Mutations in the IC138 regulatory subunit of I1 dynein alter dynein motor activity and the flagellar waveform but do not affect phototaxis.
Abstract
I1 dynein, or dynein f, is a highly conserved inner arm isoform that plays a key role in the regulation of flagellar motility. To understand how the IC138 IC/LC subcomplex modulates I1 activity, we characterized the molecular lesions and motility phenotypes of several bop5 alleles. bop5-3, bop5-4, and bop5-5 are null alleles, whereas bop5-6 is an intron mutation that reduces IC138 expression. I1 dynein assembles into the axoneme, but the IC138 IC/LC subcomplex is missing. bop5 strains, like other I1 mutants, swim forward with reduced swimming velocities and display an impaired reversal response during photoshock. Unlike mutants lacking the entire I1 dynein, however, bop5 strains exhibit normal phototaxis. bop5 defects are rescued by transformation with the wild-type IC138 gene. Analysis of flagellar waveforms reveals that loss of the IC138 subcomplex reduces shear amplitude, sliding velocities, and the speed of bend propagation in vivo, consistent with the reduction in microtubule sliding velocities observed in vitro. The results indicate that the IC138 IC/LC subcomplex is necessary to generate an efficient waveform for optimal motility, but it is not essential for phototaxis. These findings have significant implications for the mechanisms by which IC/LC complexes regulate dynein motor activity independent of effects on cargo binding or complex stability.
INTRODUCTION
The axonemal dyneins are microtubule motors necessary for production of the elegant and sophisticated movement of cilia and flagella. Axonemal dyneins consist of two main classes: two- or three-headed outer arm dyneins, which are responsible for generation of power and flagellar beat frequency (Brokaw and Kamiya, 1987), and multiple inner arm dyneins, which control the size and shape of the flagella bends (Porter and Sale, 2000; Smith and Yang, 2004; King and Kamiya, 2009). The inner dynein arms consist of more than seven different subspecies that are localized to specific regions within each 96 nm repeat (Brokaw and Kamiya, 1987; Mastronarde et al., 1992; Brokaw, 1994; Porter et al., 1996; Porter and Sale, 2000; Kamiya, 2002; Nicastro et al., 2006; Wirschell et al., 2007; Bui et al., 2008; Yagi et al. 2009). Each inner arm dynein likely plays a unique role in the formation of the flagellar bend (Kamiya, 2002; Brokaw, 1994, 2009), but the specific function of each isoform is still unclear.
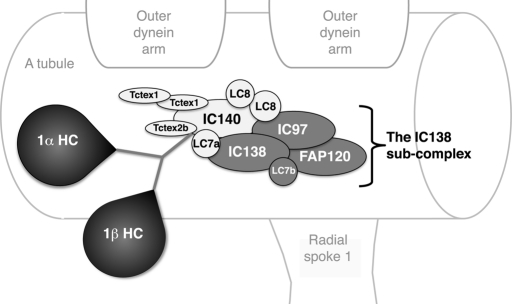
To begin to understand each inner arm dynein's role in motility, we have focused on one subspecies, known as I1 or f dynein (reviewed in Porter and Sale, 2000; Kamiya, 2002; Wirschell et al., 2007). I1 dynein is the most highly conserved and widely distributed inner arm dynein (Wickstead and Gull, 2007). The I1 complex is the only two-headed inner arm dynein, and it assembles as a trilobed structure near the base of radial spoke 1 (Figure 1). The two heads contain the motor domains of the two DHCs (dynein heavy chains), 1α and 1β, and the base consists of the DHC tail domains, intermediate chains (IC97, IC138, and IC140), light chains (LC7a, LC7b, LC8, TcTex1, and Tctex2b), and FAP120. (Goodenough and Heuser, 1985a, 1985b; Piperno et al., 1990; Smith and Sale, 1991, 1992; Porter et al., 1992; Myster et al., 1997, 1999; Harrison et al., 1998; Perrone et al., 1998, 2000; Yang and Sale, 1998; DiBella et al., 2004a, 2004b; Hendrickson et al., 2004; Bower et al., 2009; Ikeda et al., 2009; Wirschell et al., 2009). A collection of mutations in the genes encoding different I1 subunits, which results in the assembly of partial I1 dynein complexes in the axoneme, has been useful in determining the locations of and interactions between different subunits of the complex. Likewise, these mutant strains have given us insights into the function of each subunit (see Table 1).
FIGURE 1:
Diagrammatic representation of I1 dynein substructure in the axoneme. This diagram illustrates the structure of the I1 dynein and the proposed interactions between I1 subunits based on previously published work (Myster et al., 1999; Perrone et al., 2000; Porter and Sale, 2000; Ikeda et al., 2009; Wirschell et al., 2009; modified from Bower et al., 2009).
TABLE 1:
Strains used in this study.
| Strain (CC #) | Motility | Axonemal phenotype | Original study |
|---|---|---|---|
| Control strains | |||
| Wild-type mt+ (CC-125) | Fast forward | Wild type | |
| Wild-type mt– (CC-124) | Fast forward | Wild type | |
| ppr2 | Slow forward, no photoshock response | Defective CAV2 channel | Fujiu et al., 2009 |
| mia1 (CC-4265) | Slow forward, phototaxis defect | hyperphosphorylation of IC138, missing Mia1p | King and Dutcher, 1997 |
| mia2 (CC-4266) | Slow forward, phototaxis defect | Hyperphosphorylation of IC138, missing Mia2p | King and Dutcher, 1997 |
| mbo2 (CC-3664) | Backward swimming | Missing beak proteins, MBO2 | Segal et al., 1984;Tam and Lefebvre, 2002 |
| pf10 (CC-3666) | Spins in circles | Unknown | Dutcher et al., 1988 |
| uni1-2 (CC-2507) | Spins in circles | Uni-flagellate | Huang et al., 1982 |
| I1 DHC mutants | |||
| pf9-3 (CC-3913) | Slow forward | Missing I1 dynein | Myster et al., 1997 |
| pf9-3::DHC1-G41a (CC-3920) | Faster forward | Missing 1α HC motor domain | Myster et al., 1999 |
| pf28; pf9-2::DHC1 G4 (CC-3917) | Slow forward | Missing outer arms, 1α HC motor domain | Myster et al., 1999 |
| pf28; pf9-2 E8 (CC-3903) | Spins in circles | Missing outer arms, I1 dynein, full-length flagella | Porter et al., 1992 |
| ida2-6 (CC-3922) | Slow forward | Missing I1 dynein | Perrone et al., 2000 |
| ida2-7 (CC-3923) | Slow forward | Missing I1 dynein | Perrone et al., 2000 |
| ida2-7::DHC10 (CC-4401) | Faster forward | Missing 1β HC motor domain | Perrone et al., 2000 |
| pf28; ida2-7::DHC10 (CC-4079) | Slow forward | Missing outer arms, 1β motor domain | Perrone et al., 2000 |
| IC138 mutants | |||
| bop5-1 (CC-4080) | Slow forward | Truncated IC138, missing LC7b, FAP120 | Dutcher et al., 1988; Hendrickson et al., 2004; Ikeda et al., 2009 |
| bop5-2 mt+ (CC-4284) | Mixed motility | Missing IC138 subcomplex, MBO2 | Bower et al., 2009 |
| bop5-3 mt+ (CC-4389) | Slow forward | Missing IC138 subcomplex | Ikeda et al., 2009 |
| bop5-3 mt- (CC-4390) | Slow forward | Missing IC138 subcomplex | Ikeda et al., 2009 |
| bop5-4 mt-(CC-4391) | Slow forward | Missing IC138 subcomplex | Ikeda et al., 2009 |
| bop5-5 mt+ (CC-4392) | Slow forward | Missing IC138 subcomplex | Ikeda et al., 2009 |
| bop5-6 mt+ (CC-4393) | Slow forward | Reduced IC138 subcomplex | Ikeda et al., 2009 |
| bop5-1::IC138 (CC-4081) | Fast forward | Wild type | Hendrickson et al., 2004 |
| bop5-2::IC138 mt+ (CC-4285) | Mostly backward | Missing MBO2 | Bower et al., 2009 |
| bop5-3::IC138 2D1 mt+ (CC-4395) | Fast forward | Wild type | This study |
| bop5-3::IC138 2E1 mt+ | Fast forward | Wild type | This study |
| Double mutants | |||
| pf10; bop5-3 (CC-4398) | Slow forward | Missing IC138 subcomplex | This study |
| mbo2; bop5-3 (5e) (CC-4397) | Slow backward | Missing IC138 subcomplex, MBO2p | This study |
| mbo2; bop5-3 (7b) | Slow backward | Missing IC138 subcomplex, MBO2p | This study |
| uni1-2; bop5-3 (CC-4399) | Uniflagellate, spin | Missing IC138 subcomplex | This study |
| mia1; bop5-1 (CC-4394) | Slow forward | Truncated IC138, missing LC7b, FAP120, Mia1p | This study |
| mia2; bop5-3 (CC-4396) | Slow forward | Missing IC138 subcomplex, Mia2p | This study |
| mia1; bop5-4 (CC-4400) | Slow forward | Missing IC138 subcomplex, Mia1p | This study |
| uni1-2; ida3 | Uniflagellate, spin | Missing I1 dynein | Bayly et al., 2010 |
The first I1 mutant to be extensively studied was pf9-3, which affects the DHC1 gene and is null for the 1α DHC subunit (Myster et al., 1997). Loss of the 1α DHC prevents the I1 complex from being assembled in the axoneme (Myster et al., 1997). Most mutations in the 1β DHC subunit (ida2) and IC140 (ida7) also block assembly of the I1 dynein (Perrone et al., 1998, 2000; Yagi et al., 2009). These mutant strains exhibit slow, forward-swimming velocities, reduced microtubule sliding, an inability to phototax, and abnormal flagellar waveforms (Perrone et al., 2000). Notably, transformation of the mutants with gene fragments sometimes results in the partial assembly of I1 dynein in the axoneme. For example, transformation with constructs encoding the N-terminal portions of the DHCs resulted in strains (pf9::DHC1 and ida2::DHC10) that assemble all I1 subunits except the 1α or 1β motor domain and display increased swim speeds and phototaxis ability (Myster et al., 1999; Perrone et al., 2000). More recent studies have revealed the distinct functional capability of each motor domain in the I1 dynein complex (Toba et al., 2011).
