Two functions of the p24 complex are described: one connects GPI-anchored proteins to COPII proteins at ER exit sites to facilitate their incorporation into ER-derived vesicles, and the other serves in quality control of GPI-anchored proteins to retrieve unremodeled GPI-anchored proteins from the Golgi back to the ER.
Abstract
Glycosylphosphatidylinositol (GPI)-anchored proteins are secretory proteins that are attached to the cell surface of eukaryotic cells by a glycolipid moiety. Once GPI anchoring has occurred in the lumen of the endoplasmic reticulum (ER), the structure of the lipid part on the GPI anchor undergoes a remodeling process prior to ER exit. In this study, we provide evidence suggesting that the yeast p24 complex, through binding specifically to GPI-anchored proteins in an anchor-dependent manner, plays a dual role in their selective trafficking. First, the p24 complex promotes efficient ER exit of remodeled GPI-anchored proteins after concentration by connecting them with the COPII coat and thus facilitates their incorporation into vesicles. Second, it retrieves escaped, unremodeled GPI-anchored proteins from the Golgi to the ER in COPI vesicles. Therefore the p24 complex, by sensing the status of the GPI anchor, regulates GPI-anchored protein intracellular transport and coordinates this with correct anchor remodeling.
INTRODUCTION
In eukaryotic cells, the secretory pathway is initiated by the selective incorporation of correctly folded and assembled secretory proteins into vesicles that mediate transport from the endoplasmic reticulum (ER) to the Golgi apparatus. ER budding is driven by the assembly of cytosolic coat complex COPII at specific domains of the ER membrane called ER exit sites (ERES; Lee et al., 2004). For efficient ER exit, most secretory proteins are believed to be actively captured and concentrated at ERES through interactions with the cytosolic COPII coat prior to budding (Lee et al., 2004; Sato and Nakano, 2007). According to this cargo capture model, those secretory molecules that cannot interact directly with the COPII coat subunits, like soluble secretory proteins, might be selectively incorporated into ERES and COPII vesicles by interacting with a cargo receptor, which would couple cargo selection with vesicle coat assembly.
Glycosylphosphatidylinositol (GPI)-anchored proteins constitute a special category of secretory cargo, which contains a soluble protein portion attached by a glycolipid anchor to the external leaflet of the plasma membrane (Orlean and Menon, 2007). Once glycolipid anchoring has occurred in the ER lumen, GPI-anchored proteins are delivered to the Golgi apparatus via COPII vesicles. Like soluble cargoes, GPI-anchored proteins are exclusively luminal and cannot interact directly with the cytosolic COPII coat. Therefore a transmembrane cargo receptor/adaptor may be required to recognize and concentrate the GPI-anchored proteins at ERES and COPII vesicles. This possible receptor/adaptor requirement might be fulfilled by the members of the conserved p24 family, which are abundant type I transmembrane proteins assembled into heteromeric complexes that cycle between the ER and Golgi compartments (Sohn et al., 1996; Rojo et al., 1997; Fullekrug et al., 1999; Belden and Barlowe, 2001). In yeast, at least four members of the p24 family (Emp24p, Erv25p, Erp1p, and Erp2p) function in the p24 complex (Marzioch et al., 1999). Previous studies have shown a direct role of the yeast p24 complex in the ER exit of GPI-anchored proteins. First, the p24 complex is required for efficient transport of the GPI-anchored protein Gas1p to the Golgi, and it is necessary for proper targeting of other GPI-anchored proteins to the cell surface (Schimmoller et al., 1995; Belden and Barlowe, 1996; Castillon et al., 2009). Second, the p24 complex is directly and selectively required for in vitro ER budding of Gas1p (Muniz et al., 2000). Third, the p24 proteins can be cross-linked to Gas1p in purified ER-derived vesicles (Muniz et al., 2000). Finally, the tail of the p24 proteins efficiently recruits COPII coat subunits (Belden and Barlowe, 2001). On the basis of these observations, we initially proposed that the yeast p24 complex can directly promote the efficient ER exit of GPI-anchored proteins by acting as a cargo receptor. Moreover, this potential receptor function might be conserved in mammalian cells, since ER-to-Golgi transport of GPI-anchored proteins is specifically delayed after silencing of the mammalian homologues of several yeast p24 genes (Takida et al., 2008; Bonnon et al., 2010).
Although the cargo receptor model offers a mechanistic explanation for the efficient ER exit of yeast GPI-anchored proteins, their special transport requirements may suggest an alternative and more complex ER export mechanism than previously anticipated. Indeed, GPI-anchored proteins are sorted from other secretory proteins during their transport to the plasma membrane (Mayor and Riezman, 2004). In yeast, this sorting occurs initially upon exit from the ER. GPI-anchored proteins are selectively concentrated at specific ERES, from where they are incorporated into distinct transport vesicles (Muniz et al., 2001; Castillon et al., 2009). Moreover, we have shown that, in contrast to other secretory proteins, GPI-anchored proteins do not employ the COPII machinery for concentration at ERES. Instead, they use a concentrative mechanism that depends on the remodeling of their GPI anchors (Castillon et al., 2009). This process consists of inositol deacylation followed by the replacement of the primary lipid moiety by another lipid containing a highly saturated acyl chain, usually ceramide. In yeast, the entire remodeling process occurs at the ER after the anchor attachment to the protein and is proposed to lead to the association of GPI-anchored proteins with ceramide-enriched membrane domains, based on the fact that they can be biochemically isolated in a detergent-resistant membrane (DRM) fraction (Pittet and Conzelmann, 2007; Fujita and Jigami, 2008). In mammalian cells remodeling confers this property in the Golgi (Maeda et al., 2007). This COPII assembly–independent mechanism for concentration of GPI-anchored proteins at ERES may imply that the COPII function in the ER exit of GPI-anchored proteins might be restricted just to the final vesicle formation event after cargo concentration. Consequently, the p24 complex may not act in yeast, as initially expected, like a conventional cargo receptor during the ER export of GPI-anchored proteins.
Therefore the exact mode of action of the yeast p24 complex during the selective ER exit of GPI-anchored proteins is unclear. To clarify this issue, we tested several predictions of the cargo receptor hypothesis directly, including the substrate-binding capacity of the p24 complex and its influence on cargo concentration and sorting at ERES. Our results indicate that the yeast p24 complex does not behave as a typical cargo receptor but functions as an adaptor that facilitates ER exit by connecting the COPII coat with GPI-anchored proteins after their remodeling and concentration. Furthermore, we found a new function of the p24 complex in the quality control of GPI-anchored proteins. We show that the p24 complex effects the ER retention of incompletely remodeled GPI-anchored proteins by a mechanism that depends on recycling them from the Golgi back to the ER. Therefore our results support a model in which the p24 complex regulates the intracellular transport of GPI-anchored proteins by monitoring GPI anchor remodeling.
RESULTS
The p24 complex interacts specifically with GPI-anchored proteins through their GPI anchor within the ER
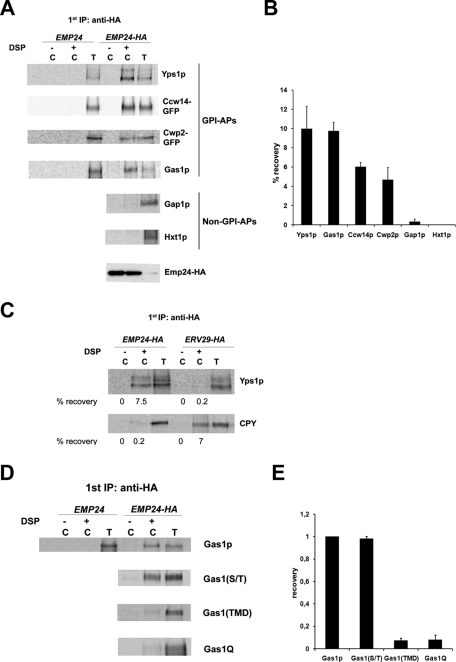
We showed previously that the yeast p24 complex binds to the GPI-anchored protein Gas1p and is directly required for its efficient ER exit (Muniz et al., 2000). These findings led to the idea that the p24 complex might constitute a conventional cargo receptor that collects GPI-anchored proteins into COPII vesicles to accelerate their transport to the Golgi. This hypothesis is reinforced by the fact that p24 complex requirement for ER exit is specific to all GPI-anchored proteins tested so far (Supplemental Figure S1; Castillon et al., 2009) but does not affect transport of carboxypeptidase Y or pro–alpha factor. To further examine the potential role of the p24 complex as a cargo receptor for GPI-anchored proteins, we investigated whether other GPI-anchored proteins interact with p24 proteins within the ER. Pulse-radiolabeled yeast cells from EMP24-HA-tagged and untagged strains were converted to perforated spheroplasts and exposed to the cleavable cross-linker DSP. After solubilization, Emp24-hemagglutinin (HA) was immunoprecipitated using anti-HA antibodies. The precipitates were denatured and subjected to a second immunoprecipitation using antibodies against different GPI-anchored proteins. The cross-linker was cleaved, and the samples were analyzed by SDS–PAGE. Emp24-HA was cross-linked to the ER form of all the GPI-anchored proteins tested with good efficiency (Figure 1, A and B). No GPI-anchored proteins were recovered if cross-linker was omitted. Moreover, GPI-anchored proteins could not be detected when the cross-linking was performed on untagged membranes, proving that the GPI-anchored proteins were recovered as part of a complex containing Emp24p. This association is specific because two unrelated transmembrane proteins, the general amino acid permease Gap1p and the glucose transporter Hxt1p, cannot be cross-linked to Emp24p (Figure 1, A and B). As an additional specificity control, the cargo-binding capacity of the p24 complex was compared with that of Erv29p, a well-characterized ER cargo receptor (Belden and Barlowe, 2001). Whereas Erv29p can bind efficiently one of its known soluble cargoes, the vacuolar carbopeptidase Y (CPY), the GPI-anchored protein Yps1p fails to be cross-linked to Erv29p (Figure 1C), and CPY is not cross-linked to Emp24p. These cross-linking experiments show that Emp24p binds specifically to GPI-anchored proteins within the ER.
