltz et al., 2009). Significantly, Claspin also interacts in a highly specific manner with replication forks, which suggests this protein helps the cell to monitor the progress of DNA replication (Lee et al., 2005; Petermann et al., 2008; Scorah and McGowan, 2009). The association of Chk1 with Claspin depends upon phosphorylation of Claspin on multiple conserved residues within its Chk1-activating domain (CKAD; Kumagai and Dunphy, 2003; Clarke and Clarke, 2005; Lee et al., 2005; Chini and Chen, 2006; Bennett et al., 2008). These sites reside in highly conserved repeats of about ten amino acids (there are two such repeats in Xenopus Claspin and three in its human homologue). During a checkpoint response, serine and threonine residues (e.g., Ser864 and Ser895 in Xenopus and Thr916, Ser945, and Ser982 in humans) within each repeat undergo phosphorylation. These phosphorylations create phosphopeptide sequences that dock with the catalytic domain of Chk1, thereby promoting an increase in its kinase activity (Jeong et al., 2003). Mutagenesis studies have indicated that both phosphorylations of Xenopus Claspin and at least two out of three phosphorylations of the human protein are critical for the ultimate activation of Chk1 (Kumagai and Dunphy, 2003; Bennett et al., 2008).
Despite the importance of these processes, the kinase responsible for these modifications has not yet been identified. These phosphorylations are dependent upon ATR, but ATR does not phosphorylate these sites directly (Kumagai and Dunphy, 2003). Although it has been reported that Chk1 can phosphorylate Thr916 of human Claspin in vitro (Chini and Chen, 2006), more recent evidence has established that Chk1 is not responsible for the phosphorylation of the CKAD in vivo (Bennett et al., 2008). Thus, the identity of the Claspin-activating kinase has remained an open question. In this study, we have adopted a multifaceted approach to identify kinase(s) responsible for phosphorylation of the CKAD of Claspin. We first performed a whole-kinome RNAi screen in Drosophila S2R+ cells to identify kinases that influence the activation of Chk1 (Bettencourt-Dias et al., 2004). We followed through with these studies by carrying out various functional tests in both Drosophila S2R+ and human cells. Our results have indicated that one or more forms of casein kinase 1 gamma 1 (CK1γ1) can phosphorylate Claspin very effectively and regulate its ability to activate Chk1. Therefore, we have identified a novel player in the regulatory mechanisms through which metazoan cells safeguard genomic integrity.
RESULTS
Kinome-wide RNAi screen in Drosophila S2R+ cells
To identify the kinase(s) responsible for phosphorylation of the CKAD of Claspin, we carried out a kinome-wide RNA interference (RNAi) screen in Drosophila S2R+ tissue culture cells. Initially, we screened for kinases whose knockdown would diminish phosphorylation of Chk1. We reasoned that this approach might identify both the Claspin-regulatory kinase and other kinases that could affect the Chk1-activating pathway. Owing to technical limitations in detecting phosphorylation of the endogenous Drosophila Chk1 (Grapes), we decided to express an inducible version of Xenopus Chk1 in the S2R+ cells (Figure 1A). There are various high-quality antibodies available that detect the whole Xenopus Chk1 protein and phosphorylation of two residues involved in its activation (Thr314 and Ser344). As shown in Figure 1B, treatment of cells with either aphidicolin (APH) or hydroxyurea (HU) resulted in robust phosphorylation of the exogenously expressed Chk1. The kinetics of this activation closely mirrored that of the endogenous Chk1 in human cells (Figure 1C). For example, in both Drosophila and human cells, strong phosphorylation of Chk1 occurred within 5 min after addition of APH. To assess whether this phosphorylation involves the same regulators as in other organisms, we treated the cells with interfering dsRNAs directed against the Drosophila versions of ATR (Mei-41) and Claspin (CG32251/CG1326). We found that both dsRNAs dramatically reduced the phosphorylation of Chk1 in response to APH and HU (Figure 1, D and E). Taken together, these tests indicate that this Chk1 reporter system recapitulates a faithful and specific checkpoint response.
FIGURE 1:
Assay of DNA damage checkpoint responses in Drosophila S2R+ cells. (A) Diagram showing the Xenopus Chk1 reporter construct used in Drosophila S2R+ cells. MT, metallothionein promoter; NLS, nuclear localization signal. (B) Expression of the Chk1-GST fusion protein was induced for 16 h and cells were treated in the absence or presence of either APH (10 μg/ml for 10 min) or HU (2 mM for 16 h). Whole-cell lysates were prepared and immunoblotted with antibodies against the Ser344-phosphorylated form of Xenopus Chk1 (Top) or GST (Bottom). (C) The Drosophila reporter cells and human U2OS cells were treated with APH for the indicated length of times. Whole-cell lysates were immunoblotted with the indicated antibodies against phospho-Chk1, GST, and human Chk1. (D and E) Phosphorylation of Chk1 in Drosophila cells is dependent on ATR and Claspin. Drosophila reporter cells were incubated with dsRNAs against Mei-41 (M) or Claspin (Cl) for 96 h. Subsequently, cells were treated with APH or HU, as in (B). Whole-cell lysates were prepared and analyzed by immunoblotting. dsRNA against firefly luciferase (F) was used as a control.
After transfecting individual dsRNAs targeting each of 228 Drosophila kinases into the reporter cells, we monitored the effect on the APH-induced phosphorylation of Chk1 by immunoblotting with antiphosphopeptide antibodies. After multiple rounds of screening, we found approximately 16 dsRNAs that elicited a moderate to strong reduction in the phosphorylation of Chk1 (unpublished data). The results of the entire screen will be reported elsewhere. One positive dsRNA was directed against the casein kinase Gish. Moreover, dsRNAs against the casein kinases CG7094 and CG12147 caused some reduction in the phosphorylation of Chk1 in the initial round of screening, although this effect was not apparent in a secondary round of screening. In general, casein kinases have a preference for acidic substrates (Cheong and Virshup, 2011). Interestingly, the sequences around the critical phosphorylation sites in the CKAD of Claspin are also quite acidic. For example, the Xenopus CKAD repeat 1 sequence is MDELLDLCSGQF.
Identification of Drosophila Gish, a homologue of casein kinase 1γ, as a kinase that directly and specifically phosphorylates Claspin
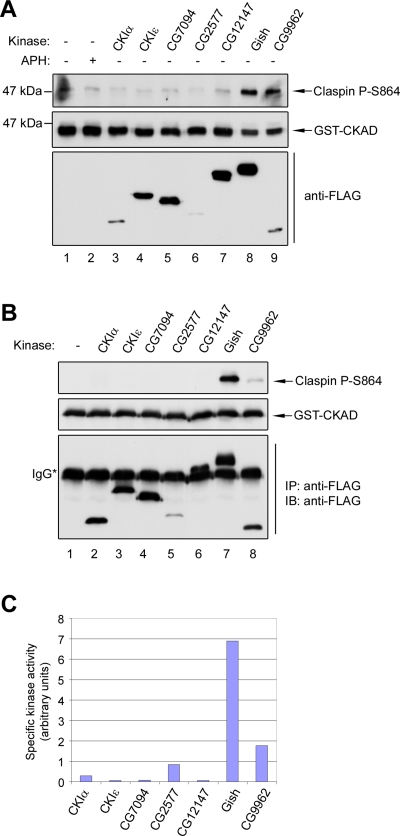
On the basis of these considerations, we decided to coexpress Gish as well as other Drosophila casein kinases in S2R+ cells along with a fragment containing the CKAD from Xenopus Claspin. We monitored phosphorylation of this fragment by using antibodies that detect the Ser864-phosphorylated form of Claspin (Kumagai and Dunphy, 2003). In these experiments, we observed that cells overexpressing Gish exhibited strong phosphorylation of Claspin, but there was no phosphorylation in cells expressing CG7094 and CG12147 (Figure 2A). There was also significant phosphorylation of Claspin in cells expressing the CG9962 kinase. However, CG9962 was negative in our dsRNA screen. To test if these kinases could phosphorylate Claspin directly, we expressed FLAG-tagged versions of these proteins in S2R+ cells. We isolated the kinases with anti-FLAG antibody beads, and then performed in vitro phosphorylation assays using the purified glutathione S-transferase (GST)-CKAD fragment as substrate. We observed that only Gish could phosphorylate the GST-CKAD fragment efficiently, whereas the CG9962 kinase was considerably less effective (Figure 2, B and C). Taken together, these results indicate that Gish can phosphorylate Claspin both in vivo and in vitro.