Another subunit thought to play a key role in the function of I1 dynein and control of flagellar bending is IC138. For example, in vitro studies revealed that changes in phosphorylation of IC138 correlate with changes in microtubule sliding velocities (Habermacher and Sale, 1997). In addition, analyses of the mia mutants, which cannot phototax and swim slowly, showed hyperphosphorylation of IC138 in the axoneme (King and Dutcher, 1997). These results suggested that IC138 is a regulatory phosphoprotein important for control of motility. The first IC138 mutation, bop5-1, was isolated as a suppressor that could restore partial motility to the paralyzed flagellar (pf) mutant pf10 (Dutcher et al., 1988). The DIC4/BOP5 gene encodes IC138 1, and the bop5-1 mutation produces a truncated protein with reduced activity, which supports the idea that IC138 is a key regulatory subunit (Hendrickson et al., 2004). When the full-length, wild-type DIC4 gene is transformed into bop5-1, the motility and protein defects are rescued (Hendrickson et al., 2004). Because bop5-1 is not a true null, however, further analysis of IC138 function was limited.
The first IC138 null to be characterized in detail was the bop5-2 strain (Bower et al., 2009). Analysis of bop5-2 showed that IC138 is required for the assembly of four subunits at the base of the I1 dynein: IC138, LC7b, IC97, and FAP120, which is named the IC138 subcomplex (Figure 1 and Bower et al., 2009). This study also showed that regulation of microtubule sliding velocities by the central pair/radial spoke phosphorylation pathway depends on the presence of the IC138 subcomplex (Bower et al., 2009; Wirschell et al., 2009). Similar to bop5-1, microtubule sliding defects in bop5-2 and bop5-2 pf17 were restored upon transformation with the wild-type IC138 gene and reassembly of the IC138 subcomplex. Because bop5-2 has additional mutations, however, it was not possible to discern how the loss of IC138 affects flagellar motility and cell behavior (Bower et al., 2009).
In this study, we used the entire complement of bop5 mutants affecting IC138 (bop5-1 through bop5-6 [ Hendrickson et al., 2004; Bower et al., 2009; Ikeda et al., 2009]) to determine the specific roles of IC138 in regulating flagellar motility. On the basis of earlier studies of I1 dynein mutants, we postulated that assembly of IC138 would be required to control swimming speed, flagellar waveform, and phototaxis. We determined that, although the I1 dynein is necessary for normal phototaxis, the IC138 subcomplex is not. In contrast, the loss of any subunit of the I1 complex is sufficient to impair cell reversal during the photoshock response. Also, we characterized the flagellar waveform of the bop5-3 mutant and compared it to those previously described for wild type and ida3 (missing the entire I1) strains (Bayly et al., 2010) to determine how waveform might be affected by loss of the IC138 subcomplex. We observed that the bop5-3 waveform is intermediate between wild type and ida3. Mutations in I1 do not affect the maximum and minimum curvature of the waveform, but they do affect the sliding velocity and shear amplitude of the wave patterns during forward swimming. Thus, we propose that the IC138 IC/LC subcomplex is critical for control of microtubule sliding and flagellar waveform, but it is not required for phototaxis. Given the high degree of I1 dynein subunit conservation, these observations have important implications for the mechanisms by which IC/LC subcomplexes regulate ciliary and flagellar motility in many species.
RESULTS
Molecular characterization of the mutations in bop5-3, bop5-4, bop5-5, and bop5-6
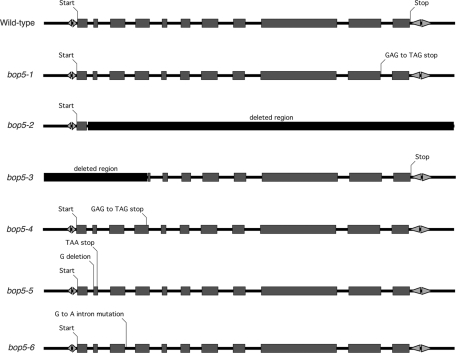
To determine the nature of the bop5 mutations that lead to decreased IC138 expression in the collection first described in Ikeda et al. (2009), the entire IC138 gene in each strain was recovered by PCR and sequenced directly (Figure 2). In bop5-3, we were unable to amplify a small region at the 5′ end of the gene, and no other mutations were found in the rest of the gene. The failure to recover a PCR product led to the conclusion that bop5-3 has a deletion that removes most of the 5′UTR and the first four exons but does not extend into the adjacent gene. The deleted region is between 1500 and 2000 base pairs (Figure 2 and unpublished data). bop5-4 contains a G-to-T mutation toward the end of the fourth exon that results in a premature stop codon (TAG) instead of a glutamic acid residue (GAG) (Figure 2). Another premature stop is created in bop5-5, from deletion of a single G that results in a frame-shift mutation and a stop codon at the end of the second exon (Figure 2). The last bop5 allele, bop5-6, contains a single G-to-A mutation in the third intron (Figure 2). This mutation does not directly alter the predicted wild-type splice site, but sequence analysis of RT-PCR products indicates that this mutation results in multiple alternatively spliced transcripts (unpublished data). Most of these transcripts contain premature stop codons and would likely not produce a functional protein. A small amount of normally spliced message is also present in bop5-6, which results in the expression of an IC138 polypeptide at lower levels than in wild type (Ikeda et al., 2009; Figure 3).
FIGURE 2:
Schematic diagram of the bop5 mutations. The molecular defects in each bop5 mutant are shown in this intron–exon diagram of the IC138 (DIC4) gene. The bop5-1 and bop5-2 mutations were characterized in Hendrickson et al. (2004) and Bower et al. (2009). bop5-3, 4, 5, 6 mutations were identified by PCR and sequencing of the IC138 gene. Bop5-3 has a deletion affecting the 5′ end of the gene; bop5-4 and bop5-5 have mutations that result in premature stops; and bop5-6 has a mutation in an intron that results in alternative splicing.
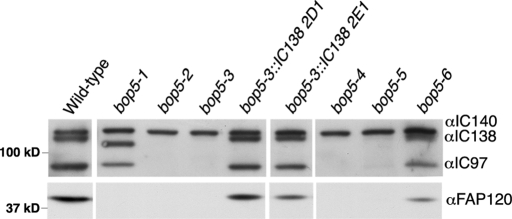
FIGURE 3:
Assembly of I1 dynein subunits in bop5 mutant axonemes. Western blots of isolated axonemes were probed with antibodies to I1 dynein subunits. IC140 serves as a loading control, as it is not affected in any bop5 mutant. IC138 is shifted in bop5-1 and missing in bop5-2 through bop5-5. FAP120 is missing in all bop5 strains. IC138, IC97, and FAP120 are reduced in bop5-6. Note the reassembly of all I1 subunits in bop5-3 strains that had been transformed with a wild-type copy of the IC138 gene (2D1 and 2E1).
The various bop5 mutations affect expression of IC138 and assembly of several other I1 subunits into the axoneme. Figure 3 shows an immunoblot that summarizes these results. As previously described (Hendrickson et al., 2004; Bower et al., 2009, Ikeda et al., 2009), bop5-1 assembles a truncated IC138 and no FAP120 into the axoneme. Assembly of IC97 is not significantly affected. Similar to bop5-2, the bop5-3, bop5-4, and bop5-5 strains lack IC138, and do not assemble IC97 or FAP120 into the axoneme. bop5-6 shows a reduction of IC138, IC97, and FAP120 expression in the axoneme. IC140 serves as a loading control, as it is an I1 component the assembly of which is not affected by bop5 mutations (Bower et al., 2009; Ikeda et al., 2009). bop5-3 transformed with a wild-type copy of the IC138 gene shows restoration of all I1 subunits missing in the bop5-3 mutant. Because bop5 mutants still assemble the two I1 dynein HCs, IC140, and other LCs (Bower et al., 2009; Ikeda et al., 2009), these mutants provide a unique opportunity to study the role of IC/LC subcomplexes in the regulation of dynein motor activity.