Figure 1:
The p24 complex associates specifically with the GPI-anchored proteins through the GPI anchor. (A) Emp24p can be cross-linked specifically to GPI-anchored proteins in the ER. Pulse-radiolabeled yeast cells from EMP24-HA-tagged and untagged strains were converted to perforated spheroplasts and incubated with (+) and without (–) DSP. The samples were denatured and immunoprecipitated with anti-HA antibody and reprecipitated with antibody against the designated protein (labeled C). Five percent of the DSP-exposed spheroplasts were immunoprecipitated with antibodies against GPI-anchored proteins or non–GPI-anchored proteins to use as a standard (labeled T) for recovery. Samples were incubated with 5% β-mercaptoethanol, analyzed by SDS–PAGE, and visualized using a Phosphorimager. Emp24-HA was detected by immunoblot. (B) Quantification of several experiments described in A. The graph plots the average percentage of the recovery of different secretory proteins. GPI-APs, GPI-anchored proteins. (C) Erv29p can be cross-linked to CPY but not to GPI-anchored proteins. Spheroplasts obtained from Emp24-HA and Erv29-HA strains were incubated with (+) or without (–) DSP, denatured, immunoprecipitated with anti-HA antibody, and then reprecipitated with antibody against the Yps1p or CPY and processed as earlier. The percentage recovery of cross-linked Yps1p and CPY is shown. (D) Emp24p can be cross-linked to GPI-anchored proteins through the GPI anchor. Spheroplasts obtained from Emp24-HA cells expressing different constructs of the Gas1p were treated with (+) or without (–) DSP, denatured, immunoprecipitated with anti-HA antibody, and then reprecipitated with antibody against the Gas1p and processed as earlier. (E) Quantification of several experiments described in D. The graph plots the average percentage of the recovery of different Gas1p mutant constructs normalized relative to the recovery of wild-type Gas1p.
All GPI-anchored proteins seem to share two conserved motifs that could potentially function as a recognition motif for the p24 complex: the GPI anchor and a serine/threonine (S/T) region. The GPI-anchor is covalently attached to a newly generated COOH-terminal residue (ω site) after cleavage of the GPI-attachment signal in the ER. The S/T region upstream of the ω site (Caro et al., 1997) is a site for O-mannosylation, which begins in the ER in yeast (Gentzsch and Tanner, 1997). To identify the region of GPI-anchored protein binding to Emp24p, we used three different mutant constructs of the Gas1p (Nuoffer et al., 1993; Watanabe et al., 2008). In one construct Gas1p is devoid of its GPI anchor and instead contains an artificial transmembrane domain (TMD) consisting of 19 leucine residues and two flanking arginine and serine residues (Gas1TMD). In a second construct Gas1p is deleted of its S/T region [Gas1(-S/T)]. In the third construct Gas1p is mutated at the ω site to prevent GPI anchoring (Gas1Q). We observed that only wild-type Gas1p and Gas1(-S/T) can be cross-linked to Emp24p (Figure 1, D and E). Therefore these results show that Emp24p recognizes and binds to the GPI-anchored proteins via the GPI anchor prior to and/or during ER exit.
The p24 complex is not required to sort and concentrate GPI-anchored proteins into ERES
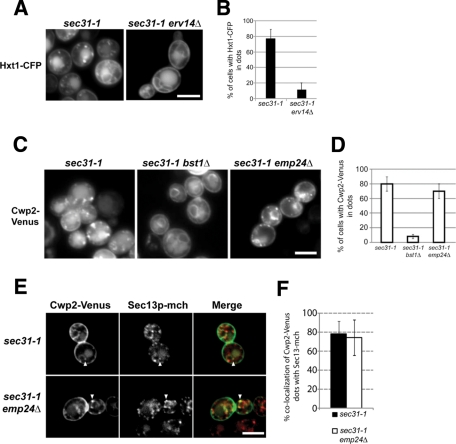
According to the cargo receptor model, our results would imply that the p24 complex interacts with the GPI moiety to concentrate GPI-anchored proteins in ERES and COPII vesicles. Therefore we assessed the requirement of the p24 complex for accumulation of GPI-anchored proteins into ERES. Cargo concentration at ERES can be visualized under the fluorescence microscope by blocking the ER exit with the temperature-sensitive sec31-1 (COPII) allele (Castillon et al., 2009). First, we assessed the reliability of this system by investigating Erv14p, which has been proposed to act as a cargo receptor of several transmembrane secretory proteins, including the glucose transporter Hxt1p (Castillon et al., 2009; Dancourt and Barlowe, 2010). If selective Hxt1p incorporation into ERES depends on Erv14p, an erv14Δ sec31-1 double mutant strain should not accumulate Hxt1p into ERES upon shift to the restrictive temperature (37°C). As shown in Figure 2, A and B, Hxt1p showed punctuate staining in sec31-1 mutant cells at 37°C. We previously showed that these dots correspond to ERES in yeast because they colocalize with the ERES marker and COPII coat component Sec13p (Castillon et al., 2009). However, in an erv14Δ sec31-1 mutant strain, Hxt1p fails to accumulate into ERES (Figure 2, A and B). To show that this observation is not the result of an ERES formation defect, we confirmed that the ERES marker Sec13p localized properly in the erv14Δ sec31-1 strain (Supplemental Figure S2). These results show that Erv14p acts as expected for a classical cargo receptor by promoting cargo concentration in ERES and confirms the reliability of the method.
Figure 2:
Emp24p is not required for GPI-anchored cargo sorting and concentration into ERES. (A) Fluorescence micrographs of live sec31-1 and sec31-1 erv14Δ cells expressing Hxt1-CFP at 37ºC. Raw images. (B) Quantification of several micrographs described in A. The graph plots the average percentage of the cells, for which Hxt1-CFP is found in dot-like structures. n, number of cells plotted; 74 ≤ n ≤ 89. (C) Fluorescence micrographs of live sec31-1, sec31-1 bst1Δ, and sec31-1 emp24Δ cells expressing Cwp2-Venus at 37ºC. Raw images. (D) Quantification of several micrographs described in C. The graph plots the average percentage of the cells, for which Cwp2-Venus is found in dot-like structures. n, number of cells plotted; 74 ≤ n ≤ 89. (E) Fluorescence micrographs of live sec31-1 and sec31-1 emp24Δ cells expressing Cwp2-Venus (green) and Sec13-mCh (red) at 37°C. White arrowheads, colocalizing dots. Deconvoluted images by 10 iterations. (F) Quantification of several micrographs described in E. The graph displays the means of the percentage of colocalization per cell of Cwp2-Venus dots with Sec13-mCh dots in sec31-1 (black bars, n = 36) and in sec31-1 emp24Δ (white bars, n = 55). Scale bar, A, C, E, 5 μm.
Next we examined whether incorporation of GPI-anchored proteins into ERES depends on the p24 complex. For this purpose, we used the deletion of EMP24 that destabilizes the other proteins of the complex, leading to a complete loss of p24 complex function (Marzioch et al., 1999). In sec31-1 mutant cells at 37°C, the GPI-anchored protein Cwp2p showed punctuate staining, which corresponds to accumulation of Cwp2p molecules into ERES as previously described (Castillon et al., 2009). Strikingly, this pattern was also reproduced in emp24Δ sec31-1 mutant cells at 37°C (Figure 2, C and D). We confirmed that these dot-like structures containing Cwp2p are ERES by colocalization with the ERES marker Sec13p (Figure 2, E and F). Therefore these data show that GPI-anchored protein concentration at ERES does not require the p24 complex.
