The spindle checkpoint protein Mad2 sets the duration of meiosis I by down-regulating APC/C activity to ensure the timely degradation of APC/C substrates. In the absence of Mad2, premature APC/C activity can cause misregulation of meiotic cell cycle events, resulting in chromosome missegregation.
Abstract
In many eukaryotes, disruption of the spindle checkpoint protein Mad2 results in an increase in meiosis I nondisjunction, suggesting that Mad2 has a conserved role in ensuring faithful chromosome segregation in meiosis. To characterize the meiotic function of Mad2, we analyzed individual budding yeast cells undergoing meiosis. We find that Mad2 sets the duration of meiosis I by regulating the activity of APCCdc20. In the absence of Mad2, most cells undergo both meiotic divisions, but securin, a substrate of the APC/C, is degraded prematurely, and prometaphase I/metaphase I is accelerated. Some mad2Δ cells have a misregulation of meiotic cell cycle events and undergo a single aberrant division in which sister chromatids separate. In these cells, both APCCdc20 and APCAma1 are prematurely active, and meiosis I and meiosis II events occur in a single meiotic division. We show that Mad2 indirectly regulates APCAma1 activity by decreasing APCCdc20 activity. We propose that Mad2 is an important meiotic cell cycle regulator that ensures the timely degradation of APC/C substrates and the proper orchestration of the meiotic divisions.
INTRODUCTION
The cell cycle is precisely controlled to ensure a specific order and timing of events. Cell cycle regulators promote the correct sequence of events, and checkpoint mechanisms monitor specific events, delaying the cell cycle if those events have not been completed. Cell cycle proteins have been characterized extensively in mitosis; however, less is known about the activity of the proteins that promote progression through meiosis. There are sequential steps required to ensure that, in meiosis I, homologous chromosomes segregate, and in meiosis II, sister chromatids separate (reviewed in Brar and Amon, 2008). Characterization of the regulation of the two meiotic divisions is necessary to understand how meiotic errors occur.
For faithful chromosome segregation in meiosis I, homologous chromosomes pair, recombine, and attach to spindle microtubules. Once chromosomes are properly attached to microtubules emanating from opposite spindle poles, a ubiquitin ligase called the anaphase-promoting complex/cyclosome (APC/C), bound by the regulatory subunit Cdc20, targets substrates for proteasomal degradation (Pesin and Orr-Weaver, 2008). One substrate, securin (Pds1 in budding yeast), sequesters separase, the protease that cleaves Rec8, the meiotic cohesin (Klein et al., 1999; Buonomo et al., 2000; Kitajima et al., 2003; Kudo et al., 2006). When securin is degraded, separase is active, and chromosomes segregate. A meiosis-specific activator of the APC/C, Ama1, also targets securin and other substrates for degradation (Cooper et al., 2000; Oelschlaegel et al., 2005; Penkner et al., 2005). Although Ama1 is not required for meiosis I, APCAma1 activity does promote the timely degradation of APC/C substrates (Cooper et al., 2000; Oelschlaegel et al., 2005; Penkner et al., 2005).
For faithful segregation of chromosomes in meiosis II, the cohesins around the centromere are protected from cleavage in metaphase I. Sgo1, an orthologue of the Drosophila MEI-S332 protein, is required for the protection of cohesins to ensure that sister chromatids stay together until meiosis II (Kerrebrock et al., 1995; Katis et al., 2004; Kitajima et al., 2004; Marston et al., 2004). After meiosis I, securin reaccumulates and sequesters separase, and Sgo1 is inactivated and degraded. The APC/C will again target securin for degradation, and in the absence of Sgo1, separase will cleave the remaining cohesins, allowing sister chromatids to separate.
In mitosis and meiosis, the attachment of chromosomes to spindle microtubules is monitored at the metaphase-to-anaphase transition by the spindle assembly checkpoint (Musacchio and Salmon, 2007). Improperly attached kinetochores send a signal to delay the cell cycle by inhibiting the APC/C, allowing the cells additional time to correct the error in attachment. In mammalian cells, the spindle checkpoint protein Mad2 also has a kinetochore-independent role as a cell cycle timer. Depletion of Mad2 results in the premature loss of APC/C substrates, a faster cell cycle, and an increase in chromosome missegregation in both mitosis and meiosis (Geley et al., 2001; Wassmann et al., 2003; Meraldi et al., 2004; Michel et al., 2004; Tsurumi et al., 2004; Homer et al., 2005a, 2005b). In contrast, in budding yeast, the spindle checkpoint proteins are not essential in mitosis (Hoyt et al., 1991; Li and Murray, 1991; Hardwick et al., 1999). Deletion of Mad2 results in only a modest increase in mitotic chromosome missegregation unless cells are challenged with genetic or chemical perturbations that disrupt microtubule–kinetochore attachments. In meiosis, however, Mad2 is essential for chromosome segregation; deletion of Mad2 results in an increase in meiosis I nondisjunction (Shonn et al., 2000). The molecular basis for this phenotype was previously unknown.
Here we investigate Mad2's role in meiotic cell cycle regulation in budding yeast to understand how Mad2 functions to promote proper chromosome segregation. We find that, similar to mammalian oocytes, in budding yeast, Mad2 serves as a “meiotic timer” to set the duration of meiosis I by regulating the activity of APCCdc20. To further understand the consequence of premature APC/C activity, we investigated the timing of other meiotic cell cycle events in mad2Δ cells. We find that Mad2 has a role in coordinating chromosome segregation with the regulation of the meiotic cell cycle. In the absence of Mad2, premature APCCdc20 can lead to premature APCAma1 activity in some cells, resulting in a single aberrant division in which sister chromatids separate inappropriately because the chromosome segregation cycle is uncoupled from other cell cycle events. Our results indicate that Mad2 has a role in meiosis I separate from its role in monitoring chromosome attachment: ensuring the timely degradation of APC/C substrates and the proper execution of the meiotic divisions.
RESULTS
Mad2 affects the timing of the meiotic cell cycle
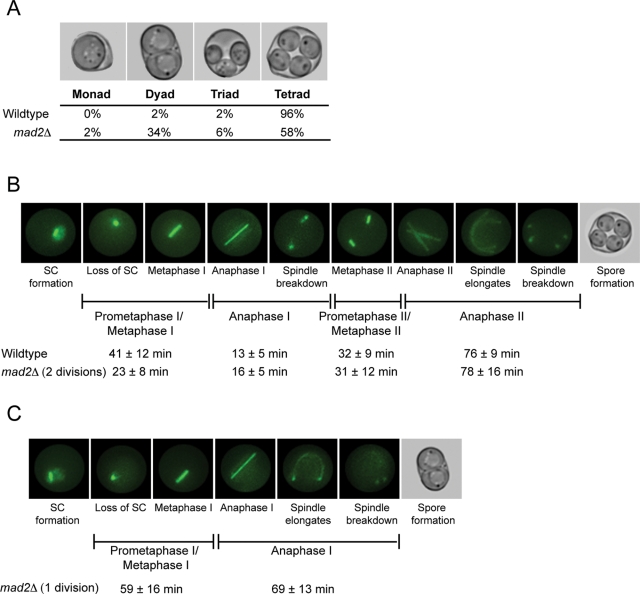
To investigate the role of Mad2 in meiosis, we analyzed sporulation in wild-type and mad2Δ cells. We used the W303 budding yeast strain because the mitotic and meiotic phenotypes of mad2Δ were previously characterized in this background (Hwang et al., 1998; Shonn et al., 2000, 2003). The process of sporulation, which includes meiosis and spore formation, can be induced through nutrient starvation of diploid budding yeast cells. Sixty-five percent of wild-type W303 cells sporulate, and of those, 96% package the four products of meiosis into four spores, forming a tetrad. Sixty-five percent of mad2Δ cells sporulate as well. However, sporulation of mad2Δ cells results in two major populations of spores: 1) 58% form tetrads, and 2) 34% form dyads, or asci containing two spores (Figure 1A). A small fraction of wild-type and mad2Δ cells form triads (2 and 6%, respectively). We reasoned that investigating the differences between meiotic cell cycle events in wild-type cells, mad2Δ cells that form dyads, and mad2Δ cells that form tetrads might uncover a role of Mad2 in regulating the meiotic divisions.