Forward swim speeds and microtubule sliding velocities are defective in bop5 mutants
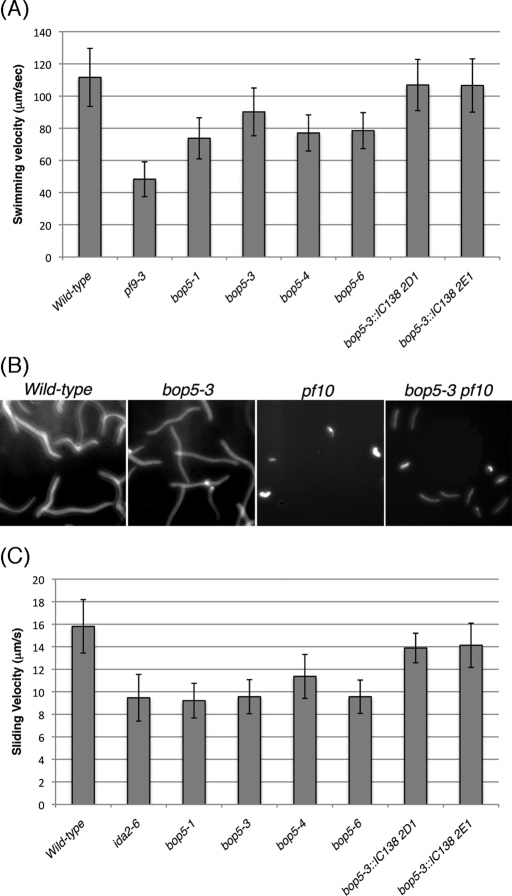
Many I1 mutants, such as pf9, ida7, and ida2, show defects in swimming (the cells swim slowly, but smoothly, in a forward direction). These defects have led to the hypothesis that slow, smooth swimming is a hallmark of strains with defects in the I1 dynein complex (Myster et al., 1997; Perrone et al., 1998; 2000). Consistent with this hypothesis, the bop5 strains, which are not complicated by second mutations, also display a slow, smooth swimming phenotype (Figure 4A). The pf9-3 mutant swims most slowly (48.3 ± 10.8 μm/s), whereas bop5-1, bop5-3, bop5-4, and bop5-6 all swim between 70 and 90 μm/s (73.8 ± 12.7, 90.2 ± 14.9, 77.1 ± 11.2, and 78.5 ± 11.2 μm/s, respectively). Neither bop5-2 nor bop5-5 was analyzed in greater detail as they both contain additional mutations that affect motility (Bower et al., 2009 and unpublished results). Transformation of bop5-3 with the wild-type IC138 gene restores forward swim speeds to near wild-type velocities (see Figure 4A).
FIGURE 4:
Forward swimming behavior and microtubule sliding in bop5 mutants. (A) Graph showing the average forward swimming velocities for wild type, pf9-3, bop5-1, bop5-3, bop5-4, bop5-6, and the bop5-3 rescued strains. At least 190–552 cells were measured for each strain. The swimming velocities of the IC138 rescued strains are significantly faster than the bop5 mutants (p < 0.005), not significantly different from one another (p = 0.832), and only slightly slower than wild type (p < 0.005). The small differences (∼5%) observed between wild type and rescued strains are typical of those seen with other transgene rescues. (B) Long exposure (1 s) images showing the forward progression of wild type, bop5-3, pf10, and bop5-3 pf10 cells. (C) Isolated axonemes from wild type and mutant strains were treated with protease and ATP, and the velocities of microtubule sliding were measuring during axoneme disintegration in vitro (n > 16 axonemes per strain). The sliding velocities of the bop5 strains are not significantly different from another or ida2-6, with the exception that bop5-4 is slightly faster than bop5-1 (p < 0.005). The sliding velocities of the IC138 rescued strains are significantly faster than all of the bop5 mutants, including bop5-3 (p < 0.005), but not significantly different from one another (p = 0.678) or wild type (p = 0.007 for 2D1 and p = 0.036 for 2E1).
We also investigated the phenotype of a bop5-3; pf10 double mutant, as the original bop5-1 mutant, which encodes a truncated IC138 polypeptide, was initially isolated as a suppressor of pf10 (Dutcher et al., 1988). Interestingly, the bop5-3; pf10 double mutant, which lacks the IC138 subcomplex, also suppresses the pf10 phenotype, as seen by tracking the forward motility of several cells in long exposure movies. Motility in the double mutants is not restored to wild-type levels, but the double mutant cells progress forward more rapidly than do pf10 cells, which circle in place (Figure 4B).
Previous studies of I1 function showed that mutations in I1 subunits cause microtubule sliding defects (DiBella et al., 2004b, Hendrickson et al., 2004; Bower et al., 2009; Wirschell et al., 2009; Toba et al., 2011). Specifically, both bop5-1 and bop5-2 have reduced microtubule sliding velocities (Hendrickson et al., 2004; Bower et al., 2009). As expected, bop5-3, bop5-4, and bop5-6 also show decreases in microtubule sliding velocity, to ∼75% of the wild-type rate and similar to that of the I1 mutant ida2-6 (Figure 4C). Microtubule sliding velocities are restored to near wild-type rates in the bop5-3 IC138 rescued strains (Figure 4C).
Analysis of bop5-3 waveforms identifies defects in shear amplitude, sliding velocities, and bend propagation
Because bop5-1 results in the assembly of a truncated IC138, and bop5-2 has other mutations that affect motility (Hendrickson et al., 2004; Bower et al., 2009), the in vivo motility phenotypes of an IC138 null mutant have never been analyzed in significant detail. To investigate the IC138 null phenotypes, we first looked at the waveforms of forward swimming cells. Because bop5 mutants swim slowly but do not show a significant reduction in beat frequency (Table 2 and data not shown), and the ida3 mutant, which lacks the entire I1 complex, has an altered waveform (Bayly et al., 2010), we hypothesized that bop5-3 would also show changes in waveform. To better illustrate the parameters assayed, two individual waveform tracings from wild type and ida3 are highlighted (Figure 5A). The principal bend is the bend with higher curvature, meaning the radius of a circle approximating the curve is smaller, and the reverse bend is the one with lower curvature. Although the waveform tracings from wild type and ida3 have similar maximum and minimum curvatures, the two waveforms look different because the regions of high curvature are of different sizes. If the curvature were imposed by two circles, as shown in Figure 5A, the ida3 flagellum would be wrapped around a shorter arc of each circle than the wild-type flagellum would be (Figure 5A). In other words, the bends (the regions of roughly constant, higher curvature) occur over smaller sections in ida3 flagella compared with wild type (Figure 5A). When the bends are shorter, the result is a narrower (“stunted”) waveform, as in ida3 compared with wild type. Reducing the length of the bends reduces shear amplitude and shortens the stroke. This idea is also illustrated in the plots of the shear angle of each waveform, in which the regions of constant slope correspond to portions of the flagella with constant curvature. In the shear angle plots of wild-type flagella, relatively long sections have either a constant negative or constant positive slope. These regions of constant slope are shorter in shear angle plots for ida3 flagella, so the curves have lower amplitude, and the distribution of shear angles is narrower in ida3 flagella (Figure 5A).
TABLE 2:
Flagellar waveform parameters.
| Wild type | bop5-3 | ida3 | |
|---|---|---|---|
| Flagellar beat frequency (Hz) | 69.45 ± 8.41 | 69.59 ± 10.35 | 64.29 ± 7.26 |
| Body revolutions per second*† | 4.52 ± 0.95 | 3.55 ± 0.91 | 2.24 ± 0.40 |
| Length (μm)*† | 12.79 ± 1.15 | 10.34 ± 2.14 | 13.87 ± 1.17 |
| Avg. root mean square shear amplitude (rad)*† | 0.68 ± 0.05 | 0.58 ± 0.05 | 0.52 ± 0.04 |
| Avg. min. (R) sliding velocity-physical (rad/s)* | –404.1 ± 50.06 | –340.2 ± 51.09 | –313.3 ± 35.73 |
| Avg. max. (P) sliding velocity-physical (rad/s)*† | 543.6 ± 54.77 | 488.7 ± 38.23 | 430.8 ± 42.04 |
| Ratio of propagation speeds (Reverse/Principal)*† | 1.03 ± 0.10 | 0.87 ± 0.16 | 0.76 ± 0.10 |
| Avg. distance between principal and reverse curvature extremes† | 0.75 ± 0.09 | 0.78 ± 0.04 | 0.72 ± 0.08 |
*Parameters for which values for bop5-3 and ida3 were significantly different from wild type.