As a control, we used the disruption of BST1, which encodes for the first anchor-remodeling enzyme that deacetylates the GPI inositol. We previously showed that in bst1Δ sec31-1 double mutant cells Cwp2p is not accumulated in ERES at 37ºC, displaying just the characteristic ER nuclear ring staining as the bst1Δ single mutant (Figure 2, C and D). This defect in concentration at ERES, which is also observed in other remodeling mutants (Castillon et al., 2009), correlates with the lack of association with DRMs of the unremodeled, ER-localized GPI-anchored proteins (Fujita et al., 2006a). Therefore we expected that since the p24 complex is not required for GPI-anchored proteins concentration at ERES, the p24 complex should not be required for anchor remodeling or DRM association of GPI-anchored proteins.
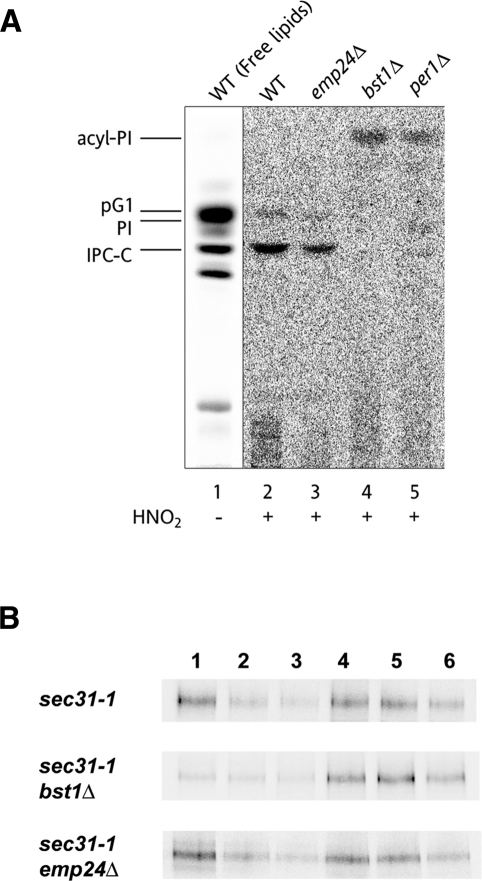
Thus we examined whether remodeling is influenced by the emp24Δ mutation. As shown in Figure 3A, we confirmed that the p24 complex is not required for anchor remodeling. The anchors of the emp24Δ strain (Figure 3A, lane 3) mainly contained a remodeled phosphatidylinositol and inositolphosphorylceramide as in wild-type strain. Next we investigated whether the GPI-anchored proteins become detergent insoluble at the ER in the absence of the p24 proteins, by using the sec31-1 thermosensitive allele and performing a pulse-chase experiment at restrictive temperature. DRM association of labeled proteins was monitored by the acquisition of detergent insolubility by subjecting cell extracts in the cold to incubation with TX-100 and density gradient centrifugations. In sec31-1 cells, the ER form of the GPI-anchored protein Gas1p was enriched in the fractions corresponding to DRMs (Figure 3B). As expected, Gas1p derived from bst1Δ sec31-1 cells was entirely located in the detergent-soluble fractions, verifying that remodeling is essential for DRM isolation of GPI-anchored proteins. However, in emp24Δ sec31-1 cells, there was no significant difference in the amount of Gas1p associated with DRM fractions compared with sec31-1 cells (Figure 3B). These results indicate that the p24 complex is not required for the isolation of GPI-anchored proteins in DRMs.
Figure 3:
The p24 complex is not required for anchor remodeling or DRM partition of GPI-anchored proteins in the ER. (A) Lipid remodeling of the GPI anchor is normal in emp24Δ cells. Wild-type, emp24Δ, bst1Δ, and per1Δ strains were labeled with [3H]myo-inositol for 2 h at 25C. The labeled PI moieties were prepared from GPI-anchored proteins and analyzed by TLC using the solvent system 55:45:10 chloroform/methanol/0.25% KCl. Lipids extracted from wild-type cells (lane 1) were used as a standard. pG1, phosphatidylinositol with a C26:0 fatty acid in sn-2 position; PI, phosphatidylinositol; IPC-C, inositolphosphorylceramide consisting of 4-hydroxysphinganine and a hydroxylated C26:0 fatty acid (Fujita et al., 2006a); acyl-PI, inositol-acylated PI (Ghugtyal et al., 2007) (B) GPI-anchored proteins are associated with DRMs at the ER level in emp24Δ cells. DRM association of the Gas1p in the ER was analyzed using sec31-1, sec31-1 bst1Δ, and sec31-1 emp24Δ cells, which were previously pulse labeled and chased at 37ºC. The cells were broken with glass beads and subjected to TX-100 extraction and density gradient centrifugation. Six fractions were collected and analyzed by immunoprecipitation with antibodies against Gas1p.
Because cargo receptors are believed to target secretory proteins to the ERES and GPI-anchored proteins are sorted from non–GPI-anchored proteins into different ERES upon cargo concentration, we next investigated whether the p24 complex is required for GPI-anchored protein sorting and targeting into their specific ERES. To test this possibility, we analyzed the colocalization of Cwp2p and the transmembrane protein Hxt1p contained in ERES in sec31-1 and emp24Δ sec31-1 cells at 37°C (Supplemental Figure S3, A and B). No significant differences could be observed, suggesting that sorting upon cargo concentration at ERES is not affected by the emp24Δ mutation.
Efficient ER exit of GPI-anchored proteins involves p24 protein–specific interaction with the specialized form of COPII coat subunit Lst1p
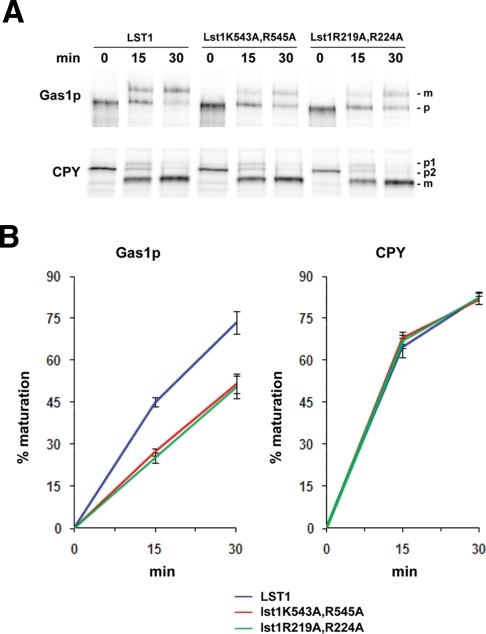
Taken together, the data presented earlier show that sorting and concentration of GPI-anchored proteins at ERES do not depend on p24 proteins, supporting the idea that the p24 complex does not act as a conventional cargo receptor. Therefore the export function by which the p24 complex directly promotes the ER exit of GPI-anchored proteins must be subsequent to their concentration at ERES. One possibility is that p24 complexes facilitate COPII vesicle budding from ERES containing concentrated GPI-anchored proteins. Given the lack of a cytosolic domain on GPI-anchored proteins and the ability of the p24 proteins to interact with the COPII components, p24 complexes could link the COPII coat on these specific ERES to GPI-anchored proteins and ensure their incorporation into COPII vesicles. In line with this hypothesis, the Sec24p homologue Lst1p holds a selective binding site for the p24 proteins (Miller et al., 2003) and, in addition, it is specifically required for the efficient ER-to-Golgi transport of the GPI-anchored protein Gas1p (Peng et al., 2000). Therefore the p24 complex might connect Lst1p with GPI-anchored proteins to stimulate their export from the ER. To investigate this possibility, we analyzed by pulse chase the influence on Gas1p transport of two Lst1p mutant forms (Lst1K543A,R545A and Lst1R219,224A) that disrupt specifically the binding site for p24 proteins, impairing its packaging into COPII vesicles (Miller et al., 2003). As shown in Figure 4, these mutations decreased the transport of Gas1p from the ER to the Golgi. This transport defect was specific for the GPI-anchored protein Gas1p, because the transport of the non–GPI-anchored protein CPY was not affected. It should be noted that the Gas1p transport delay in the lst1 point mutants is similar to the defect observed in lst1Δ (Peng et al., 2000). We conclude from this experiment that the efficient ER exit of GPI-anchored proteins requires the interaction of p24 proteins with the specialized form of COPII coat subunit Lst1p. Therefore our data suggest that the p24 complex, rather than being a conventional cargo receptor, acts as an adaptor that links COPII coat to GPI-anchored proteins in ERES. In agreement with this, p24 proteins are present in GPI-anchored protein containing ERES (Supplemental Figure S4, C and D) and exit the ER in the same vesicles (Muniz et al., 2000).