Figure 1:
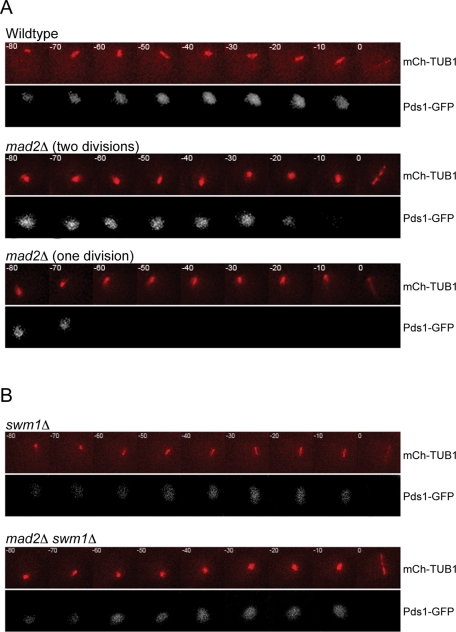
Mad2 affects the duration of the meiotic cell cycle. (A) Wild-type and mad2Δ/mad2Δ sporulated cells were counted for the number of spores in each ascus. Nine hundred sporulated cells were counted in three biological replicates. (B, C) Synaptonemal complex (SC) formation and loss, and spindle assembly and disassembly, were visualized by expressing Zip1-GFP and TUB1-GFP and monitored using time-lapse fluorescence microscopy. The stages of meiosis were determined based on loss of Zip1 and spindle morphology, and the time of each stage was calculated (in minutes ± SD). (B) Still images from a representative movie of wild-type cells. One hundred cells of each genotype were counted. (C) Still images from a representative movie of mad2Δ cells that form dyads. Fifty mad2Δ cells that formed dyads were counted.
We first asked whether Mad2 is required for the proper timing of the meiotic divisions. Past studies did not detect a difference in timing of mad2Δ cells, but these studies analyzed fixed cells and, due to asynchrony in meiotic induction, may not have detected small changes in specific phases of the meiotic cell cycle (Shonn et al., 2000). To analyze more carefully the timing of the meiotic divisions, we used time-lapse microscopy to measure the duration of meiotic phases in individual wild-type and mad2Δ cells. We expressed two green fluorescent protein (GFP)–tagged proteins, ZIP1-GFP and TUB1-GFP, to follow progression through meiosis. Zip1 is a component of the synaptonemal complex that assembles in zygotene and disassembles in diplotene, and it serves as a visual marker for prophase I (Sym et al., 1993; Scherthan et al., 2007). TUB1-GFP encodes a tagged α-tubulin, permitting observation of spindle formation and breakdown in meiosis I and meiosis II (Carminati and Stearns, 1997; Straight et al., 1997). Although we used the same fluorescent tag for both proteins, they are distinguishable because they are both morphologically and temporally different during the meiotic cell cycle (Figure 1B). Time-lapse images taken during sporulation allow us to measure the duration of each stage. We define the cell cycle stages based on disappearance of Zip1 and spindle morphology (Figure 1B).
The time-lapse microscopy indicates that the mad2Δ cells that form dyads undergo only one meiotic division, but the mad2Δ cells that form tetrads undergo both meiotic divisions (Figure 1, B and C). We will refer to the mad2Δ cells that undergo one meiotic division and form dyads as “1 division mad2Δ cells” and the mad2Δ cells that undergo both divisions and form tetrads as “2 division mad2Δ cells.” The small fraction (6%) of mad2Δ cells that form triads also undergo two divisions; however, there is an error in spore formation, and only three of the four products of meiosis are packaged into spores. Therefore, 64% of sporulating cells undergo both divisions; in our analysis of the 2 division mad2Δ cells, we only include those that form tetrads since we are unsure about the underlying cause of the spore formation error resulting in a triad.
Observation of the 2 division mad2Δ cells shows that these cells have a faster prometaphase I/metaphase, 23 ± 8 min, compared with 41 ± 12 min in wild-type cells (Figure 1B). This difference is highly significant (p < 0.0001; unpaired Student's t test). This change in cell cycle timing only occurs in meiosis I; the duration of each stage in meiosis II of 2 division mad2Δ cells is similar to that of wild-type cells. The shorter duration of prometaphase I/metaphase I may cause the increase in chromosome missegregation that occurs in 2 division mad2Δ cells.
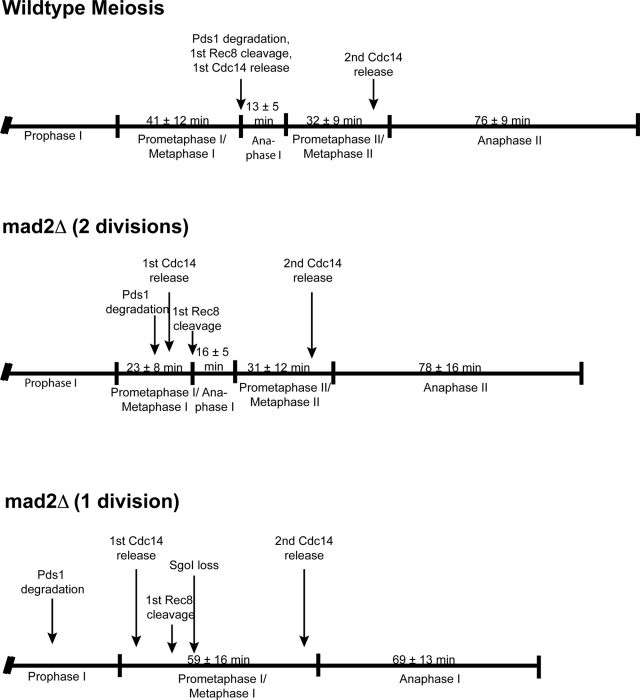
In contrast, the 1 division mad2Δ cells have an extended prometaphase I/metaphase I at 59 ± 16 min, compared with 41 ± 12 min in wild-type cells (Figure 1C). The difference in the duration of prometaphase I/metaphase I between wild-type and 1 division mad2Δ cells is highly statistically significant (p < 0.0001; unpaired Student's t test). The duration of anaphase I in 1 division mad2Δ cells is 69 min ± 13 min, substantially greater than the 13 ± 5 min in wild-type cells but much more similar to anaphase II in wild-type cells (76 ± 9 min). In the 1 division mad2Δ cells, the anaphase I spindle elongates and bends around the cell, which is a characteristic of anaphase II spindles but not anaphase I spindles in wild-type cells (Figure 1, B and C). In summary, the 2 division mad2Δ cells have a faster prometaphase I/metaphase I, and the 1 division mad2Δ cells have a longer prometaphase I/metaphase I, but the entire meiotic cell cycle is shorter than in wild-type cells.
The 1 division mad2Δ cells undergo an aberrant meiotic division in which the homologous chromosomes pair and recombine, but sister chromatids separate inappropriately
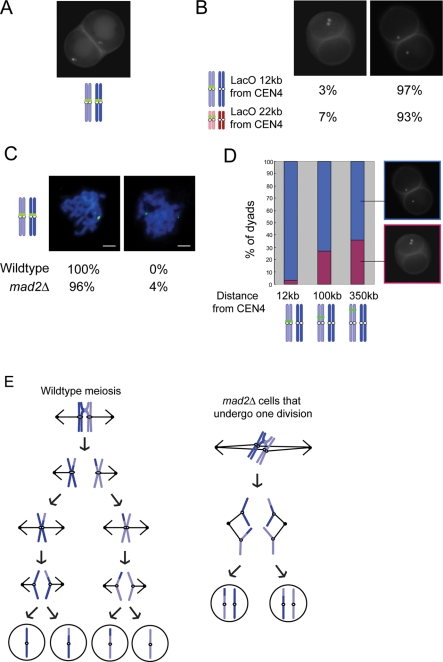
To determine the role of Mad2 in meiosis, we further analyzed the 1 division mad2Δ cells. We monitored the segregation of chromosome IV by placing a lactose operator (LacO) array near the centromere and expressing a GFP–lactose repressor fusion protein (GFP-LacI), targeting GFP to the chromosome (Straight et al., 1996; Shonn et al., 2000). When both homologous chromosomes have GFP targeted near CEN4, 97% of the mad2Δ dyads have two GFP-marked chromosomes in each spore, suggesting that the spores are diploid (Figure 2A). As a further confirmation that the spores are diploid, we dissected the dyad spores and found that 78% were viable and able to sporulate.