†Parameters for which values for bop5-3 and ida3 were significantly different from each other (p < 0.05).The average minimum and maximum sliding velocities are spatial averages of the minimum (maximum) sliding velocity at each point on the flagellum, averaged over the middle 80% of its length. The minimum sliding velocity occurs in the reverse (R) bend and characterizes the effective stroke; the maximum sliding velocity occurs in the principal (P) bend and describes the recovery stroke.
FIGURE 5:
Asymmetric waveforms of I1 mutant strains during forward swimming. (A) Illustration of flagellar waveforms (left) and shear angle curves (right) plotted vs. normalized distance along the flagellum (s/L). The effects of bend curvature and bend arc length on the amplitude of flagellar waveform are illustrated on the left. Either reduced curvature (larger radius of curvature) or shorter curved regions (less wrapping) will lead to narrower waveforms. The effective stroke is dominated by the reverse bend, and the recovery stroke is dominated by the principal bend. (B) Flagellar waveforms of five representative cells of wild type, bop5-3, and ida3, showing every 1/12th cycle. (C) Spatial distribution of shear angle of the same five representative cells of wild type, bop5-3, and ida3, showing every 1/12th of a cycle. Data on wild type and ida3 are reprinted with modification from Bayly et al. (2010).
The differences between bop5-3, ida3, and wild type are evident in the traces of the overall waveform beat pattern (Figure 5B): bop5-3 waveforms are narrower than wild type, but not as severely affected as ida3. These differences can also be seen in the supplemental movies (Supplemental Movies S1–S3). To characterize the waveforms quantitatively, shear angle curves were plotted (Figure 5C), and various parameters of the curves were calculated, as described in Bayly et al., 2010. Qualitatively, wild type exhibits larger oscillations in shear angle than does bop5-3, and the bop5-3 shear angle curves have larger oscillations than those of ida3 (Figure 5C); the bop5-3 and ida3 graphs of shear angle distribution are narrower than wild type. These oscillations in shear angle are represented quantitatively by the root mean square shear amplitude. Wild-type values are higher than those of bop5-3, and bop5-3 values are higher than those of ida3 (0.68 ± 0.05, 0.58 ± 0.05, and 0.52 ± 0.04 rad, respectively, p < 0.05; Table 2).
The sliding velocities in the principal and reverse bends (average minimum, or R, sliding velocity and average maximum, or P, sliding velocity) in bop5-3 and ida3 also differ significantly from their values in wild type. These parameters describe the average minimum (or maximum) interdoublet sliding velocity at each point over the middle 80% of the flagellar length. The minimal sliding velocity at a given point occurs as the reverse bend propagates past that point; the maximum sliding velocity at a point occurs as the principal bend propagates. Both the bop5-3 and ida3 display reduced sliding velocities in the reverse bend relative to wild type, but the values for bop5-3 and ida3 are not significantly different from each other. The sliding velocity of the principal bend in bop5-3 is significantly lower than in wild type but significantly higher than in ida3 (Table 2). These reductions in the calculated microtubule sliding velocities are similar to the reductions measured in the in vitro sliding assays (Figure 4C).
The final parameter that exhibits differences between bop5-3 and wild type is the propagation speed of the reverse bend (Table 2). As this value is also dependent on flagellar length, we compensated for minor variations in length by analyzing the ratio of the reverse propagation speed to the principal propagation speed. The values of this ratio in bop5-3 and ida3 are 15–20% lower than those of wild type. These lower values result primarily from the lower propagation speed of the reverse bend, suggesting that the effective stroke is affected most strongly. Speed of bend propagation is closely related to (although not completely determined by) the speed of microtubule sliding, which is significantly lower during the effective strokes of bop5-3 and ida3 (see preceding paragraph and Discussion).
I1 dynein mutants exhibit defects in photoshock behavior
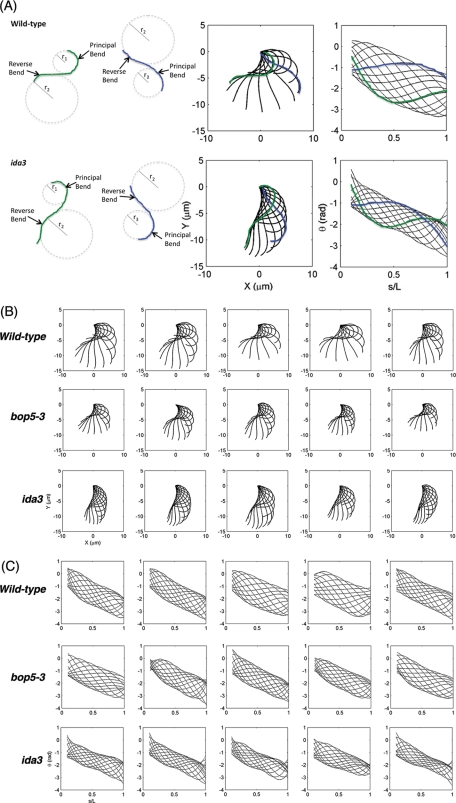
To further characterize the effect of the missing IC138 subcomplex, we compared the photoshock response in the I1 mutant strains to that of wild type and other mutants (Table 3). Wild-type cells normally swim forward using an asymmetric ciliary-type waveform, but abruptly halt upon sudden exposure to bright light and convert their waveforms to a more symmetric flagellar-type beat in response to elevated intracellular calcium. This calcium increase results in a brief period (∼0.5 s) of backward movement (photoshock), but then, as intracellular calcium levels drop, the cells switch back to the more asymmetric ciliary waveform and resume normal forward motility (Witman, 1993; see Table 3 and Figure 6). In contrast, mutant cells with defects in the flagellar membrane voltage activated calcium channel (ppr2) or mutant cells with defects in outer doublet beak structures (mbo2) do not alter their motility in response to sudden bright light (Segal et al., 1984; Matsuda et al., 1998; Fujiu et al., 2009; see Table 3). All of the I1 mutants, however, displayed an altered photoshock response, where the cells paused upon bright light exposure (suggesting that they detected the light stimulus), but the cells did not appear to swim backward any significant distance (Figure 6 and Table 3). The mia1 and mia2 mutants, which exhibit altered IC138 phosphorylation (King and Dutcher, 1997) also did not appear to reverse during the photoshock response. Interestingly, backward movement during photoshock can be observed in bop5-3, bop5-1, pf9-3, and ida2-7 strains that have been rescued by transformation with genes encoding IC138, 1α HC, or 1β HC, respectively (Table 3).
TABLE 3:
Photoshock responses in wild type and mutant strains.
| Strain | Reversal (%) | Pause/direction change (%) | No change (%) | Number of cells scored |
|---|---|---|---|---|
| Wild-type + | 92 | 8 | 0 | 50 |
| Wild-type − | 90 | 10 | 0 | 30 |
| mia1-1 | 0 | 100 | 0 | 30 |
| mia2-1 | 0 | 100 | 0 | 30 |
| mbo2 | 0 | 5 | 95 | 20 |
| ppr2 | 0 | 12 | 88 | 25 |
| pf9-3 | 0 | 79 | 21 | 53 |
| pf9-3::DHC1 | 90 | 10 | 0 | 30 |
| pf28; pf9-2::DHC1 | 0 | 45 | 55 | 78 |
| pf28; pf9-2 E8 | 0 | 52 | 48 | 66 |
| ida2-7 | 0 | 81 | 19 | 67 |
| ida2-7::DHC10 | 16 | 66 | 18 | 85 |
| pf28; ida2-7::DHC10 | 0 | 57 | 43 | 23 |
| bop5-1 | 0 | 80 | 20 | 50 |
| bop5-1::IC138 | 70 | 27 | 3 | 30 |
| bop5-3 mt+ | 0 | 100 | 0 | 54 |
| bop5-3::IC138 (2D1) | 63 | 37 | 0 | 30 |
| bop5-3::IC138 (2E1) | 67 | 33 | 0 | 30 |
| bop5-3; mbo2 (5e) | 0 | 0 | 100 | 30 |
| bop5-3; mbo2 (7b) | 0 | 0 | 100 | 20 |
| bop5-4 | 0 | 97 | 3 | 30 |
| bop5-5 | 0 | 68 | 32 | 30 |
| bop5-6 | 0 | 97 | 3 | 30 |
Actively moving cells were exposed to a brief flash of bright light and then scored for reversal, pausing, and/or change in forward direction, or no change in direction of movement. The most prevalent phenotypes are highlighted in bold type, with the exception of ida2-7::DHC10 (see text for details).