FIGURE 2:
The CKAD sites of Claspin are specifically phosphorylated by a Drosophila homologue of CK1γ. (A) The GST-tagged Claspin CKAD fragment was expressed in Drosophila S2R+ cells in the absence (lanes 1 and 2) or presence (lanes 3–9) of individual FLAG-tagged Drosophila casein kinases. After 48 h, total cell lysates were prepared and immunoblotted with antibodies against Ser864-phosphorylated Claspin, GST, or the FLAG epitope. In lane 2, cells were treated with APH for 10 min prior to lysis. (B) In vitro kinase assay. Purified GST-CKAD peptide was incubated with the indicated FLAG-tagged Drosophila casein kinases immunoprecipitated from S2R+ cell lysates. The reactions were subjected to SDS–PAGE and immunoblotted with antibodies against Ser864-phosphorylated Claspin, GST, or the FLAG epitope. (C) Quantitation of the data from (B), expressed as a ratio of Ser864-phosphorylated Claspin to the total amount of each kinase.
Human CK1γ1, but not CK1γ2 or CK1γ3, phosphorylates Claspin in human cells
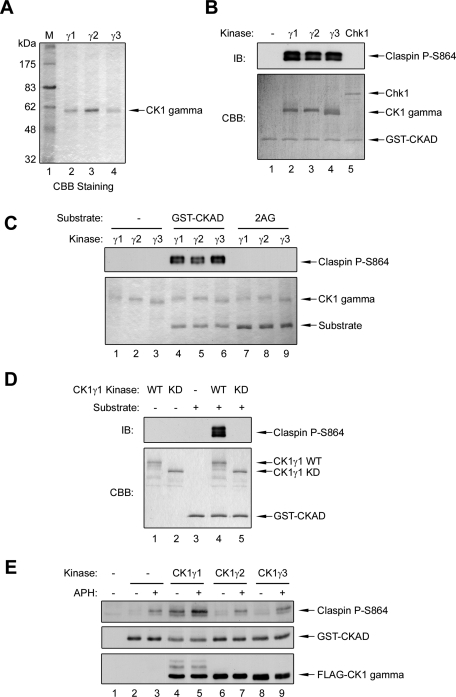
To investigate further the functional significance of these findings, we extended our studies to human cells. Among human proteins, Drosophila Gish is most similar to the CK1 gamma family of kinases (CK1γ), which includes three subfamilies (CK1γ1, CK1γ2, and CK1γ3; Zhai et al., 1995; Kusuda et al., 2000; Knippschild et al., 2005). Although all three kinases exhibit a high degree of homology within the kinase domain, there are significant differences in their N- and C-terminal extensions, which suggests these proteins are functionally distinct. To test whether these kinases can directly phosphorylate Claspin, we prepared recombinant versions of CK1γ1, CK1γ2, and CK1γ3 in Sf9 insect cells and then performed in vitro kinase assays (Figure 3A). In these experiments, we observed that all three kinases could phosphorylate the GST-CKAD fragment well (Figure 3B). In control experiments, we observed that recombinant Chk1 could not phosphorylate this fragment significantly (Figure 3B). Moreover, we demonstrated that the CK1γ kinases could not phosphorylate a mutant form of the CKAD (2AG) in which the two critical phosphorylation sites (Ser864 and Ser895) had been changed to alanine (Figure 3C). Finally, we also produced wild-type and kinase-dead (N169A mutant) forms of GST-tagged CK1γ1 in bacteria. We observed that the wild-type GST-CK1γ1 could phosphorylate Ser864 of Claspin efficiently, whereas the kinase-dead mutant was inactive (Figure 3D).
FIGURE 3:
Human CK1γ1 phosphorylates the Claspin CKAD sites both in vitro and in human cells. (A) Tagged versions of human CK1γ1, CK1γ2, and CK1γ3 proteins were purified from Sf9 insect cells with nickel agarose beads, resolved by SDS–PAGE, and stained with Coomassie Brilliant Blue (CBB). (B) In vitro kinase assays. Purified GST-CKAD was incubated in vitro with no added kinase (lane 1), the indicated forms of CK1γ (lanes 2–4), or Xenopus Chk1-His6-GST (lane 5), as described in Materials and Methods. The samples were subjected to SDS–PAGE and immunoblotted with antibodies against Ser864-phosphorylated Xenopus Claspin (Top) or stained with Coomassie Brilliant Blue (Bottom). (C) In vitro kinase assays were performed as described in (B) with no substrate (lanes 1–3), wild-type GST-CKAD (lanes 4–6), or a mutant of the substrate in which Ser864 and Ser895 were changed to alanine (2AG; lanes 7–9). (D) In vitro kinase assays were performed as described in (B). Purified wild-type GST-CK1γ1 (WT) and its kinase-dead (KD) mutant were incubated in the absence (lanes 1–2) or presence (lanes 4–5) of the GST-CKAD substrate. For lane 3, the incubation contained substrate, but not any recombinant kinase. (E) Only CK1γ1 phosphorylates Claspin well in human cells. CK1γ1, CK1γ2, and CK1γ3 were coexpressed with the GST-CKAD in human U2OS cells. Cells were incubated in the absence or presence of APH for 10 min as indicated. Cell lysates were prepared and immunoblotted with anti-P-Ser864, anti-GST, and anti-FLAG antibodies.
We pursued these findings by comparing the abilities of these kinases to phosphorylate Claspin in human cells. We proceeded to express recombinant forms of CK1γ1, CK1γ2, and CK1γ3 at similar levels in human U2OS cells. Concomitantly, we also introduced the GST-CKAD fragment into the cells. We observed that only cell lines expressing CK1γ1 could phosphorylate the fragment of Claspin above the background level found in cells containing no recombinant kinase (Figure 3E, lanes 2, 4, 6, and 8). Moreover, cells expressing exogenous CK1γ1 also displayed a modest increase in phosphorylation of the CKAD in response to APH (Figure 3E, lanes 3, 5, 7, and 9).
CK1γ1 is required for the checkpoint-induced phosphorylation of Claspin and activation of Chk1 in human cells
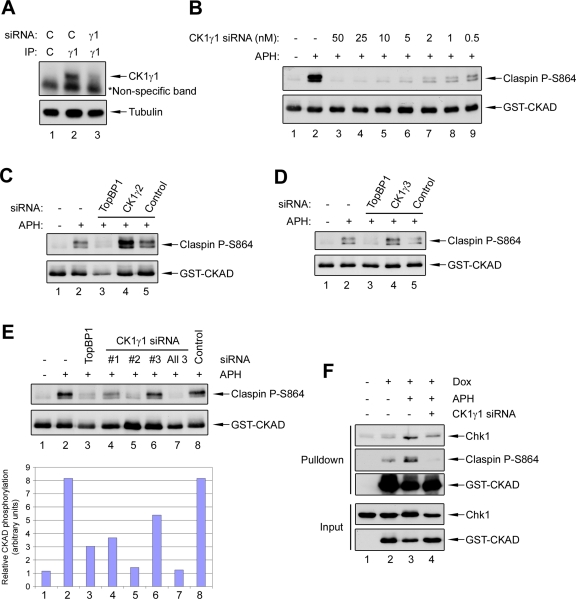
To further evaluate the role of CK1γ1 in the phosphorylation of Claspin, we utilized small interfering RNAs (siRNAs) to knock down the expression of the various CK1γ kinases in human cells. We monitored the efficiency of siRNA knockdown at both the mRNA and protein level for CK1γ1 and CK1γ2 (Figure 4A and Supplemental Figure S1, A, B, and D–F). Due to the fact that a suitable antibody for detection of CK1γ3 is not available, we were able to monitor only the mRNA level for this kinase (Figure S1C). As an initial test for the effect of the CK1γ1 knockdown, we examined phosphorylation of the transiently expressed form of the Xenopus CKAD in human U2OS cells. For this purpose, we generated a line of U2OS cells in which expression of this fragment is under control of a doxycycline-inducible promoter. We observed a large increase in the phosphorylation of the CKAD fragment within 5 min after addition of APH, which correlated well with the activation of Chk1 (Figure S1G). We found that this phosphorylation could be abolished almost completely if the cells had been transfected with CK1γ1 siRNA (Figure 4B). In contrast, when the cells were transfected with siRNAs against CK1γ2 or CK1γ3, there was no such reduction (Figure 4, C and D). We could also observe reduced phosphorylation of Claspin with multiple CK1γ1 siRNAs, which suggests the results are not due to off-target effects (Figure 4E). Finally, depletion of CK1γ1 (as well as that of CK1γ2 or CK1γ3) did not affect the cellular levels of other regulators of Chk1, such as ATR, Claspin, TopBP1, and Rad17 (Figure S1, H and I). Thus, removal of CK1γ1 does not generally affect components of the ATR-mediated regulatory pathway.
