Abstract
Aims
Myo-inositol levels are frequently altered in several brain disorders. Myo-inositol 3-phosphate synthase, encoded by the Isyna1 gene, catalyzes the synthesis of myo-inositol in cells. Very little is known about the mechanisms regulating Isyna1 expression in brain and other tissues. In this study, we have examined the role of DNA methylation in regulating Isyna1 expression in rat tissues.
Materials & methods
Transfection analysis using in vitro methylated promoter constructs, Southern blot analysis of genomic DNA from various tissues digested with a methylation-sensitive enzyme and CpG methylation profiling of genomic DNA from different tissues were used to determine differential methylation of Isyna1 in tissues. Transfection analysis using plasmids harboring mutated CpG residues in the 5’-upstream region of Isyna1 was used to identify critical residues mediating promoter activity.
Results
The −700 bp to −500 bp region (region 1) of Isyna1 exhibited increased methylation in brain cortex compared with other tissues; it also exhibited sex-specific methylation differences between matched male and female brain cortices. Mutation analysis identified one CpG residue in region 1 necessary for promoter activity in neuronal cells. A tissue-specific differentially methylated region (T-DMR) was found to be localized between +450 bp and +650 bp (region 3). This DMR was comparatively highly methylated in spleen, moderately methylated in brain cortex and poorly methylated in testis, consistent with mRNA levels observed in these tissues.
Conclusion
Rat Isyna1 exhibits tissue-specific DNA methylation. Brain DNA was uniquely methylated in the 5’-upstream region and displayed gender specificity. A T-DMR was identified within the gene body of Isyna1. These findings suggest that Isyna1 is regulated, in part, by DNA methylation and that significant alterations in methylation patterns during development could have a major impact on inositol phosphate synthase expression in later life.
Keywords: CpG island, CpG methylation, epigenetics, Isyna1, mean methylation index, myo-inositol, myo-inositol 3-phosphate synthase, T-DMR, tissue-specific differentially methylated region
Myo-inositol 3-phosphate synthase (E.C. 5.5.1.4; IP synthase) is a rate-limiting enzyme that catalyzes the first step in the biosynthesis of all inositol containing compounds. It converts glucose 6-phosphate to Myo-inositol 3-phosphate. Free Myo-inositol (MI) is generated when the phosphate moiety is removed by inositol monophosphatase 1 (IMPase1). IP synthase was first purified by Maeda and Eisenberg from rat testis, the richest source of this enzyme [1]. The native mammalian enzyme is considered to be a homotrimer made up of approximately 68-kDa subunits [1]. In rats, these subunits arise by translation of a full-length mRNA, spliced from 11 exons, encoded by the Isyna1 gene (ISYNA1 in human). Recently, we and others have identified and characterized a plethora of shorter isoforms [2,3], arising by alternative splicing and intron retention mechanisms. The identification of novel isoforms in different tissues, in addition to the full-length isoform (now termed the α-isoform), suggests that the regulation of IP synthase gene expression is complex.
Myo-inositol is an ubiquitous six-carbon cyclic sugar which is an important component of membrane phospholipids and a key precursor for the phosphoinositide (PI) signaling pathway. MI can be phosphorylated at multiple positions in its ring to yield an array of inositol phosphates (IPs) and PIs that can participate in a multitude of cellular processes. Brain exhibits high levels of MI [4] which may, in part, be attributed to MI’s active role in the PI signaling pathway, which also generates the two second messengers – diacylglycerol and inositol triphosphate [5]. MI plays a critical role in the developing axons of sympathetic neurons, as IMPA1 transcripts encoding IMPase1 constitute the single most abundant transcript in these neurons [6,7]. A plethora of data indicate that alterations in brain MI levels can lead to severe behavioral problems and neurological deficits. Altered MI levels have been observed in the brains of Alzheimer’s [8], obsessive compulsive disorder [9,10] and autism spectrum disorder patients [11], as well as in suicide [12] and stroke [13] victims. High fetal MI concentrations in the cerebrospinal fluid have been attributed to the pathogenesis of Down Syndrome [14], while a deficiency is known to cause neural tube defects [15,16]. Oral administration of MI has been reported to be therapeutic for panic disorder [17]. The role of MI in bipolar disorder is an area that has garnered considerable attention and research focus. Bipolar patients in the manic phase of the disorder are often treated with lithium or valproate, drugs that are presumed to decrease free MI levels by inhibiting IMPase1 and IP synthase, respectively [18], while oral administration of MI has been reported to alleviate the depressive phase of bipolar disorder [19]. IP synthase, therefore, is a potential drug target for effecting mood stabilization.
Insights into MI metabolism and its role in brain function can be obtained through a comprehensive understanding of the processes that regulate both Isyna1 and IMPA1 expression. We have previously demonstrated that ISYNA1 is upregulated by E2F1 [20], an observation that has been confirmed in yeast [21]. More recently, we demonstrated that Isyna1 generates a number of alternatively spliced isoforms, one of which was shown to negatively modulate enzyme activity [2]. Studies presented here clearly implicate epigenetic mechanisms (DNA methylation) in the tissue-specific expression of Isyna1. These observations provide fundamental new insights into the complexity of IP synthase regulation and add a new dimension to the understanding of MI’s role in neuropsychiatric and neurological disorders.
Materials & methods
Materials
Sodium bisulfite and hydroquinone were obtained from Sigma-Aldrich Corp. (MO, USA); cell culture media were obtained from ATCC (Manassas, VA, USA) or Invitrogen (CA, USA); P32-dCTP (3000 Ci/mmol) and Microspin G-25 columns were from Amersham Biosciences (NJ, USA); PCR primers were synthesized by Integrated DNA Technologies, Inc. (IA, USA). The plasmid pFRL2 was a kind gift from Dr Richard Hodin, Massachusetts General Hospital, MA, USA [22]. Unless otherwise specified, all chemicals and reagents were of analytical grade procured from established manufacturers.
Cell culture
Cell lines SK-N-AS (CRL-2137), HEK (CRL-1573) and NTERA-2cl.D1 (CRL-1973) were obtained from ATCC and cultured according to their protocols. All cell lines were grown in the presence of 10 U/ml penicillin and 10 µg/ml streptomycin.
Animals
Adult Sprague-Dawley rats, obtained from Harlan Bioproducts for Science Inc. (IN, USA) were fed standard rat chow (Purina rat chow 5001) and water ad libitum. Animal use was approved by the Institutional Review Board of the Veterans Affairs Medical Center, KY, USA, and were conducted as per NIH guidelines. Rats were housed according to Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC) recommendations.
