The lectin Yos9p is a subunit of the HRD ligase that partakes in the degradation of aberrant glycoproteins from the ER. We demonstrate that a variant of Yos9p that has lost its ability to bind glycans enhances the elimination of non-glycosylated substrates indicating that Yos9p has an additional role in the degradation of non-glycosylated proteins.
Abstract
The HRD ubiquitin ligase recognizes and ubiquitylates proteins of the endoplasmic reticulum that display structural defects. Here, we apply quantitative proteomics to characterize the substrate spectrum of the HRD complex. Among the identified substrates is Erg3p, a glycoprotein involved in sterol synthesis. We characterize Erg3p and demonstrate that the elimination of Erg3p requires Htm1p and Yos9p, two proteins that take part in the glycan-dependent turnover of aberrant proteins. We further show that the HRD ligase also mediates the breakdown of Erg3p and CPY* engineered to lack N-glycans. The degradation of these nonglycosylated substrates is enhanced by a mutant variant of Yos9p that has lost its affinity for oligosaccharides, indicating that Yos9p has a previously unrecognized role in the quality control of nonglycosylated proteins.
INTRODUCTION
The ubiquitin-proteasome system regulates the degradation of numerous proteins that control cellular events ranging from cell-cycle progression to apoptosis. Ubiquitin-mediated protein breakdown has an additional role in protein quality control, as it eliminates polypeptides that deviate from their native fold. If not properly disposed of, misfolded proteins likely threaten the cellular homeostasis, primarily by forming toxic aggregates. Accordingly, all cellular compartments that support protein synthesis also harbor protein quality control systems to degrade unwanted polypeptides (Hirsch et al., 2006). The quality control system of the endoplasmic reticulum (ER) retains nonnative proteins in this organelle and directs polypeptides that cannot fold productively to the cytosol for proteasomal degradation. This degradation pathway is referred to as ER-associated degradation or ERAD (Hirsch et al., 2009).
Which polypeptides are typically eliminated by ER quality control? The removal of damaged proteins after stress conditions is of vital importance, as cells with defects in protein quality control are less likely to survive such insults (Casagrande et al., 2000; Friedlander et al., 2000; Travers et al., 2000). Even under normal conditions, proteins may deteriorate in the ER due to oxidative processes (Chakravarti and Chakravarti, 2007). Another source of aberrant polypeptides is protein synthesis itself, because imperfections in this process yield faulty proteins as unwanted by-products (Drummond and Wilke, 2009). Owing to size or intrinsic complications in the folding pathway, some proteins are more likely to attain aberrant conformations. Presumably, a fraction of all newly synthesized proteins fails to fold and is therefore degraded. Complete breakdown of a protein species occurs if mutations in the corresponding gene prevent productive folding. Such mutant proteins have proven to be valuable tools to investigate protein quality control. Prominent examples are the G255R derivative of the yeast carboxypeptidase Y (termed CPY*) or the ΔF508 variant of the mammalian cystic fibrosis conductance regulator (Ward et al., 1995; Knop et al., 1996). In addition, truncated proteins and orphan subunits of multiprotein complexes are also used as substrates.
The ER protein quality control system screens a highly diverse range of clients for irreversible structural defects. This pursuit is complicated by the fact that the ER is populated by nascent polypeptides that also deviate from their native fold. These immature proteins must not be degraded, as they will fold in time. Is there a feature that distinguishes aberrant proteins from nascent polypeptides? Newly synthesized glycoproteins appear to be protected from degradation by an oligosaccharide structure made of nine mannose and two N-acetyl glucosamine residues (Man9GlcNAc2) to allow their maturation (Helenius and Aebi, 2001). The slow acting ER mannosidase I voids the protective function of the sugar moiety by trimming Man9-oligosaccharides to Man8 on virtually all ER resident glycoproteins (Gemmill and Trimble, 1999). Proteins that bear a Man8GlcNAc2 glycan and display features characteristic of misfolded proteins (such as hydrophobic patches) are presumably eliminated by the ER quality control system. The first characterized step that prompts the disposal of a glycoprotein entails an additional mannosidase that processes Man8GlcNAc2 oligosaccharides, yielding Man7GlcNAc2 glycans. Although direct evidence is missing, it appears that Htm1p catalyzes this reaction (Clerc et al., 2009). How Htm1p recognizes its substrates is still enigmatic, but the notion that Htm1p interacts with Pdi1p suggests that Htm1p may process glycoproteins bound by the oxidoreductase.
Proteins that have been flagged with a Man7GlcNAc2 sugar are ubiquitylated by the HRD ligase. Five subunits of this complex have been identified: Yos9p, Hrd3p, Hrd1p, Usa1p, and Der1p. The lectin Yos9p recognizes the Man7GlcNAc2 structure generated by Htm1p (Quan et al., 2008; Hosokawa et al., 2009; Satoh et al., 2010). Yos9p interacts with Hrd3p and, apparently, both subunits cooperate to select misfolded glycoproteins for ubiquitylation (Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006a, 2006b). Current data suggest that Hrd3p interacts with misfolded domains on substrate proteins and Yos9p scans the bound polypeptide for the appropriate Man7GlcNAc2 structure (Denic et al., 2006; Gauss et al., 2006a; Carvalho et al., 2010). Polypeptides that display such a bipartite signal are ubiquitylated by Hrd1p (Bays et al., 2001; Deak and Wolf, 2001). Usa1p organizes the structure of the complex and recruits Der1p to the ligase (Carvalho et al., 2006; Horn et al., 2009). Der1p is required for the breakdown of soluble luminal substrates and few membrane proteins, but its function in this process is unknown (Knop et al., 1996; Taxis et al., 2003; Vashist and Ng, 2004; Willer et al., 2008).