Figure 2:
The mad2Δ dyad spores are the result of an aberrant meiotic division in which homologous chromosomes pair and recombine and then sister chromatids separate. (A) mad2Δ/mad2Δ cells with both chromosome IV's marked with a LacO array near CEN4 were analyzed for the segregation of the marked chromosome. Three hundred dyads were counted in three experiments. (B) Dyads from mad2Δ/mad2Δ cells with only one of the two homologous chromosome marked with a LacO array were analyzed for the segregation of the marked chromosome. Strains contain a LacO array either 12 kb from CEN4 or 22 kb from CEN3. Three hundred dyads from each genotype were counted in three experiments. (C) Wild-type and mad2Δ/mad2Δ cells with both homologous chromosome IV's marked with a LacO array 12 kb from CEN4 were prepared for chromosome spreads. Fifty spreads from each genotype were counted. Scale bar, 2 μm. (D) mad2Δ/mad2Δ dyads with only one of the two homologous chromosomes marked with a LacO array were analyzed for the segregation of the marked chromosome. Strains contain the LacO array 12, 100, or 350 kb from CEN4. The percentage of dyads with one marked chromosome in each spore is shown in blue. The percentage of dyads with two marked chromosomes in one spore is shown in pink. Three hundred dyads from each genotype were counted in three experiments. (E) Diagram showing a wild-type meiosis compared with the aberrant meiosis in mad2Δ cells that results in the separation of sister chromatids in a single division.
The formation of two diploid spores after a single division in mad2Δ cells could be the result of either 1) segregating homologous chromosomes and ending the cell cycle after meiosis I or 2) separating sister chromatids inappropriately. To determine how the chromosomes separated in the 1 division mad2Δ cells, we labeled one of the two homologous chromosomes with a LacO array near the centromere of chromosome IV. If homologous chromosomes separate, one of the two spores will have two copies of the marked chromosome, and the other spore will have two copies of the unmarked chromosome. If sister chromatids separate, each spore will have one marked chromosome. When the LacO array is placed at the TRP1 locus, approximately 12 kb from CEN4, 97% of the dyads contain one marked chromosome in each spore (Figure 2B). We verified that chromosome III also segregates sister chromatids by placing the LacO array at the LEU2 locus, approximately 22 kb away from CEN3. Ninety-three percent of mad2Δ dyads contained one marked chromosome in each spore (Figure 2B). Surprisingly, the data show that sister chromatids separate inappropriately in the single meiotic division. These results suggest that in the 1 division mad2Δ cells, meiosis II events occur in the single meiotic division.
To investigate whether other events of meiosis I were perturbed in the 1 division mad2Δ cells, we examined whether the cells initiate meiosis correctly by pairing and recombining homologous chromosomes in prophase I. To monitor pairing, we examined spread meiotic nuclei of mad2Δ cells in the pachytene stage of prophase. In pachytene, homologous chromosomes have paired, synapsed, and initiated recombination. We marked both homologous chromosomes with a LacO array near CEN4 and expressed GFP-LacI. Chromosomes that are paired will have two GFP marked chromosomes in close proximity. One hundred percent of wild-type and 96% of mad2Δ cells have paired homologous chromosomes (Figure 2C). Using this assay, we cannot determine which cells will form dyads, but the proportion of cells with paired chromosomes is so great that it must include the mad2Δ cells that undergo one division, showing that there is not a defect in pairing in mad2Δ cells.
To determine whether crossovers occur in the 1 division mad2Δ cells, we monitored the segregation pattern of a LacO array placed at different locations along one of the homologous chromosome IVs. As shown earlier, if a LacO array is placed only 12 kb from the centromere on one of the homologous chromosomes, the sister centromeres split, and 97% of the dyads have one chromosome with the array in each spore. Only 3% of the dyads have two chromosomes with the array in one spore. We figured that if an array is placed further from the centromere, a crossover could occur between the array and the centromere, and the dyads would inherit two marked chromosomes in one spore. Indeed, with an array located approximately 100 kb from the centromere, 27% of the dyads have two marked chromosomes in one spore. With an array located 350 kb from the centromere, 36% of the dyads have two marked chromosomes in one spore (Figure 2D). In summary, the mad2Δ dyads undergo a single meiotic division in which homologous chromosomes pair, recombine, and separate sister chromatids inappropriately (Figure 2E).
In the 1 division mad2Δ cells, kinetochores are not clamped together, and cohesin does not remain protected around the centromere
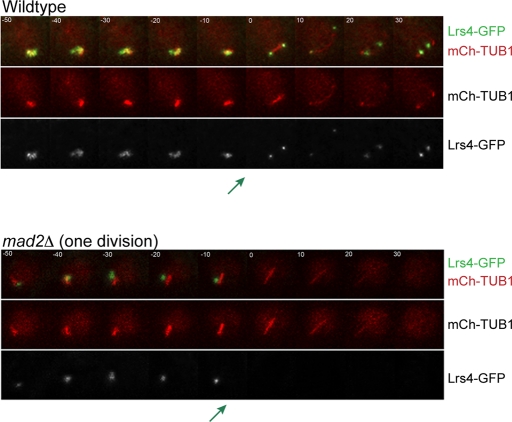
Our results suggest that in the 1 division mad2Δ cells, the paired homologous chromosomes attach sister chromatids to opposite spindle poles in metaphase I instead of homologous chromosomes (Figure 2E). In wild-type cells, sister chromatids do not attach to opposite spindle poles because the monopolin complex holds the sister chromatids' kinetochores together, assembling one microtubule-binding site (Toth et al., 2000; Rabitsch et al., 2003; Winey et al., 2005; Petronczki et al., 2006; Monje-Casas et al., 2007). To determine whether monopolin localization is disrupted in 1 division mad2Δ cells, we monitored the localization of the monopolin component Lrs4 tagged with GFP, using time-lapse microscopy (Figure 3). In wild-type cells, Lrs4 normally resides in the nucleolus until the end of prophase I. Then Lrs4 leaves the nucleolus and binds to the kinetochores until the end of anaphase I (Rabitsch et al., 2003). In 2 division mad2Δ cells, Lrs4-GFP behaves similarly to wild-type cells (unpublished data). In 1 division mad2Δ cells, Lrs4-GFP leaves the nucleolus but does not bind to kinetochores (Figure 3).
Figure 3:
In the mad2Δ cells that undergo one meiotic division, the monopolin complex does not bind sister kinetochores. (A) Time lapse images of meiosis in wild-type and 1 division mad2Δ/mad2Δ cells. Both strains are expressing Lrs4-GFP and Tub1-mCherry. The green arrow shows the point in which Lrs4-GFP leaves the nucleolus. One hundred of the wild-type cells, 100 of the 2 division mad2Δ cells, and 50 of the 1 division mad2Δ cells were analyzed.
The chromosome segregation pattern of 1 division mad2Δ cells is quite distinct from that in monopolin mutants. In monopolin mutants, sister chromatids cannot separate in meiosis I due to the protected centromeric cohesins. However, the spindle poles will separate, and the cells will undergo meiosis II, making four mostly inviable spores (Toth et al., 2000; Rabitsch et al., 2003). However, in the 1 division mad2Δ cells, sister chromatids separate in the single division, making two viable diploid spores, suggesting that centromeric cohesins are not protected.