Figure 4:
The disruption of the p24 protein–binding site on the specialized COPII subunit Lst1p specifically impairs the efficient ER-to-Golgi transport of Gas1p. (A) Pulse-chase analysis of the ER-to-Golgi transport in the deletion strain lst1Δ expressing wild-type Lst1p or the mutant forms Lst1K543A,R545A and Lst1R219,224A. Proliferating cells were radiolabeled for 5 min, chased for the indicated times at 24°C, and lysed. Gas1p and CPY were immunoprecipitated, resolved by SDS–PAGE, and analyzed by Phosphorimager. Gas1p-p, ER-precursor form; -m, Golgi form. CPY-p2, ER-precursor form; -p1, Golgi precursor form; -m, mature form. (B) Quantification of several experiments described in A. The graph plots the average percentage of Gaslp and CPY maturation in lst1Δ strain expressing wild-type Lst1p or Lst1p mutants.
The p24 complex recognizes both remodeled and unremodeled GPI-anchored proteins
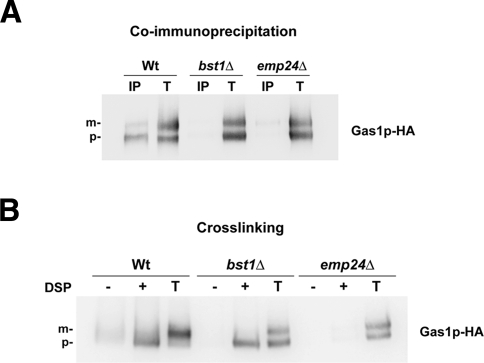
We have shown that Emp24p binds specifically GPI-anchored proteins through their GPI anchors. Therefore, if the p24 complex promotes vesicle budding by connecting the COPII coat with the GPI-anchored proteins after their concentration at ERES, we expected that the p24 proteins recognize GPI-anchored proteins once their GPI anchors have been completely remodeled. We addressed this issue by examining the extent of association between Emp24p and GPI-anchored proteins in wild-type strain and in strains with mutations in genes encoding remodeling enzymes, including the inositol deacylase Bst1p, the GPI phospholipase Per1p, or the acyltransferase Gup1p. For this purpose, cells expressing the GPI-anchored protein Gas1p-HA were analyzed by two different methods—native coimmunoprecipitation and chemical cross-linking. As seen in Figure 5A, immunoprecipitation in 1% digitonin-solubilized extracts revealed that in the wild-type strain Emp24p coprecipitated mainly with the 105-kDa ER precursor form of Gas1p, suggesting that GPI-anchored proteins dissociate from Emp24p upon arrival to the Golgi. By using this method, we also found that Emp24p did not coprecipitate with Gas1p in the remodeling mutant strains bst1Δ (Figure 5A), per1Δ, and gup1Δ (Supplemental Figure S5). For the cross-linking study, the cross-linked material was immunoprecipitated with antibody against Emp24p and blotted against anti-HA antibody. In this case, Gas1p molecules could be specifically cross-linked to Emp24p in both wild-type and bst1Δ mutant strains (Figure 5B). Thus association of Emp24p with unremodeled GPI-anchored proteins can be detected by cross-linking but not by native coimmunoprecipitation. Because chemical cross-linking is well known to stabilize weak or transient protein–protein interactions (Tatu and Helenius, 1997), these results suggest that Emp24 binding to unremodeled GPI-anchored proteins is weaker than that to remodeled GPI-anchored proteins. This could imply that Emp24p preferentially recognizes remodeled GPI-anchored proteins, which would be consistent with the idea that the p24 complex promotes the efficient export of GPI-anchored proteins after their remodeling and concentration at ERES. Nevertheless, the cross-linking data show that unremodeled GPI-anchored proteins are also specifically recognized by Emp24p. This result prompted us to evaluate whether the interaction between Emp24p and unremodeled GPI-anchored proteins plays any physiological role in the transport of GPI-anchored proteins.
Figure 5:
Emp24p can bind both remodeled and unremodeled GPI-anchored proteins. (A) Native coimmunoprecipitation assay between Emp24p and Gas1p. Enriched ER fractions (wild-type, bst1Δ, and emp24Δ mutant cells expressing Gas1-HA) were solubilized in 1% digitonin and analyzed by native immunoprecipitation (IP) with anti-Emp24p antibody followed by immunoblotting with anti-HA peroxidase antibody. Totals (T) represent a fraction of the solubilized input material. (B) Cross-linking assay between Emp24p and Gas1p. Spheroplasts from wild-type, bst1Δ, and emp24Δ mutant cells were incubated with (+) and without (−) DSP, denatured, and immunoprecipitated with anti-Emp24p antibody, followed by immunoblotting with anti-HA peroxidase antibody. Totals (T) represent a fraction of the solubilized input material. Gas1p: p, ER-precursor form; m, Golgi form.
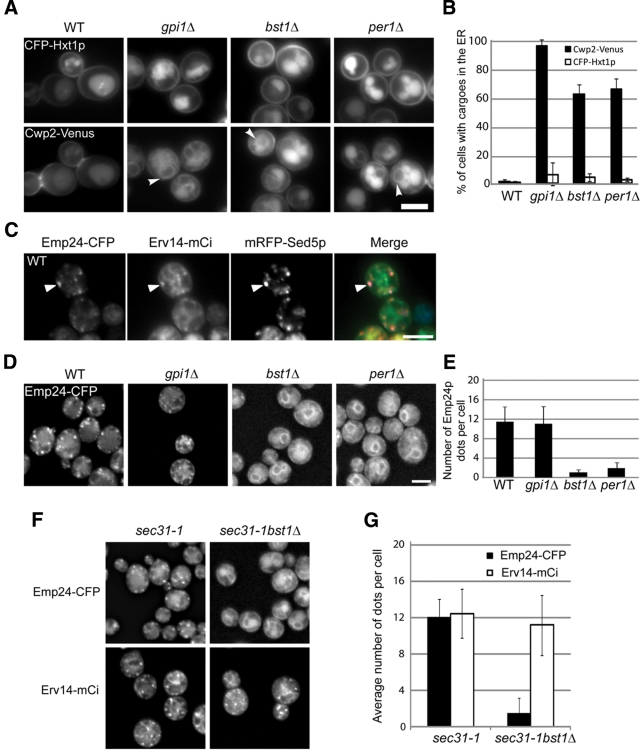
A defect in GPI anchor remodeling relocalizes the p24 complex from the Golgi to the ER
We showed previously that anchor remodeling is required for the efficient ER exit of GPI-anchored proteins (Castillon et al., 2009). Indeed, bst1Δ or per1Δ mutations lead to the ER accumulation of unremodeled GPI-anchored proteins. This accumulation is specific since the unrelated transmembrane cargo Hxt1p is properly targeted to the plasma membrane in the absence of remodeling (Figure 6, A and B; Castillon et al., 2009). Because Emp24p is able to interact with unremodeled GPI-anchored proteins (Figure 5B), we decided to investigate whether the ER accumulation of unremodeled GPI-anchored proteins observed in the remodeling mutants could influence the intracellular localization of Emp24p, which normally cycles between Golgi and ER compartments (Belden and Barlowe, 2001). To address this possibility, we analyzed the localization of functional, fluorescent protein–tagged Emp24p (Supplemental Figure S6B) in the absence of remodeling by using the bst1Δ and per1Δ mutant strains. In wild-type cells Emp24p is preferentially found in dot-like structures that colocalize with the cis-Golgi SNARE Sed5p (Figure 6C), whereas in both bst1Δ and per1Δ mutant cells it is completely relocated to the ER (Figure 6, D and E). This relocalization phenotype could be explained if the p24 complex is unstable in the absence of remodeling, and thus the p24 proteins would be subsequently retained in the ER. However, we examined this possibility and found that the p24 complex is indeed stable and presumably functional in the remodeling mutants (Supplemental Figure S7). Therefore we assume that the relocation and ER retention of Emp24p observed in remodeling mutants are due to the interaction of Emp24p with the anchor of the GPI-anchored proteins. If this hypothesis is correct, Emp24p localization should not be affected in the absence of GPI anchor synthesis even though unanchored proteins are retained in the ER (Figure 6, A and B). We tested this assumption by analyzing the localization of Emp24p in the gpi1Δ mutant, which is defective in the synthesis of the GPI anchor (Leidich and Orlean, 1996). As predicted, in the gpi1Δ mutant such precursor accumulation does not affect the localization of Emp24p, which is not trapped at the ER and can be properly sent off to the Golgi. (Figure 6, D and E). This shows that an unremodeled GPI anchor is required to cause redistribution of Emp24p to the ER.