How are aberrant polypeptides that lack N-glycans detected by the ER quality control system? The first soluble HRD substrate of this type was identified in mammals (Okuda-Shimizu and Hendershot, 2007). A nonglycosylated variant of the immunoglobulin (Ig) light chain interacts with the mammalian orthologues of Der1p and Usa1p, suggesting that these factors take part in the breakdown of this substrate. In yeast, nonglycosylated substrates of the HRD ligase are also coming to light. A mutant variant of the vacuolar proteinase A (ngPrA*Δ295-331) is degraded via the HRD ligase (Kanehara et al., 2010). How the ligase interacts with ngPrA*Δ295-331 is currently unknown. More information is available on the recognition of membrane-anchored proteins with structural flaws in the lipid bilayer. Such defects are recognized by the intramembrane region of Hrd1p, which queries the lipid bilayer for membrane domains with folding defects (Sato et al., 2009). This recognition mode governs the breakdown of mutant proteins such as Pdr5*, Sec61-2p, and the HMG CoA reductase Hmg2p (3-hydroxy-3-methyl-glutaryl-CoA reductase).
Polypeptides that were ubiquitylated by the HRD ligase are mobilized from the ER membrane with the help of the AAA ATPase Cdc48p, which is recruited to the HRD complex by the adapter protein Ubx2p (Neuber et al., 2005; Schuberth and Buchberger, 2005). After being released from the membrane, the aberrant proteins are proteolyzed by the proteasome. A second pathway that degrades certain unwanted transmembrane proteins and some cytosolic and nuclear polypeptides involves the membrane-embedded ubiquitin ligase Doa10p (Swanson et al., 2001).
So far, only a small number of endogenous ER quality control substrates has been reported. Therefore, ER quality control is mainly investigated using proteins that carry engineered folding defects. The limited number of substrates restricts the investigation of the ERAD pathway, because other clients may follow branches of this system that are not yet characterized. To establish additional ERAD substrates that may allow the elucidation of novel aspects in this pathway, we examined the endogenous protein clientele of the HRD complex. To this end, we identified proteins that are stabilized in yeast cells that lack Hrd1p. Among the proteins that are more abundant in this background is Erg3p, a component of the sterol biosynthesis pathway.
The yeast Saccharomyces cerevisiae harnesses more than 20 genes for the synthesis of ergosterol, the end product of the sterol pathway (Daum et al., 1998). Key steps include the initial reaction that converts AcetylCoA to HMG-CoA, which is reduced to mevalonate by the Hmg2p. Because this reaction is irreversible and rate limiting, Hmg2p activity is tightly regulated by several mechanisms (Flury et al., 2005). As described earlier in this text, Hmg2p is a target of the HRD ligase and belongs to the few characterized endogenous ER quality control substrates. Elimination of Hmg2p is triggered, however, by a physiological event and not by irreversible structural flaws. If sterols are abundantly available, Hmg2p assumes a conformation that prompts its degradation via the HRD ligase, and mevalonate synthesis subsides (Gardner and Hampton, 1999). The next step of the sterol pathway converts mevalonate to isopentenyl-pyrophosphate, the building block of the terpenoids and steroids. To produce sterol, six isoprene units are condensed to form a squalene molecule, which is cyclized into sterol. The sterol C5-desaturase Erg3p introduces a double bond into the B ring of ergosterol. This reaction depends on cytochrome b5 reductase and requires molecular oxygen (Osumi et al., 1979). The catalytically active domain of Erg3p is highly conserved. It contains a cytosolically exposed iron-binding motif composed of eight histidines, which are required for the activity of the enzyme (Shanklin and Cahoon, 1998).
RESULTS
Erg3p is an endogenous target of the HRD ligase
With the aim to extend the known client spectrum of the HRD ligase, we identified proteins that are stabilized in Δhrd1 cells by performing a SILAC analysis (stable isotope labeling by amino acids in cell culture; de Godoy et al., 2006). The strains used in the assay expressed CPY* as endogenous control and lacked Ire1p, the sensor of the unfolded protein response (UPR), to exclude the detection of proteins that are induced in response to ER stress (Ron and Walter, 2007). To reduce the sample complexity after the isotope labeling, we separated the combined cell lysates into a soluble and a membrane-associated fraction by centrifugation and subjected both fractions to SDS-gel electrophoresis, tryptic in-gel digestion, and mass spectrometric analysis. We queried the identified proteins against the gene-ontology database and selected only constituents of the secretory pathway for further analysis. In total, we found 85 proteins identified by at least two unique peptides that were significantly increased in their abundance in the Δhrd1 strain (Supplemental Table 1). Among the stabilized proteins was CPY*, confirming that the assay was functional. Another protein that displayed enhanced levels in the Δhrd1 stain was Erg3p, a constituent of the sterol pathway.
Generation of epitope-tagged Erg3p variants
At present, little is known about the properties of Erg3p. To facilitate the characterization of endogenously expressed Erg3p, we chromosomally modified ERG3 either with a C-terminal myc-tag or an N-terminal HA-epitope. We verified that the constructs integrate into the ER membrane like their wild-type (wt) counterpart (Nishino et al., 1981) by performing high-speed centrifugation of cell lysates, which confirmed that both variants reside exclusively in the particulate fraction (Supplemental Figure 1A). To analyze if the tagged Erg3p variants are biologically active, we grew the generated yeast strains in the presence of caffeine, which impairs the growth of cells that lack functional Erg3p (Dudley et al., 2005). The strain expressing C-terminally myc-tagged Erg3p behaved like control cells whereas the N-terminal HA-tag resulted in a phenotype comparable to the Δerg3 strain (Supplemental Figure 1B). On the basis of these results, we conclude that both epitope tags allow integration of Erg3p into the ER membrane, whereas only the C-terminally myc-tagged Erg3p is functional.