FIGURE 6:
Photoshock behavior in I1 mutant strains. The movement of wild type, ppr2, and bop5-3 cells were recorded before, during, and after exposure to a bright light stimulus. Six frames from each movie are shown here, illustrating the movement of an individual cell for ∼0.90 s. Forward motion is toward the top of the figure, and reverse motion is toward the bottom. On sudden exposure to bright light, the wild-type cell stops forward motion, moves backward briefly, and then resumes forward motion. The ppr2 cell moves forward at a constant rate throughout the assay. The bop5-3 cell stops briefly, but does not reverse before recovering and swimming forward again.
One possible interpretation is that cells with an altered I1 complex might not be capable of converting their waveforms to the more symmetric flagellar-type beat. Previous analysis of an I1 mutant has shown, however, that waveform conversions are possible (Brokaw and Kamiya, 1987). In addition, we constructed double mutants containing both bop5-3 and mbo2 mutations, and we observed vigorous backward swimming when the double mutant cells were viewed with conventional light microscopy (Table 1). These results strongly suggest that cells lacking the IC138 subcomplex can alter their waveforms, but may not generate enough power to swim backward efficiently during photoshock (see Discussion).
I1 null mutants, but not bop5 mutants, have impaired phototaxis
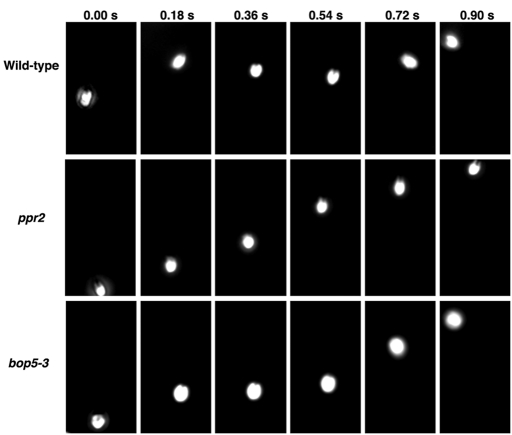
Because I1 mutants such as pf9/ida1, ida2, ida3, mia1, and mia2 exhibit phototaxis defects (King and Dutcher, 1997; Okita et al., 2005), we assayed phototactic ability in the bop5 mutants using both photoaccumulation and quantitative phototaxis assays (Table 4, Figure 7, and Supplemental Movie S4). As the phototaxis measurements showed similar results to the photoaccumulation assays (Table 4), phototaxis diagrams are illustrated for only a subset of the strains (Figure 7B). Wild-type mating-type plus (CC-125) and mating-type minus (CC-124) cells showed positive and negative taxis, as reported previously (Pazour et al., 1995; Okita et al., 2005). The phototactic sign (positive or negative) depends on the genetic background (AGG1 or agg1) of each strain (Smyth and Ebersold, 1985; Iomini et al., 2006). As expected, the I1 DHC mutants (pf9-3 and ida2-7) did not show significant phototaxis in either direction (Myster et al., 1999; Perrone et al., 2000). The DHC-rescued strains (pf9-3::DHC1 and ida2-7::DHC10), however, were able to phototax, although the strain with the full-length 1β DHC (pf9-3::DHC1) displayed more robust photoaccumulation than the strain with the full-length 1α DHC (ida2-7::DHC10) (Table 4).
TABLE 4:
Phototaxis responses in wild-type and mutant strains.
| Strain | Photoaccumulation | Phototaxis | Direction |
|---|---|---|---|
| Wild-type mt+ | Yes | Yes | Positive |
| Wild-type mt- | Yes | Yes | Negative |
| mia1 | No | No | NA |
| mia2 | No | No | NA |
| ppr2 | Yes | NT | Positive |
| pf9-3 | No | NT | NA |
| pf9-3::DHC1 G4 1a | Yes | NT | Negative |
| pf28; pf9-2::DHC1 G4 | No | NT | NA |
| pf28; pf9-2 E8 | No | NT | NA |
| ida2-7 | No | NT | NA |
| ida2-7::DHC10 | Yes, weak | NT | Positive |
| pf28; ida2-7::DHC10 | No | NT | NA |
| bop5-1 | Yes | Yes | Positive |
| bop5-1::IC138 | Yes | NT | Positive |
| bop5-3 mt+ | Yes | Yes | Negative |
| bop5-3::IC138 (2D1) | Yes | Yes | Negative |
| bop5-3::IC138 (2E1) | Yes | Yes | Negative |
| bop5-4 | Yes | Yes | Positive |
| bop5-6 | Yes | Yes | Variable |
| mia1; bop5-1 | No | NT | NA |
| mia2; bop5-3 | No | No | NA |
| mia1; bop5-4 | No | No | NA |
NT, not tested in this study; NA, not applicable.
FIGURE 7:
Phototaxis behavior in wild type and I1 mutant strains. (A) Photographs of selected strains showing their photoaccumulation behavior in a Petri dish after 10 min in front of a light box. The light box is located at the right of the page. (B) Polar histograms showing the swimming direction of individual cells ∼5 s after the phototactic light source was turned on at 0 degrees (right side).
The mia1 and mia2 mutants, which contain the entire I1 complex but exhibit hyperphosphorylation of IC138, are not able to phototax (King and Dutcher 1997; Okita et al., 2005). Surprisingly, all of the bop5 mutant strains were able to phototax, in either the positive or negative direction, nearly as well as wild type (Figure 7 and Table 4). Because previous studies have suggested that hyperphosphorylation of IC138 might be the basis of the phototaxis defects in the mia mutants, we wondered if loss of the IC138 subcomplex could suppress the mia phenotype. To test this hypothesis, we constructed double mutant strains containing bop5-1, bop5-3, or bop5-4 with either mia1 or mia2. Interestingly, the bop5; mia double mutants were not able to phototax, and their forward swimming velocities were similar to those displayed by the single mia mutants (Table 4, Figure 7, and unpublished data).
DISCUSSION
The IC138 subcomplex is necessary for normal flagellar motility
Although the IC138 subcomplex is not required for the stability, targeting, and/or docking of I1 dynein in the axoneme (Bower et al., 2009), the IC138 subcomplex is essential for control of the size and shape of the flagellar waveform during forward swimming. All four bop5 mutant strains (bop5-1, bop5-3, bop5-4, bop5-6) show similar motility defects, suggesting not only that the IC138 subcomplex is necessary for normal flagellar motility, but that either a reduction in IC138 subcomplex subunits (bop5-6) or a truncated IC138 and missing LC7b (bop5-1) can also cause abnormal motility (Hendrickson et al., 2004; Bower et al., 2009). All four mutants swim at ∼2/3 the velocity of wild-type strains (Figure 4A). The reduction in swimming velocity is caused by altered and inefficient flagellar waveforms, not reduced beat frequency, similar to other I1 mutants (Brokaw and Kamiya, 1987; Perrone et al., 2000; Myster et al., 1997, 1999; Bayly et al., 2010). The reduced swim speeds are further correlated with reduced microtubule sliding velocities in bop5 axonemes (Figure 4C). In addition, the bop5 mutations partially suppress the motility defects seen in the pf10 mutant (Figure 4B). These observations suggest that the IC138 subcomplex and the protein product encoded by the PF10 gene interact directly or indirectly to regulate motility. Further insight into this interaction will require identification and localization of the PF10 gene product.
Loss of the IC138 subcomplex changes the shape and amplitude of the flagellar waveform
Comparisons of waveform parameters between wild-type, bop5-3, and ida3 mutants have provided new insights into the role of both the I1 dynein and the IC138 subcomplex in controlling motility. First, the overall distances between principle and reverse curvature extremes are approximately equal in all three strains (Table 2 and Figure 5B). Strains lacking the IC138 subcomplex or the entire I1 dynein are still able to generate bends with curvatures similar to wild type, suggesting that the net forces bending the flagella are similar to those in wild type (i.e., that the loss of the I1 dynein or IC138 subcomplex does not dramatically affect the total dynein force summed over the length of the flagellum).
In contrast, the calculated microtubule sliding velocities in mutants lacking I1 or the IC138 subcomplex significantly differ from the sliding velocities in wild-type flagella (Table 2). Moreover, calculated sliding velocity and shear amplitude are affected more severely in ida3 than in bop5-3 flagella, suggesting that the I1 dynein contributes to control of microtubule sliding and shear, even in the absence of the IC138 subcomplex (Table 2). Consistent with the calculated reduction in waveform sliding velocity, direct measurement in isolated axonemes also reveals reductions in the microtubule sliding velocity when the IC138 subcomplex fails to assemble (Figure 4C). This reduction in sliding velocity suggests that, even if the total force summed along the length of the flagellum is similar to the corresponding cumulative force in wild type, the local force generated by dyneins at any one point on the flagellum is reduced in I1 mutants.