Figure 6:
Emp24p is relocalized from the Golgi to the ER in remodeling mutants. (A) Selective defect in the ER export of GPI-anchored proteins in remodeling mutants. Live images of wild-type, gpi1Δ, bst1Δ, and per1Δ expressing Hxt1-CFP and Cwp2-Venus at 30°C. (B) Quantification of several micrographs described in A. The graph plots the average percentage of cells displaying Cwp2-Venus (black bars) and Hxt1-CFP (white bars) in the ER. n, number of cells plotted; 37 ≤ n ≤ 53. (C) Live images of wild-type cells expressing Emp24-CFP, Erv14-mCi, and mRFP-Sed5 at 30°C. (D) Emp24p localization depends on remodeling of GPI-anchored proteins. Live images of wild-type, gpi1Δ, bst1Δ, and per1Δ cells expressing Emp24-CFP at 30°C. (E) Quantification of several micrographs described in D. The graph plots the average number of Emp24-CFP dots per cell seen in the different strains. n, number of cells plotted; 46 ≤ n ≤ 63. (F) Emp24p is not incorporated into ERES in remodeling mutants. Fluorescence micrographs of live sec31-1 and sec31-1 bst1Δ cells expressing Emp24-CFP and Erv14-mCi at 37ºC. (G) Quantification of several micrographs described in F. The graph plots the average percentage of the sec31-1 and sec31-1 bst1Δ cells for which Emp24-CFP and Erv14-mCi are found in dot-like structures. n, number of cells plotted. 74 ≤ n ≤ 89. A, C, D, Raw images. Scale bar, 5 μm.
Next we investigated whether Emp24p also fails to be concentrated into ERES in remodeling mutants. To assess this possibility, we analyzed the distribution of Emp24p in the bst1Δ sec31-1 mutant strain at 37ºC. As shown in Figure 6, F and G, Emp24p is preferentially found in ERES in sec31-1 cells. Nevertheless, in a bst1Δ sec31-1 cells Emp24p did not show the typical ERES punctuate pattern, displaying only the ER-characteristic nuclear ring staining. The defect in ERES association is specific for Emp24p because Erv14p, a transmembrane protein that also cycles continuously between ER and Golgi (Powers and Barlowe, 1998), is properly accumulated at ERES in the bst1Δ sec31-1 mutant cells (Figure 6, F and G). Therefore this result indicates that the ER accumulation of unremodeled GPI-anchored proteins prevents the p24 complex from being incorporated into ERES.
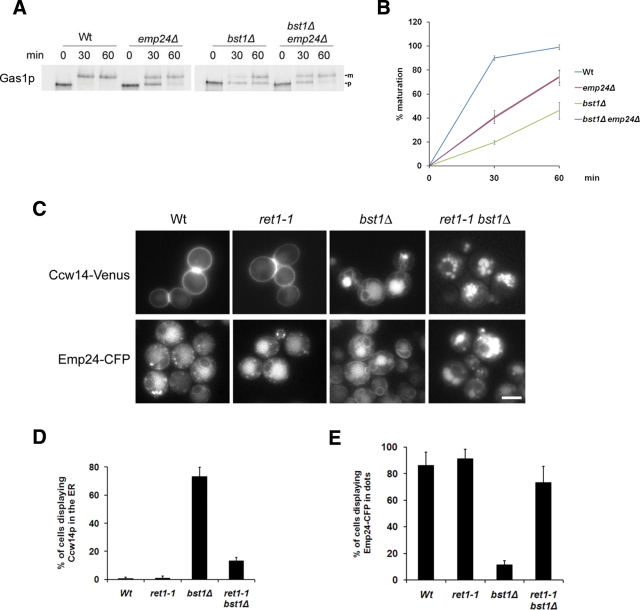
The p24 complex contributes to the effective ER retention of unremodeled GPI-anchored proteins by recycling them from the Golgi
Taken together, these data suggest a role of the p24 complex in the quality control of GPI-anchored proteins. The p24 complex might contribute to the retention of GPI-anchored proteins in the ER until they become properly remodeled. We addressed this possibility by checking whether the unremodeled GPI-anchored proteins can be transported faster in the absence of the p24 complex. To do this, we analyzed by a pulse-chase experiment the ER-to-Golgi transport of Gas1p in remodeling mutants lacking Emp24p (Figure 7, A and B). Compared to the wild-type strain, the Golgi maturation kinetics of Gas1p is delayed in the emp24Δ mutant and the remodeling mutant bst1Δ, as observed previously (Schimmoller et al., 1995; Tanaka et al., 2004). Nevertheless, the delay is stronger in the remodeling mutant than in the emp24Δ mutant strain. Remarkably, in the double mutant emp24Δ bst1Δ the transport rate is improved with respect to the remodeling mutant. To discard the possibility that the increase of the ratio of Gas1p mature form observed in the double mutant is due to a higher protein turnover in the ER, we quantified the loss of total signal remaining after 60 min of chase. The percentage of signal remaining is 82% for wild type, 78% for emp24Δ, 87% for bst1Δ, and 83% for the bst1Δ emp24Δ double mutant, indicating that there are no significant differences among strains. Therefore we conclude that the p24 complex contributes to the ER retention of unremodeled GPI-anchored proteins, supporting the idea that the p24 complex is involved in a quality control mechanism that regulates the intracellular transport of GPI-anchored proteins. Of interest, the transport rate in the double mutant is only recovered to the transport level of the emp24Δ mutant strain. This partial recovery was what we expected because in the double-mutant strain the ER exit function of the p24 complex is still lost, preventing a recovery to wild-type levels.
Figure 7:
Efficient ER retention of unremodeled GPI-anchored proteins requires their recycling from the Golgi to the ER by the p24 complex. (A) The emp24Δ mutation partially suppresses the GPI-anchored protein transport defect in remodeling mutants. Pulse-chase analysis to follow the transport from ER to Golgi of Gas1p in wild-type and deletion strains. Proliferating cells were radiolabeled for 5 min, chased for the indicated times at 24°C, and lysed. Gas1p was immunoprecipitated, resolved by SDS–PAGE, and analyzed by Phosphorimager. ER (p) and Golgi (m) Gas1p forms are indicated. (B) Quantification of several experiments described in A. The graph plots the average percentage of Gas1p maturation in wild-type and deletion strains. (C) An active retrograde transport is required for ER retention of GPI-anchored proteins and ER redistribution of Emp24p in remodeling mutants. Live images of wild-type, ret1-1, bst1Δ, and ret1-1 bst1Δ expressing Ccw14-Venus or Emp24-CFP at 24°C. Raw images. Scale bar, 5 μm. (D) Quantification of several micrographs described in C. The graph plots the average percentage of cells displaying Ccw14-Venus in the ER. n, number of cells plotted; n ≥ 100. (E) Quantification of several micrographs described in D. The graph plots the average number of Emp24-CFP dots per cell seen in the different strains. n, number of cells plotted; n ≥ 100.
Because the p24 proteins are normally cycling between ER and Golgi, the ER retention of unremodeled GPI-anchored proteins mediated by the p24 complex might require retrieval from the Golgi. The p24 complex could bind unremodeled GPI-anchored proteins in the ER, travel with them, inefficiently, to the cis-Golgi, and mediate their efficient recycling to the ER via COPI-dependent retrograde transport pathway. We investigated this possibility by testing whether retrograde transport is responsible for the ER retention of unremodeled GPI-anchored proteins. To do this, we further examined the ER accumulation of unremodeled GPI-anchored proteins in the coatomer mutant ret1-1. The temperature-sensitive ret1-1 mutant has a lesion in the α subunit of coatomer and, at permissive temperature (24ºC), shows a defect in the Golgi-to-ER retrograde transport of dilysine-harboring proteins but not in the anterograde transport of GPI-anchored proteins (Letourneur et al., 1994). If the ER retention of unremodeled GPI-anchored proteins depends on retrograde transport, a ret1-1 bst1Δ double mutant should fail to massively accumulate unremodeled GPI-anchored proteins in the ER. As shown in Figure 7, C and D, in wild-type and ret1-1 mutant cells, the GPI-anchored protein Ccw14p showed a cell surface staining, whereas in bst1Δ, the ER-characteristic nuclear ring staining was observed. In contrast, when Ccw14p was expressed in ret1-1 bst1Δ double-mutant cells, no ER staining was observed and Ccw14p was mainly localized at the vacuole (see also Supplemental Figure S8). In addition, the ER staining for Emp24p exhibited in the bst1Δ mutant was not detected in ret1-1 bst1Δ double-mutant cells, and Emp24p was present in dot-like structures, perhaps Golgi or endosomes (Figure 7, C and E). Therefore these results show that an active retrograde transport is required for the ER retention of both GPI-anchored proteins and Emp24p in the remodeling mutant bst1Δ, which strongly suggests that one of the p24 complex functions in the early secretory pathway is to retrieve escaped unremodeled GPI-anchored proteins from the Golgi back to the ER.