Erg3p is a glycoprotein with an ER luminal N terminus and a cytosolic C terminus
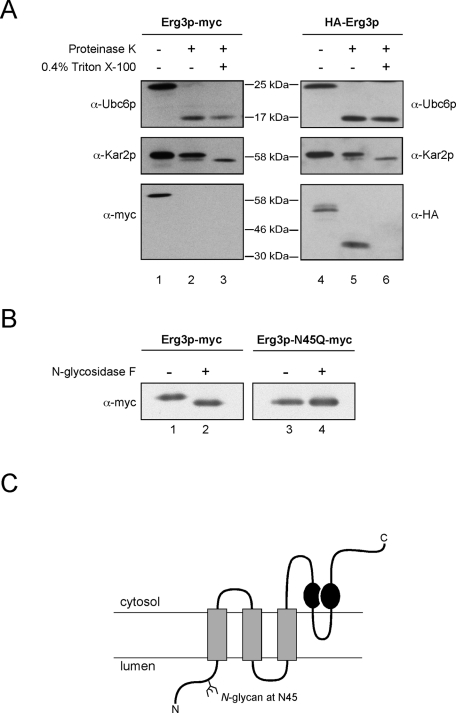
To determine the membrane topology of Erg3p, we analyzed the tagged variants in protease protection assays (Figure 1A). Addition of proteinase K to crude cell extracts efficiently removed the cytosolic domain of the control protein Ubc6p (Figure 1A, lane 2). Likewise, the myc-tag of Erg3p was also proteolyzed in the assay (Figure 1A, lane 2), demonstrating that the C terminus of Erg3p is cytosolic. The luminal control protein Kar2p remained protease-protected, unless detergent was included in the assay to solubilize the microsomes (Figure 1A, compare lanes 2 and 3). The N-terminal HA-tag of Erg3p behaved like Kar2p and resides therefore in the ER lumen.
FIGURE 1:
Membrane topology and posttranslational modifications of Erg3p. (A) A protease protection assay was carried out with extracts from yeast cells expressing either N- or C-terminally tagged Erg3p. The extracts were either left untreated or incubated with 0.3 mg/ml proteinase K. Additionally, detergent was added where indicated. Subsequently, samples were separated by SDS–PAGE and subjected to immunoblotting with the indicated antibodies. Integrity of the vesicles and activity of the protease were controlled by immunoblotting against luminal Kar2p and cytosolically exposed Ubc6p. Protease treatment increased the electrophoretic mobility of HA-Erg3p (A, lane 5). Consequently, the C terminus of Erg3p is protease accessible. (B) Erg3p-myc was immunoprecipitated from solubilized membranes expressing either Erg3p-myc or a variant termed Erg3p-N45Q-myc that carried a mutation in the potential glycosylation site. Precipitated Erg3p was treated with N-glycosidase F where indicated and analyzed by immunoblotting. (C) Predicted membrane topology of Erg3p. Gray boxes illustrate transmembrane segments; black circles indicate the positions of the histidines required for Erg3p function.
Erg3p has two potential N-glycosylation sites at position N45 and N283 (Supplemental Figure 1C). To assess whether Erg3p is N-glycosylated, we digested immunoprecipitated Erg3p-myc with N-glycosidase F. This procedure increased the electrophoretic mobility of Erg3p, demonstrating that Erg3p is a glycoprotein (Figure 1B, compare lanes 1 and 2). We generated an N45Q variant of Erg3p-myc, which migrated faster than the wt protein. Exposure to N-glycosidase did not further increase the electrophoretic mobility of Erg3p-N45Q, indicating that Erg3p is monoglycosylated (Figure 1B, compare lanes 3 and 4). Is the glycan required for Erg3p function? In the presence of caffeine, cells carrying an N45Q mutation in ERG3 grew like wt cells, demonstrating that nonglycosylated Erg3p is biologically active (Supplemental Figure 1B). We cannot rule out that the N-terminal HA-tag altered the membrane topology of Erg3p, as this variant is not functional. Our data on the glycosylation sites of Erg3p confirm the predicted membrane topology, however, because N45 is near the N terminus, which we regard as luminal, whereas N283 is in the vicinity of the cytosolic catalytic center of Erg3p (Shanklin and Cahoon, 1998). A hydropathy prediction based on a Kyte-Doolittle plot indicated that Erg3p contains three transmembrane segments ahead of the first histidine-rich motif and an additional loop between the two histidine-rich motifs (Figure 1C).
Erg3p is a substrate of the HRD ligase
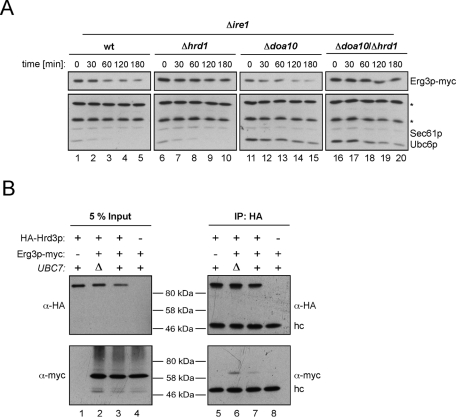
Our SILAC analysis revealed that Erg3p is more abundant in cells that lack HRD1. To exclude the possibility that transcription of ERG3 was induced in response to the HRD1 deletion, we analyzed Erg3p mRNA levels by real-time quantitative PCR, which were unchanged in Δhrd1 cells (Supplemental Figure 2). To confirm that Erg3p is a substrate of the HRD ligase, we assessed the stability of Erg3p by cycloheximide decay assays in cells that lack either Hrd1p or Doa10p and a Δhrd1/Δdoa10 strain (Figure 2A). Because the SILAC analysis was performed with IRE1-deficient cells, the strains used in this experiment also lacked IRE1 to ensure that both experiments were carried out in a similar genetic background. Consistent with the SILAC data, we observed breakdown of Erg3p-myc in the wt strain and in cells carrying a DOA10 deletion, and Erg3p turnover was severely reduced in the Δhrd1 strain (Figure 2A). Conversely, Δdoa10 cells displayed enhanced stability of the Doa10p-dependent substrate Ubc6p (Swanson et al., 2001).
FIGURE 2:
Erg3p is short-lived in the presence of Hrd1p. (A) Cells of the indicated strains expressing Erg3p-myc were treated with cycloheximide, and equal aliquots were removed at the specified time points. To analyze Erg3p-myc stability, protein extracts were separated by SDS–PAGE followed by immunoblotting. Sec61p served as loading control. The asterisk (*) indicates background bands that cross-react with the a-Ubc6 antibody. (B) Microsomes were isolated from cells expressing HA-Hrd3p and Erg3p-myc as indicated, and solubilized with NP40 lysis buffer, and HA-Hrd3p was immunoprecipitated. Samples from the total lysates (left panel) and the precipitates (right panel) were separated by SDS–PAGE followed by immunoblotting using the indicated antibodies. (Δ) denotes deletion of the respective gene. (hc) refers to the heavy chain of the a-HA antibody used for precipitation.