In vitro promoter methylation analysis
The minimal promoter of ISYNA1 in pGL3 luciferase reporter vector (Promega, WI, USA) has been extensively characterized in our laboratory [20]. A plasmid harboring the minimal promoter region (PstI/pGL-3) was methylated in vitro using HhaI, HaeIII and SssI methylases according to the supplier’s (New England BioLabs, Inc., MA, USA) protocol [20]. Methylation was ascertained by digestion with HaeIII (for plasmids methylated by HaeIII and SssI methylases) and HhaI (for the plasmid methylated by HhaI methylase) and resolving them on gels. Absence of digested products seen with the methylated plasmids and their presence with the unmethylated plasmid indicated completion of methylation. Equal amounts (2 µg/ml) of methylated and unmethylated promoter plasmids were transfected into SK-N-AS human neuroblastoma cells along with 50 ng/ml of a control vector (pRL-TK; Promega) expressing Renilla luciferase. Transfection was performed in 2 ml media containing 3 × 105 cells using FuGENE6® Reagent (Roche, IN, USA) in 6-well plates. Cells were harvested after 48 h, and firefly and Renilla luciferase activities were assayed using the dual luciferase® reporter assay system (Promega) in a Turner 20/20 luminometer. Normalized luciferase activities were then determined. The experiment was repeated thrice with duplicates and expressed as mean ± standard error of the mean. The reporter activity of the unmethylated parent plasmid was taken as 100%.
Southern blot analysis
A total of 20 µg of genomic DNA from various rat tissues (testis, brain cortex, spleen, heart and pancreas) was digested with EcoRI and HindIII, followed by digestion with SmaI and resolved on 1% agarose gels. Gels were Southern blotted onto Zeta-Probe GT membranes (BioRad, CA, USA) and probed with a full-length Isyna1 cDNA labeled with α-P32dCTP using the Prime-A-Gene (Promega) system and purified through Microspin™ G-25 columns (GE healthcare, WI, USA). Membranes were hybridized overnight and washed first in 1XSSC/0.1% sodium dodecyl sulfate for 10 min at room temperature, followed by another wash at 68°C, and finally with two washes in 0.1X SSC/0.1% sodium dodecyl sulfate for 10 min at 68°C. Membranes were placed in Saran Wrap and exposed to Fuji Medical X-Ray film (Super HR-G30) overnight in −80°C.
Bisulfite conversion, sequencing & determination of CpG methylation profiles
Approximately 10–15 µg of genomic DNA was digested with EcoRI (an enzyme that does not digest the amplicons) and subjected to the bisulfite reaction in the presence of 3.1 M sodium bisulfite, pH 5.0, and 0.5 mM hydroquinone at 55°C for 16 h under mineral oil. DNA was purified using the QIAquick PCR purification kit (Qiagen, CA, USA). DNA, after denaturation in 0.3M NaOH, was ethanol precipitated and dissolved in Tris-ethylene diamine tetra acetate buffer and used for PCR amplification using primers specific for the bisulfite-modified DNA. Primers (Table 1) were designed using the BiSearch program [23]. Each amplicon was ligated into pGEM-T Easy vector (Promega) and transformed into Escherichia coli DH5α competent cells. Approximately 10 random colonies were selected for mini-prep plasmid DNA preparation (Qiagen), sequenced at the University of Louisville Sequencing Core facility (KY, USA), and methylated CpG residues were identified and their methylation levels determined.
Table 1.
PCR primers used for CpG methylation profiling of rat Isyna1.
| Region | Primer name | CpG† | Sequence‡ |
|---|---|---|---|
| Region 1 | rM-Isyn 313 | 1–9 | GATGAGTTTAGTTGGGTTGG |
| rM-Isyn 553 | ACCTCCCCCTAAACAA | ||
| Region 2§ | rM-Isyn 611 | 11–31 | ATTAGGAGTAGGTAGGATAAT |
| rM-Isyn 930 | CTTACCRACTAAAAATTAAATCAAAC | ||
| rM-Isyn 905 | 31–43 | GTTTGATTTAATTTTTAGTY | |
| rM-Isyn 1046 | RAACTATCCACCAAAATCTC | ||
| rM-Isyn 1027 | 44–70 | GAGATTTTGGTGGATAGTTY | |
| rM-Isyn 1203 | AAAAATAAATAAAAAATAAATCR | ||
| Region 3 | rM-Isyn 1415 | 89–106 | GTGGTTGTTAGGAGGA |
| rM-Isyn 1770 | AACACCAAACCCATCC | ||
Refers to the position of CpG residues in the 1.35-kb fragment (see Figure 1).
Primers used on bisulfite-modified DNA.
CpG profile for region 2 was derived from three overlapping amplicons.
R: A/G; Y: C/T.
Mutation analysis of CpG sites in region 1 of the 5’ flanking region
To determine the effect of differential methylation in region 1 (−700 bp–−500 bp) on promoter activity, each CpG residue (CpG 1–9; Figure 1) in region 1 was subjected to mutation analysis. A 758-bp amplicon from the 5´-flanking region of rat Isyna1, terminating 72 bp upstream of the ATG start site, and containing CpG 1–9, was cloned into pGEM®-T Easy vector (Promega). CpG residues were mutated by conversion of the CG residue to AG using the QuikChange® site-directed mutagenesis kit (Stratagene, CA, USA). The mutated insert, after confirmation by sequencing, was recovered and recloned into the pFRL2 vector [22]. This vector is unique in that it carries both the firefly luciferase gene for determining the cloned promoter activity and a Renilla luciferase gene that serves as a control to normalize transfection efficiency in the same vector, obviating the need for cotransfections. Only one CpG residue was mutated per plasmid thus permitting the assessment of the effect of each specific residue on total promoter activity. The mutated and unmutated parent plasmids were transfected (as described earlier) into neuroblastoma (SK-N-AS), human embryonic kidney (HEK) and human testicular embryonic carcinoma (NTerra-2) cell lines and normalized promoter activity was determined. Cell lines were grown as described previously [20], or following ATCC protocols. Transfection was carried out at least three-times, in duplicate wells, and relative promoter activity was expressed as mean ± standard error of the mean. The promoter activity of each mutant construct was expressed as a ratio of the unmutated construct (taken as 100%). Statistical significance was computed using the unpaired two-tailed Student’s t-test using software from GraphPad Software, Inc. (CA, USA).
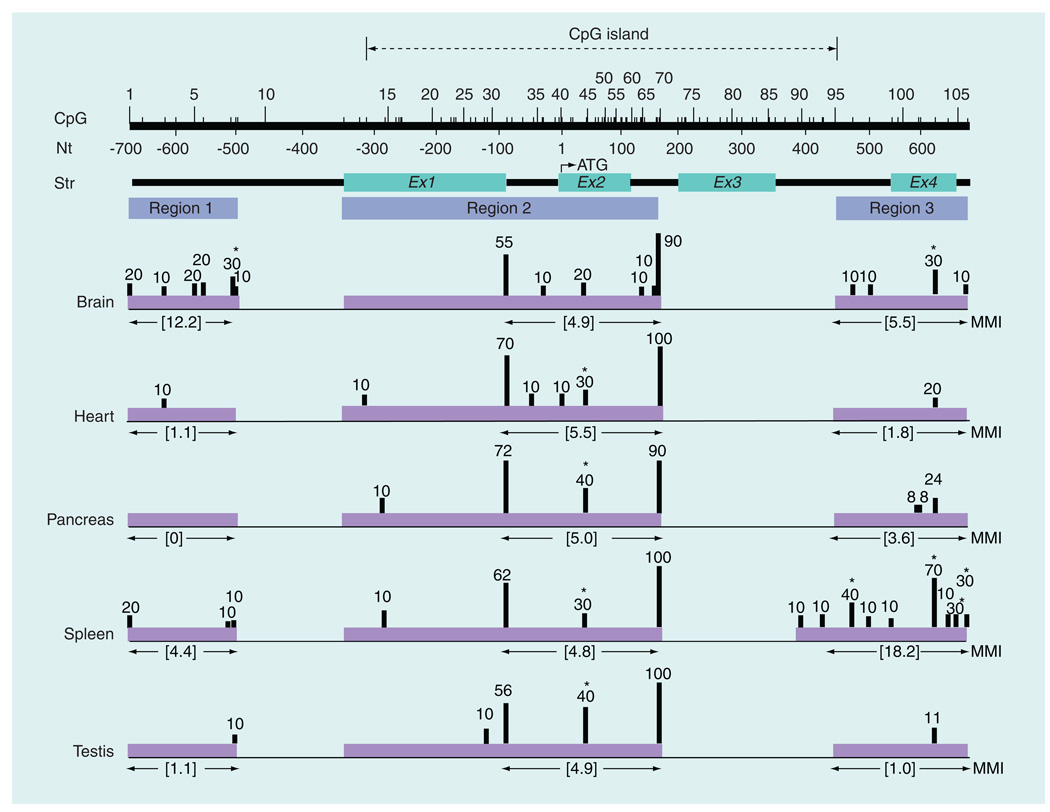
Figure 1. CpG methylation profile of rat Isyna1 in various tissues.