FIGURE 4:
siRNA knockdown of CK1γ1, but not CK1γ2 or CK1γ3, abolishes phosphorylation of Claspin in human cells. (A) Reduction of CK1γ1 protein levels by siRNA. U2OS cells were transfected with control or CK1γ1 siRNA for 48 h. Whole-cell lysates were prepared and subjected to immunoprecipitation with control (lane 1) or anti-CK1γ1 antibodies (lanes 2–3). The immunoprecipitates were subjected to immunoblotting with anti-CK1γ1 antibodies (Top). The asterisk denotes a nonspecific background band. The total level of α-tubulin in each lysate was also determined by immunoblotting (Bottom). (B) U2OS cells harboring an inducible form of the GST-CKAD peptide were treated with various amounts of CK1γ1 siRNA (0.5–50 nM) for 48 h. Cells were incubated in the absence (lane 1) or presence (lanes 2–9) of APH for 10 min. Expression of the GST-CKAD was induced with doxycycline 16 h before addition of APH. Whole-cell lysates were immunoblotted as indicated. (C) U2OS cells were incubated with no siRNA (lanes 1 and 2), TopBP1 siRNA (lane 3), CK1γ2 siRNA #1 (lane 4), or control siRNA for 48 h, and incubated in the absence (lane 1) or presence of APH for 10 min (lanes 2–5). Expression of the GST-CKAD was induced as described in (B). Whole-cell lysates were immunoblotted as indicated. (D) Same as (C), except CK1γ3 siRNA #2 was used for lane 4. (E) U2OS cells were treated for 48 h with no siRNA (lanes 1 and 2), TopBP1 siRNA (lane 3), CK1γ1 siRNA #1 (lane 4), CK1γ1 #2 (lane 5), CK1γ1 #3 (lane 6), all three CK1γ1 siRNAs (lane 7), or control siRNA (lane 8). Cells were incubated in the absence (lane 1) or presence (lanes 2–8) of APH for 10 min. Expression of the GST-CKAD was induced as described in (B). Total cell lysates were prepared and immunoblotted with the indicated antibodies. Band signals for phospho-Claspin and Claspin were detected and quantitated by using the Odyssey Infrared Imaging System (Bottom). Blotting membranes were also processed for standard ECL with secondary anti-rabbit antibodies (Top). (F) GST pulldown experiments. U2OS cells were treated with no siRNA (lanes 1 and 2), control siRNA (lane 3), or CK1γ1 siRNA #2 (lane 4) for 48 h and then incubated in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of APH for 10 min. Expression of the GST-CKAD was either not induced (lane 1) or induced with doxycycline (lanes 2–4) 16 h before addition of APH. Whole-cell lysates were prepared and incubated with glutathione beads. Bead-associated proteins (top three panels) and whole-cell lysates (bottom two panels) were immunoblotted with the indicated antibodies.
Since phosphorylation of Claspin is necessary for binding to Chk1, we also examined whether knockdown of CK1γ1 would affect the Claspin–Chk1 interaction. For this purpose, we performed pulldown experiments with the GST-CKAD fragment, which is known to interact well with Chk1. As expected, there was increased binding of Chk1 to the GST-CKAD in cells treated with APH (Figure 4F). By contrast, this increase was abrogated in cells transfected with CK1γ1 siRNA. Altogether, our data indicate CK1γ1, but not CK1γ2 or CK1γ3, is a specific kinase responsible for the checkpoint-induced phosphorylation of Claspin in human cells.
To pursue these findings further, we examined the effect of ablation of CK1γ1 on the ultimate activation of Chk1. We observed that removal of CK1γ1 by siRNA treatment from U2OS cells substantially diminished the phosphorylation of Chk1 (Figure 5, A and B). The reduction was similar to that observed in cells treated with siRNA directed against TopBP1, the upstream activator of ATR–ATRIP. By contrast, depletion of CK1γ2 or CK1γ3 in the same experiments had no effect on phosphorylation of Chk1. We observed a reduction in the phosphorylation of Chk1 following treatment with three different siRNAs against CK1γ1 (Figure 5B). Moreover, we obtained very similar results in a different cell line, namely, HeLa cells (Figure S2A–C). Finally, as a complementary means to confirm the effect of the siRNAs, we also generated a series of endoribonuclease-prepared siRNAs (esiRNAs), which are a mixture of siRNA oligonucleotides generated in vitro by cleavage of long double-stranded RNA with RNase III. As shown in Figure S2D, esiRNAs directed against CK1γ1 strongly inhibited phosphorylation of Chk1 but comparably generated esiRNAs against CK1γ2 and CK1γ3 had no effect.
FIGURE 5:
CK1γ1 is required for the damage-induced phosphorylation of Chk1. (A) U2OS cells were incubated for 48 h with no siRNA (lanes 1 and 2), control siRNA (lanes 3, 4, and 9), TopBP1 siRNA (lane 5), CK1γ1 siRNA #2 (lane 6), CK1γ2 siRNA #1 (lane 7), and CK1γ3 siRNA #2 (lane 8). Cells were incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4–9) of APH for 10 min. Whole-cell lysates were immunoblotted with antibodies that detect phospho-Chk1 Ser345, phospho-Chk1 Ser317, Chk1 protein, and tubulin. (B) U2OS cells were treated for 48 h with no siRNA (lanes 1 and 2), TopBP1 siRNA (lane 3), CK1γ1 siRNA #1 (lane 4), CK1γ1 #2 (lane 5), CK1γ1 #3 (lane 6), all three CK1γ1 siRNAs (lane 7), or control siRNA (lane 8). Cells were incubated in the absence (lane 1) or presence (lanes 2–8) of APH for 10 min. Whole-cell lysates were prepared and immunoblotted with antibodies that detect phospho-Chk1 Ser317, Chk1 protein, and tubulin. Band signals for phospho-Chk1 and Chk1 were detected and quantitated by using the Odyssey Infrared Imaging System (Bottom). Blotting membranes were also processed for standard ECL, with secondary anti-rabbit or anti-mouse antibodies (Top).
Among the various substrates of ATR–ATRIP, Chk1 appears to be unique in its need for assistance by Claspin (Liu et al., 2006; Smits et al., 2010). Accordingly, we asked whether ablation of CK1γ1 might affect other substrates of ATR–ATRIP. For this purpose, we examined the ATR-dependent phosphorylation of Rad17 on Ser645 (Bao et al., 2001; Post et al., 2001). As anticipated, depletion of TopBP1 diminished phosphorylation of Rad17 in response to APH (Figure S2E). On the other hand, removal of CK1γ1 (or CK1γ2 and CK1γ3) had no effect on this phosphorylation. This result lends further support to the hypothesis that CK1γ1 acts specifically at the level of the Claspin–Chk1 interaction. This finding also indicates depletion of CK1γ1 does not generally perturb the ATR-mediated signaling pathway.