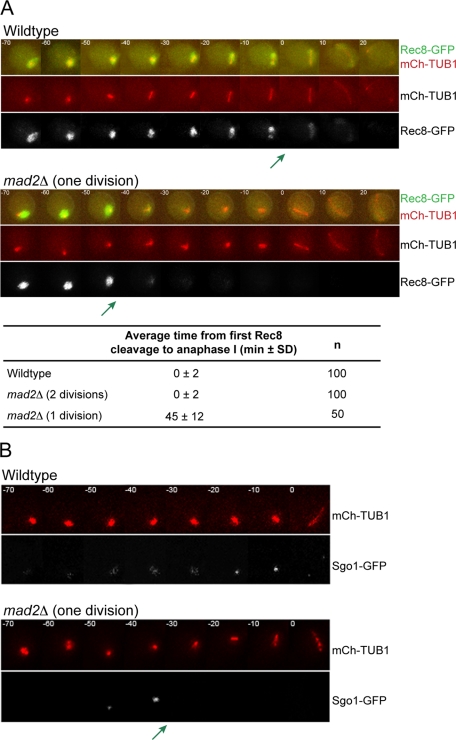
We monitored the meiotic cohesin Rec8 to determine whether centromeric cohesin is lost prematurely in the 1 division mad2Δ cells. In wild-type cells, previous reports showed that there is stepwise cleavage of Rec8; the Rec8 along chromosome arms is cleaved in meiosis I, and the centromeric Rec8 is cleaved in meiosis II (Klein et al., 1999; Buonomo et al., 2000; Kitajima et al., 2003, 2004). To monitor cohesin cleavage during the meiotic cell cycle, we tagged Rec8 with GFP in cells also expressing mCherry-TUB1. As expected, wild-type and 2 division mad2Δ cells show a stepwise loss of Rec8. The majority of Rec8 is cleaved concurrently with anaphase I spindle assembly, leaving a fraction of Rec8 until meiosis II (Figure 4A). In the 1 division mad2Δ cells, there is still a stepwise loss of cohesin, except that the first cleavage occurs prematurely, 45 ± 12 min prior to anaphase I.
Figure 4:
In the mad2Δ cells that undergo one meiotic division, protection of the meiotic cohesin Rec8 is lost prematurely. (A) Time lapse images of meiosis in wild-type and 1 division mad2Δ/mad2Δ cells. Cells are expressing Tub1-mCherry and Rec8-GFP. A green arrow marks the time of the first Rec8 cleavage. The time from first cleavage to anaphase I was calculated (± SD). (B) Time lapse images of meiosis in wild-type and 1 division mad2Δ/mad2Δ cells. Cells are expressing Tub1-mCherry and Sgo1-GFP. A green arrow marks the time at which Sgo1 is lost in the 1 division mad2Δ cells. One hundred wild-type cells and 50 mad2Δ cells that formed dyads were analyzed.
Our results suggest that in the 1 division mad2Δ cells, centromeric cohesins do not remain protected in anaphase I, allowing the cells to separate sister chromatids. We used time-lapse microscopy to monitor Sgo1, one of the factors required for protection of centromeric cohesins (Kerrebrock et al., 1995; Katis et al., 2004; Kitajima et al., 2004; Marston et al., 2004). We made an Sgo1-GFP fusion protein and expressed mCherry-TUB1 in wild-type and mad2Δ cells. As expected, Sgo1-GFP associates with chromosomes throughout meiosis I in wild-type and in 2 division mad2Δ cells (Figure 4B). In contrast, in 1 division mad2Δ cells, Sgo1 is lost 38 ± 7 min prior to anaphase I. The loss of Sgo1 most likely results in the cleavage of centromeric Rec8 (Kitajima et al., 2004; Marston et al., 2004). Therefore, our data show that in the 1 division mad2Δ cells, sister chromatids separate because sister kinetochores are not clamped together by monopolin, Sgo1 is lost prematurely, and centromeric cohesins are cleaved in the single meiotic division. Clamping of sister kinetochores by monopolin is independent of cohesin and cohesin protection (Monje-Casas et al., 2007), suggesting that the phenotype seen in mad2Δ cells is due to pleiotropic misregulation of the meiotic cell cycle. We diagram the timing of cell cycle events with respect to the stages of meiosis (as defined by spindle morphology) in Figure 5.
Figure 5:
Schematic representing the timing of different cell cycle events with respect to the stage of the cell cycle in wild-type, 2 division mad2Δ, and 1 division mad2Δ cells.
In the absence of Mad2, the APC/C is prematurely active in prometaphase of meiosis I
We considered that Mad2 regulates the meiotic cell cycle by modulating APCCdc20 activity. During spindle checkpoint signaling, Mad2, together with other checkpoint proteins, inhibits APCCdc20 activity to delay the cell cycle in metaphase I (Musacchio and Salmon, 2007) To determine whether APCCdc20 is prematurely active in the absence of Mad2, we investigated the timing of the degradation of the APCCdc20 substrate securin/Pds1. Using time-lapse microscopy, we monitored Pds1-GFP and mCherry-TUB1. In wild-type cells, Pds1-GFP is degraded, and the cells enter anaphase I (Figure 6A). We were surprised to find that in the 2 division mad2Δ cells, Pds1 is degraded on average 13 ± 12 min prior to anaphase I spindle assembly (Figure 6A and Table 1). Because prometaphase I/metaphase I is ∼18 min faster in 2 division mad2Δ cells than in wild-type cells (Figure 1B), Pds1 is in fact degraded ∼31 min early (Figure 5). It is not clear why the cells do not enter anaphase I immediately after loss of Pds1, but there may also be a misregulation of other cell cycle events preventing cohesin cleavage. It is striking that in the 1 division mad2Δ cells, Pds1 is degraded even more prematurely: 81 min prior to anaphase I spindle assembly (Figure 6A and Table 1).
Figure 6:
The APC is prematurely active in mad2Δ cells. (A) Time-lapse images of meiosis in wild-type and mad2Δ/mad2Δ cells. Both strains are expressing Pds1-GFP and Tub1-mCherry. One hundred wild-type cells, 100 mad2Δ cells that formed tetrads, and 50 mad2Δ cells that formed dyads were analyzed. (C) Time lapse images of meiosis in swm1Δ/swm1Δ and swm1Δ/swm1Δ mad2Δ/mad2Δ cells. Both strains are expressing Pds1-GFP and Tub1-mCherry. No single meiotic division in swm1Δ/swm1Δ mad2Δ/mad2Δ was observed. One hundred cells of each genotype were analyzed.
TABLE 1:
Average time from securin/Pds1 degradation to anaphase I spindle formation.
| Average time from Pds1 degradation to anaphase I (min) | SD | Range (min) | n | |
|---|---|---|---|---|
| Wild type | 1 | 2 | 0–10 | 100 |
| mad2Δ (2 divisions) | 13 | 12 | 0–60 | 100 |
| mad2Δ (1 division) | 81 | 35 | 35–190 | 50 |
| swm1Δ | 0 | 0 | 0 | 100 |
| mad2Δ swm1Δ | 1 | 2 | 0–10 | 100 |
| mad3Δ | 1 | 2 | 0–10 | 100 |
| ama1Δ | 1 | 2 | 0–10 | 100 |
| mad2Δ ama1Δ | 11 | 8 | 0–30 | 100 |
If prematurely active APC is indeed the cause of premature Pds1 degradation, then decreasing APC/C activity should prevent the early Pds1 degradation. We measured the timing of the loss of Pds1-GFP in mad2Δ cells that also have a deletion of Swm1, a nonessential component of the APC/C. Cells that lack Swm1 target substrates for ubiquitination less efficiently than wild-type cells (Hall et al., 2003; Schwickart et al., 2004; Oelschlaegel et al., 2005), but Pds1 is degraded at anaphase I in swm1Δ cells (Figure 6B and Table 1). In mad2Δ swm1Δ cells, we do not see premature degradation of Pds1; the degradation of Pds1 occurs within 1 ± 2 min of anaphase I spindle formation (Figure 6B and Table 1). Using time-lapse microscopy, we find that no mad2Δ swm1Δ cells undergo only one meiotic division. Therefore, down-regulating APC/C activity prevents premature degradation of Pds1 in mad2Δ cells and rescues the single-division phenotype. We conclude that in mad2Δ cells, the APC/C is prematurely active.