CpG methylation profile was determined in five (male) rat tissues – brain cortex, heart, pancreas, spleen and testis. The CpG island (dotted lines with arrowheads), number and position of each CpG residue, nucleotide position and gene structure are indicated on the top of the figure. The CpG island, determined using European Molecular Biology Open Software Suite (EMBOSS) CpGPlot [102], extends from CpG13–CpG95. Regions 1, 2 and 3 represent amplicons used for CpG analysis; region 2 is composed of three overlapping amplicons. Analyzed regions are shown in purple shaded horizontal blocks with the MMI values directly below each block. Methylation levels (%) are denoted by the height of the vertical bars and are based on the percentage of clones analyzed. For CpG31, the methylation level was averaged from two overlapping amplicons. CpG sites that are significantly differentially methylated in tissues are marked by an asterisk. Note, in particular, the pronounced differences in methylation patterns in the brain cortex in region 1 and in spleen in region 3. Except for CpG10 and CpG71–94, the status of all CpG residues spanning a 1.35-kb region from nucleotides −700 bp of the promoter to +650 bp in intron 4 was examined.
MMI: Mean methylation index; Nt: Nucleotide; Str: Structure.
Results
In vitro methylation of human ISYNA1 promoter decreases reporter activity
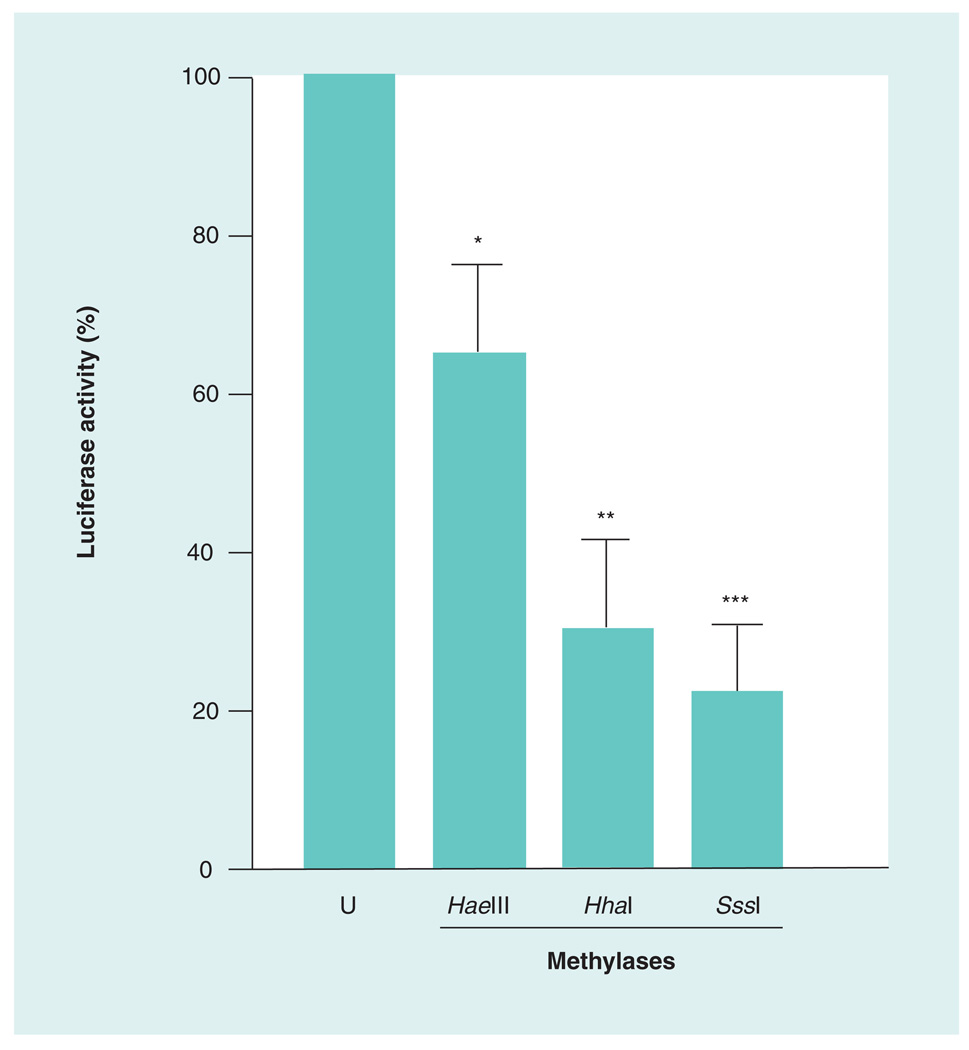
Because the rat Isyna1 gene promoter was uncharacterized, a fully characterized human ISYNA1 minimal promoter construct, available in our laboratory, was used [20]. The rat and human genes are highly conserved, including in the 5´-flanking region. Transfection analysis with the methylated and unmethylated promoter constructs indicated that promoter activity was significantly decreased upon methylation (Figure 2). The greatest decrease (~78%) was observed with SssI methylase, followed by HhaI (~70%) and HaeIII (~36%) methylases. SssI methylase methylates C residues at all CG dinucleotides whereas the HhaI and HaeIII methylases are sequence-specific and methylate the internal Cs at GCGC and GGCC motifs, respectively. The decrease in reporter activity observed with the SssI methylase is consistent with more CpG sites being available for methylation. These results not only indicate that the promoter activity of ISYNA1 can be suppressed by methylation but also that a number of CpG sites participate in the regulation of gene expression.
Figure 2. Promoter activity of ISYNA1 can be suppressed by methylation.
The minimal promoter of ISYNA1 [20] in pGL3 luciferase reporter vector (Promega, WI, USA) was methylated in vitro using HaeIII, HhaI and SssI methylases. Equal amounts of the methylated and U promoter plasmids were transfected into SK-N-AS human neuroblastoma cells along with a control vector (pRL-TK) expressing Renilla luciferase and normalized luciferase activities were determined. The experiment was repeated three-times with duplicates and the results expressed as mean ± standard error of the mean. The reporter activity of the U parent plasmid was taken as 100%. Statistical significance was computed using the unpaired two-tailed Student’s t-test using GraphPad software (GraphPad Software, Inc., CA, USA).