If dynein force generation is reduced at any one point, but similar overall, we wondered how dynein force generation is being altered in the I1 mutants. The spatial distribution of dynein activity must be different, and this difference affects the overall shape of the mutant waveforms, as seen by the reduction in shear amplitude (Table 2). It is clear that ida3 and bop5-3 flagella exhibit shorter regions of constant curvature than do wild type. Once again, the effect is most severe in ida3, and intermediate in bop5-3. This may mean that fewer dynein cross-bridges are active per unit length of the flagellum than in wild type or, alternatively, that the mechanisms that coordinate the selective activation and inactivation of different dyneins during bending are disrupted in I1 mutants. The latter may be consistent with the notion that I1 dynein plays a role in the local inhibition of sliding driven by other dyneins (Porter et al., 1992; Smith, 2002; Kotani et al., 2007; Toba et al., 2011). Moreover, multiple connections between I1 dynein and both outer and inner arm dyneins have been observed by cryoelectron tomography of isolated axonemes (Nicastro et al., 2006; Bui et al., 2008; Heuser et al., 2009).
Another parameter that differs significantly between wild type and the I1 mutants is the ratio of propagation speeds. This ratio represents the speed of the reverse bend compared with the speed of the principal bend and reveals if the dyneins in one type of bend are activated/inactivated differently than in another type of bend. The ratio of propagation speed is reduced more severely in ida3 flagella than in bop5-3 flagella, but both ratios are lower than the ratio for wild-type flagella. The reduction is caused by the reduced speed of the reverse bend (see Table 2). One possible explanation is that dyneins are not being properly inactivated during the effective stroke. The lack of inactivation may limit curve progression, which could slow down the overall speed of the bend. The reduction of both shear amplitude and reverse bend propagation speed in bop5 mutants leads to the hypothesis that dyneins are not being properly activated and inactivated temporally and spatially along the flagella, which is consistent with the model that the IC138 subcomplex is important in regulating shape of the flagellar bend (Hendrickson et al., 2004; Bower et al., 2009). The defects in waveform also explain the slow swimming phenotype seen in both bop5 and other I1 mutants.
The IC138 subcomplex is necessary for an efficient photophobic response
One important feature of Chlamydomonas behavior is the photoshock response. All I1 mutants showed a similar defect in the photoshock response: They paused, but did not reverse (Figure 6 and Table 3). This includes the mia mutants, which have altered phosphorylated IC138 levels, and bop5-1, which is only missing part of IC138, LC7b, and FAP120 (Table 3). The defects in the photoshock response are partially restored in the rescued strains. Interestingly, in mutant cells missing the 1α motor domain, the photoshock response is more effectively restored compared with mutants lacking only the 1β motor domain (Table 3). These results suggest that the 1β motor domain is more important in regulating the photoshock response. This hypothesis is in line with recent work that suggests that the 1β HC is a more effective motor that the 1α HC (Toba et al., 2011).
The basis of the “no reversal” phenotype is not well understood, but it is consistent with the phenotype seen in other dynein mutants (Kamiya and Okamoto, 1985; King and Kamiya, 2009). One hypothesis is that the cells may not be able to generate a backward, more symmetric, flagellar-type waveform without assembly of I1 dynein. bop5-3; mbo2 mutant cells, however, swim backward efficiently (Table 3), and waveform conversion has been detected in the I1 mutant, ida98, now known as ida1-1 (Brokaw and Kamiya, 1987), so this seems unlikely. It is more probable that, although bop5 cells can switch their waveforms, these waveforms do not generate enough power to result in the net backward movement of the cell body during the brief time period (∼0.5 s) of the photoshock response.
The IC138 subcomplex is not required for phototaxis
Phototaxis is a sensitive measure of the coordination and precise control of bending for each flagellum in Chlamydomonas (Witman, 1993). I1 mutant cells display defects in phototaxis when measured using simple photoaccumulation assays (King and Dutcher, 1997) or more quantitative phototaxis measurements (Okita et al., 2005). We have confirmed these observations using the I1 DHC mutants (Table 4; Myster et al., 1999; Perrone et al., 2000). Moreover, assembly of only one of the two motor head domains is sufficient to restore phototaxis ability, as the loss of either the 1α or 1β motor domain does not impair phototaxis (Table 4; Myster et al., 1999; Perrone et al., 2000). Cells missing the 1α motor domain show better phototaxis than do cells lacking the 1β motor domain (see Table 4). These results correlate with recent observations showing that the 1β dynein has higher motor activity than the 1α dynein (Toba et al., 2011). Although the I1 motor domain mutants and outer arm mutants (pf28) can each undergo phototaxis, double mutant cells missing both the outer arms and an I1 motor domain cannot (Table 4). This observation suggests a functional interaction between the motor domains in I1 dynein and the outer dynein arms.
Because nearly all of the bop5 mutant strains can phototax, we conclude that the IC138 subcomplex is not absolutely required for phototaxis (Table 4). These observations suggest that the remaining subunits of the I1 complex must play an essential role in phototaxis, even in the absence of the IC138 subcomplex. The bop5; mia double mutant strains, however, are all defective in phototaxis (Table 4 and Figure 7). This observation is significant because it has been assumed that phototaxis defects in the mia mutants were primarily the result of hyperphosphorylation of IC138, leading to inhibition of I1 dynein. Because the rest of the I1 dynein is present, but the IC138 subcomplex is missing in the double mutants, we postulate that defects in other, unknown axonemal proteins may be the real cause of the phototaxis defects observed in the mia mutants.
The C-terminal region of IC138 plays a critical role in I1 dynein function
The motility defects of the bop5-1 (truncated IC138) mutant are similar to those of the bop5 null strains. The forward swimming velocities of bop5-1, bop5-3, bop5-4, and bop5-6 are ∼80 μm/s, which suggests that their waveform parameters may be similar (Figure 4A). The bop5-1 strain also displays a similar defect in the photoshock response (Table 3). These results indicate that the C-terminal region of IC138, which binds LC7b and possibly FAP120, plays a critical role in I1 dynein function (Hendrickson et al., 2004; Ikeda et al., 2009).
Our study of the bop5 mutant alleles provides new insights into the function of the IC138 subcomplex. Loss of the IC138 subcomplex has several effects in vivo: reduced swimming velocities, altered waveforms, and inefficient reversal responses during photoshock. Microtubule sliding velocities are reduced both in vivo and during sliding disintegration in vitro. The net result is that the amplitude of the flagellar waveform is reduced: bop5 cells are less efficient at moving forward during normal motility and less effective at moving backward during photoshock. Contrary to expectation, however, loss of the IC138 subcomplex does not result in phototaxis defects. More work is needed to determine exactly how the I1 dynein contributes to control of phototaxis without the IC138 subcomplex. Nonetheless, evidence presented here demonstrates that the IC138 subcomplex plays an important role in controlling both the shape of the flagellar waveform and the response of the cell to environmental stimuli. Given the high degree of conservation of I1 dynein subunits (Wickstead and Gull, 2007), the IC138 subcomplex likely plays a critical role in regulating flagellar motility in many species. In addition, the bop5 mutants demonstrate that an IC/LC complex can modulate dynein motor activity independent of any effects on complex stability and/or cargo binding.
MATERIALS AND METHODS
Strains, culture conditions, and genetic analyses
All strains discussed in this study are shown in Table 1. Strains were grown on Tris acetate phosphate (TAP) medium (Harris, 1989). For most assays, a pea-sized amount of cells was resuspended in minimal medium lacking nitrogen (Harris, 1989) and rocked overnight under constant light to encourage maximum flagellar assembly and motility. Photobehaviors, especially phototaxis, were more robust in gametic versus vegetative cells. For flagellar waveform analysis, cells were cultured overnight in R medium (Harris, 1989).
Rescued bop5-3 strains were generated by cotransformation of bop5-3 mt+ with the pSI103 plasmid containing the aphVIII gene (Sizova et al., 2001) and the pHX plasmid containing the gene that encodes IC138 (Hendrickson et al., 2004). Transformants that grew on TAP plus 10 μg/ml paromomycin were screened for rescue of the motility and protein defects. To generate double mutants, nonparental ditype tetrads were dissected and analyzed by light microscopy, PCR, and/or Western blots to identify the appropriate double mutant combination.
PCR analyses
PCR and sequencing of the IC138 gene in each mutant were performed using primers designed in the MacVector program (MacVector, Cary, NC) to the IC138 genomic sequence (GenBank accession AY743342). The PCR products were sequenced directly (Genewiz, South Plainfield, NJ), and specific mutations or deletions were identified in bop5-3, bop5-4, bop5-5, and bop5-6.
Isolation of axonemes, SDS–PAGE, and Western blot analysis
To isolate axonemes, cells were subjected to pH shock, and then isolated flagella were demembranated by 0.1% NP-40 treatment as described previously (Witman, 1986). The resulting axonemes were resuspended in HMEEN (10 mM HEPES, pH 7.4, 5 mM MgSO4, 1 mM EGTA, 0.1 mM EDTA, 30 mM NaCl) with 0.1 μg/ml protease inhibitors (pepstatin, leupeptin, aprotinin) and 1 mM dithiothreitol (Bower et al., 2009).