To assess whether Erg3p physically interacts with the HRD ligase, we precipitated HA-tagged Hrd3p and checked for copurifying Erg3p-myc. Immunoblotting revealed a weak interaction between ligase and Erg3p (Figure 2B). Because binding of substrates to the HRD complex likely triggers their degradation, interactions between ligase and client proteins are difficult to detect. To stabilize the complex between ligase and Erg3p, we deleted UBC7. In this background, substrates of the HRD complex are not ubiquitylated and interactions with the ligase are stabilized. Indeed, the signal of coprecipitated Erg3p-myc increased in Δubc7 cells (Figure 2B, compare lanes 6 and 7). Our observation that Erg3p binds to the HRD complex and the notion that this interaction is enhanced in Δubc7 cells underscore the conclusion that Erg3p is an endogenous substrate of the HRD ligase.
Degradation of Erg3p depends on Htm1p and Yos9p
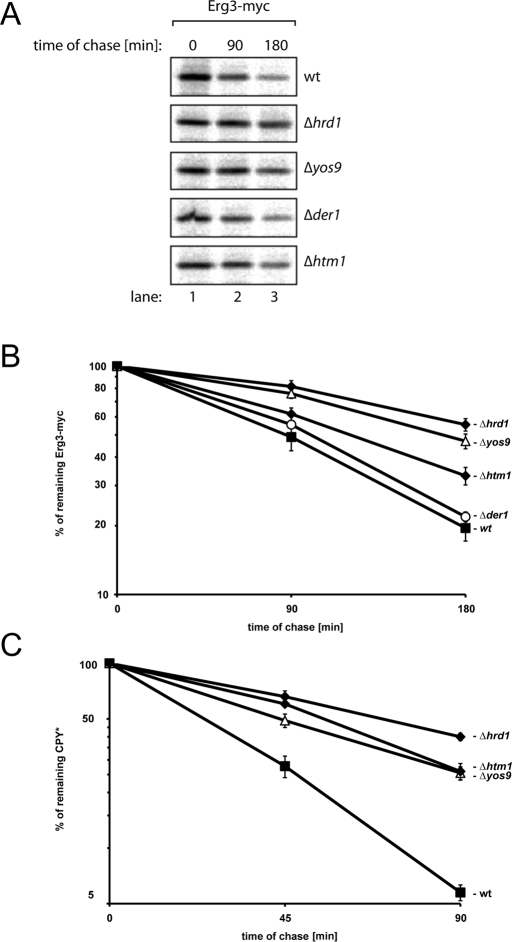
The core complex of the HRD ligase consists of Hrd1p and Hrd3p. This configuration suffices to ubiquitylate membrane-anchored proteins like Hmg2p and Sec61-2p, whereas the degradation of soluble or glycosylated substrates entails additional subunits, such as Der1p or Yos9p (Vashist and Ng, 2004; Carvalho et al., 2006). Because Erg3p is an integral membrane protein, we tested if Hrd1p directly recognizes this substrate by analyzing the stability of Erg3p-myc in Δhrd1 cells carrying plasmids that encode Hrd1p mutants that stabilize Hmg2p, Pdr5*, or Sec61-2p (Sato et al., 2009). These Hrd1p mutants were proficient in the elimination of Erg3p (Supplemental Figure 3A). Therefore we investigated whether additional factors partake in Erg3p turnover by performing pulse-chase experiments in cells lacking HTM1, YOS9, DER1, or HRD1 (Figure 3, A and B). In agreement with the notion that breakdown of most membrane proteins occurs independently of Der1p, we observed that this factor is also dispensable for Erg3p disposal. Deletion of HTM1 or YOS9 impaired Erg3p turnover, indicating that Erg3p degradation is glycan dependent. Knockout of HTM1, however, did not delay Erg3p breakdown to the extent seen in Δyos9 cells. This result was unexpected, because Htm1p and Yos9p act in the same pathway. Consequently, each deletion should stabilize Erg3p to the same extent. Furthermore, the half-life of Erg3p was highest in Δhrd1 cells. The differential effect of the individual deletions suggests that the Man7 signal generated by Htm1p is not obligatory for Erg3p breakdown and that the HRD ligase ubiquitylates Erg3p even in the absence of Yos9p. We also analyzed the stability of the established model substrate CPY* in the same setting (Figure 3C). Deletion of either HTM1 or YOS9 stabilized CPY* to the same extent, indicating that Yos9p-mediated breakdown of CPY* requires the glycan signal generated by Htm1p. Deletion of HRD1, however, stabilized CPY* further, suggesting that the HRD ligase recognizes CPY* also by an Htm1p/Yos9p- independent mechanism.
FIGURE 3:
Degradation of Erg3p requires Htm1p and Yos9p but not Der1p. (A) Exponentially growing cells of the indicated yeast strains were pulse-labeled with 35S for 15 min, and equal aliquots were removed at the specified time points. Erg3p-myc was immunoprecipitated from cell lysates, and samples were subsequently separated by SDS–PAGE. A PhosphorImager scan of a typical gel is shown. (B) Erg3p-myc degradation was analyzed in the indicated yeast strains. Five independent experiments were quantified using a PhosphorImager, and the results were averaged. The errors bars indicate the SE of the experiments. (C) Stability of CPY* was analyzed in the indicated yeast strains. At least four independent experiments were quantified for each strain using a PhosphorImager, and the results were averaged. The errors bars indicate the SE of the experiments.