*p < 0.05; **p < 0.005; ***p < 0.001.
U: Unmethylated.
Differential methylation of rat Isyna1 in various tissues
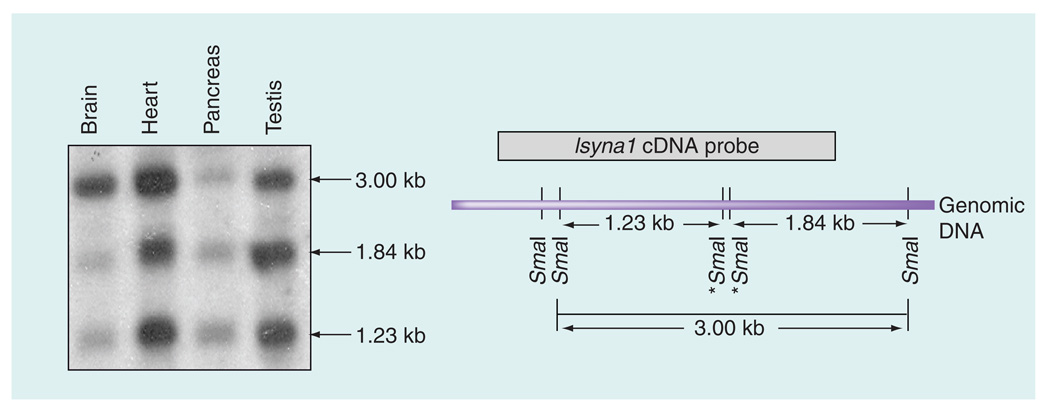
To examine if the rat Isyna1 gene is differentially methylated in various tissues, genomic DNA from four (adult male) tissues – brain cortex, heart, pancreas and testis – was isolated, digested with SmaI (which does not cleave DNA when the 3´C in its recognition sequence – CCCGGG – is methylated) and analyzed by Southern blot using a rat Isyna1 cDNA probe (Figure 3). Three major SmaI fragments were identified – a 3.0 kb partially undigested fragment and the 1.23-kb and 1.84-kb fragments derived from the complete digestion of the 3.0-kb fragment. The 3.0-kb fragment can be detected if either or both internal SmaI sites (marked by asterisks in Figure 3) are methylated and the flanking SmaI sites are unmethylated. Thus, the extent of methylation can be assessed by visual comparison of the intensity of the 3.0-kb band with that of its digested products. It is evident from the Southern blot that the internal SmaI sites are methylated to varying degrees in the four tissues analyzed. This is exemplified in the brain cortex where the internal SmaI sites appeared to be relatively highly methylated because the intensity of the two completely digested bands were barely evident compared with the 3.0-kb band. Conversely, in testis and pancreas, the digested bands were comparatively more intense than the 3.0-kb band suggesting that the internal SmaI sites are poorly methylated. The observation with the testis is consistent with this tissue expressing high levels of IP synthase [2]. In the heart, all three bands appeared equally pronounced with the 3.0-kb band slightly more intense than the two digested bands. Comparison of the intensities of the 3.0-kb fragment with that of its digested products suggests the following order of decreasing Isyna1 methylation: first, brain cortex; then the heart; followed by the pancreas/testis. These results imply that methylation of rat Isyna1 may contribute to differential gene expression in various tissues.
Figure 3. Southern blot analysis of SmaI-digested Isyna1 genomic DNA in different rat tissues.
A total of 20 µg of genomic DNA from four rat tissues (brain cortex, heart, pancreas and testis) was first digested with EcoRI and HindIII, followed by SmaI, resolved on 1% agarose gels, Southern blotted onto a Zeta-Probe GT membrane (BioRad, CA, USA) and probed with a P32-labeled Isyna1 cDNA. The SmaI restriction map of the rat Isyna1 genome is shown on the right. The 1.23-kb and 1.84-kb bands represent completely digested SmaI fragments while the 3.0-kb fragment is due to partial digestion resulting when one or both internal SmaI sites (marked by asterisks) are methylated. The probe detects 1.23-, 1.84- and the 3.00-kb fragments. Differential methylation was inferred by comparing the intensity of the 3.00 kb fragment with that of the other two restriction digest fragments on the blot. Note the striking difference in band intensities between the brain cortex (more methylated) and testis (less methylated).
Mean methylation index
We next sought to characterize the CpG methylation profiles of Isyna1 in different tissues. To compare methylation of identical CpG regions amongst different tissues, we determined the mean methylation index (MMI), a value that describes the average methylation level of CpG residues within a defined region. It is computed by dividing the sum of the methylation levels of all CpG sites present within a region by the number of CpG sites present in that region. The MMI can have a maximum value of 100 (complete methylation) or a minimum of 0 (no methylation). The methylation level of a CpG residue is the percentage of analyzed clones (~10) that harbor a methylated residue at that site. Methylation levels more than or equal to 30% were considered significant. Significant methylation levels were further characterized as being highly methylated (>60%) or moderately methylated (30–60%). Methylation levels less than 30% were considered poorly methylated and insignificant.
CpG methylation profile of rat Isyna1 in various tissues
We examined the CpG methylation status of a 1.35-kb region extending from nucleotide −700 bp in the 5´-flanking region (relative to the ATG start site) to approximately +650 bp in intron 4 in genomic DNA from five (adult male) tissues – brain cortex, heart, pancreas, spleen and testis (Figure 1). Over 250 clones were analyzed to decipher the overall CpG methylation profiles of Isyna1 in DNA from these tissues. Specifically, methylation of all but 25 (CpG10 and CpG71–94) of the 106 CpG residues spanning this region were examined. The 25 CpG residues probably reside in a region of poor sequence context generated after bisulfite conversion making them refractory to PCR amplification, a typical problem encountered in bisulfite sequencing [24]. Upon bisulfite conversion, there are a large number of C to T conversions, which end up creating long stretches of Ts. Hence, primer design programs do not always identify appropriate primer pairs for amplifying the genomic region of interest. Second, amplicons larger than approximately 300 bp are typically difficult to obtain as the harsh bisulfite treatment results in large-scale degradation [25]. Unique tissue-specific methylation signatures were apparent in the 5´-flanking region between −700 bp and −500 bp (hereafter, region 1) and in the intron 3–intron 4 region between +450 bp and +650 bp (region 3); no significant changes were observed between −350 bp and +200 bp (region 2) which, interestingly, is located within a CpG island. Methylation differences in region 1 of Isyna1 were most prominent in the brain cortex while the most pronounced differences in region 3 were apparent in spleen (see the ‘Discussion’ section for more details).