Depletion of CK1γ1 leads to multiple checkpoint defects
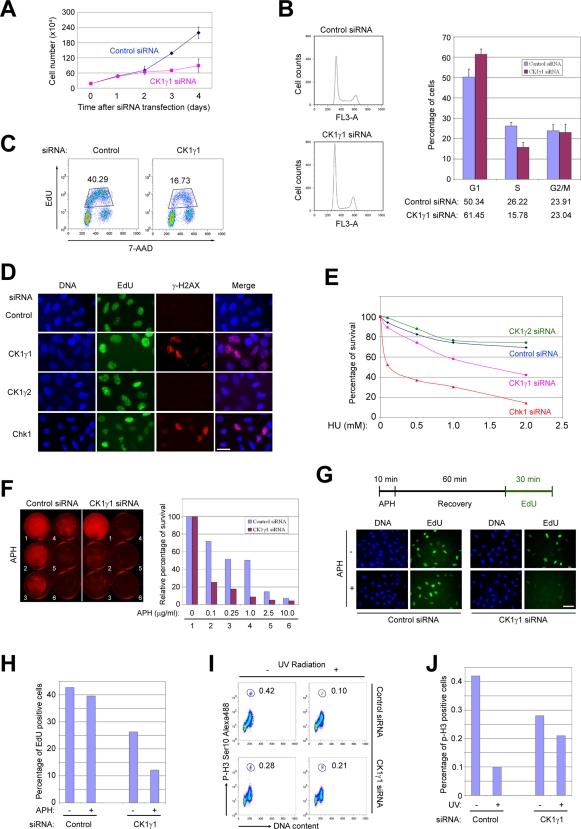
Once we established that CK1γ1 is responsible for the phosphorylation of Claspin and activation of Chk1, we sought to extend our studies by examining the roles of CK1γ1 in various aspects of cell physiology, including checkpoint responses. We noticed depletion of CK1γ1 eventually led to a decrease in cellular proliferation (Figures 6A and S3A). The effect was relatively mild within 48 h after siRNA transfection, but became more noticeable by 72 h. To analyze this effect in more detail, we examined cell cycle progression in the siRNA-treated cells. By performing flow cytometry, we observed that the percentage of cells in G2/M remained about the same upon depletion of CK1γ1, but there appeared to be a significant drop in the percentage of S phase cells as well as an increase in G1 cells (Figure 6B). As another means to examine the effect on S phase, we labeled cells with 5-ethynyl-2′-deoxyuridine (EdU) and detected the EdU-positive cells by flow cytometry in order to assess DNA replication. In these experiments, we observed an approximately 50–60% reduction of incorporated EdU in CK1γ1-depleted cells compared with control cells (Figure 6C).
FIGURE 6:
Depletion of CK1γ1 results in abnormal cell growth, increased DNA damage, and failure in checkpoint responses. (A) Cell numbers in cultures of U2OS cells were determined at the indicated times following transfection with control siRNA or CK1γ1 siRNA #2. (B) Cell cycle distribution. Asynchronous culture of U2OS cells treated with control siRNA or CK1γ1 siRNA #2 for 48 h were stained with 7-AAD, and DNA content was analyzed by flow cytometry. Percentages of cells in each phase of the cell cycle were quantified with the FlowJo program (Ashland, OR). (C) DNA replication. U2OS cells treated with control siRNA or CK1γ1 siRNA #2 for 48 h, pulse-labeled with EdU for 45 min, stained with 7-AAD, and analyzed by flow cytometry. Percentages of EdU-positive cells are indicated. (D) Assay for damaged DNA. U2OS cells were treated with control siRNA, CK1γ1 siRNA #2, CK1γ2 siRNA #1, or Chk1 siRNA for 48 h. Cells were pulse-labeled with EdU for 45 min. Immunofluorescence staining was performed by incubating cells with Hoechst dye to detect DNA, Click-iT reagent to detect incorporated EdU, and antibodies against γ-H2AX to detect DNA damage foci. Scale bar: 30 μm. (E) Colony survival assay. U2OS cells were treated with control siRNA, CK1γ1 siRNA #2, CK1γ2 siRNA #1, or Chk1 siRNA for 48 h, and then incubated with the indicated doses of HU for 24 h. The cells were washed, trypsinized, and then replated at low density. Surviving colonies were stained with crystal violet and counted after 8 d. (F) Killing curve. U2OS cells treated with control siRNA or CK1γ1 siRNA #2 were grown in the presence of different doses of APH. Cells were stained with crystal violet after 6 d. Staining intensities were measured with the Odyssey Imaging System, and the relative percentages of surviving cells were plotted. (G) S-phase checkpoint. In the experimental scheme summarized on top, U2OS cells transfected with control siRNA or CK1γ1 siRNA #2 were first treated with APH for 10 min, allowed to recover for 60 min, and finally labeled with EdU for 30 min. Representative images showing the incorporation of EdU before and after treatment with APH are shown in the bottom. Scale bar: 50 μm. (H) Percentages of EdU-positive cells in (G) were counted and plotted. (I) G2/M checkpoint. U2OS cells transfected with control siRNA or CK1γ1 siRNA #2 were irradiated with UV (20 J/m2), and then incubated for 1 h before fixation. Cells were stained with anti-phospho-histone H3 antibodies conjugated with Alexa Fluor 488 dye and with 7-AAD. (J) Percentages of mitotic cells from (I) were determined by flow cytometry.
To examine potential causes for the decrease in DNA replication, we asked whether depletion of CK1γ1 might lead to the occurrence of DNA damage. For this purpose, we stained cells with antibodies directed against Ser139-phosphorylated H2AX, known as γ-H2AX, which is a well-known marker for the presence of damaged DNA (Paull et al., 2000). We observed the highly elevated presence of nuclear foci containing γ-H2AX in CK1γ1-depleted cells compared with control cells (Figures 6D and S3B). Thus, lack of CK1γ1 apparently leads to the accumulation of damage without exposure to any exogenous damaging agent. As expected from published results, depletion of Chk1 also resulted in elevated staining for γ-H2AX (Paulsen et al., 2009). On the other hand, removal of CK1γ2 did not have this effect. It is well known that the presence of damaged DNA suppresses DNA replication. Therefore, chronic DNA damage in CK1γ1-depleted cells may at least partially explain the reduced levels of DNA synthesis. However, we do not have direct evidence for this possibility.
We next examined various physiological responses to genotoxic stress following ablation of CK1γ1. Toward this end, we examined cell survival upon treatment with replication inhibitors. In one type of assay, we treated cells with HU for 24 h, and then examined the ability of the cells to form colonies upon removal of the drug. We also monitored survival of cells incubated continuously with various doses of APH. In both types of assays, we observed depletion of CK1γ1 significantly compromised the ability of cells to withstand replication stress (Figure 6, E and F). In another line of experiments, we monitored recovery of replication forks after stalling. For this purpose, we used the experimental design shown in Figure 6G. We first challenged cells with a transient (10 min) treatment with APH to block DNA replication. Subsequently, we removed the drug, allowed the cells to recover for 60 min, and finally performed pulse-labeling with EdU to assess the replicative capacity of the cells. We observed control cells exhibited nearly complete recovery from the challenge to DNA replication, whereas the CK1γ1-depleted cells were not able to recover effectively (Figure 6, G and H). This result further supports a role for CK1γ1 in mediating a proper DNA damage response in the S phase of the cell cycle.
Finally, we examined whether CK1γ1 also plays a role in mediating the G2/M checkpoint. To test this possibility, we irradiated control or CK1γ1-depleted cells with ultraviolet (UV) light, labeled the cells with antibodies against the M-phase marker phospho-histone H3, and quantitated the staining by flow cytometry. In these experiments, we observed the expected large reduction in the mitotic marker in the irradiated control cells, but this response was substantially defective in the CK1γ1-depleted cells (Figure 6, I and J). As part of these experiments, we also verified that depletion of CK1γ1 also compromised activation of Chk1 in response to UV (Figure S3C). Together, our results indicate that CK1γ1 plays an important role in regulating DNA damage checkpoint responses in both the S and G2/M phases.
Multiple CK1γ1 isoforms contribute to the phosphorylation of Claspin
As part of these studies, we sought to rescue the effects of depletion of CK1γ1 by introducing an siRNA-resistant form of CK1γ1 into human cells. However, as shown in Figure S4A, we were able to obtain only a partial rescue of these phenotypes in stable lines of U2OS cells harboring an inducible, siRNA-resistant version of the published form of CK1γ1 (Zhai et al., 1995; Kusuda et al., 2000). An important consideration for these experiments is that there appear to be multiple isoforms of CK1γ1 in human cells. After performing a systematic search of the human Expressed Sequence Tags database, we could readily identify numerous potentially distinct forms of CK1γ1. After aligning the sequences, we noticed that all of them contained the same sequence at their 5′ end (which encodes the kinase domain), but that their 3′ ends were substantially different. After carefully analyzing possible polyadenylation signal sites and stop codons and performing comparisons with genomic DNA data, we identified at least four potential new isoforms of CK1γ1. On the basis of these sequences, we designed primers specific to each isoform and were able to amplify three out of the four isoforms from human U2OS cells by reverse transcription PCR (RT-PCR; see sequence alignments in Figure S4B). For clarity of presentation, we have designated the published version of CK1γ1 as isoform A, and the remaining three new forms that we were able to isolate as B, C, and D.