HRD-dependent degradation of nonglycosylated Erg3p and CPY*
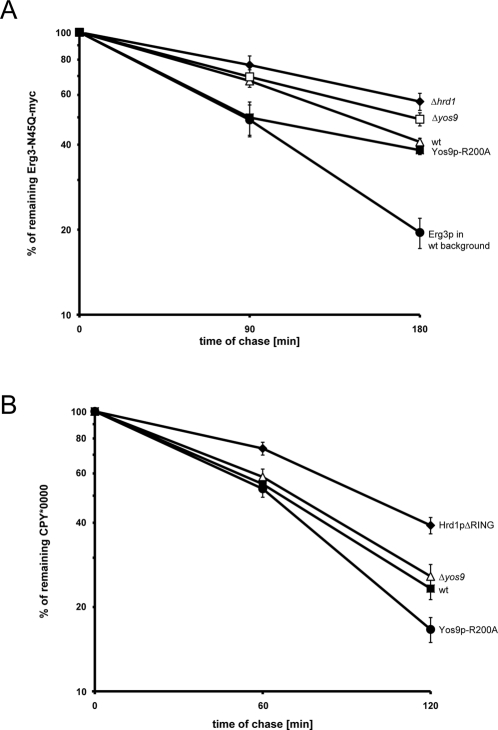
Our results imply that the Htm1p/Yos9p-dependent quality control is supported by an N-glycan–independent mechanism that also delivers Erg3p and CPY* to Hrd1p for ubiquitylation. If this were the case, the nonglycosylated variants of Erg3p and CPY* should also undergo HRD-dependent degradation. To test this hypothesis, we analyzed the stability of Erg3p-N45Q-myc by pulse-chase experiments. We observed that a considerable fraction of the expressed Erg3p-N45Q-myc was degraded in control cells (wt), whereas breakdown of this substrate was significantly delayed in the Δhrd1 strain (Figure 4A). Thus, nonglycosylated Erg3p is also a client of the HRD ligase. Because Erg3p-N45Q-myc is biologically active, we suppose that its structure is not markedly distinct from wt Erg3p. This assumption is further supported by the fact that the Hrd1p mutants, which stabilize Sec61p-2, Pdr5*, or Hmg2p, have no effect on the turnover of Erg3p-N45Q (Supplemental Figure 3B). As a consequence, the glycan-independent pathway that partakes in the turnover of glycosylated Erg3-myc should also eliminate Erg3p-N45Q. Does the lectin Yos9p have a role in the selection of nonglycosylated Erg3p, as implied by our previous findings? We compared the stability of Erg3p-N45Q-myc in a Δyos9 strain and cells expressing Yos9p-R200A chromosomally from the endogenous YOS9 promoter. The R200A mutant selectively disables the lectin function of Yos9p (Bhamidipati et al., 2005; Szathmary et al., 2005; Quan et al., 2008). Therefore, glycan-dependent protein quality control is disrupted in cells expressing Yos9p-R200A, whereas other functions of Yos9p are unaffected. Expression of Yos9p-R200A had almost no discernible effect on the turnover of Erg3p-N45Q-myc when compared with control cells. We observed a considerable stabilization when YOS9 was deleted, however. This finding suggests that Yos9p also takes part in the breakdown of nonglycosylated Erg3p.
FIGURE 4:
Elimination of nonglycosylated substrates by the ER quality control. (A) Pulse-chase experiments were performed to analyze the stability of Erg3p-N45Q-myc in the indicated strain backgrounds. Five independent experiments were quantified using a PhosphorImager, and the results were averaged. For comparison, a degradation curve of glycosylated Erg3p in control cells is included. The errors bars indicate the SE of the experiments. (B) Same as in (A), but CPY*0000 was used as substrate.
To support these results with a different substrate, we extended our investigations to CPY*0000, a variant of the mutant CPY* that lacks all four N-linked glycans found on CPY*. To avoid overexpression of the substrate, we chromosomally exchanged PRC1, the gene that encodes CPY, for the CPY*0000 variant. Although the most C-terminal glycan of CPY* is required for efficient disposal of this substrate, CPY*0000 was degraded in control cells as judged from our pulse-chase analysis (Figure 4B). Furthermore, we observed a markedly increased half-life of this substrate in cells expressing Hrd1p-ΔRING, a catalytically inactive variant of Hrd1p, indicating that CPY*0000 is also a client of the HRD ligase. Next we analyzed the stability of CPY*0000 in Δyos9 cells, which displayed a phenotype comparable to control cells. Although this finding suggested that CPY*0000 breakdown is independent of Yos9p, we also tested the stability of CPY*0000 in cells expressing Yos9p-R200A. Surprisingly, the Yos9p-R200A mutant accelerated the turnover of CPY*0000 when compared with control cells. Because Yos9p-R200A enhances the degradation of both Erg3p-N45Q and CPY*0000, we conclude that Yos9p participates in the turnover of these nonglycosylated proteins.
We also tested whether Yos9p contributes to the interaction between the substrates used in this study and the HRD ligase; however, the main binding partner for both substrates appears to be Hrd3p, as the interaction between substrate and ligase persists upon deletion of YOS9 and is most prominent in Δhrd1 cells (Supplemental Figure 4, A and B). In agreement with this observation, we have previously reported that the binding of glycosylated CPY* to the HRD ligase occurs independently of Yos9p (Gauss et al., 2006a). Because the interaction of free Man7 oligosaccharides with Yos9p is weak and best detected by frontal affinity chromatography (Quan et al., 2008; Hosokawa et al., 2009), the influence of Yos9p on the ability of the HRD complex to bind substrates is probably not detectable by coimmunoprecipitation.