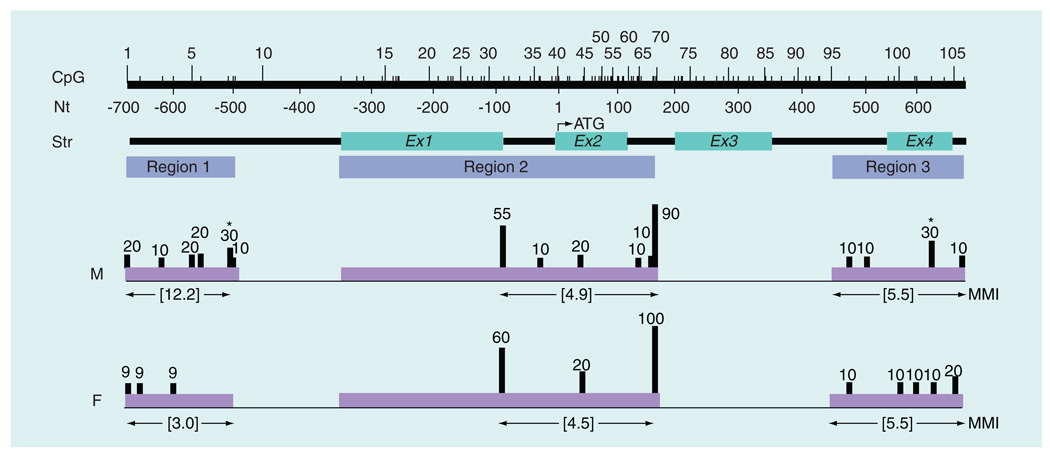
CpG methylation profile of rat Isyna1 in male & female brain cortex
As a preliminary step to identify possible gender bias (sex differences) in brain genomic DNA methylation patterns, the CpG methylation profiles of Isyna1 in age-matched male and female rat brain cortex were compared. The CpG profile of the 1.35-kb Isyna1 region from male brain cortex DNA (as shown in Figure 1) was compared with that from an age-matched adult female (Figure 4). The most pronounced difference in methylation was in region 1 of the gene. In DNA from female brain cortex, region 1 of the gene was relatively unmethylated (MMI: 3.0) compared with DNA from the male cortex (MMI: 12.2) – indeed, all the methylated residues in region 1 of the gene from female cortex were found in only one out of 11 clones analyzed. Methylation levels were essentially similar in regions 2 (MMI: 4.9 for males vs 4.5 for females) and 3 (MMI: 5.5 for both sexes) of the Isyna1 gene. CpG103 in region 3 of the gene was the lone exception being moderately methylated in DNA from male cortex (30%) but essentially unmethylated (10%) in the DNA from female cortex. Interestingly, CpG103 was highly methylated (70%) in DNA from spleen (Figure 1), the only other tissue showing significant overall methylation. These preliminary data, though speculative, indicate that the Isyna1 gene in male brain cortex is comparatively more methylated than the gene in female brain cortex in region 1 and, therefore, might harbor putative sex-specific regulatory element(s) required for IP synthase expression in this tissue.
Figure 4. Comparison of Isyna1 methylation profiles between male and female rat brain cortex.
Details are the same as in Figure 1. Methylation levels were relatively higher in region 1 in the male compared with the female, with no significant differences in regions 2 or 3. The male brain cortex also harbored two significantly methylated CpG residues (asterisks) at CpG8 and CpG103 (each 30%). CpGs31 and 70, although highly methylated, exhibited no major sex-specific differences. The CpG profile for the male brain cortex is derived from Figure 1.
F: Female; M: Male; MMI: Mean methylation index; Nt: Nucleotide; Str: Structure.
Mutation analysis of CpG residues in the 5’ flanking region of rat Isyna1
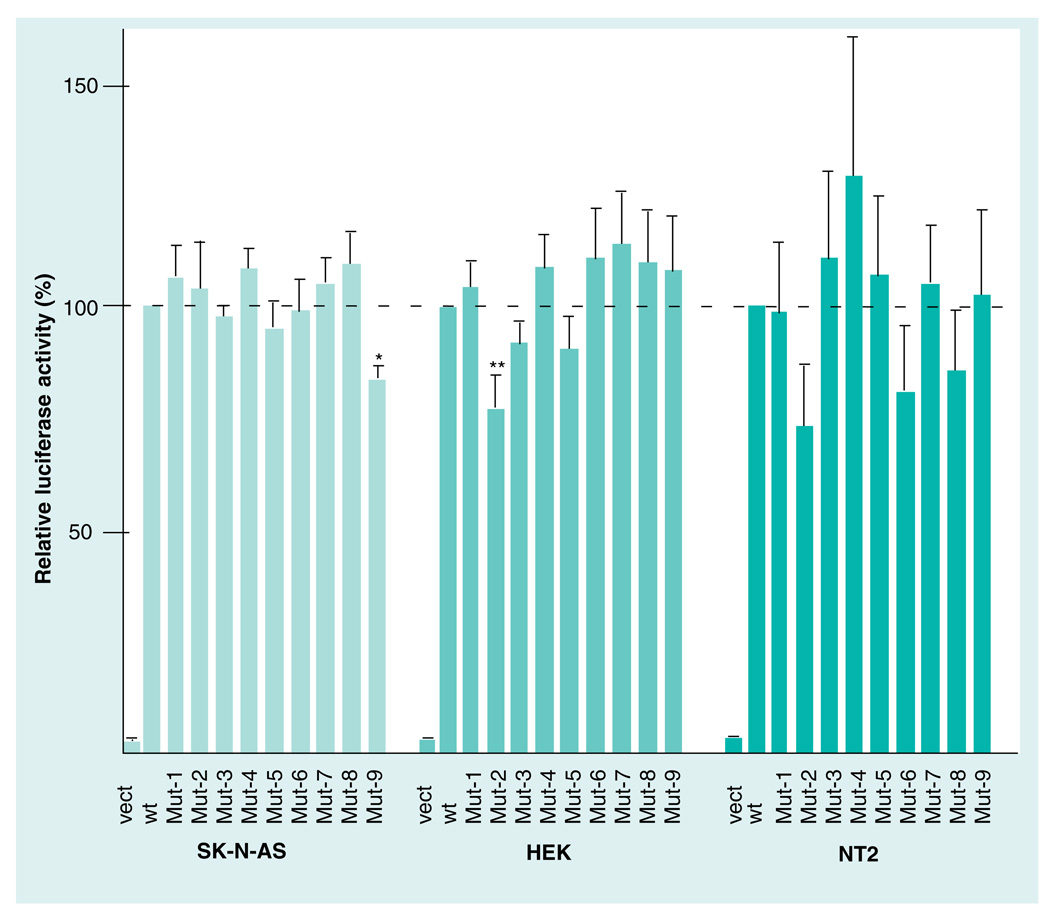
To ascertain if the increased methylation observed in region 1 of Isyna1 in male brain cortex had any functional consequences, all CpG residues in region 1 (i.e., CpG1–9; Figure 1) were individually altered by site-directed mutagenesis (CG>AG) and the effect of each mutation (Mut1–9, respectively) was examined in three different cell lines: SK-N-AS, HEK and NTerra-2 cells (Figure 5). Two CpG sites exhibited significant downregulation of promoter activity when mutated: Mut9 decreased promoter activity in the neuroblastoma cell line by 17% (p = 0.002) and Mut2 decreased activity by 23% (p = 0.01) in the kidney cell line. These results indicate that CpG9 and CpG2 may be components of distinct regulatory elements required for gene expression in neuronal and kidney cells, respectively. None of the nine mutations had any significant effect on Isyna1 promoter activity in the testis cell line.