The p24 complex alleviates the ER stress caused by the accumulation of unremodeled GPI-anchored proteins
Under normal growth conditions, a defect in remodeling causes the ER accumulation of unremodeled GPI-anchored proteins, which results in constitutive and moderate activation of the multifaceted unfolded protein response (UPR) (Jonikas et al., 2009). The UPR has been shown to alleviate the stress produced by the accumulation of aberrant secretory cargo in the ER (Travers et al., 2000). Because the p24 complex binds unremodeled GPI-anchored proteins, we explored the possibility that this binding contributes to reduce the ER stress in a remodeling mutant. If this is the case, the absence of the p24 proteins in a remodeling mutant should further activate the UPR. We measured the UPR induction from a reporter construct (pJC31) that contains the 22–base pair UPRE (unfolded protein responses element) of KAR2 fused to LacZ (Cox and Walter, 1996). β-Galactosidase activity was increased in the emp24Δ bst1Δ double mutant compared with the single mutants (Table 1), reflecting a synergistic activation of the UPR. This result suggests that p24 complex bound to unremodeled GPI-anchored proteins in the ER helps to shield them from components of the UPR pathway. Therefore, although we have shown that unremodeled GPI-anchored proteins exit the ER faster in the absence of the p24 proteins, the remaining protein fraction could be more exposed and thus increase the UPR activation. This result supports a role of the p24 complex in protecting the cell from the stress induced by the accumulation of unremodeled GPI-anchored proteins in the ER.
TABLE 1:
Activation of the UPR in wild-type and mutant strains.
| Strain | β-Galactosidase units |
|---|---|
| Wild type | 1.5 ± 0.009 |
| emp24Δ | 6.3 ± 0.005 |
| bst1Δ | 10 ± 0.021 |
| bst1Δ emp24Δ | 15.4 ± 0.021 |
β-Galactosidase assays were performed on strains harboring the reporter construct (pJC31). Activity is given in β-galactosidase units and represents the average of four independent determinations with SE.
DISCUSSION
In this study we describe the mechanisms by which the yeast p24 complex operates in the intracellular transport of GPI-anchored proteins. In yeast, GPI-anchored proteins are selectively concentrated at specific ERES, from where they are incorporated into COPII vesicles that are distinct from those carrying other secretory proteins (Muniz et al., 2001; Castillon et al., 2009). A direct role in this sorting process was suggested by our in vitro experiment showing that preincubation of wild-type membranes with antibodies against the cytosolic tail of Emp24p selectively inhibited the ER budding of a GPI-anchored protein. Even though p24 complex binds specifically to GPI-anchored proteins in the ER in an anchor-dependent manner, it is not required to sort and concentrate them into their specific ERES. This implies that the p24 complex does not behave like a conventional cargo receptor, and that its ER export function takes place downstream of cargo concentration, consistent with our previous results that GPI-anchored proteins do not require COPII machinery for their concentration in ERES. The direct role of the p24 complex in the ER exit of yeast GPI-anchored proteins seems to be to link the GPI-anchored proteins to the COPII coat. This function is required since GPI-anchored proteins are completely luminal and need an adaptor to bind sites on the COPII coat. This binding is likely to ensure an efficient incorporation of the GPI-anchored proteins into the vesicles during their formation and might influence vesicle formation itself. In strong support of the “linker” function is our observation that the disruption of the p24 protein–binding site on the specialized COPII subunit Lst1p specifically impairs the efficient ER-to-Golgi transport of Gas1p. Therefore the p24 complex may act as an adaptor that facilitates COPII vesicle formation by stabilizing COPII components on the GPI-anchored protein containing ERES. A similar mechanism is believed to be used by the p24 complex for the generation of COPI vesicles, in which the yeast p24 proteins can promote budding by acting as a primer to induce COPI coat polymerization onto the Golgi membrane (Aguilera-Romero et al., 2008). Furthermore, p24 tails from animal cells can stimulate in vitro the formation of COPI-coated vesicles when they are displayed on liposomes (Bremser et al., 1999).
Because in mammalian cells lipid remodeling is not terminated at the ER (Fujita and Jigami, 2008), it is possible that concentration of mammalian GPI-anchored proteins upon ER exit does not depend on this process as it does in yeast. It seems that the mammalian p24 complex acts in this case as expected for a conventional cargo receptor by concentrating GPI-anchored proteins at ERES prior vesicle budding (Fujita et al., 2011).
Our study reveals a novel role of the yeast p24 complex in quality control of GPI-anchored proteins by monitoring the completion of anchor remodeling. In the absence of the remodeling machinery, Emp24p can bind unremodeled GPI-anchored proteins. Furthermore, unremodeled GPI-anchored proteins are transported more efficiently in the absence of Emp24p. These findings indicate that the p24 complex helps retain incompletely remodeled GPI-anchored proteins at the ER when the remodeling machinery is absent or inactive. Our results also indicate that this ER retention requires retrieval from Golgi. We observed a complete redistribution of Emp24p from Golgi to ER when remodeling is defective. Likewise, the mammalian KDEL receptor, whose steady-state localization is in the cis-Golgi compartment, also redistributes to the ER upon overexpression of KDEL-bearing secretory proteins (Lewis and Pelham, 1992). Moreover, we have shown that an active Golgi-to-ER retrograde transport is required for the ER accumulation of both unremodeled GPI-anchored proteins and Emp24p in a remodeling mutant. These results strongly suggest that the p24 complex contributes to ER retention of unremodeled GPI-anchored proteins by recycling them from the Golgi via COPI-dependent retrograde transport. Consistent with a retrieval action in a post-ER quality control, the p24 complex is continuously cycling between ER and Golgi (Stamnes et al., 1995; Sohn et al., 1996; Belden and Barlowe, 2001) and facilitates COPI vesicle formation from the Golgi membrane (Aguilera-Romero et al., 2008).
Both export and retention mechanisms seem to involve GPI anchor recognition by the p24 complex. Consistently, Emp24p can bind Gas1p but not to Gas1-TMD or Gas1Q, for which the GPI attachment signal has been exchanged for a transmembrane domain or mutated in the ω site to prevent anchoring, respectively. Emp24p localization is strongly affected by the presence of unremodeled GPI-anchored protein but not by unanchored proteins. One possibility is that Emp24p recognizes the lipid moiety on the GPI anchor. Our observation that both remodeled and unremodeled GPI-anchored proteins can be cross-linked to Emp24p but only remodeled GPI-anchored proteins are coprecipitated in detergent solutions might suggest that lipid remodeling allows higher binding affinity to p24 proteins in the ER. This would lead to a preferential ER export of remodeled GPI-anchored proteins by the p24 complex. Even though unremodeled GPI-anchored protein binding to Emp24p could be weaker, it is still sufficient for their retrieval from Golgi, as shown by the relocalization of Emp24p to the ER in remodeling mutants. Another possible explanation for the difference between the cross-linking and coprecipitation experiments might be that the interaction of the p24 complex with unremodeled GPI anchored proteins depends more on the lipid structure than for remodeled proteins. This hydrophobic interaction is more likely to be inhibited by the presence of the detergent in the coprecipitation experiment.
Our binding studies also indicate that remodeled GPI-anchored proteins dissociate from the p24 complex in the Golgi, since Emp24p binds only to the ER form of Gas1p. As proposed for cargo receptors, dissociation could be caused by a decreasing pH in a later Golgi subcompartment that would induce conformational changes in the p24 proteins to lower affinity for bound ligand (Dancourt and Barlowe, 2010). However, the fact that unremodeled GPI-anchored proteins are recycled from Golgi to the ER suggests that they do not release from the p24 complex in this organelle. We speculate that either the lower-pH subcompartment is not reached by unremodeled GPI-anchored proteins because of their physical properties or the hydrophobic nature of the interaction of unremodeled proteins with p24 renders the interaction insensitive to pH. Perhaps the short length of their unremodeled anchors precludes them from proper insertion into the thicker Golgi membrane, leading to their retrieval by the p24 complex.
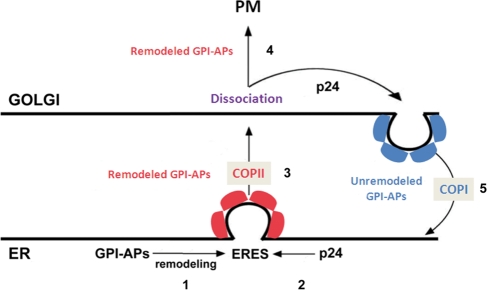
We present the following model for p24 function in yeast (Figure 8). GPI-anchored proteins are concentrated and sorted into their specific ERES upon anchor remodeling. Then p24 complexes are recruited to these ERES due to its efficient interaction with fully remodeled GPI-anchored proteins. This late binding would avoid an unproductive competition with the remodeling machinery. Once recruited to the GPI-anchored protein ERES, the p24 complex links the GPI-anchored proteins to COPII components on these ERES, which ensures the formation of COPII vesicles containing correctly remodeled GPI-anchored proteins. During or after arrival to the Golgi, remodeled GPI-anchored proteins dissociate from the p24 complex. On their release, remodeled GPI-anchored proteins can progress through the secretory pathway to be finally delivered to the plasma membrane. By contrast, unremodeled GPI-anchored proteins that have escaped to the Golgi will be recycled back to the ER in COPI vesicles by the p24 complex. Once in the ER, unremodeled GPI-anchored proteins would have another opportunity to acquire the proper remodeled anchor.