Axoneme samples were run on 5–15% polyacrylamide gradient gels and transferred to Immobilon P (Millipore, Billerica, MA) as described previously (Bower et al., 2009). Antibodies were used at the following dilutions (vol/vol): IC138, 1:10,000 (Hendrickson et al., 2004); IC97, 1:10,000 (Wirschell et al., 2009); IC140, 1:10,000 (Yang and Sale, 1998); FAP120, 1:10,000 (Ikeda et al., 2009); IC69, 1:50,000 (Sigma-Aldrich, St. Louis, MO). Signal was detected with alkaline-phosphatase–conjugated secondary antibodies and the Tropix detection system (Bower et al., 2009).
Analysis of swimming speed and motility assays
To measure forward swimming velocity, gametic cells were viewed on an Axioscope (Carl Zeiss, Thornwood, NY) with 20× phase-contrast optics and a halogen light source. Short (30 s – 1 min) movies were recorded with a Rolera-MGi EM-CCD camera (Q-Imaging, Tucson, AZ), and velocities were calculated using MetaMorph software (version 7.6.5.0; Molecular Devices, Sunnyvale, CA), as described previously (Bower et al., 2009). All data are expressed as mean ± SD. F tests and t tests were used to determine the significance of differences between means (Supplemental Table S1). To assay motility in a population of cells, long (1 s) exposures were taken with the Rolera-MGi EM-CCD camera. Final images are cropped regions of the field of view that show several motile cells.
Microtubule sliding assays
Microtubule sliding velocity was determined using the methods developed by Okagaki and Kamiya (1986) with minor modifications (Howard et al., 1994; Habermacher and Sale, 1996, 1997; Hendrickson et al., 2004; Toba et al., 2011). Flagella were resuspended in buffer lacking protease inhibitors, demembranated with 0.5% NP-40, and placed in perfusion chambers. Sliding was started by adding buffer with 1 mM ATP and 3 μg/ml subtilisin A type VIII protease (Sigma-Aldrich). Recordings of sliding were made on an Axiovert 35 microscope (Carl Zeiss) with dark field optics and a Silicon intensified camera (VE-1000; Dage-MTI, Michigan City, IN). The analogue videos were digitized, and velocity was determined manually on the flat computer screen by measuring displacement of microtubules calibrated with a stage micrometer. All data are expressed as mean ± SD, and F tests and t tests were used to determine the significance of differences between means (Supplemental Table S2).
Flagellar waveform analysis of forward swimming cells
Analysis of the flagellar waveform during forward swimming was done using bop5-3; uni1 double mutant cells grown overnight in minimal medium. The acquisition and analysis of flagella waveforms were conducted as described in Bayly et al. (2010). Briefly, 600-frame videos (350 frames per second, 2.88 ms shutter speed) were taken on a Zeiss Universal Microscope (Carl Zeiss) at 100× phase contrast with a Dragonfly Express IEEE-1394b Digital Camera System and FlyCapture software (Point Grey Research, Scottsdale, AZ) and saved as avi files. Videos were analyzed using custom Matlab programs (The Mathworks, Natick, MA), which include correction of rigid-body motion, identification of flagellar point clouds, sorting of the point clouds, fitting of curves to the data, and characterization and statistical analysis of the final waveforms (Bayly et al., 2010). To determine if a waveform parameter is dependent on flagellar length, SAS 9.2 software (SAS Institute, Cary, NC) was used to create a general linear model of the data with the strain as a class variable (see also Supplemental Table S3).
High-speed movies of biflagellate cells were taken on an Axioscope (Carl Zeiss) with 63× DIC optics and a halogen light source. Movies were recorded with a 1024 PCI high-speed camera (Motion Engineering Company, Indianapolis, IN) at 500 frames per second. To facilitate viewing the movies, ImageJ (National Institutes of Heatlh, Bethesda, MD) was used to compile every fourth frame into a QuickTime movie with a playback speed of 15–30 frames per second, depending on the strain. Movies of wild type, bop5-3, and pf9-3 are Supplemental Movies S1, S2, and S3.
Photoaccumulation, phototaxis, and photoshock assays
For photoaccumulation assays, gametic cells at a concentration of 2.5 × 106 cells/ml were transferred to 35 mm Petri dishes, and dark adapted for 10 min. Cells were then exposed to a fluorescent light source located 30 cm away (2 W/m2) for 10 min. Cell densities on both sides of the dish were determined at various time points and recorded by digital photography to determine relative photoaccumulation ability (Supplemental Movie S4). Each strain was independently assayed at least three times or tested separately for phototaxis to confirm the results. For phototaxis, cells were dark adapted in red light before illumination by unidirectional blue-green light. The cell behavior was recorded, and the angle between the light direction and the swimming direction of each cell was measured and pooled into 30° bins, as described previously (Okita et al., 2005). Where applicable, the direction of the movement relative to the light source is noted: positive if toward the light source, and negative if away from the light source.
To assay for the photoshock response, we noted whether forward swimming cells exhibited any change in behavior upon brief exposure to bright light. Cell movement was recorded using phase- contrast microscopy at 20× and a red filter. The cells were then briefly exposed to bright light using a green fluorescent protein excitation filter on the epi-illuminescence port (λ = 450–500 nm). The position of each cell (n > 30) before and after light exposure was traced and scored as no change, paused, changed in forward direction only, or reversed.
Supplementary Material
Acknowledgments
Thanks to Claire Parker and Jaimee Reck for assistance with the analysis of swimming velocities, photo-accumulation, and photoshock; Shota Mochiji (University of Tokyo) for assistance with the phototaxis assays; Kimberly VanderWaal (University of California at Davis) for assistance with statistical analysis of quantitative waveform data; and members of the Dutcher lab (Washington University) for providing the equipment, instruction, and guidance for the recording and analysis of flagellar waveforms. Thanks also to Stuart Goldstein (University of Minnesota) for helpful discussions on waveform parameters. K. VanderWaal was supported by a predoctoral fellowship from the American Heart Association (Fellowship 0715799Z) and a Doctoral Dissertation Fellowship from the University of Minnesota. R. Yamamoto is supported by a long-term fellowship from the TOYOBO Biotechnology Foundation. K. Wakabayashi is supported by a Grant-in-Aid for Young Scientists (B) from Japan Society for Promotion of Sciences (22770189). R. Kamiya is supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (20051007). P.V. Bayly is supported by the Children's Discovery Institute. S.K. Dutcher is supported by NIH (GM032843); W.S. Sale by NIH (GM051173), and M.E. Porter by NIH (GM056677) and a Grant-in-Aid from the University of Minnesota.
Abbreviations used:
- BOP
bypass of paralysis
- DHC
dynein heavy chain
- FAP
flagellar associated protein
- HC
heavy chain
- IC
intermediate chain
- IDA
inner dynein arm
- LC
light chain
- MBO
move backward only
- MIA
modifier of inner arms
- PF
paralyzed flagellar
- TAP
Tris acetate phosphate
Footnotes
Using the new nomenclature for dynein genes in Chlamydomonas (Hom et al., submitted), the IC138 subunit of the I1 inner arm dynein is encoded by the DIC4 gene (previously known as the BOP5 gene), and mutations in DIC4 are designated bop5 alleles.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0270) on June 22, 2011.