The role of Mad2 in down-regulating APC/C activity in metaphase I is distinct from its role in delaying the cell cycle if a chromosome is not attached to the spindle
Because the activity of the APC/C is inhibited during spindle checkpoint signaling when a chromosome is not attached to spindle microtubules, we wanted to determine whether another protein required for the spindle checkpoint signaling in meiosis, Mad3, is also required to decrease APC/C activity during prometaphase I and metaphase I (Shonn et al., 2000; Musacchio and Salmon, 2007). We monitored loss of Pds1-GFP with respect to anaphase I spindle assembly in mad3Δ cells. Pds1-GFP is degraded within 1 ± 2 min of anaphase I spindle formation in mad3Δ cells, similar to what occurs in wild-type cells (Table 1). We also do not see the formation of dyad spores or the single-division phenotype. Our results indicate that in the absence of Mad3, the APC/C is not prematurely active, suggesting that Mad2 functions independently of Mad3 to down-regulate APC/C activity during prometaphase I/metaphase I. This is consistent with previous studies showing that mad3Δ cells do not have an increase in meiosis I nondisjunction (Shonn et al., 2003).
Both APCCdc20 and APCAma1 are prematurely active in 1 division mad2Δ cells
The meiosis-specific cofactor of the APC/C, Ama1, also targets Pds1 for ubiquitination and subsequent degradation and, therefore, may be prematurely active in mad2Δ cells (Cooper et al., 2000; Oelschlaegel et al., 2005; Penkner et al., 2005). Ama1 is not essential for meiosis but does function in meiosis I to promote the rapid degradation of APC/C substrates (Oelschlaegel et al., 2005). To determine whether APCAma1 is prematurely active in mad2Δ cells, we deleted Ama1 in wild-type and mad2Δ cells and followed the degradation of Pds1-GFP with respect to spindle assembly by live-cell imaging. In ama1Δ cells, Pds1 is degraded within 1 ± 2 min of anaphase I spindle assembly (Table 1). In ama1Δ mad2Δ cells, Pds1 is degraded ∼11 ± 8 min prior to anaphase I spindle assembly, similar to 2 division mad2Δ cells. We do not see any cells that undergo a single meiotic division. We conclude that premature APCAma1 activity results in the very premature Pds1 degradation and the single-division phenotype in mad2Δ cells.
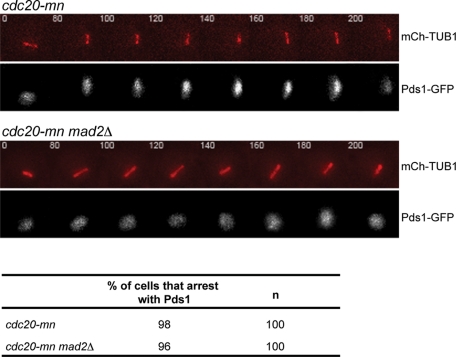
Our results suggest that Mad2 may have a role in preventing premature APCAma1 activity. However, Mad2 could directly or indirectly inhibit APCAma1. In wild-type cells, Cdc20 is required for Pds1 degradation, suggesting that APCAma1 is only active after APCCdc20 targets substrates for degradation (Salah and Nasmyth, 2000; Oelschlaegel et al., 2005). We reasoned that Mad2 may not directly inhibit APCAma1, but might instead inhibit APCCdc20 activity, and that this inhibition prevents APCAma1 activity. To determine whether Cdc20 is required for the premature activity of APCAma1, we analyzed whether Pds1 can be degraded in the absence of Cdc20 in mad2Δ cells. We replaced the Cdc20 promoter with the mitosis-specific Clb2 promoter to make a Cdc20 meiotic null (cdc20-mn; Lee and Amon, 2003) and monitored Pds1 degradation with respect to metaphase I spindle assembly. The cdc20-mn and cdc20-mn mad2Δ cells do not degrade Pds1 prematurely. Ninety-eight percent of the cdc20-mn and 96% of cdc20-mn mad2Δ cells that enter meiosis are blocked in metaphase I with Pds1-GFP present for at least 200 min (Figure 7). Our results demonstrate that APCCdc20 activity is required for the activation of APCAma1 in 1 division mad2Δ cells. We conclude that in prometaphase I, Mad2 indirectly prevents premature APCAma1 activity by inhibiting APCCdc20.
Figure 7:
APCCdc20 activity is required for the premature APCAma1 activity in mad2Δ cells. (A) Time-lapse images of meiosis in cdc20-mn/cdc20-mn and mad2Δ/mad2Δ cdc20-mn/cdc20-mn cells. All strains are expressing Pds1-GFP and Tub1-mCherry. The percentage of cells that arrest with Pds1 is shown.
We decided to further explore the regulation of APCAma1 by APCCdc20. A previous study demonstrates that APCAma1 activity is inhibited during metaphase I by cyclin-dependent kinase (CDK; Oelschlaegel et al., 2005). However, an allele of AMA1, Ama1-m8, with all eight putative CDK phosphorylation sites mutated to alanine does not result in premature APCAma1 activity, suggesting that the inhibition of APCAma1 by CDK is not direct or that there is redundancy in the regulatory pathway. To determine whether Ama1 phosphorylation regulates APCAma1 activity in mad2Δ cells, we analyzed mad2Δ Ama1-m8 cells expressing mCherry-TUB1 by time-lapse microscopy. Surprisingly, 87% of sporulated mad2Δ Ama1-m8 cells undergo a single meiotic division (Table 2). The percentage sporulation of mad2Δ Ama1-m8 cells was similar to that in wild-type cells. In accordance with previous observations, we did not see a phenotype of Ama1-m8 in the wild-type background (Oelschlaegel et al., 2005). Our results suggest that there is redundancy in the regulatory network to prevent premature activity of APCAma1 in metaphase I. We propose that Ama1 is inhibited by phosphorylation and by another activity of CDK. In the absence of Mad2, premature APCCdc20 activity could lead to less CDK activity and, in some cells, the dephosphorylation of Ama1, resulting in the one-division phenotype.
TABLE 2:
A nonphosphorylatable form of Ama1 increases the percentage of mad2Δ cells that undergo 1 meiotic division.
| Genotype | Percentage sporulation | Percentage of sporulated cells that undergo one meiotic division | n |
|---|---|---|---|
| Wild-type | 65 | 0 | 100 |
| Ama1-m8 | 67 | 0 | 100 |
| mad2Δ | 65 | 34 | 100 |
| mad2Δ + Ama1-m8 | 60 | 87 | 100 |
Premature APCAma1 activity results in the premature release of the Cdc14 phosphatase
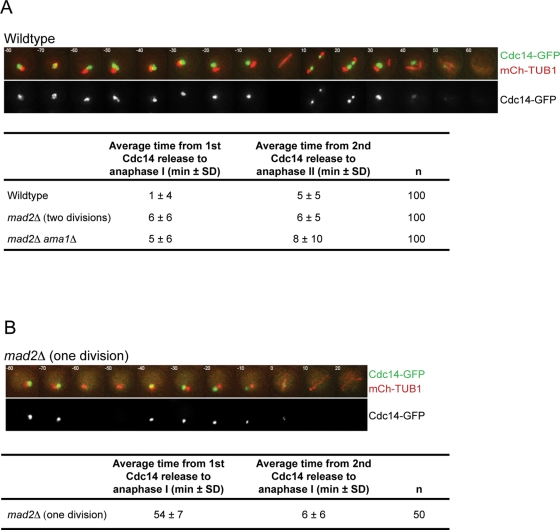
Our results demonstrate that in the absence of Mad2, securin/Pds1 is degraded prematurely. The degradation of securin results in the release of the protease separase, which cleaves sister chromatid cohesion. Separase also functions in the Cdc14 early anaphase release (FEAR) network, promoting the release of the Cdc14 phosphatase from the nucleolus (Stegmeier et al., 2002; Buonomo et al., 2003; Marston et al., 2003). Once released, Cdc14 counteracts CDK activity by dephosphorylating CDK substrates. Because the 1 division mad2Δ cells degrade securin/Pds1 prematurely, we analyzed whether Cdc14 is released prematurely. We monitored the timing of release and resequestration of Cdc14-GFP in wild-type and mad2Δ cells using time-lapse microscopy. In wild-type cells, Cdc14-GFP is released from the nucleolus 1 ± 4 min before anaphase I, is resequestered into the nucleolus in metaphase II, and then is released again 5 ± 5 min before anaphase II (Figure 8A). The 2 division mad2Δ cells have similar timing of Cdc14 release; Cdc14 is released 6 ± 6 min before anaphase I, resequestered, and released again 6 ± 5 min before anaphase II. Surprisingly, in 1 division mad2Δ cells, Cdc14 is also released twice, but both releases occur before the first division. Cdc14 is first released 54 ± 7 min before anaphase I spindle formation, resequestered, and released again 6 ± 6 min before anaphase I spindle formation (Figure 8B). These results support our conclusion that meiosis II cell cycle events are uncoupled from the chromosome segregation cycle in the 1 division mad2Δ cells (Figure 5).