Figure 5. Mutation analysis of CpG residues in region 1 (−700 bp– −500 bp) of rat Isyna1.
A 758-bp PCR fragment from the 5’ flanking region of Isyna1 containing CpG residues 1–9 of region 1 was cloned into the pFRL2 vector [22]. CpG residues were mutated (CG>AG) to yield the cognate single mutants, Mut-1–9. The mutants, the wt parent and the empty pFRL2 vect were transfected into SK-N-AS (human neuroblastoma), HEK and NT2 cell lines and their relative luciferase activities were determined after normalization with Renilla luciferase. The normalized promoter activity of the parent vector was set at 100% (dotted line). Mut-9 in SK-N-AS and Mut-2 in HEK cell lines decreased promoter activity to 83 and 77%, respectively. The mutations did not affect the promoter activity in the NT2 cell line. Transfections were carried out at least three-times using duplicate wells. Values are expressed as mean ± standard error of the mean. Statistical significance was computed using the unpaired two-tailed student’s t-test using GraphPad software (GraphPad Software, Inc., CA, USA).
*p = 0.002; **p = 0.01.
HEK: Human embryonic kidney; Mut: Mutation; NT2: NTerra-2 human testicular embryonic carcinoma; vect: Vector; wt: Wild-type.
Discussion
In this study, we provide evidence that the differential expression of rat Isyna1 in tissues is regulated, in part, by DNA methylation. Evidence for the role of DNA methylation comes from three different experimental approaches. First, in vitro methylation of the cognate human (ISYNA1) promoter, cloned into a luciferase reporter vector and transfected into a neuroblastoma cell line, significantly decreased reporter activity. Of the three methylases used for in vitro methylation – SssI, HaeIII and HhaI methylases – the greatest decrease (78%) was observed with SssI methylase, an enzyme that methylates all CpG residues. The remaining two enzymes which act in a sequence-specific context caused decreases of 70 and 36%, respectively (Figure 2). Because all three enzymes significantly decreased Isyna1 reporter activity to different extents, a number of CpG residues in the promoter are probably required for gene expression. Given that the rat and human genes are highly conserved, including in the 5´-flanking region (data not shown), it is likely that CpG methylation of the upstream region may also play an important role in rat Isyna1 gene expression.
Second, Southern blot analysis of SmaI-digested rat genomic DNA from four different tissues, probed with P32-labeled Isyna1 cDNA, revealed differing band intensities. As the CG dinucleotide in the SmaI recognition sequence – CCCGGG – is sensitive to methylation, the amount of digested/undigested DNA depends on the extent of methylation of the SmaI sites that are located within the gene segment detectable by the probe (Figure 3). Comparison of the intensities of the undigested and digested fragments clearly suggests that the SmaI sites located within Isyna1 are differentially methylated in the four tissues. When tissues are ranked by decreasing methylation levels, based on the degree of digestion of the 3.0-kb fragment (see ‘Results’ section), the order is: first brain cortex, second the heart, and third the pancreas/testis.
Third, CpG methylation profiling of rat Isyna1 provided direct evidence for differential methylation in tissues. CpG methylation profiling was undertaken on a 1.35-kb region, extending from −700 bp to +650 bp of the rat Isyna1 gene in genomic DNA from five different tissues isolated from a single male rat. Because CpG methylation patterns can be influenced by extraneous factors such as diet, maternal health during gestation and growth environment, we did not pool tissues from different animals; rather, we chose to compare CpG methylation patterns among different tissues from a single animal. The strength of this study, therefore, is that it allows direct comparison of tissue-specific Isyna1 methylation patterns in the same genetic background with little or no contribution from confounding factors. It also allows the identification of subtle methylation changes in the gene that may not be evident in DNA from pooled samples, especially for a housekeeping gene such as Isyna1, which is anticipated to exhibit low methylation levels overall. This is also a drawback, as the data, though derived from an apparently healthy rat, may not be representative of the population. Nevertheless, the present study provides novel insights into the epigenetic regulation of Isyna1. The analyzed region encompasses 106 CpG residues, of which the methylation levels of 81 CpG residues were successfully determined through the use of overlapping amplicons. For CpG methylation profiling, the Isyna1 gene was arbitrarily divided into three regions, based on amplicon contiguity: region 1 (−700 bp– −500 bp in the 5´-flanking region), region 2 (−350 bp–+200 bp flanking the ATG start site) and region 3 (+450 bp–+650 bp in the gene body). As anticipated, overall methylation levels for Isyna1 were expectedly low, consistent with the housekeeping nature of this gene. Tissue-specific differentially methylated regions (T-DMRs) were, however, apparent in regions 1 and 3.
For region 1, the most intriguing difference pertained to the (male) brain cortex where a relatively higher level of methylation (MMI: 12.2) of Isyna1 was observed, compared with that in all other tissues (MMI: 0–4.4; Figure 1) examined. A total of six CpG sites in region 1 were methylated in genomic DNA from brain cortex compared with three in DNA from spleen, one each in DNA from testis/heart and none in DNA from pancreas (see also Table 2). In addition to having more methylated sites, genomic DNA from brain cortex also had the only significantly methylated site (CpG8; 30%) in region 1 of the gene. The unique gene methylation pattern in DNA from the brain cortex could imply the probable presence of regulatory element(s) required for fine-tuning Isyna1 gene expression in the brain. To test this hypothesis, substitution mutations (CG>AG) were introduced into each of the 9 CpG residues present in region 1 (CpG 1–9; Figure 1) of Isyna1. The effect of each of these altered CpG sites on Isyna1 promoter activity was examined in SK-N-AS, HEK and NTerra2 cell lines (Figure 5). A significant decrease in promoter activity was observed with Mut9 in the neuronal cell line and Mut2 in the HEK cell line. None of the Isyna1 mutations had any significant effect in the testis cell line. This result is consistent with the data on the recently characterized human methylome where cell lines were observed to have different methylation and expression levels [26]. The data suggest that CpG9 may be important for brain/neural gene expression and likely defines a putative brain-specific regulatory element. CpG9, however, is poorly methylated in DNA from the brain cortex (10%; Figure 1), but is located one nucleotide away from CpG8, which is significantly methylated (30%). Methylation of CpG8 of Isyna1, therefore, could sterically hinder the binding of a transcription factor/complex to CpG9, or confer a more compact and less accessible DNA conformation [27], thereby accounting for a decrease in Isyna1 expression in this tissue. In line with this argument, elimination of CpG8 of Isyna1 by substitution mutation (Mut8) did not affect promoter activity (Figure 5), presumably because the putative transcription factor/complex can now access its binding site at CpG9 to drive promoter activity. CpGs 8 and 9 are located within a putative Sp1 binding site, CCTGCCCGCC, that matches the consensus Sp1 sequence, MSNNCCCSSC (inferred by AliBaba2.1 [101]). The methylation of region 1 of Isyna1, and CpG8 in particular, observed in normal brain cortex likely contributes to decreased MI levels. However, unlike the other tissues analyzed, regulation in the brain may be more complex because region 3 is also methylated (see below). How these two regions contribute to Isyna1 expression in brain cortex remains to be determined. On a cautionary note, we cannot rule out that the observed decrease in gene expression may be owing to the mutations disrupting a transcription factor binding site.
Table 2.