REFERENCES
- Bayly P, Lewis BL, Kemp PL, Pless RB, Dutcher SK. Efficient spatiotemporal analysis of the flagellar waveform of Chlamydomonas reinhardtii. Cytoskeleton. 2010;67:56–69. doi: 10.1002/cm.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, VanderWaal KE, O'Toole E, Fox L, Perrone CA, Mueller J, Wirschell M, Kamiya R, Sale WS, Porter ME. IC138 defines a subdomain at the base of the I1 dynein that regulates microtubule sliding and flagellar motility. Mol Biol Cell. 2009;20:3055–3063. doi: 10.1091/mbc.E09-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw C. Simulation of the cyclic dynein-driven sliding, splitting and reassociation in an outer doublet pair. Biophys J. 2009;97:2939–2947. doi: 10.1016/j.bpj.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw C, Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ. Control of flagellar bending: a new agenda based on dynein diversity. Cell Motil Cytoskeleton. 1994;23:199–204. doi: 10.1002/cm.970280303. [DOI] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J Cell Biol. 2008;183:923–932. doi: 10.1083/jcb.200808050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella L, Sakato M, Patel-King R, Pazour G, King S. The LC7 light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol Biol Cell. 2004a;15:4633–4646. doi: 10.1091/mbc.E04-06-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella L, Smith E, Patel-King R, Wakabayashi K, King S. A novel Tctex2-related light chain is required for stability of inner dynein arm I1 and motor function in the Chlamydomonas flagellum. J Biol Chem. 2004b;279:21666–21676. doi: 10.1074/jbc.M313540200. [DOI] [PubMed] [Google Scholar]
- Dutcher SK, Gibbons W, Inwood WB. A genetic analysis of suppressors of the pf10 mutation in Chlamydomonas reinhardtii. Genetics. 1988;120:965–976. doi: 10.1093/genetics/120.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiu K, Nakayama Y, Yanagisawa A, Sokabe M, Yoshimura K. Chlamydomonas CAV2 encodes a voltage-dependent calcium channel required for the flagellar waveform conversion. Curr Biol. 2009;19:133–139. doi: 10.1016/j.cub.2008.11.068. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Outer and inner dynein arms of cilia and flagella. Cell. 1985a;41:341–342. doi: 10.1016/s0092-8674(85)80003-9. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J Cell Biol. 1985b;100:2008–2016. doi: 10.1083/jcb.100.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermacher G, Sale W. Regulation of flagellar dynein by an axonemal type-1 phosphatase in Chlamydomonas. J Cell Biol. 1996;109:1899–1907. doi: 10.1242/jcs.109.7.1899. [DOI] [PubMed] [Google Scholar]
- Habermacher G, Sale W. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J Cell Biol. 1997;136:167–176. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: CA: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Harrison A, Olds-Clarke P, King S. Identification of the t complex-encoded cytoplasmic dynein light chain tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J Cell Biol. 1998;140:1137–1147. doi: 10.1083/jcb.140.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson TW, Perrone CA, Griffin P, Wuichet K, Mueller J, Yang P, Porter ME, Sale WS. IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol Biol Cell. 2004;15:5431–5442. doi: 10.1091/mbc.E04-08-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol. 2009;187:921–933. doi: 10.1083/jcb.200908067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DR, Habermacher G, Glass DB, Smith EF, Sale WS. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J Cell Biol. 1994;127:1683–1692. doi: 10.1083/jcb.127.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Dutcher S, Luck D. Uniflagellar mutants of Chlamydomonas: evidence for the role of basal bodies in the transmission of positional information. Cell. 1982;29:745–753. doi: 10.1016/0092-8674(82)90436-6. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Yamamoto R, Wirschell M, Yagi T, Bower R, Porter ME, Sale WS, Kamiya R. A novel ankyrin-repeat protein interacts with the regulatory proteins of inner arm dynein f (I1) of Chlamydomonas reinhardtii. Cell Motil Cytoskeleton. 2009;66:448–456. doi: 10.1002/cm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C, Li L, Mo W, Dutcher SK, Piperno G. Two flagellar genes, AGG2 and AGG3, mediate orientation to light in Chlamydomonas. Curr Biol. 2006;16:1147–1153. doi: 10.1016/j.cub.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 2002;219:115–155. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Okamoto M. A mutant of Chlamydomonas reinhardtii that lacks the flagellar outer arm but can swim. J Cell Sci. 1985;74:181–191. doi: 10.1242/jcs.74.1.181. [DOI] [PubMed] [Google Scholar]
- King S, Dutcher SK. Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J Cell Biol. 1997;136:177–191. doi: 10.1083/jcb.136.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Kamiya R. The Chlamydomonas Sourcebook. 2nd Edition. GB Witman, San Diego: CA: Academic Press; 2009. Axonemal dyneins: assembly, structure, and force generation; pp. 131–208. Volumn 3. [Google Scholar]
- Kotani N, Sakakibara H, Burgess SA, Kojima H, Oiwa K. Mechanical properties of inner-arm dynein-f (dynein I1) studied with in vitro motility assays. Biophys J. 2007;93:886–894. doi: 10.1529/biophysj.106.101964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D, O'Toole E, McDonald K, McIntosh J, Porter M. Arrangement of inner dyenin arms in wild-type and mutant flagella of Chlamydomonas. J Cell Biol. 1992;118:1145–1162. doi: 10.1083/jcb.118.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A, Yoshimura K, Sineshchekov OA, Hirono M, Kamiya R. Isolation and characterization of Chlamydomonas mutants that display phototaxis but not photophobic response. Cell Motil Cytoskeleton. 1998;41:353–362. doi: 10.1002/(SICI)1097-0169(1998)41:4<353::AID-CM7>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Myster S, Knott JA, O'Toole E, Porter ME. The Chlamydomonas Dhc1 gene encodes a dynein heavy chain subunit required for assembly of the I1 inner arm complex. Mol Biol Cell. 1997;8:607–620. doi: 10.1091/mbc.8.4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster SH, Knott JA, Wysocki KM, O'Toole E, Porter ME. Domains in the 1-alpha dynein heavy chain required for inner arm assembly and flagellar motility in Chlamydomonas. J Cell Biol. 1999;146:801–818. doi: 10.1083/jcb.146.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- Okagaki T, Kamiya R. Microtubule sliding in mutant Chlamydomonas axonemes devoid of outer or inner dynein arms. J Cell Biol. 1986;103:1895–1902. doi: 10.1083/jcb.103.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita N, Isogai N, Hirono M, Kamiya R, Yoshimura K. Phototactic activity in Chlamydomonas “non-phototactic” mutants deficient in Ca2+-dependent control of flagellar dominance or in inner-arm dynein. J Cell Sci. 2005;118:529–537. doi: 10.1242/jcs.01633. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Sineshchekov OA, Witman GB. Mutational analysis of the phototransduction pathway in Chlamydomonas reinhardtii. J Cell Biol. 1995;131:427–440. doi: 10.1083/jcb.131.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Myster S, Bower R, O'Toole E, Porter ME. Insights into the structural organization of the I1 inner arm dynein from a domain analysis of the 1β dynein heavy chain. Mol Biol Cell. 2000;11:2297–2313. doi: 10.1091/mbc.11.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Yang P, O'Toole E, Sale WS, Porter ME. The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol Biol Cell. 1998;9:3351–3365. doi: 10.1091/mbc.9.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Ramanis Z, Smith EF, Sale WS. Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J Cell Biol. 1990;110:379–389. doi: 10.1083/jcb.110.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Knott JA, Myster SH, Farlow SJ. The dynein gene family in Chlamydomonas reinhardtii. Genetics. 1996;144:569–585. doi: 10.1093/genetics/144.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Power J, Dutcher SK. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol. 1992;188:1163–1176. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Sale WS. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol. 2000;151:37–42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal RA, Huang B, Ramanis Z, Luck DJ. Mutant strains of Chlamydomonas reinhardtii that move backwards only. J Cell Biol. 1984;98:2026–2034. doi: 10.1083/jcb.98.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P. A Streptomyces rimosus aphVIII gene coding for a new type of phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 2001;277:221–229. doi: 10.1016/s0378-1119(01)00616-3. [DOI] [PubMed] [Google Scholar]
- Smith E, Sale W. Microtubule binding and translocation by inner dynein arm subtype I1. Cell Motil Cytoskeleton. 1991;18:258–268. doi: 10.1002/cm.970180403. [DOI] [PubMed] [Google Scholar]
- Smith E, Sale W. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 1992;257:1557–1559. doi: 10.1126/science.1387971. [DOI] [PubMed] [Google Scholar]
- Smith E, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF. Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil Cytoskeleton. 2002;52:33–42. doi: 10.1002/cm.10031. [DOI] [PubMed] [Google Scholar]
- Smyth RD, Ebersold WT. Genetic investigation of a negatively phototactic strain of Chlamydomonas reinhardtii. Genet Res. 1985;46:133–148. doi: 10.1017/s001667230002262x. [DOI] [PubMed] [Google Scholar]
- Tam L, Lefebvre PA. The Chlamydomonas MBO2 locus encodes a conserved coiled-coil protein important for flagellar waveform conversion. Cell Motil Cytoskeleton. 2002;51:197–212. doi: 10.1002/cm.10023. [DOI] [PubMed] [Google Scholar]
- Toba S, Fox LA, Sakakibara H, Porter ME, Oiwa K, Sale WS. Distinct roles of the 1α and 1β heavy chains of the inner arm dynein I1 of Chlamydomonas flagella. Mol Biol Cell. 2011;22:342–353. doi: 10.1091/mbc.E10-10-0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–1721. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Hendrickson T, Sale WS. Keeping and eye on I1: I1 dynein as a model for flagellar dynein assembly and regulation. Cell Motil Cytoskeleton. 2007;64:569–579. doi: 10.1002/cm.20211. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Yang C, Yang P, Fox L, Yanigasawa H, Kamiya R, Witman GB, Porter ME, Sale WS. IC97 is a novel intermediate chain of I1 dynein that interacts with tubulin and regulates interdoublet sliding. Mol Biol Cell. 2009;20:3044–3054. doi: 10.1091/mbc.E09-04-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Witman GB. Chlamydomonas phototaxis. Trends Cell Biol. 1993;3:403–408. doi: 10.1016/0962-8924(93)90091-e. [DOI] [PubMed] [Google Scholar]
- Yagi T, Uematsu K, Liu Z, Kamiya R. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J Cell Sci. 2009;122:1306–1314. doi: 10.1242/jcs.045096. [DOI] [PubMed] [Google Scholar]
- Yang P, Sale WS. The Mw 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol Biol Cell. 1998;9:3335–3349. doi: 10.1091/mbc.9.12.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.