Figure 8:
The phosphatase Cdc14 is released from the nucleolus prematurely in the 1 division mad2Δ cells, and the premature release is dependent on APCAma1 activity. (A) Time-lapse images of meiosis in wild-type, mad2Δ/mad2Δ, and mad2Δ/mad2Δ ama1Δ/ama1Δ cells. All strains are expressing Cdc14-GFP and Tub1-mCherry. Average time of Cdc14 release from the nucleolus before anaphase I and anaphase II spindle formation is shown (± SD). (B) Time-lapse images of meiosis in 1 division mad2Δ/mad2Δ cells. All strains are expressing Cdc14-GFP and Tub1-mCherry. Average times of the two Cdc14 releases from the nucleolus before anaphase I are shown.
Our results suggest that the premature degradation of Pds1 leads to separase activation of the FEAR network and Cdc14 release. Because Ama1 is required for the premature degradation of Pds1 in mad2Δ cells that undergo one division, Ama1 should also be required for the premature release of Cdc14. Analysis of Cdc14 release in mad2Δ ama1Δ cells reveals that it occurs with timing similar to that of wild-type cells during both meiotic divisions (Figure 8A). These data show that the activity of AMA1 is required for the premature release of Cdc14, most likely through targeting Pds1 for ubiquitination and subsequent degradation.
DISCUSSION
Mad2 regulates the timing of the degradation of APC/C substrates
In this study, we observed that a deletion of Mad2 can disrupt the normal timing of meiotic cell cycle events. In the absence of Mad2, cells execute one of two alternative meiotic programs: 1) 64% of cells undergo two meiotic divisions, but prometaphase I/metaphase I is shorter in duration, and 2) 34% of cells undergo a single meiotic division in which sister chromatids separate inappropriately. Both pathways are the result of the APC/C prematurely targeting substrates such as securin/Pds1 for ubiquitination. Down-regulation of APC/C activity, by deleting the APC/C component Swm1, rescues the premature securin/Pds1 degradation and the single-division phenotype. Our data indicate that premature APCCdc20 activity causes the faster prometaphase I/metaphase I in the 2 division mad2Δ cells, and premature APCCdc20 and APCAma1 result in the single-division phenotypes. We propose that Mad2 restrains APCCdc20 activity, setting the duration of prometaphase I and metaphase I to prevent a misregulation of cell cycle events and the missegregation of chromosomes.
In the 2 division mad2Δ cells, the premature APCCdc20 activity is likely to lead to the increase in meiosis I nondisjunction by targeting substrates for degradation and transitioning into the next cell cycle stage too rapidly. By using time-lapse microscopy to monitor cell cycle proteins in individual cells instead of a population of cells, we find that securin/Pds1 is degraded ∼30 min earlier in 2 division mad2Δ cells and the duration of prometaphase I/metaphase I is ∼18 min shorter when compared with wild-type cells. This change in duration of prometaphase I/metaphase I in mad2Δ cells shortens the time allotted for chromosomes to properly attach to the bipolar spindle. Because the disruption of Mad2 in mammalian oocytes, worms, fission yeast, and budding yeast results in an increase in meiosis I nondisjunction, regulation of cell cycle timing may be a conserved role of Mad2 in meiosis (Kitagawa and Rose, 1999; Shonn et al., 2000, 2003; Bernard et al., 2001; Tsurumi et al., 2004; Homer et al., 2005a, 2005b; Stein et al., 2007). Similarly, in Drosophila oocytes, disruption of the spindle checkpoint component Ald/Mps1 leads to an advance in anaphase I onset and an increase in meiosis I nondisjunction (Gilliland et al., 2007).
Can premature APCAma1 and APCCdc20 activity result in all of the phenotypes seen in 1 division mad2Δ cells to produce two viable spores? The premature activity of both APCAma1 and APCCdc20 could lead to the early degradation of substrates, such as Pds1, Sgo1, Dbf4, and Spo13 (Cooper et al., 2000; Ferreira et al., 2000; Salah and Nasmyth, 2000; Oelschlaegel et al., 2005; Penkner et al., 2005; Sullivan and Morgan, 2007). Pds1 regulates the meiotic divisions by inhibiting separase to ensure the timely segregation of chromosomes and release of Cdc14 phosphatase (Rock and Amon, 2009). Sgo1 protects centromeric cohesins (Katis et al., 2004; Kitajima et al., 2004; Marston et al., 2004). Dbf4, as a component of the DDK kinase, regulates the localization of monopolin to kinetochores (Matos et al., 2008). Finally, Spo13 has a role in monopolin localization and protection of sister chromatid cohesion. The premature degradation of these substrates could result in the phenotypes of the 1 division mad2Δ cells: 1) Cdc14 phosphatase is prematurely released from the nucleolus; 2) cohesin does not remain protected in metaphase I; 3) kinetochores on sister chromatids are not clamped together; and 4) sister chromatids separate in a single division. Therefore, the production of viable spores after a single meiotic division in mad2Δ cells is likely the consequence of the combined loss of many APC/C substrates.
Our results show that APCAma1 activity at the end of prophase I results in an uncoupling of cell cycle events from the chromosome segregation cycle, and meiosis II events occur in a single meiotic division. The cell has evolved many different mechanisms to prevent premature APCAma1 activity. AMA1 is transcribed and spliced only in meiosis (Cooper et al., 2000). An inhibitor of Ama1, Mnd2, prevents APCAma1 activity in the early meiotic stages (Oelschlaegel et al., 2005; Penkner et al., 2005). In addition, CDK/M-phase cyclin also inhibits APCAma1 activity (Oelschlaegel et al., 2005). In this study, we show that the premature APCAma1 activity in mad2Δ cells requires APCCdc20 activity. Therefore, we do not believe that Mad2 directly inhibits APCAma1 but is more likely to indirectly inhibit APCAma1 through the regulation of APCCdc20 activity. We are unsure why only 34% of mad2Δ cells have premature APCAma1 activity and why this phenotype is uncovered in the W303, but not SK1 strain background (S. Lacefield, unpublished data). We suspect that the regulation of Ama1 activity in W303 and SK1 is slightly different. In the mad2Δ cells that have premature activity of APCAma1, we predict that the timing of the meiotic cell cycle was altered such that APCCdc20 was activated early, leading to activation of APCAma1 prior to the inhibition by CDK/M-phase cyclin activity. Indeed, in mad2Δ cells with a version of Ama1 with all putative CDK phosphorylation sites mutated to alanine, 87% of cells undergo a single division.