The HRD ligase interacts with nonglycosylated, folding-competent CPY
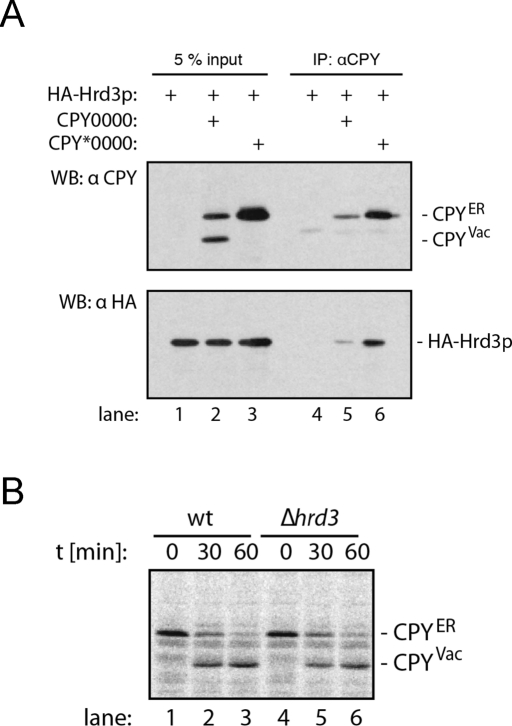
Presumably, both immature and irreversibly misfolded glycoproteins interact with Hrd3p. Proteins that display oligosaccharides other than Man7 glycans are thought to return to the ER lumen for further refolding. Thus, binding of glycoproteins to the HRD complex does not inevitably trigger their destruction. Are immature proteins that lack glycans also recruited to the HRD ligase, and, if so, what is their fate? To address these questions, we first analyzed whether CPY0000, the folding proficient variant of CPY*0000, interacts with the HRD ligase (Figure 5A). We included CPY*0000 as control protein in the experiment, because this substrate strongly binds to Hrd3p (Gauss et al., 2006a). We prepared lysates from cells expressing CPY0000 or CPY*0000 and immunoprecipitated CPY. When we tested the precipitates for the presence of HA-Hrd3p, we observed that both CPY*0000 and CPY0000 specifically interact with the HRD complex. Does binding of CPY0000 to the HRD ligase trigger its degradation? We performed pulse-chase experiments to compare the stability of CPY0000 in control cells and a strain that lacks HRD3 (Figure 5B). In control cells, we observed the classical maturation of the ER-luminal proCPY into the active protease that resides in the vacuole. Deletion of HRD3 did not increase the amount of CPY0000 that reached the vacuole, indicating that binding of CPY0000 to the HRD complex does not trigger its degradation. Therefore we conclude that the HRD ligase ubiquitylates only misfolded proteins, even if the glycan signal that informs the HRD complex about the folding state of the bound substrate is missing.
FIGURE 5:
Interaction of CPY0000 with the HRD ligase does not prompt its disposal. (A) Microsomes from cells expressing HA-Hrd3p and either CPY*0000 or CPY0000 as indicated were lysed in NP40 buffer, and CPY was precipitated. Samples of the total lysates (lanes 1–3) and the precipitates (lanes 4–6) were separated by SDS–PAGE and tested for the presence of CPY or HA-Hrd3 by immunoblotting. CPYER denotes the ER form of CPY, whereas the faster migrating vacuolar form is labeled CPYVac. (B) Stability of CPY0000 in control and Δhrd3 cells. Exponentially growing cells of the indicated yeast strains expressing CPY0000 were pulse-labeled with 35S for 10 min, and equal aliquots were removed at the specified time points. CPY was immunoprecipitated from cell lysates. Precipitates were separated by SDS–PAGE, and the gel was scanned using a PhosphorImager.
DISCUSSION
In this study we identified cellular targets of the HRD ligase. On the basis of the rationale that substrates of the HRD complex are stabilized in Δhrd1 cells, we performed a SILAC analysis to detect proteins that display increased abundance in cells that lack Hrd1p. The accumulation of aberrant proteins activates the UPR, which induces the synthesis of proteins that maintain the fidelity of the ER. To exclude UPR targets from the screen, we also deleted IRE1, the gene encoding the stress sensor that triggers the UPR (Ron and Walter, 2007). Furthermore, all strains expressed CPY*, which is constitutively degraded via the HRD ligase as endogenous control. Indeed, we identified CPY* in the screen, confirming that the approach was functional. In addition, 85 proteins of the secretory pathway exhibited protein levels that were increased. The large majority of these proteins was increased by 1.5- to 2-fold, implying that the folding machinery of the ER may be more efficient than anticipated. Alternatively, but not mutually exclusively, Δhrd1 cells may degrade aberrant proteins by different mechanisms, such as enhanced autophagy. This process is linked to ER stress and may represent a less specific alternative to the ERAD pathway (Yorimitsu and Klionsky, 2007).
Among the proteins that were more abundant in Δhrd1 cells was the C-5 sterol-desaturase Erg3p. This enzyme participates in the synthesis of an ergosterol precursor as it introduces a C-5(6) double bond into episterol in a reaction that depends on molecular oxygen (Osumi et al., 1979). From our characterization, we conclude that Erg3p has three transmembrane domains and a membrane-inserted loop. Furthermore, the C terminus is cytosolic, whereas the N terminus resides in the ER lumen. Additionally, Erg3p is posttranslationally modified by a single N-glycan that is located near the N terminus at position N45.
Based on a quantification of the pulse-chase experiments shown here, Erg3p has a half-life of 80 min, whereas CPY* is degraded with a half-life of 20 min. Because Erg3p is not misfolded by default, a high turnover rate such as that observed for CPY* and other mutant substrates would be unexpected. What causes the turnover of Erg3p? Pharmacological or genetic inhibition of the ergosterol biosynthesis pathway did not influence the stability of Erg3p (unpublished data), suggesting that the enzyme is not degraded to regulate sterol metabolism. Instead, we speculate that the oxygen-dependent reaction catalyzed by Erg3p inflicts oxidative damage on the enzyme that demands its disposal.
Degradation of Erg3p is promoted by the HRD ligase, but Doa10p does not contribute to this process. The initial finding that Erg3p is a substrate of the HRD complex is backed by our coimmunoprecipitation experiments, which demonstrate that Erg3p engages in a complex with the HRD ligase that is enhanced upon deletion of UBC7 or HRD1. Additionally, a proteomic screen found ubiquitylated peptides of Erg3p, lending further support to the conclusion that Erg3p is a substrate of the ubiquitin-proteasome pathway (Peng et al., 2003).