To evaluate the differences between these forms, we expressed these proteins as FLAG-tagged fusions in human cells and examined their intracellular localization by indirect immunofluorescence (Figure 7A). Interestingly, their localizations appeared to be quite distinct. We observed that immunofluorescence staining for isoform A covered the entire cell with a prominent staining of the plasma membrane. This distribution is consistent with the fact that isoform A contains a lipid attachment sequence near the C-terminal end of the protein that would confer membrane localization. By contrast, isoforms B and C exhibited both nuclear and cytoplasmic staining, whereas isoform D appeared to reside only in the cytoplasm. None of the isoforms B, C, and D appeared to exhibit membrane localization, which is consistent with the fact that all three proteins lack the lipid attachment sequence.
FIGURE 7:
Contribution of multiple CK1γ1 isoforms to the phosphorylation of Claspin. (A) Distinct localizations of various isoforms of CK1γ1 in human cells. U2OS cells expressing FLAG-tagged CK1γ1 isoforms were processed for indirect immunofluorescence with anti-FLAG antibodies. Scale bar: 30 μm. (B) Differential phosphorylation of Claspin by CK1γ1 isoforms. The CKAD fragment of Claspin was coexpressed with different isoforms of CK1γ1 in human U2OS cells. Whole-cell lysates were prepared and analyzed by immunoblotting. (C) Isoform-specific siRNAs. U2OS cells expressing the FLAG-tagged CK1γ1 isoform C were treated with a control siRNA, CK1γ1 siRNA #2, or two isoform-specific siRNAs (#71 and #156). Note that siRNA #156, which targets the coding/3′-UTR junction region of CK1γ1 isoform C, only partially pairs with the FLAG-tagged version of CK1γ1 isoform C in the expression plasmid. (D and E) Knockdown of CK1γ1 isoform C expression only partially compromises phosphorylation of Claspin and activation of Chk1. U2OS cells, transfected with the different siRNAs as indicated, were treated with APH, and whole-cell lysates were then prepared and analyzed by immunoblotting. (F and G) Rescue experiment. U2OS cells were concurrently infected with three lentiviruses encoding siRNA-resistant forms of CK1γ1 isoforms A, B, and C. After siRNA transfection and treatment with APH as indicated, whole-cell lysates were prepared and analyzed by immunoblotting. Quantitation of the band intensities was performed with the Odyssey Imaging System. Relative levels of phosphorylation of Claspin and Chk1 were calculated and plotted in (G). (H) Model for the role of CK1γ1 in mediating the activation of Chk1.
We then compared the activities of these different proteins in phosphorylating the GST-CKAD fragment of Claspin upon coexpression in human cells (Figure 7B). In this in vivo assay, phosphorylation of Claspin by isoform A was comparatively weak. Significantly, however, both isoforms B and C were very effective in phosphorylating S864 of Claspin. We could not observe good phosphorylation with isoform D, but it was difficult to express this protein at high levels. As another assessment of specificity, we also coexpressed the GST-CKAD with other members of the casein kinase family (namely, CK1α , two different forms of CK1δ, and CK1ε). As shown in Figure S4C, none of these kinases could phosphorylate S864 of Claspin significantly in a side-by-side comparison with CK1γ1 (isoform C).
In an attempt to dissect the relative contributions of these isoforms to phosphorylation of Claspin, we designed isoform-specific siRNAs against one of the nuclear isoforms (isoform C). Unfortunately, the unique sequence of isoform B is not long enough for design of multiple nonoverlapping siRNAs. We observed that depletion of isoform C by two distinct siRNAs resulted in an approximately 50% inhibition of the checkpoint-dependent phosphorylation of Claspin and Chk1 (Figure 7, C–E). These findings suggest isoform C is partially responsible for the phosphorylation of Claspin, and imply that one or more of the other isoforms would carry out the remainder of the phosphorylation.
Finally, we sought to rescue the decreased phosphorylation of Claspin and Chk1 in CK1γ1-depleted cells by introducing siRNA-resistant versions of one or more isoforms of CK1γ1 into the cells. For this purpose, we generated lentiviruses encoding siRNA-resistant versions of isoforms A, B, and C. As shown in Figure 7, F and G, coinfection of siRNA-treated cells with three lentiviruses encoding all three isoforms resulted in good rescue of both the phosphorylation of Claspin (monitored with the exogenously expressed GST-CKAD) and the phosphorylation of Chk1 in response to treatment with APH. In various experiments, we obtained the best rescue with this combination of viruses. We obtained a similar rescue of the phosphorylation of Chk1 in cells lacking the exogenously expressed GST-CKAD (unpublished data). Furthermore, these rescued cells also exhibited a significantly restored cell cycle profile and G2/M checkpoint (Figure S5, A and B). Taken together, these results suggest that multiple isoforms of CK1γ1 are necessary for the normal phosphorylation of Claspin.
DISCUSSION
In this study, we sought to resolve the molecular basis of a key step in the checkpoint-dependent activation of Chk1 in response to genomic stress. The activation of Chk1 involves phosphorylation-dependent docking of Chk1 with its cognate mediator protein (Claspin) and recognition of the resulting Claspin–Chk1 complex by ATR–ATRIP. In particular, phosphorylation of Claspin on multiple residues in its CKAD mediates the binding of Chk1. Direct biochemical evidence has indicated the presence of Claspin in this complex enhances the ability of ATR–ATRIP to phosphorylate Chk1 on key residues necessary for the ultimate activation of Chk1 (Kumagai et al., 2004; Lindsey-Boltz et al., 2009). Thus, the kinase that phosphorylates Claspin on the CKAD plays a key role in this pathway.
To address the identification of this kinase(s) systematically, we initially performed a kinome-wide RNAi screen in Drosophila S2 cells. Various RNAi screens in these cells have been used to identify numerous regulators in diverse cellular pathways (Bettencourt-Dias et al., 2004; Bjorklund et al., 2006; Boutros and Ahringer, 2008). For our purposes, the fact that Drosophila contains approximately one-third as many kinases as humans was a considerable advantage. S2 cells appear to possess checkpoint-signaling pathways generally similar to those in other metazoan cells (de Vries et al., 2005; Yi et al., 2009). In particular, the Drosophila Chk1 homologue Grapes exhibits a conserved checkpoint function in S2 cells. It functions downstream of Mei-41 (the Drosophila homologue of ATR) and is critical for a proper checkpoint response to genotoxic stress.
Nonetheless, the reagents available for the study of checkpoint responses in Drosophila S2 cells are still relatively limited. To circumvent this problem, we introduced the Xenopus version of Chk1 into the Drosophila cells as a more readily traceable marker to monitor the checkpoint response. We were able to establish a system in which phosphorylation of this reporter could be induced following treatment with a variety of replication inhibitors. Moreover, this response seems to have molecular features similar to those present in vertebrate cells. For example, RNAi-mediated knockdown of Drosophila homologues of ATR and Claspin abolished checkpoint-dependent phosphorylation of the exogenously introduced Chk1 reporter molecule.
The results of our screen indicated that knockdown of several casein kinases led to the reduced phosphorylation of Chk1. Following this lead, we next employed a candidate-based cDNA overexpression approach to further pinpoint a specific kinase that can directly phosphorylate the CKAD of Claspin. From these tests, we eventually found that Drosophila Gish, a homologue of CK1γ, could phosphorylate the CKAD of Claspin quite effectively both in vitro and in the Drosophila tissue culture cells. At this juncture, we chose to extend our studies to human cells. Interestingly, humans possess three different versions of CK1γ, namely, CK1γ1, CK1γ2, and CK1γ3. All three proteins have very similar central kinase domains but are significantly different in their N- and C-terminal extensions. We observed that all three kinases could phosphorylate the CKAD relatively well in vitro. However, our further characterizations established that CK1γ1 appears to be the form primarily responsible for the phosphorylation of CKAD in human cells. Upon expression in human cells, only CK1γ1 could phosphorylate the CKAD of Claspin in vivo. More importantly, siRNA-mediated knockdown of CK1γ1 resulted in greatly reduced phosphorylation of Claspin, whereas knockdowns of CK1γ2 and CK1γ3 did not have a significant effect. As expected from the known function of the phosphorylation of Claspin, cells with diminished levels of CK1γ1 were greatly compromised in their ability to carry out the activation of Chk1 in response to a variety of genotoxic agents, including APH, HU, and UV. Furthermore, these cells had the physiological defects characteristic of cells with impairment of the Chk1-mediated signaling pathway. In particular, these cells showed reduced survival following treatment with genotoxic agents, impaired recovery of stalled replication forks, a defective G2/M checkpoint response, and spontaneous DNA damage in the absence of exogenous stress. Taken together, our results indicated CK1γ1 is an important regulator in Chk1-mediated cellular checkpoint responses.