Distribution of methylated CpG residues in Isyna1.
| Tissue | Total methylated sites† |
Region 1 | Region 2 | Region 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total |
Significant sites‡ |
MMI | Total |
Significant sites |
MMI | Total |
Significant sites |
MMI | ||
| Brain M§ | 16 | 6 | CpG8 | 12.2 | 6 | CpG31, 70 | 4.9 | 4 | CpG103 | 5.5 |
| Brain F§ | 11 | 3 | 3.0 | 3 | CpG31, 70 | 4.5 | 5 | 5.5 | ||
| Heart | 8 | 1 | 1.1 | 6 | CpG31, 45, 70 | 5.5 | 1 | 1.8 | ||
| Pancreas | 7 | 0 | 0 | 4 | CpG31, 45, 70 | 5.0 | 3 | 3.6 | ||
| Spleen | 16 | 3 | 4.4 | 4 | CpG31, 45, 70 | 4.8 | 7¶ | CpG96, 103, 105, 106 | 18.2 | |
| Testis | 6 | 1 | 1.1 | 4 | CpG31, 45, 70 | 4.9 | 1 | 1.0 | ||
Number of methylated CpG sites identified in the analyzed 1.35-kb region: regions 1–3.
CpG residues with ≥30% methylation.
Brain cortex.
Nine CpGs, if the longer amplicon is considered (see Figure 1).
F: Female; M: Male; MMI: Mean methylation index.
The brain is a sexually dimorphic organ as the male and female brain follow different developmental trajectories [28]. The human male brain is masculinized and defeminized by estrogens during development, a process that is countered by α-fetoprotein in the developing female brain [28]. Male/female brain patterning could involve selective methylation of target genes to fine-tune gene expression. As an exploratory step, we sought to determine if the CpG profile of the Isyna1 gene in male brain cortex and that from an age-matched female exhibited differences in methylation signatures. As seen in Figure 4, the most salient difference was observed only in region 1, which was essentially unmethylated in the female brain cortex (MMI: 3 for female vs 12.2 for male); regions 2 and 3 were identical. Indeed, the CpG profile of region 1 in DNA from a female brain resembled that in DNA from other tissues more than it did in DNA from the male brain cortex. This indicates that the regulatory element(s) putatively identified in region 1 (discussed above) is likely associated with male-specific gene expression in the brain cortex. This intriguing observation, though highly speculative, warrants more detailed follow-up analyses.
None of the five tissues analyzed displayed notable differences in Isyna1 CpG profiles for region 2 (MMI: 4.8–5.5 for all tissues within the methylated segment used for comparison; Figure 1). Region 2 also had two of the most methylated CpG residues in Isyna1 – CpG31, bordering the exon 1/intron 1 junction, was methylated 55–72% and CpG70, in intron 2, was methylated 90–100% in all tissues. The only exception appeared to be CpG45, which was moderately methylated in all tissues (30–40%), but poorly methylated in both male and female brain cortex (20%) (Figures 1, 4 & Table 2). It is also noteworthy that region 2 of the gene, located within a CpG island, did not demonstrate tissue-specific DNA methylation differences. Interestingly, the CpG islands in rat and human genes, as determined by CpGPlot [102], are located in identical regions, extending from exon 1 to intron 3 (data not shown). Although sequence divergence is expected in the upstream regions of both genes, it would be interesting to see if similar features – namely, the lack of tissue-specific methylation differences in the CpG island and the presence of a brain-specific methylated region upstream of the CpG island – are also present in human tissues.
CpG methylation of region 3 of Isyna1 showed pronounced differences among tissues. When ranked according to decreasing levels of DNA methylation (based on region 3 MMI values), the order is: first spleen, second the brain cortex, third the pancreas, fourth the heart and last the testis. The Isyna1 gene from spleen was comparatively the most methylated (MMI: 18.2) based not only on the number of sites methylated, but also on the number of sites significantly methylated – a total of nine CpG residues (seven within the region of comparison) were methylated with four residues showing significant methylation (Figure 1 & Table 2). Indeed, the most highly methylated residue in region 3 of the gene was CpG103 (70%) in DNA from spleen, which implicates this residue in regulating expression in this tissue. By contrast, region 3 of the Isyna1 gene from testis, the tissue expressing highest levels of Isyna1 was hardly methylated (MMI: 1.0) with methylation observed at only one site, CpG103 (11%). Region 3 of Isyna1 from male brain cortex (MMI: 5.5) was ranked second in levels of DNA methylation and showed an intermediate methylation pattern with four methylated CpGs; it was also the only other tissue exhibiting significant DNA methylation (30%) at CpG103. While the total number of significantly methylated (≥30%) sites in region 3 of the Isyna1 gene was highest in DNA from spleen (four CpG sites), followed by brain cortex (one CpG site), no significant methylated residue in region 3 of the gene was observed in DNA from the remaining tissues. The CpG methylation profiles in region 3 of the gene in DNA from spleen, testis and brain cortex appear to be inversely correlated to Isyna1 mRNA expression levels in both rat and human tissues [2,29], which suggests that methylation in this region of the gene may dictate tissue-specific expression. Northern blot analysis of Isyna1 mRNA indicates that expression is highest in testis [2], moderate in brain, and poor in spleen, a pattern also seen in humans [29], which correlates extremely well with increasing MMI values of 1.0, 5.5 and 18.2, respectively, for the gene in these tissues. Interestingly, male and female brain cortex exhibited the same methylation levels in region 3 of Isyna1 (MMI: 5.5 for both sexes; Figure 4), in keeping with their brain-specific expression, as any significant deviation in methylation levels will negate the argument that these differentially methylated regions (DMRs) are associated with tissue-specificity. This observation presupposes that the region 3 T-DMR likely harbors positive regulatory element(s) necessary for increased gene expression. Methylation of these putative element(s), as seen in DNA from spleen, and to a lesser extent in that from brain cortex, may prevent a positively acting factor (activator) from binding to these elements, thereby, downregulating Isyna1 gene expression. On the other hand, in testis, in which region 3 of the Isyna1 gene is relatively unmethylated, the putative activator can presumably bind facilitating increased gene expression. However, as noted before, regulation of Isyna1 in the brain may be more complex as region 1 also appears to contribute to the moderate levels seen in this tissue. Many of these promoter-distal DMRs are also typically associated with enhancer elements [30].
Alternatively, the methylation of the T-DMR in region 3 of Isyna1 may be associated with alternative splicing. The full-length transcript of Isyna1 encodes a polypeptide (the α-isoform) that is 557 amino acids long. Recently, we identified a number of Isyna1 splice variants, one of which – a short γc-isoform – was shown to modulate IP synthase enzyme activity negatively [2]. This mRNA initiates at the normal ATG codon, in exon 2, but terminates in intron 4, encoding a polypeptide of only 148 amino acids. Interestingly, the termination site for this isoform coincides with the T-DMR in region 3 of the gene. Increased methylation of region 3, therefore, could affect splicing at the intron 4 splice donor site, facilitating the expression of the γc-isoform which would result in the negative modulation of Isyna1 expression in spleen. This observation is concordant with recent observations on the human methylome where sites of significant methylation differences were found to occur at splice junctions, suggesting an association between transcription and splicing [31]. Mutational analysis of individual CpG residues in region 3 of the Isyna1 gene cloned in the correct positional context (i.e., cloned downstream of a promoter) should permit a better understanding of the role of the T-DMR in region 3. Of note, our observations on the methylation of the region 3 DMR is in contrast with the prevailing view that increased gene body methylation correlates with increased gene expression [32]. Testis, a tissue that expresses high levels of Isyna1, shows negligible methylation in the gene body.