Mad2 functions as a meiotic cell cycle regulator to promote faithful chromosome segregation
The activity of Mad2 in meiosis in budding yeast may be similar to its essential activity in meiosis in mammals. As in budding yeast, depletion of Mad2 in mouse oocytes causes a shortened duration of metaphase I, premature degradation of securin, and chromosomal missegregation (Wassmann et al., 2003; Tsurumi et al., 2004; Homer et al., 2005a, 2005b). We showed that Mad2 prevents premature APC/C substrate degradation in prometaphase I/metaphase I in budding yeast. This activity is independent of the spindle checkpoint component Mad3, although both Mad2 and Mad3 are required to delay the cell cycle at the metaphase I–to–anaphase I transition if chromosomes are not attached to microtubules. Mad3 was previously implicated in regulating the duration of prophase I, an earlier meiotic stage (Cheslock et al., 2005). In mammalian oocytes, BubR1 (which shares a homology domain with Mad3) was also shown to have a role in prophase I, and depletion of BubR1 does not cause the same metaphase I phenotypes as depletion of Mad2 (Homer et al., 2009). We note that the consequences of loss of checkpoint components are different in meiosis I than in mitosis in both budding yeast and mammalian cells (Shonn et al., 2003; Meraldi et al., 2004; Homer et al., 2009). In budding yeast, the absence of Mad2 does not cause a difference in cell cycle timing in mitosis and only has a modest effect on chromosome segregation. The APC/C may be differentially regulated in meiosis I compared with mitosis to ensure that the substrates that have meiotic functions are degraded at the proper time. In the absence of Mad2, cell cycle events are uncoupled from the chromosome segregation cycle. Our results demonstrate that Mad2 is an important meiotic cell cycle regulator, preventing premature APC/C activity in prometaphase I and metaphase I and ensuring the proper orchestration of meiotic cell cycle events.
MATERIALS AND METHODS
Strains and manipulations
All strains are W303 derivatives and are described in Table 3. Deletions were made using standard methods (Longtine et al., 1998). Chromosomes were tagged with GFP as described (Straight et al., 1996; Shonn et al., 2000). Lrs4, Rec8, Sgo1, Pds1, and Cdc14 C-terminal GFP-fusion proteins were made by targeting GFP to the endogenous locus, as described (Wach et al., 1997; Sheff and Thorn, 2004). Zip1-GFP with GFP located at the end of the second coiled-coil domain at position 700 replaced Zip1 at the endogenous locus as described (Scherthan et al., 2007). To visualize tubulin, constructs containing PTUB1TUB1-GFP were integrated into the URA3 locus. Constructs containing PHIS3mCherry-TUB1 were integrated into the URA3 or ADE2 locus. The Clb2 promoter replaced the Cdc20 promoter using a PCR-based method described in Longtine et al. (1998). Strains were sporulated in liquid culture by growing in peptone, yeast extract, and dextrose at 30°C to saturation, diluted into 1% yeast extract, 2% bactopeptone, and 1% potassium acetate for 12–15 h at 30°C, washed with water, and resuspended in 1% potassium acetate at 25°C.
TABLE 3:
Strains used in this study.
| Strain name | Genotype |
|---|---|
| LY56 | MATa/α, LacO:TRP1/LacO:TRP1, PTUB1-GFP-TUB1-URA3/PCYC1-GFP-LacI2-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15 |
| LY7 | MATa/α, mad2::LEU2/mad2::LEU2, LacO:TRP1/LacO:TRP1, PCYC1-GFP-LacI2-URA3/ura3-1, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15 |
| LY9 | MATa/α, mad2::LEU2/mad2::LEU2, LacO:TRP1/trp1-1, PCYC1-GFP-LacI2-URA3/ura3-1, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15 |
| LY806 | MATa/α, mad2::LEU2/mad2::kanMX4, PTUB1-GFP-TUB1-URA3/PCYC1-GFP-LacI2-URA3, ZIP1-GFP-700/ZIP1, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY838 | MATa/α, PTUB1-GFP-TUB1-URA3/ura3-1, ZIP1-GFP-700/ZIP1, LacO:TRP1/trp1-1, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, |
| LY849 | MATa/α, mad2:: kanMX4/mad2::kanMX4, PCYC1-GFP-LacI2-URA3/ura3-1, LacO:LEU2/leu2-3112, ade2-1/ade2-1, can1-100/can1-100, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY850 | MATa/α, mad2:: kanMX4/mad2::kanMX4, PCYC1-GFP-LacI2-URA3/ura3-1, LacO:TRP1(at chromIV genomic fragment 550857)/+, leu2-3112/leu2-3112, ade2-1/ade2-1, can1-100/can1-100, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY851 | MATa/α, mad2:: kanMX4/mad2::kanMX4, PCYC1-GFP-LacI2-URA3/ura3-1, LacO:TRP1(at chromIV genomic fragment 793927)/+, leu2-3112/leu2-3112, ade2-1/ade2-1, can1-100/can1-100, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY578 | MATa/α, mad2::LEU2/mad2::LEU2, LRS4-GFP-HIS3/LRS4-GFP-HIS3, LacO:TRP1/LacO:TRP1, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15 |
| LY579 | MATa/α, LRS4-GFP-HIS3/LRS4-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY500 | MATa/α, mad2::kanMX4/mad2::kanMX4, REC8-GFP-kanMX4/REC8-GFP-kanMX4, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY527 | MATa/α, REC8-GFP-kanMX4/REC8-GFP-kanMX4, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY685 | MATa/α, mad2::kanMX4/mad2::kanMX4, SGO1-GFP-HIS3/SGO1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY684 | MATa/α, SGO1-GFP-HIS3/SGO1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY389 | MATa/α, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY358 | MATa/α, mad2::kanMX4/mad2::kanMX4, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY768 | MATa/α, mad2::LEU2/mad2::LEU2, swm1::kanMX4/swm1::kanMX4, LacO:TRP1/LacO:TRP1, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY774 | MATa/α, swm1::kanMX4/swm1::kanMX4, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY528 | MATa/α, CDC14-GFP-HIS3/CDC14-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, PHIS3-mCherry-TUB1-ADE2/PHIS3-mCherry-TUB1-ADE2, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY411 | MATa/α, mad2::LEU2/mad2::LEU2, CDC14-GFP-HIS3/CDC14-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY698 | MATa/α, mad2::LEU2/mad2::LEU2, ama1::kanMX4/ama1::kanMX4, CDC14-GFP-HIS3/CDC14-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY274 | MATa/α, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, PCLB2-3HA-CDC20-kanMX6/PCLB2-3HA-CDC20-kanMX6, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY720 | MATa/α, mad2::LEU2/mad2::LEU2, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3 (at mad2)/PHIS3-mCherry-TUB1-URA3 (at mad2), PCLB2-3HA-CDC20-kanMX6/PCLB2-3HA-CDC20-kanMX6, PCYC1-GFP-LacI2-URA3/, PCYC1-GFP-LacI2-URA3, LacO:TRP1/trp1-1 ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15 |
| LY752 | MATa/α, mad3::kanMX4/mad3::kanMX4, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, PHIS3-mCherry-TUB1-ADE2/PHIS3-mCherry-TUB1-ADE2, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY689 | MATa/α, cdc20-127/cdc20-127, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY657 | MATa/α, ama1::kanMX4/ama1::kanMX4, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY643 | MATa/α, mad2::LEU2/mad2::LEU2, ama1::kanMX4/ama1::kanMX4, LacO:TRP1/LacO:TRP1, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY829 | MATa/α, mad2::kanMX/mad2::kanMX, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, PAma1-Ama1-m8-LEU2/PAma1-Ama1-m8-LEU2 ade2-1/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
| LY802 | MATa/α, PDS1-GFP-HIS3/PDS1-GFP-HIS3, PHIS3-mCherry-TUB1-URA3/PHIS3-mCherry-TUB1-URA3, ama1::kanMX/ama1::kanMX PAma1-Ama1-m8-LEU2/PAma1-Ama1-m8-LEU2, PHIS3-mCherry-TUB1-ADE2/ade2-1, can1-100/can1-100, leu2-3112/leu2-3112, his3-11,15/his3-11,15, trp1-1/trp1-1 |
All strains are derivatives of W303 (ade2-1 his3-11,15 leu2-3112 trp1-1 ura3-1 can1-100).