The casein kinase 1 family of serine/threonine kinases is highly conserved and ubiquitously expressed. The functions of CK1 encompass a wide variety of processes, including cell proliferation, cell division, apoptosis, circadian rhythms, and others (Gross and Anderson, 1998; Knippschild et al., 2005; Cheong and Virshup, 2011). In mammals, this family contains at least seven members (α, β, δ, ε, γ1, γ2, and γ3) with multiple splicing variants. All of the CK1 proteins share significant homology in the central kinase domain (53–98% identical), but differ significantly in the flanking N- and C-terminal sequences, which most likely confer unique properties to the various kinases. In the case of CK1γ1, we identified and isolated multiple splicing variants of this kinase from human U2OS cells. These isoforms contain distinct C-terminal sequences (ranging from 3 to 106 amino acids), display discrete subcellular localizations, and exhibit differences in their abilities to phosphorylate the CKAD. A similar phenomenon has also been observed in various organisms in the case of CK1α, which contains four types of isoforms with distinct localizations and kinase activities (Zhang et al., 1996; Green and Bennett, 1998; Burzio et al., 2002). These findings strongly suggest various isoforms of CK1 may participate in distinct cellular activities due to differences in access to and/or affinity for substrates.
Interestingly, we initially found it difficult to achieve good rescue of CK1γ1-depleted cells with vectors encoding an siRNA-resistant version of the major published form of CK1γ1. However, as described in the last part of Results, there are multiple forms of CK1γ1 in human cells. We were able to clone and express three additional versions of CK1γ1, which we designated as isoforms B, C, and D to distinguish them from the published form (isoform A). In cellular localization studies, we found that isoform A had the expected prominent localization to cell membranes (Davidson et al., 2005). However, isoforms B and C resided in both the nucleus and cytoplasm, whereas isoform D was mainly cytoplasmic. In coexpression studies, we found that isoforms A, B, and C (but not D) could phosphorylate the CKAD of Claspin, although isoforms B and C were more effective than isoform A in this in vivo assay. Ultimately, we were able to rescue depletion of CK1γ1 by introducing siRNA-resistant forms of isoforms A, B, and C into the cells. It is straightforward to understand how the nuclear isoforms could regulate Claspin, but it is intriguing that a membrane-bound enzyme is also partially responsible. Conceivably, some fraction of this isoform could be absent from the membrane. It is also possible that Claspin could shuttle between the nucleus and cytoplasm and thus be subject to regulation by enzymes in both locations. In this regard, a recent study has shown that perturbed cell-surface signaling through the Sonic Hedgehog (Shh) pathway inhibits ATR-mediated signaling by disrupting the interaction between Claspin and Chk1 (Leonard et al., 2008).
It will be important to understand the mechanisms that control the phosphorylation of Claspin by CK1γ1 (see Figure 7H). It is known that phosphorylation of the CKAD in Xenopus egg extracts is dependent upon ATR. However, ATR itself is unable to phosphorylate the critical sites in the CKAD directly. One possibility would be that ATR might somehow regulate the activity of CK1γ1. However, we have been able to detect only a subtle increase in the activity of CK1γ1 in response to genomic stress. Moreover, when we mutated the only apparent potential target site for ATR (Ser361) to Ala in CK1γ1, we could not observe a change in its kinase activity toward the CKAD upon coexpression in U2OS cells (unpublished data). Another possibility is that ATR might regulate the accessibility of Claspin to CK1γ1. Further studies will be required to understand the dynamics of this phosphorylation.
In summary, we identified a conserved casein kinase, Gish/CK1γ1, from Drosophila and humans as a specific enzyme that controls phosphorylation of the CKAD of Claspin. Functional studies have revealed that this kinase is critical for mediating activation of Chk1 and ensuring a proper checkpoint response under conditions of genotoxic stress. Further studies of its regulation and function should help us gain more insight into the molecular basis of checkpoint responses.
MATERIALS AND METHODS
Drosophila cell culture
Drosophila S2R+ cells were purchased from the Drosophila Genomics Resource Center (Bloomington, IN) and cultured at room temperature (23°C) in Schneider's Drosophila culture medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 100 U/ml penicillin and 100 μg/ml streptomycin. Stable S2R+ cell lines expressing the pMT-Chk1-NLS-GTS reporter were generated by cotransfection of the reporter plasmid with the pCoHygro plasmid (19:1) and selection with 300 μg/ml hygromycin for ∼21 d. Expression of the reporter gene was induced with CuSO4 (500 μM) for 16 h. DNA replication blocks were induced by treating cells with APH (10 μg/ml) for 10 min or HU (2 mM) for 16 h.
Plasmid transfection and dsRNA-mediated RNA interference in Drosophila cells
Plasmid DNA transfections were performed using FuGENE 6 reagent (Roche, Indianapolis, IN) according to the manufacturer's instructions. Briefly, S2R+ cells (0.6 × 106) were seeded in 12-well plates and grown overnight. On the following day, a mixture of plasmid DNA (0.6 μg) and FuGENE 6 reagent (5 μl) was diluted in 100 μl H2O and incubated with cells in serum-free medium for 4 h. Next, medium containing FBS was added, and cells were incubated for a total of 48 h.
dsRNA-mediated RNA interference in S2R+ cells was performed essentially as described previously (Clemens et al., 2000). Briefly, dsRNAs (∼500 base pairs long) were produced by in vitro transcription using DNA templates containing T7 promoters at both ends. The quality of dsRNAs was monitored by gel electrophoresis to ensure that only single bands were present. dsRNAs (15 μg) were added directly to 0.5 ml of serum-free medium and incubated with cells for 60 min. Subsequently, 1 ml complete medium with 15% FBS was added, and cells were incubated for another 96 h to allow for turnover of the proteins of interest.
Whole-cell lysates were prepared in TEB150 buffer (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 5 mM ethylene glycol tetraacetic acid [EGTA], 0.5% Triton X-100, and 10% glycerol) containing freshly added 1 mM dithiothreitol (DTT), 1 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 10 μg/ml each of pepstatin, chymostatin, and leupeptin. Lysates were briefly sonicated, diluted in 2× protein sample buffer, and resolved by SDS–PAGE.
Human cell culture and transfection
HeLa cells, U2OS cells, and U2OS T-REx cells (a kind gift from Thomas C. Spelsberg, Mayo Clinic, Rochester, MN) were cultured in DMEM containing 10% FBS, penicillin, and streptomycin. Plasmid DNA transfections were performed using the FuGENE 6 reagent, and siRNA (50 nM) transfections were carried out with Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer's instructions. For cotransfection experiments with the U2OS T-Rex cells, a 2:1 ratio of GST-CKAD reporter plasmid to the kinase expression plasmid was used. Transfections were carried out for a total of 48 h, and kinase expression was induced by treatment with doxycycline (100 ng/ml) for 16 h.