Figure 8:
Model of the specific roles of the p24 complex in the trafficking of GPI-anchored proteins along the early secretory pathway in yeast. The p24 complex promotes the efficient ER export of fully remodeled GPI-anchored proteins by linking the proteins to the COPII coat at their specific ERES and prevents the progression of incompletely remodeled GPI-anchored proteins along the secretory pathway by recycling them back from Golgi to the ER in COPI vesicles. 1) GPI anchored proteins are concentrated and sorted into their specific ERES upon anchor remodeling. 2) p24 complex is efficiently recruited to these ERES due to its binding to fully remodeled GPI-anchored proteins. 3) The p24 complex acts as an adaptor by linking COPII components to the GPI-anchored protein at ERES, which might facilitate vesicle biogenesis. 4) On arrival to the Golgi, GPI-anchored proteins dissociate from the p24 complex. On their release, correctly remodeled GPI-anchored proteins can progress through the secretory pathway to be delivered to the plasma membrane. 5) Escaped unremodeled GPI-anchored proteins are retrotransported from Golgi to the ER by the p24 complex. GPI-APs, GPI-anchored proteins.
Therefore the yeast p24 complex senses the status of the GPI anchor, regulates GPI-anchored protein intracellular transport, and coordinates this with correct anchor remodeling.
MATERIALS AND METHODS
Yeast strains and plasmids
Strains of Saccharomyces cerevisiae used for this work are listed in Supplemental Table S1. The erv14-mCi-SpHIS5 allele was obtained after the PCR of the mCi-SpHIS5 cassette from the EUROSCARF pKT211 plasmid and after homologous recombination of the PCR product at the locus in the 3′ of ERV14 open reading frame (ORF) excluding the stop codon (Longtine et al., 1998). In this strain CFP-Hxt1p is normally found at the plasma membrane and vacuole, similar to what is observed in wild-type yeast, whereas in erv14Δ cells a fraction of CFP-Hxt1p accumulated in the ER (Supplemental Figure S6A). This result confirms that Erv14-mCi is functional.
The plasmids expressing Cwp2-Venus (pRS416ADH-CWP2-VENUS), Ccw14-Venus (pRS416ADH-CCW14-VENUS), and CFP-Hxt1p (pRS415ADH-CERULEAN-HXT1) were made in a previous study (Castillon et al., 2009). The plasmid expressing mRFP-Sed5p was kindly provided by A. Nakano (Matsuura-Tokita et al., 2006). Other plasmids used in this study are pGAS1, pGAS1TMD, pGAS1(-S/T) (Watanabe et al., 2008), pGAS1Q (Nuoffer et al., 1993), pTKY12 (LST1), pLM218 (Lst1K543A,R545A), and pLM219 (Lst1R219,224A) (Miller et al., 2003). To construct the plasmid expressing Sec13-mCh, we first integrated by homologous recombination the PCR product containing mCherry-KanMX from pBS34 (Yeast Resource Center, http://depts.washington.edu/yeastrc/index.html) at the 3′ end of the SEC13 ORF excluding the stop codon. From the resultant strain we amplified by PCR a fragment with a forward primer (gggatatcaggaggcttccgagattttgg) hybridizing 500 base pairs upstream of the SEC13 ORF and a reverse primer (gggatatcctcgcaggtctgcagcgaggcgcc) recognizing the downstream sequence of mCherry. The PCR product was inserted in YCplac22 after enzymatic digestion by EcoRV to produce YCplac22-Sec13-mCh. YCplac111-Emp24-CFP was produced after amplification of the region named arbitrarily Emp24-1 upstream of codon 128 coding for asparagine of the EMP24 ORF using the primers F1, gctctagacgataatggtcttgctcttggtaacc, and R1, gcggatcctgggtcgtccaaatccacatata, and the region named Emp24-2 downstream of codon 127 coding for proline of the EMP24 ORF using the primers F2, gcggatccaacaccaatacattggatagtgc, and R2, gccccgggccactagtgtatgcgactgcgattca. Next the PCR product of Emp24-1 was digested by XbaI and BamHI, and the PCR product of Emp24-2 was digested by XmaI and BamHI. Both digested fragments were ligated into YCplac111 predigested by XbaI and XmaI in order to obtain YCplac111-Emp24. We then amplified the sequence coding for the fluorescent protein Cerulean (Rizzo et al., 2004) by PCR with a forward primer containing a BamHI site and a reverse primer excluding the stop codon of Cerulean and a BamHI site. The fragment containing Cerulean was inserted into BamHI-digested YCplac111-Emp24 plasmid to obtain YCplac111-Emp24-CFP. This fluorescent fusion protein is functional because it is able to rescue Cwp2p localization in emp24Δ cells (Supplemental Figure S6B).
Cross-linking assay
EMP24-HA-tagged and untagged strains were first grown in SUD medium [0.16% yeast nitrogen base without amino acids and without (NH4)2SO4, 2% glucose, 0.1% urea] supplemented with the required amino acids at 24°C to express Gap1p and then grown overnight in SDYE medium (0.67% yeast nitrogen base without amino acids, 2% glucose, and 0.2% yeast extract) supplemented with the required amino acids and nutrients at 24ºC. A total of 200 ml of cells was harvested at 5 × 106 cell/ml, washed twice with SD medium (0.67% yeast nitrogen base, 2% glucose supplemented with the required nutrients), resuspended in 4 ml, and incubated 15 min at 24ºC. Cells were then pulse labeled for 3 min with EasyTag Express Protein Labeling Mix (PerkinElmer Life Sciences, Waltham, MA) in SD medium at 24°C. Metabolic activity was stopped by the addition of NaN3 (20 mM final) and incubation on ice for 10 min. Spheroplasting and lysis was performed as described (Muniz et al., 2000). Before the cross-linking reaction, permeabilized spheroplasts were resuspended in 2.5 M urea in B88 (20 mM HEPES, pH 6.8, 150 mM KOAc, 250 mM sorbitol, 5 mM Mg(OAc)2) incubated for 10 min on ice, and washed twice with B88. A total of 25 × 107 permeabilized spheroplasts was incubated with 0.5 mM dithiobis(succinimidylpropionate) (DSP; Pierce, Thermo Fisher Scientific, Rockford, IL; 20ºC, 20 min). The cross-linking reaction was quenched by addition of glycine (50 mM final, 5 min, 20ºC). A portion of the sample was removed for analysis (total), and the remaining aliquot was dissolved with 1% SDS in TEPI (50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM protease inhibitor mix; 5 min, 95ºC for GPI-anchored proteins and CPY or 55ºC for Gap1p and Hxt1p), and immunoprecipitated with anti-HA antibody, 12AC5 (Roche Diagnostics, Indianapolis, IN), and Protein G Sepharose 4 Fast Flow (GE Healthcare Bio-Sciences, Piscataway, NJ). Precipitated material was eluted from the Sepharose beads by incubation with 1% SDS in TEPI (5 min, 95 or 55ºC), reimmunoprecipitated with anti-Gas1p, anti-CPY, anti-Gap1p, or anti-GFP antibody, was incubated with 5% 2-β-mercaptoethanol, and was analyzed by SDS–PAGE with subsequent exposure and quantitation using a Phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Native coimmunoprecipitation
The native coimmunoprecipitation experiments were performed on enriched ER fractions as described (Fujita et al., 2006b).
GPI anchor remodeling and DRM partitioning
Phosphatidylinositol (PI) moieties of GPI anchor were isolated from GPI-anchored proteins labeled with [3H]myo-inositol as described previously (Guillas et al., 2000; Fujita et al., 2006a). The lipids were separated by TLC using solvent system 55:45:10 chloroform/methanol/0.25% KCl and visualized using FLA-7000 (Fujifilm, Tokyo, Japan). DRM partitioning was performed as in Bagnat et al. (2000).
β-Galactosidase assay
Assays of β-galactosidase activity in extracts of yeast cells containing the UPRE-LacZ fusion construct pJC31 were performed as described by Cox and Walter (1996).
Pulse-chase analysis
Radiolabeling and immunoprecipitations were performed as described by Watanabe et al. (2002) with some modifications, and cells were grown in SDYE medium supplemented with the required amino acids at 24°C to 0.5–2 × 107/ml, harvested, and resuspended in SD medium without methionine and cysteine. A total of 3 × 107 cells was used for each time point, preincubated at 24°C for 15 min, and labeled with 100 μCi of EasyTag Express Protein Labeling Mix, [35S], for 5 min.
Coimmunoprecipitation and stability of Emp24p
The coimmunoprecipitation experiments were performed as described (Marzioch et al., 1999). Emp24-CFP was precipitated with a monoclonal anti-GFP antibody (Roche).The protein levels of Emp24p and Erv25p after addition or not of cycloheximide (35 μg/ml for 90 min) from log-phase growing cultures were revealed after trichloroacetic acid precipitation by Western blot.