Recognition of Erg3p involves the glycan-specific ERAD factors Htm1p and Yos9p. Because Htm1p, Yos9p, and Hrd1p act in the same pathway, single deletions of the corresponding genes should delay the turnover of Erg3p to the same extent. By contrast, we observed three degrees of substrate stabilization: Breakdown of Erg3p was least affected by the deletion of HTM1. In Δyos9 cells, we observed an intermediate phenotype, whereas Erg3p stabilization was most prominent in cells lacking Hrd1p. These data reveal a hierarchical order among the components that facilitate the degradation of Erg3p. The modest delay resulting from the HTM1 deletion implies that the oligosaccharide structure generated by Htm1p is not mandatory for Erg3p breakdown, perhaps because Yos9p admits Erg3p for ubiquitylation even in the absence of a Man7 signal. The intermediate phenotype seen in Δyos9 cells demonstrates that the HRD complex recognizes Erg3p independently of Yos9p, because deletion of HRD1 stabilizes Erg3p even further. The pathways that operate independent of Htm1p or Yos9p are rather inefficient when compared with the glycan-mediated destruction, yet they allow the elimination of unwanted polypeptides. Because Erg3p is degraded in the absence of factors that mediate glycan-dependent quality control, we assumed that nonglycosylated Erg3p is also a target of the HRD ligase. Indeed, Erg3p-N45Q is proteolyzed in an HRD-dependent manner. Erg3p-N45Q, however, has a half-life of 140 min, indicating that removal of the N-glycan impairs Erg3p breakdown significantly.
To underscore this result with a different substrate, we included the nonglycosylated form of CPY* (termed CPY*0000) in our studies. When we expressed this protein in wt cells, we observed degradation of CPY*0000 with a calculated half-life of 90 min. Turnover of this substrate was significantly reduced upon deletion of HRD1, demonstrating that CPY*0000 is also a suitable substrate for the HRD complex.
Next we analyzed whether we could separate the Yos9p-mediated recognition of nonglycosylated substrates from its lectin function. To this end, we included a yeast strain chromosomally expressing Yos9p-R200A as the only source of Yos9p in our study. Surprisingly, cells that express Yos9p-R200A degraded Erg3p-N45Q or CPY*0000 more efficiently than did control cells. Why is the mutant form of Yos9p more efficient than wt protein? We assume that Yos9p-R200A interacts with a reduced number of client proteins, because the lectin function of Yos9p is disabled. As a result, the protein traffic at the HRD ligase may be diminished. With fewer potential substrates competing for binding to the ligase, degradation of nonglycosylated proteins is likely to become more efficient. This effect may be particularly prominent for substrates like CPY* that contact the ligase from the lumen of the ER.
How does Yos9p-R200A assist in the turnover of CPY*0000? A structural role seems plausible, assuming that Yos9p maintains the HRD ligase in an active conformation. The notion that deletion of YOS9 does not affect the elimination of HRD substrates such as ngPrA*Δ295-331 and Hmg2p argues against this possibility (Kanehara et al., 2010). Importantly, we observe that cells expressing Yos9p-R200A degrade CPY*0000 and Erg3-N45Q faster than wt cells. Therefore we suggest that Yos9p does not only recognize Man7 glycans, but also other structural cues indicative of protein misfolding. If Yos9p interacts with nonnative polypeptides independent of its lectin function must be investigated in a cell-free setting, because the epitope tagged form of Yos9p is unstable in the absence of Hrd3p (Gauss et al., 2006a).
Unlike glycoproteins, which can be flagged by a Man7 structure to indicate that they have exhausted their chances to mature, nonglycosylated proteins are not endowed with such a feature. What commits proteins that lack N-glycans for degradation? Perhaps a stochastic process selects these substrates. If the HRD complex continuously engages in transient interactions with putative substrates and only small number of these events trigger substrate degradation, newly synthesized polypeptides have sufficient time to mature. Yet, aberrant proteins are eventually degraded because they remain confined to the ER. The turnover rate we observe for CPY*0000 and Erg3p-N45Q demonstrates that the elimination of nonglycosylated proteins is indeed a rather slow process, perhaps to allow these proteins a maximum amount of time to acquire their native fold. Exemplified by CPY0000, a substrate that folds unusually slowly (Winther et al., 1991), we demonstrate that the process that selects nonglycosylated proteins for degradation is carefully fine-tuned to prevent loss of polypeptides that require more time to mature.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast-rich and minimal media were prepared as described. Gene deletions and epitope tagging in yeast were generated by homologous recombination as described (Gauss et al., 2006a). Genomic mutations were introduced by exchanging the gene of interest with a URA marker. Subsequently, the URA marker was replaced by a sequence carrying the desired mutation following selection against 5-fluoroorotic acid. All sequences were verified by DNA sequencing. Yeast strains used in this study are listed in Supplemental Table 2. Standard genetic methods were used to generate the plasmids. The pRS416-ERG3-MYC plasmid was generated by PCR amplification of the ERG3 locus from yLJ001. Plasmid mutations were introduced by a QuikChange Site Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA), yielding pRS416-ERG3-N45Q-MYC. All DNA sequences were confirmed by sequence analysis.
The plasmids used to express CPY* and CPY0000 in the coimmunoprecipitation experiment were generous gifts from Dieter Wolf (Stuttgart, Germany) and Morten Kielland-Brandt (Copenhagen, Denmark).
SILAC analysis
In yeast, Pro3p metabolizes arginine to proline at the convergent step of proline synthesis and arginine breakdown. To allow a double labeling by isotopic lysine and arginine supplementation, we generated Δpro3 strains that are auxotrophic for proline in addition to lysine and arginine (Δlys2, Δarg4). Based on mass spectrometric analysis, this strategy eliminated the Arg-to-Pro–derived isotopic peak scattering. The analysis was performed on a nanoLC-Electrospray-LTQ-Orbitrap system as described (de Godoy et al., 2006). Identification and quantification of peptides with the MASCOT and MaxQuant software packages resulted in 65,000 unique peptides distributed over 3237 protein groups with a false positive rate of <1%. To detect significant outlier ratios, the p-values “Significance A” were filtered with a threshold of p < 0.05.