Time-lapse microscopy
To monitor meiosis using live-cell microscopy, cells were induced to sporulate, and after 8 h in potassium acetate they were transferred on a concanavalin A–treated (1 mg/ml) cover glass bottom chamber containing 1% potassium acetate and imaged using a Nikon Ti-E inverted microscope (Melville, NY) equipped with a 60× objective (PlanApo, numerical aperture 1.4, oil), a Lambda 10-3 optical filter changer and SmartShutter (Sutter Instrument, Novato, CA), GFP and mCherry filters (Chroma Technology, Bellows Falls, VT), and a CoolSNAP HQ2 charge-coupled device camera (Photometrics, Tucson, AZ). Z-stacks of four to eight sections were acquired in 5- to 10-min intervals for 12–15 h using a 12.5% ND filter and exposure times of 50–300 ms. Z-stacks were combined into a single maximum-intensity projection with NIS-Elements software (Nikon).
Meiotic chromosome spreads
Meiosis-induced cells were cultured in 1% potassium acetate for 10–12 h at 25°C and fixed with 4% paraformaldehyde overnight at 4°C. Cells were washed twice in phosphate-buffered saline, resuspended in 1 M sorbitol, and digested in 1–1.5 mg/ml Zymolyase (Zymo Research, Irvine, CA) buffered with 1 M sorbitol for 20–30 min at 30°C. After washing twice with 1 M sorbitol, protoplasts were resuspended in 1 M sorbitol and 50 mM EDTA, pH 8.0, and put on ice for 10 min. Protoplasts were placed on a cold, positively charged slide, 100–200 μl of cold hypotonic solution (7.5 mM KCl, 0.05% Triton X) was added, and the slides were then air dried overnight. The specimens were stained with 4′,6-diamidino-2-phenylindole (1 μg/ml) and observed under an epifluorescence microscope.
Acknowledgments
We are grateful to D. Kaback, A. Neiman, and E. Schiebel for constructs. We thank F. Solomon and M. Zolan for helpful discussions. We thank B. Calvi, S. Mukhopadhyay, F. Solomon, and members of the Lacefield lab for their critical reading of the manuscript. This work was supported in part by a Basil O'Connor Award from the March of Dimes Foundation to S.L.
Abbreviations used:
- APC/C
anaphase-promoting complex/cyclosome
- cdc20-mn
Cdc20 meiotic null
- GFP-LacI
green fluorescent protein–lactose repressor fusion protein
- LacO
lactose operator
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0378) on June 22, 2011.
REFERENCES
- Bernard P, Maure JF, Javerzat JP. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat Cell Biol. 2001;3:522–526. doi: 10.1038/35074598. [DOI] [PubMed] [Google Scholar]
- Brar GA, Amon A. Emerging roles for centromeres in meiosis I chromosome segregation. Nat Rev Genet. 2008;9:899–910. doi: 10.1038/nrg2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Rabitsch KP, Fuchs J, Gruber S, Sullivan M, Uhlmann F, Petronczki M, Toth A, Nasmyth K. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev Cell. 2003;4:727–739. doi: 10.1016/s1534-5807(03)00129-1. [DOI] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheslock PS, Kemp BJ, Boumil RM, Dawson DS. The roles of MAD1, MAD2 and MAD3 in meiotic progression and the segregation of nonexchange chromosomes. Nat Genet. 2005;37:756–760. doi: 10.1038/ng1588. [DOI] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Egeland DB, Jarnik M, Strich R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci USA. 2000;97:14548–14553. doi: 10.1073/pnas.250351297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MF, Santocanale C, Drury LS, Diffley JF. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol Cell Biol. 2000;20:242–248. doi: 10.1128/mcb.20.1.242-248.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland WD, Hughes SE, Cotitta JL, Takeo S, Xiang Y, Hawley RS. The multiple roles of mps1 in Drosophila female meiosis. PLoS Genet. 2007;3:e113. doi: 10.1371/journal.pgen.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Torres MP, Schroeder GK, Borchers CH. Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J Biol Chem. 2003;278:16698–16705. doi: 10.1074/jbc.M213109200. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Li R, Mistrot C, Chen RH, Dann P, Rudner A, Murray AW. Lesions in many different spindle components activate the spindle checkpoint in the budding yeast Saccharomyces cerevisiae. Genetics. 1999;152:509–518. doi: 10.1093/genetics/152.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer H, Gui L, Carroll J. A spindle assembly checkpoint protein functions in prophase I arrest and prometaphase progression. Science. 2009;326:991–994. doi: 10.1126/science.1175326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer HA, McDougall A, Levasseur M, Murdoch AP, Herbert M. Mad2 is required for inhibiting securin and cyclin B degradation following spindle depolymerisation in meiosis I mouse oocytes. Reproduction. 2005a;130:829–843. doi: 10.1530/rep.1.00856. [DOI] [PubMed] [Google Scholar]
- Homer HA, McDougall A, Levasseur M, Yallop K, Murdoch AP, Herbert M. Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev. 2005b;19:202–207. doi: 10.1101/gad.328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Rose AM. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat Cell Biol. 1999;1:514–521. doi: 10.1038/70309. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Miyazaki Y, Yamamoto M, Watanabe Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 2003;22:5643–5653. doi: 10.1093/emboj/cdg527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Kudo NR, et al. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell. 2006;126:135–146. doi: 10.1016/j.cell.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Lee BH, Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marston AL, Lee BH, Amon A. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev Cell. 2003;4:711–726. doi: 10.1016/s1534-5807(03)00130-8. [DOI] [PubMed] [Google Scholar]
- Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell. 2008;135:662–678. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Michel L, Benezra R, Diaz-Rodriguez E. MAD2 dependent mitotic checkpoint defects in tumorigenesis and tumor cell death: a double edged sword. Cell Cycle. 2004;3:990–992. [PubMed] [Google Scholar]
- Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Oelschlaegel T, Schwickart M, Matos J, Bogdanova A, Camasses A, Havlis J, Shevchenko A, Zachariae W. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell. 2005;120:773–788. doi: 10.1016/j.cell.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Penkner AM, Prinz S, Ferscha S, Klein F. Mnd2, an essential antagonist of the anaphase-promoting complex during meiotic prophase. Cell. 2005;120:789–801. doi: 10.1016/j.cell.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Rock JM, Amon A. The FEAR network. Curr Biol. 2009;19:R1063–1068. doi: 10.1016/j.cub.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah SM, Nasmyth K. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma. 2000;109:27–34. doi: 10.1007/s004120050409. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickart M, Havlis J, Habermann B, Bogdanova A, Camasses A, Oelschlaegel T, Shevchenko A, Zachariae W. Swm1/Apc13 is an evolutionarily conserved subunit of the anaphase-promoting complex stabilizing the association of Cdc16 and Cdc27. Mol Cell Biol. 2004;24:3562–3576. doi: 10.1128/MCB.24.8.3562-3576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Shonn MA, McCarroll R, Murray AW. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- Shonn MA, Murray AL, Murray AW. Spindle checkpoint component Mad2 contributes to biorientation of homologous chromosomes. Curr Biol. 2003;13:1979–1984. doi: 10.1016/j.cub.2003.10.057. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Stein KK, Davis ES, Hays T, Golden A. Components of the spindle assembly checkpoint regulate the anaphase-promoting complex during meiosis in Caenorhabditis elegans. Genetics. 2007;175:107–123. doi: 10.1534/genetics.106.059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. A novel destruction sequence targets the meiotic regulator Spo13 for anaphase-promoting complex-dependent degradation in anaphase I. J Biol Chem. 2007;282:19710–19715. doi: 10.1074/jbc.M701507200. [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Tsurumi C, Hoffmann S, Geley S, Graeser R, Polanski Z. The spindle assembly checkpoint is not essential for CSF arrest of mouse oocytes. J Cell Biol. 2004;167:1037–1050. doi: 10.1083/jcb.200405165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wassmann K, Niault T, Maro B. Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr Biol. 2003;13:1596–1608. doi: 10.1016/j.cub.2003.08.052. [DOI] [PubMed] [Google Scholar]
- Winey M, Morgan GP, Straight PD, Giddings TH, Jr, Mastronarde DN. Three-dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol Biol Cell. 2005;16:1178–1188. doi: 10.1091/mbc.E04-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]