Microscopy
Acquisitions were made as previously described (Castillon et al., 2009). The micrographs were acquired under a 100× 1.4–numerical aperture oil objective with the AXIOZ1 microscope (Zeiss, Thornwood, NY) and the Zeiss AxioCam MRm charge-coupled device camera controlled by the software AxioVision, release 4.6. If colocalization was required, then the acquisitions were deconvoluted by 10 iterations using the Diffraction PSF 3D and the Iterative Deconvolve 3D plug-ins for ImageJ (http://www.optinav.com/Diffraction-PSF-3D.htm). The data were quantified and processed with Image J.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministerio de Ciencia e Innovación BFU2008-04119/BMC (M.M.), the Swiss National Science Foundation (H.R., R.W.), the National Centre of Competence in Research in Chemical Biology, funded by the Swiss National Science Foundation (H.R.), a Swiss National Science Foundation Assistant Professorship (R.W.), and University of Seville fellowships (A.A.R and J.M.L). Imaging was carried out at the Imaging Platform of the Swiss National Science Foundation Frontiers in Genetics. We thank Christoph Bauer for assistance with imaging, Brigitte Bernadets for technical assistance, and Veit Goder, Anne Spang, and Randy Schekman for materials and advice.
Abbreviations used:
- DRM
detergent-resistant membrane
- DSP
dithiobis(succinimidylpropionate)
- ERES
ER exit sites
- GPI
glycosylphosphatidylinositol
- UPR
unfolded protein response
- UPRE
unfolded protein response element
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-04-0294) on June 16, 2011.
REFERENCES
- Aguilera-Romero A, Kaminska J, Spang A, Riezman H, Muniz M. The yeast p24 complex is required for the formation of COPI retrograde transport vesicles from the Golgi apparatus. J Cell Biol. 2008;180:713–720. doi: 10.1083/jcb.200710025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M, Keranen S, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci USA. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J Biol Chem. 1996;271:26939–26946. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C. Distinct roles for the cytoplasmic tail sequences of Emp24p and Erv25p in transport between the endoplasmic reticulum and Golgi complex. J Biol Chem. 2001;276:43040–43048. doi: 10.1074/jbc.M108113200. [DOI] [PubMed] [Google Scholar]
- Bonnon C, Wendeler MW, Paccaud JP, Hauri HP. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J Cell Sci. 2010;123:1705–1715. doi: 10.1242/jcs.062950. [DOI] [PubMed] [Google Scholar]
- Bremser M, Nickel W, Schweikert M, Ravazzola M, Amherdt M, Hughes CA, Sollner TH, Rothman JE, Wieland FT. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96:495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- Caro LH, Tettelin H, Vossen JH, Ram AF, van den Ende H, Klis FM. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1477–1489. doi: 10.1002/(SICI)1097-0061(199712)13:15<1477::AID-YEA184>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Castillon GA, Watanabe R, Taylor M, Schwabe TM, Riezman H. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 2009;10:186–200. doi: 10.1111/j.1600-0854.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- Fujita M, Jigami Y. Lipid remodeling of GPI-anchored proteins and its function. Biochim Biophys Acta. 2008;1780:410–420. doi: 10.1016/j.bbagen.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Fujita M, Umemura M, Yoko-o T, Jigami Y. PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol Biol Cell. 2006a;17:5253–5264. doi: 10.1091/mbc.E06-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Watanabe R, Jaensch N, Romanova-Michaelides M, Satoh T, Kato M, Riezman H, Yamaguchi Y, Maeda Y, Kinoshita T. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J Cell Biol. 2011;194:61–75. doi: 10.1083/jcb.201012074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Yoko OT, Jigami Y. Inositol deacylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol Biol Cell. 2006b;17:834–850. doi: 10.1091/mbc.E05-05-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullekrug J, Suganuma T, Tang BL, Hong W, Storrie B, Nilsson T. Localization and recycling of gp27 (hp24gamma3): complex formation with other p24 family members. Mol Biol Cell. 1999;10:1939–1955. doi: 10.1091/mbc.10.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentzsch M, Tanner W. Protein-O-glycosylation in yeast: protein-specific mannosyltransferases. Glycobiology. 1997;7:481–486. doi: 10.1093/glycob/7.4.481. [DOI] [PubMed] [Google Scholar]
- Ghugtyal V, Vionnet C, Roubaty C, Conzelmann A. CWH43 is required for the introduction of ceramides into GPI anchors in Saccharomyces cerevisiae. Mol Microbiol. 2007;65:1493–1502. doi: 10.1111/j.1365-2958.2007.05883.x. [DOI] [PubMed] [Google Scholar]
- Guillas I, Pfefferli M, Conzelmann A. Analysis of ceramides present in glycosylphosphatidylinositol anchored proteins of Saccharomyces cerevisiae. Methods Enzymol. 2000;312:506–515. doi: 10.1016/s0076-6879(00)12935-0. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Leidich SD, Orlean P. Gpi1, a Saccharomyces cerevisiae protein that participates in the first step in glycosylphosphatidylinositol anchor synthesis. J Biol Chem. 1996;271:27829–27837. doi: 10.1074/jbc.271.44.27829. [DOI] [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, Demollière C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the ER. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. Ligand-induced redistribution of a human KDEL receptor from the Golgi complex to the endoplasmic reticulum. Cell. 1992;68:353–364. doi: 10.1016/0092-8674(92)90476-s. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Tashima Y, Houjou T, Fujita M, Yoko-o T, Jigami Y, Taguchi R, Kinoshita T. Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol Biol Cell. 2007;18:1497–1506. doi: 10.1091/mbc.E06-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzioch M, Henthorn DC, Herrmann JM, Wilson R, Thomas DY, Bergeron JJ, Solari RC, Rowley A. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol Biol Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A. Live imaging of yeast Golgi cisternal maturation. Nature. 2006;441:1007–1010. doi: 10.1038/nature04737. [DOI] [PubMed] [Google Scholar]
- Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Muniz M, Morsomme P, Riezman H. Protein sorting upon exit from the endoplasmic reticulum. Cell. 2001;104:313–320. doi: 10.1016/s0092-8674(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Muniz M, Nuoffer C, Hauri HP, Riezman H. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J Cell Biol. 2000;148:925–930. doi: 10.1083/jcb.148.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C, Horvath A, Riezman H. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J Biol Chem. 1993;268:10558–10563. [PubMed] [Google Scholar]
- Orlean P, Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- Peng R, De Antoni A, Gallwitz D. Evidence for overlapping and distinct functions in protein transport of coat protein Sec24p family members. J Biol Chem. 2000;275:11521–11528. doi: 10.1074/jbc.275.15.11521. [DOI] [PubMed] [Google Scholar]
- Pittet M, Conzelmann A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:405–420. doi: 10.1016/j.bbalip.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Powers J, Barlowe C. Transport of axl2p depends on erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J Cell Biol. 1998;142:1209–1222. doi: 10.1083/jcb.142.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Rojo M, Pepperkok R, Emery G, Kellner R, Stang E, Parton RG, Gruenberg J. Involvement of the transmembrane protein p23 in biosynthetic protein transport. J Cell Biol. 1997;139:1119–1135. doi: 10.1083/jcb.139.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Nakano A. Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 2007;581:2076–2082. doi: 10.1016/j.febslet.2007.01.091. [DOI] [PubMed] [Google Scholar]
- Schimmoller F, Singer-Kruger B, Schroder S, Kruger U, Barlowe C, Riezman H. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K, Orci L, Ravazzola M, Amherdt M, Bremser M, Lottspeich F, Fiedler K, Helms JB, Wieland FT. A major transmembrane protein of Golgi-derived COPI-coated vesicles involved in coatomer binding. J Cell Biol. 1996;135:1239–1248. doi: 10.1083/jcb.135.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamnes MA, Craighead MW, Hoe MH, Lampen N, Geromanos S, Tempst P, Rothman JE. An integral membrane component of coatomer-coated transport vesicles defines a family of proteins involved in budding. Proc Natl Acad Sci USA. 1995;92:8011–8015. doi: 10.1073/pnas.92.17.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takida S, Maeda Y, Kinoshita T. Mammalian GPI-anchored proteins require p24 proteins for their efficient transport from the ER to the plasma membrane. Biochem J. 2008;409:555–562. doi: 10.1042/BJ20070234. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Maeda Y, Tashima Y, Kinoshita T. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J Biol Chem. 2004;279:14256–14263. doi: 10.1074/jbc.M313755200. [DOI] [PubMed] [Google Scholar]
- Tatu U, Helenius A. Interactions between newly synthesized glycoproteins, calnexin and a network of resident chaperones in the endoplasmic reticulum. J Cell Biol. 1997;136:555–565. doi: 10.1083/jcb.136.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Castillon GA, Meury A, Riezman H. The presence of an ER exit signal determines the protein sorting upon ER exit in yeast. Biochem J. 2008;414:237–245. doi: 10.1042/BJ20080715. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Funato K, Venkataraman K, Futerman AH, Riezman H. Sphingolipids are required for the stable membrane association of glycosylphosphatidylinositol-anchored proteins in yeast. J Biol Chem. 2002;277:49538–49544. doi: 10.1074/jbc.M206209200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.