Antibodies
Antibodies specific for CPY and PGK were purchased from Molecular Probes (Eugene, OR). For detection of the c-myc or HA epitopes, monoclonal antibodies 9E10 and 12CA5 were used. Maturation of CPY was followed using the antibody ab34636 from Abcam (Cambridge, UK). Antibodies against Ubc6 and Sec61 are described elsewhere (Biederer et al., 1997; Walter et al., 2001).
Membrane preparation
Established protocols were used to prepare membrane fractions from cell extracts (Biederer et al., 1996). Briefly, microsomes were separated from the cytosolic fraction by centrifugation (20,000 × g for 20 min), and the supernatants were considered as cytosolic fractions.
Protein degradation assays
Pulse-chase experiments were performed as described (Biederer et al., 1996). Analysis was performed using a Fujifilm FLA-3000 PhosphorImager. Cycloheximide decay assays were performed as described (Walter et al., 2001).
Protease-protection assay
Yeast cell extracts were prepared and incubated on ice for 10 min; 0.4% Triton-X100 was added where indicated. Subsequently, proteinase K was added to a final concentration of 0.3 mg/ml, and samples were incubated for 15 min on ice when indicated. Addition of phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 1 mM stopped the reaction, and samples were analyzed by immunoblotting.
Immunoprecipitation and immunoblotting
Precipitation and detection of proteins was performed as described (Gauss et al., 2006a).
Glycosidase F digestion
Precipitated proteins were incubated either in the presence or absence of 0.4 U N-glycosidase F (Roche, Basel, Switzerland) and 1% β-mercaptoethanol for 1 h at 37°C and analyzed by immunoblotting.
Supplementary Material
Acknowledgments
We thank Corinna Volkwein for excellent technical support and Thomas Sommer and the members of the Sommer lab for fruitful discussions. Ernst Jarosch kindly provided several yeast strains used in this study. This work was supported by a DFG (Deutsche Forschungsgemeinschaft) grant (HI 1441/3-1 to C.H.) and the EU Network of Excellence, RUBICON. H.B. is a fellow of the MDC-HU (Max Delbrück Center-Humboldt Universität) International PhD Program.
Abbreviations used:
- CPY*
carboxypeptidase Y
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- SILAC
stable isotope labeling by amino acids in cell culture
- UPR
unfolded protein response
- wt
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0832) on July 7, 2011.
REFERENCES
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande R, Stern P, Diehn M, Shamu C, Osario M, Zúñiga M, Brown PO, Ploegh H. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell. 2000;5:729–735. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- Chakravarti B, Chakravarti DN. Oxidative modification of proteins: age-related changes. Gerontology. 2007;53:128–139. doi: 10.1159/000097865. [DOI] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- de Godoy LMF, Olsen JV, de Souza GA, Li G, Mortensen P, Mann M. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 2006;7:R50. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J Biol Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol. 2005;1:2005.0001. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury I, Garza R, Shearer A, Rosen J, Cronin S, Hampton RY. INSIG: a broadly conserved transmembrane chaperone for sterol-sensing domain proteins. EMBO J. 2005;24:3917–3926. doi: 10.1038/sj.emboj.7600855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- Gardner RG, Hampton RY. A highly conserved signal controls degradation of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase in eukaryotes. J Biol Chem. 1999;274:31671–31678. doi: 10.1074/jbc.274.44.31671. [DOI] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006a;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006b;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill TR, Trimble RB. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim Biophys Acta. 1999;1426:227–237. doi: 10.1016/s0304-4165(98)00126-3. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Sommer T. Coping with stress: cellular relaxation techniques. Trends Cell Biol. 2006;16:657–663. doi: 10.1016/j.tcb.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Horn SC, Hanna J, Hirsch C, Volkwein C, Schütz A, Heinemann U, Sommer T, Jarosch E. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell. 2009;36:782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K. Human OS-9, a lectin required for glycoprotein endoplasmic reticulum-associated degradation, recognizes mannose-trimmed N-glycans. J Biol Chem. 2009;284:17061–17068. doi: 10.1074/jbc.M809725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Xie W, Ng DTW. Modularity of the Hrd1 ERAD complex underlies its diverse client range. J Cell Biol. 2010;188:707–716. doi: 10.1083/jcb.200907055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- Nishino T, Hata S, Taketani S, Yabusaki Y, Katsuki H. Subcellular localization of the enzymes involved in the late stage of ergosterol biosynthesis in yeast. J Biochem. 1981;89:1391–1396. doi: 10.1093/oxfordjournals.jbchem.a133330. [DOI] [PubMed] [Google Scholar]
- Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T, Nishino T, Katsuki H. Studies on the delta 5-desaturation in ergosterol biosynthesis in yeast. J Biochem. 1979;85:819–826. [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Chen Y, Hu D, Hanashima S, Yamamoto K, Yamaguchi Y. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol Cell. 2010;40:905–916. doi: 10.1016/j.molcel.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- Shanklin J, Cahoon EB. Desaturation and related modifications of fatty acids1. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Taxis C, Hitt R, Park S-H, Deak PM, Kostova Z, Wolf DH. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J Biol Chem. 2003;278:35903–35913. doi: 10.1074/jbc.M301080200. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Vashist S, Ng DTW. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Urban J, Volkwein C, Sommer T. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. EMBO J. 2001;20:3124–3131. doi: 10.1093/emboj/20.12.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- Willer M, Forte GMA, Stirling CJ. Sec61p is required for ERAD-L: genetic dissection of the translocation and ERAD-L functions of Sec61P using novel derivatives of CPY. J Biol Chem. 2008;283:33883–33888. doi: 10.1074/jbc.M803054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther JR, Stevens TH, Kielland-Brandt MC. Yeast carboxypeptidase Y requires glycosylation for efficient intracellular transport, but not for vacuolar sorting, in vivo stability, or activity. Eur J Biochem. 1991;197:681–689. doi: 10.1111/j.1432-1033.1991.tb15959.x. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Eating the endoplasmic reticulum: quality control by autophagy. Trends Cell Biol. 2007;17:279–285. doi: 10.1016/j.tcb.2007.04.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.