Plasmids
The pMT-Chk1-NLS-GST reporter plasmid was generated by a two-step cloning method. First, the NLS-GST fragment was generated by PCR and inserted into the Drosophila expression vector pMT/V5-His B plasmid (Invitrogen). Next, the sequence encoding the full-length Xenopus Chk1 was ligated in-frame to the 5′ end of the NLS-GST sequence. The pUAST Drosophila casein kinase expression plasmids were kind gifts from Jin Jiang (University of Texas Southwestern Medical School, Dallas, TX). The full-length cDNA clones of human CK1γ1, CK1γ2, and CK1γ3 were purchased from Open Biosystems (Thermo Scientific, Lafayette, CO). We also obtained cDNAs encoding CK1α, a short and long form of CK1δ (409 and 415 amino acids in length, respectively), and CK1ε from the same source. For expression in human cells, these cDNAs were cloned into the tetracycline-inducible pcDNA5/TO (Invitrogen) vector, which encodes an N-terminal FLAG tag. siRNA-resistant versions of CK1γ1 were created through introduction of silent mutations into three nucleotides in the middle of siRNA-targeting regions by site-directed mutagenesis.
siRNAs and esiRNAs
Stealth siRNA duplexes specific for CK1 gamma kinases and a control siRNA (low GC) were purchased from Invitrogen. The sense strand sequences of these siRNAs are listed in Supplemental Table S1. These CK1γ1 siRNAs are directed against common regions present in all isoforms. esiRNAs were produced in vitro essentially by following the protocol kindly provided by Eric Lau and Wei Jiang (Burnham Institute, La Jolla, CA; Lau et al., 2006).
Immunoblotting and antibodies
Immunoblotting and enhanced chemiluminescence (ECL) were performed by standard methods. For detection of phosphorylation signals, 2 mM Na3VO4 and 20 mM NaF were included in all immunoblotting solutions to block potential phosphatase activities. For quantification of immunoblotting signals, membranes were stripped and blotted with IRDye-800- or IRDye-680-conjugated secondary antibodies and scanned with the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
For generation of anti-CK1γ1 antibodies, the sequence encoding the full-length human CK1γ1 isoform A was cloned into the pET30a vector and expressed in bacterial cells. Recombinant protein was solubilized in 6 M urea and purified with nickel agarose beads. This protein was used to immunize rabbits, and the generated antibody was affinity purified. Because the expression level of CK1γ1 in U2OS cells is low, we carried out immunoprecipitation and then immunoblotting on a relatively large scale for detection of endogenous CK1γ1. Whole-cell lysates were prepared in TEB150 buffer with phosphatase and protease inhibitors as described above. Next, whole-cell lysates were incubated with anti-CK1γ1 antibodies coupled to the protein-A Dynabeads (Invitrogen) for 90 min at 4°C. Finally, the beads were washed two times with Buffer A (10 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 2.5 mM EGTA, and 20 mM β-glycerolphosphate) and two times with HEPES-buffered saline (HBS; 10 mM HEPES-KOH, pH 7.5, and 150 mM NaCl). Finally, samples were boiled in protein sample buffer and subjected to SDS–PAGE.
Antibodies against the following targets were obtained from the indicated sources: Chk1 P-Ser345 and Chk1 P-Ser317 (Cell Signaling Technology, Beverly, MA); Chk1 (G-4), CK1γ2 (N-20), GST (Z-5), and Rad17 (H-3; Santa Cruz Biotechnology, Santa Cruz, CA); Claspin P-Ser864 (Kumagai and Dunphy, 2003); γ-H2AX (Millipore, Billerica, MA); ATR (Affinity BioReagents, Golden, CO); TopBP1 (Bethyl Laboratories, Montgomery, TX), Rad17 P-Ser645 (a kind gift from Eva Y. Lee, University of California, Irvine, CA); FLAG epitope (M2, Stratagene, Agilent, Santa Clara, CA); and α-tubulin (Calbiochem, San Diego, CA).
Production of recombinant proteins
Recombinant CK1 gamma kinases with FLAG, hemagglutinin (HA), and His6 tags were produced in Sf9 insect cells and purified by following a previously described protocol (Kumagai and Dunphy, 2000). Briefly, the full-length cDNAs of CK1 gamma kinases were cloned into the pFastBac-HA-HTa vector. Recombinant baculoviruses were generated with the Bac-to-Bac system (Invitrogen), and recombinant proteins were purified using nickel agarose beads. For production of recombinant wild-type or kinase-dead (N169A) versions of GST-tagged CK1γ1, the appropriate cDNAs were cloned into the pGEX-4T-3 vector. Recombinant proteins were produced in Escherichia coli cells and purified using the Glutathione Sepharose 4 Fast Flow beads (GE Healthcare, Waukesha, WI), according to the manufacturer's instructions.
Reverse transcription PCR
Total RNA was isolated using the TRIzol RNA isolation reagent (Invitrogen). Reverse transcription PCR (RT-PCR) reactions were carried out using the SuperScript III One-Step RT-PCR System (Invitrogen) according to the manufacturer's instructions. Specific primers were designed to span exon–intron boundaries of respective genes, ensuring the results were not affected by genomic DNA contamination. The linear ranges of amplification cycle for each gene were predetermined by electrophoresis and quantification of the PCR products. The sequences of RT-PCR primers are listed in Table S2.
Immunoprecipitation and in vitro kinase assays
FLAG-tagged Drosophila casein kinases were expressed in S2R+ cells for 48 h. Whole-cell lysates were prepared and incubated with anti-FLAG antibody-conjugated Dynabeads at 4°C for 60 min. Beads were washed two times with buffer A and three times with HBS. The bead-bound kinases were incubated with the GST-CKAD fragment (500 ng) in kinase buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 1 mM DTT) containing 10 mM ATP at room temperature for 30 min. Finally, the reactions were stopped by addition of protein sample buffer and boiling. In the case of in vitro kinase assays using recombinant proteins, 250 ng of purified CK1γ1, CK1γ2, CK1γ3, or Xenopus Chk1-GST-His6 (Kumagai and Dunphy, 2000) protein, instead of the immunoprecipitated kinases, were added to the reactions.
GST pulldown experiments
U2OS cells expressing GST-CKAD were lysed in TEB150 buffer as described above. Whole-cell lysates were incubated with Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) at 4°C for 1 h. Beads were washed two times with buffer A and three times with HBS. Finally, the beads were boiled in an equal volume of 2× protein sample buffer and subjected to SDS–PAGE.
Immunofluorescence staining
For immunofluorescence staining, cells grown on coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) and permeabilized with 0.5% Triton X-100 in PBS. After undergoing blocking with 5% goat serum in PBS for 30 min, cells were incubated with primary antibodies at room temperature for 60 min. Following three washes with PBS, cells were incubated with Alexa Fluor 594-conjugated secondary antibodies for 60 min. Finally, cells were washed, counterstained with 5 μg/ml Hoechst 33258, and mounted in VECTASHIELD mounting medium. For detection of EdU incorporation to assess DNA replication, the Click-iT system (Invitrogen) was used as described previously (Kumagai et al., 2010). For immunofluorescence staining of the FLAG-tagged CK1γ1 isoforms, U2OS cells were first transfected with pcDNA5-FLAG-CK1γ1 plasmids for 48 h. Cells were then fixed and stained with anti-FLAG (M2) antibody and Alexa Fluor 488–conjugated anti–mouse secondary antibodies.
Flow-cytometry analysis
For detection of replicating cells in S phase, cells were labeled with 10 μM EdU for 45 min and stained with the Click-iT system. Briefly, cells were fixed with 70% ethanol and incubated overnight. After undergoing washing with PBS containing 1% bovine serum albumin (BSA), cells were stained with freshly prepared Click-iT reaction cocktail containing Alexa Fluor 488 azide for 30 min at room temperature. Finally, cells were washed again, treated with RNase A (50 μg/ml), and counterstained with 7-amino-actinomycin D (7-AAD). Flow-cytometry analyses were performed with a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). For G2/M checkpoint assays, cells were treated with UV (20 J/m2) and incubated for 1 h. Cells were then fixed and stained with anti–phospho-histone H3 (Ser10) antibody conjugated to Alexa Fluor 488.
Cell survival assays
U2OS cells transfected with control or CK1γ1 siRNAs were seeded in 6-well plates at low density (∼200 cells/well). Cells were treated with HU (0, 0.1, 0.5, 1.0, and 2.0 mM) for 24 h. After removal of HU and addition of fresh medium, cells were continuously incubated for an additional 8 d and stained with crystal violet. Surviving colonies were counted by scanning plates with the Odyssey Imaging System.
Rescue experiments
Rescue experiments with lentiviruses were essentially performed as described previously (Qin et al., 2003). Briefly, lentiviruses expressing siRNA-resistant versions of CK1γ1 isoforms were produced by cotransfecting 293T cells with four lentivector plasmids (pFUW, pRSV-Rev, pMDLg/pRRE, and pVSV) in a 2:1:1:1 ratio using the FuGENE 6 reagent. Viral supernatants were harvested 48 h later and filtered. To rescue the effect of CK1γ1 siRNA, U2OS cells were infected with viruses in the presence of 10 μg/ml of Polybrene (Millipore) for 8 h. After an additional incubation of 16 h, cells were transfected with control or CK1γ1 siRNA. Two days after siRNA transfection, cells were treated with aphidicolin for 10 min, and whole-cell lysates were then immediately prepared, resolved on SDS–PAGE, and processed for immunoblotting.
Supplementary Material
Acknowledgments
We are grateful to laboratory members for helpful advice and comments on the manuscript. We also thank Rochelle Diamond for assistance with flow cytometry and J. Jiang (University of Texas Southwestern Medical School, Dallas, TX) for Drosophila casein kinase expression vectors. This work was supported by National Institutes of Health grants GM-043974 and GM-070891 and an Ellison Senior Scholar in Aging award to W.G.D. Work in the laboratory of D.M.G. was supported by Cancer Research UK and the Biotechnology and Biological Sciences Research Council (United Kingdom).
Abbreviations used:
- γ-H2AX
Ser139-phosphorylated H2AX
- 7-AAD
7-amino-actinomycin D
- APH
aphidicolin
- ATR
ATM- and Rad3-related kinase
- ATRIP
ATR-interacting protein
- BSA
bovine serum albumin
- Cdk
cyclin-dependent kinase
- Chk1
checkpoint kinase 1
- CK1γ
casein kinase 1 gamma
- CKAD
Chk1-activating domain
- DTT
dithiothreitol
- ECL
enhanced chemiluminescence
- EdU
5-ethynyl-2′-deoxyuridine
- EGTA
ethylene glycol tetraacetic acid
- esiRNAs
endoribonuclease-prepared siRNAs
- FBS
fetal bovine serum
- GST
glutathione S-transferase
- HA
hemagglutinin
- HBS
HEPES-buffered saline
- HU
hydroxyurea
- NLS
nuclear localization signal
- PBS
phosphate-buffered saline
- PMSF
phenylmethylsulfonyl fluoride
- RT-PCR
reverse transcription PCR
- Shh
Sonic Hedgehog
- TopBP1
topoisomerase IIb-binding protein 1
- UV
ultraviolet
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-01-0048) on June 16, 2011.
REFERENCES
- Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A, Chen SM, Abraham RT, Wang XF. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature. 2001;411:969–974. doi: 10.1038/35082110. [DOI] [PubMed] [Google Scholar]
- Bennett LN, Larkin C, Gillespie DA, Clarke PR. Claspin is phosphorylated in the Chk1-binding domain by a kinase distinct from Chk1. Biochem Biophys Res Commun. 2008;369:973–976. doi: 10.1016/j.bbrc.2008.02.154. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, Taipale J. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- Burzio V, Antonelli M, Allende CC, Allende JE. Biochemical and cellular characteristics of the four splice variants of protein kinase CK1alpha from zebrafish (Danio rerio) J Cell Biochem. 2002;86:805–814. doi: 10.1002/jcb.10263. [DOI] [PubMed] [Google Scholar]
- Cheong JK, Virshup DM. Casein kinase 1: complexity in the family. Int J Biochem Cell Biol. 2011;43:465–469. doi: 10.1016/j.biocel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Chini CC, Chen J. Human Claspin is required for replication checkpoint control. J Biol Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- Chini CC, Chen J. Repeated phosphopeptide motifs in human Claspin are phosphorylated by Chk1 and mediate Claspin function. J Biol Chem. 2006;281:33276–33282. doi: 10.1074/jbc.M604373200. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CA, Clarke PR. DNA-dependent phosphorylation of Chk1 and Claspin in a human cell-free system. Biochem J. 2005;388:705–712. doi: 10.1042/BJ20041966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- de Vries HI, Uyetake L, Lemstra W, Brunsting JF, Su TT, Kampinga HH, Sibon OC. Grp/DChk1 is required for G2-M checkpoint activation in Drosophila S2 cells, whereas Dmnk/DChk2 is dispensable. J Cell Sci. 2005;118:1833–1842. doi: 10.1242/jcs.02309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CL, Bennett GS. Identification of four alternatively spliced isoforms of chicken casein kinase I alpha that are all expressed in diverse cell types. Gene. 1998;216:189–195. doi: 10.1016/s0378-1119(98)00291-1. [DOI] [PubMed] [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Jeong S-Y, Kumagai A, Lee J, Dunphy WG. Phosphorylated Claspin interacts with a phosphate-binding site in the kinase domain of Chk1 during ATR-mediated activation. J Biol Chem. 2003;278:46782–46788. doi: 10.1074/jbc.M304551200. [DOI] [PubMed] [Google Scholar]
- Knippschild U, Gocht A, Wolff S, Huber N, Lohler J, Stoter M. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat Cell Biol. 2003;5:161–165. doi: 10.1038/ncb921. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Kim S-M, Dunphy WG. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J Biol Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–359. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuda J, Hirai M, Tanuma R, Hashimoto K. Cloning, expression analysis and chromosome mapping of human casein kinase 1 gamma1 (CSNK1G1): identification of two types of cDNA encoding the kinase protein associated with heterologous carboxy-terminal sequences. Cytogenet Cell Genet. 2000;90:298–302. doi: 10.1159/000056792. [DOI] [PubMed] [Google Scholar]
- Lau E, Zhu C, Abraham RT, Jiang W. The functional role of Cdc6 in S-G2/M in mammalian cells. EMBO Rep. 2006;7:425–430. doi: 10.1038/sj.embor.7400624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Gold DA, Shevchenko A, Shevchenko A, Dunphy WG. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol Biol Cell. 2005;16:5269–5282. doi: 10.1091/mbc.E05-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Leonard JM, Ye H, Wetmore C, Karnitz LM. Sonic Hedgehog signaling impairs ionizing radiation-induced checkpoint activation and induces genomic instability. J Cell Biol. (2008;183:385–391. doi: 10.1083/jcb.200804042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Li K, Stewart GS, Elledge SJ. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc Natl Acad Sci USA. 2004;101:6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey-Boltz LA, Sercin O, Choi JH, Sancar A. Reconstitution of human Claspin-mediated phosphorylation of Chk1 by the ATR (ataxia telangiectasia-mutated and rad3-related) checkpoint kinase. J Biol Chem. 2009;284:33107–33114. doi: 10.1074/jbc.M109.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Helleday T, Caldecott KW. Claspin promotes normal replication fork rates in human cells. Mol Biol Cell. 2008;19:2373–2378. doi: 10.1091/mbc.E07-10-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post S, Weng YC, Cimprich K, Chen LB, Xu Y, Lee EY. Phosphorylation of serines 635 and 645 of human Rad17 is cell cycle regulated and is required for G(1)/S checkpoint activation in response to DNA damage. Proc Natl Acad Sci USA. 2001;98:13102–13107. doi: 10.1073/pnas.231364598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorah J, McGowan CH. Claspin and Chk1 regulate replication fork stability by different mechanisms. Cell Cycle. 2009;8:1036–1043. doi: 10.4161/cc.8.7.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits VA, Warmerdam DO, Martin Y, Freire R. Mechanisms of ATR-mediated checkpoint signalling. Front Biosci. 2010;15:840–853. doi: 10.2741/3649. [DOI] [PubMed] [Google Scholar]
- Yi X, et al. Stwl modifies chromatin compaction and is required to maintain DNA integrity in the presence of perturbed DNA replication. Mol Biol Cell. 2009;20:983–994. doi: 10.1091/mbc.E08-06-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L, Graves PR, Robinson LC, Italiano M, Culbertson MR, Rowles J, Cobb MH, DePaoli-Roach AA, Roach PJ. Casein kinase I gamma subfamily. Molecular cloning, expression, and characterization of three mammalian isoforms and complementation of defects in the Saccharomyces cerevisiae YCK genes. J Biol Chem. 1995;270:12717–12724. doi: 10.1074/jbc.270.21.12717. [DOI] [PubMed] [Google Scholar]
- Zhang J, Gross SD, Schroeder MD, Anderson RA. Casein kinase I alpha and alpha L: alternative splicing-generated kinases exhibit different catalytic properties. Biochemistry. 1996;35:16319–16327. doi: 10.1021/bi9614444. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.