CpG methylation profiling of the entire analyzed region (regions 1–3) of the Isyna1 gene indicates that the gene in spleen and male brain cortex exhibits comparatively higher overall methylation as evidenced by the presence of 16 methylated sites each (Table 2), followed by the gene in female brain cortex (eleven CpG sites), heart (eight CpG sites), pancreas (seven CpG sites) and testis (six CpG sites). The Isyna1 gene in heart, pancreas and testis show identical, and insignificant, methylation levels overall. Methylation is most pronounced in region 3 of the gene in spleen, and to a lesser extent in this region in the male brain cortex. Increased methylation of region 1 of the gene is unique to male brain cortex. The comparatively higher level of methylation of region 1 of the gene in brain cortex is consistent with Southern blot data (Figure 3). While MMIs have been used to identify regions of differential methylation, it should be noted that different CpG methylation profiles could also yield identical MMI values. Therefore, the role of a specific CpG residue playing a key functional role in Isyna1 expression cannot be over-looked. Specific CpG residues in Isyna1 exhibiting significant methylation levels (>30%) in the various tissues analyzed are also identified and listed in Table 2.
Myo-inositol is found in high levels in the brain probably due to its active role in the PI signaling pathway [4]. As described elsewhere, altered MI levels are found in a range of behavioral and neurological disorders, thereby underlining its importance in brain function and maintenance. Some of these disorders manifest a gender bias that is not fully accounted for by changes in the sex chromosomes [33]. Autism [34,35], alcohol dependence [36], antisocial personality disorder [37] and attention deficit disorder [38], for instance, predominantly affect males, whereas, females are more prone to disorders such as anorexia nervosa [39], major depression [40], multiple sclerosis [41], panic disorder [42] and post-traumatic stress disorder [43]. Preliminary data from the present study indicate the possible existence of sex-specific DNA methylation patterns in regulatory regions of the Isyna1 gene, which may help elucidate some of the sex-specific differences observed in the etiology of these disorders. Significant alterations in the DNA methylation patterns of the developing pre- and post-natal brain in either sex could have occurred when exposed to environmental toxins that could conceivably explain some of these etiologies in later life. Such sex differences in brain DNA methylation patterns in genes have recently been reported [44]. This area of investigation, therefore, deserves greater in depth analyses.
The prevailing view regarding T-DMRs is that a significant majority are not likely to be located in CpG islands but at the outer edges of these islands in regions called ‘CpG island shores’ [45]. Consistent with this view, we find no significant change in DNA methylation levels in the CpG island of Isyna1 (CpG13–70; Figure 1). Regions of differential DNA methylation occur outside the island in region 1 of the gene in male brain cortex and in region 3 in all other tissues analyzed. We cannot discount the possibility that additional, and perhaps, more pronounced DMRs may exist downstream of exon 4 and/or outside the gene body in the 3´ regions. Other factors may also contribute to tissue specificity. For example, although genomic DNA from rat heart is poorly methylated in region 3 of the Isyna1 gene (Figure 1), this tissue does not manifest high mRNA levels [2,29]. It is possible that regulation in the heart may be dependent on nonepigenetic mechanisms. Tissues get inositol through three basic mechanisms: by de novo synthesis via Isyna1, by inositol transport, or by recycling mechanisms which generate free MI from the breakdown of inositol polyphosphates. It is not clear which mechanism predominates in the heart. Perhaps, DNA methylation plays a key role only in tissues that are solely dependent on the synthetic mechanism, as opposed to those that rely more on the other (recycling or transport) mechanisms. Future studies should resolve this issue.
Future perspective
This study opens up several interesting avenues for further investigation and emphasizes the use of animal models to unravel subtle changes in methylation profiles which are especially relevant for housekeeping genes that manifest low methylation levels. Future studies should examine:
-
▪
Whether sex-specific factors regulate gene expression in the brain;
-
▪
The significance of the T-DMR in region 3 of the gene in spleen;
-
▪
Whether methylation of the region 3 T-DMR promotes the synthesis of alternatively-spliced regulatory isoforms;
-
▪
Whether similar features of CpG methylation exist in the human ISYNA1 gene.
This study should provide a useful blueprint to address those questions.
Executive summary.
-
▪
Understanding Myo-inositol 3-phosphate synthase regulation can contribute to our understanding of why Myo-inositol levels are frequently altered in several brain disorders.
-
▪
This study examines whether the expression of rat Isyna1, the gene encoding Myo-inositol 3-phosphate synthase, is regulated by DNA methylation in different tissues.
Materials & methods
-
▪
Transfection analysis, Southern blot analysis, CpG methylation profiling and mutation analysis were used to determine if Isyna1 is differentially methylated in rat tissues.
In vitro methylation of human ISYNA1 promoter decreases reporter activity
-
▪
Transfection analysis utilizing plasmids that harbor in vitro methylated CpG residues in the human promoter indicate that CpG methylation decreases ISYNA1 promoter activity.
Differential methylation of rat Isyna1 in various tissues
-
▪
Southern blot analysis of genomic DNA, digested with a methylation-sensitive enzyme, indicates that the gene is differentially methylated in the brain cortex, heart, pancreas and testis in the rat. Ranked by decreasing methylation levels, the order is first the brain cortex, second heart and third pancreas/testis.
CpG methylation profile of rat Isyna1 in various tissues
-
▪
CpG methylation profiling was undertaken on a 1.35-kb region extending from −700 bp to +650 bp of the Isyna1 gene in rat tissues. Analysis was divided into three regions: region 1 (− 700 bp to −500 bp, in the 5’ upstream region), region 2 (−350 bp to +200 bp spanning the transcriptional start site) and region 3 (+450 bp to +650 bp, in the gene body).
-
▪
Region 1 of Isyna1 was comparatively highly methylated in the male brain cortex when compared with that from other tissues.
-
▪
Region 2 of Isyna1, localized within a CpG island, showed no distinctive DNA methylation changes among the various tissues.
-
▪
Region 3 of Isyna1, located within the gene body, was comparatively highly methylated in the spleen, moderately methylated in brain cortex and poorly methylated in testis, a tissue that expresses high levels of the enzyme. The reciprocal relationship observed between the methylation pattern found in this region and mRNA expression in these tissues suggests that region 3 could be a tissue-specific differentially methylated region.
CpG methylation profile of rat Isyna1 in male & female brain cortex
-
▪
A preliminary analysis of matched male and female brain cortex suggests the putative presence of a sex-specific regulatory element within region 1.
Mutation analysis of CpG residues in the 5’ flanking region of rat Isyna1
-
▪
Mutation analysis of each of the nine CpG residues present in region 1 of Isyna1 identified one CpG residue as significantly contributing to promoter activity in neuronal cells.
Discussion
-
▪
Rat Isyna1 exhibits tissue-specific DNA methylation patterns. Brain DNA was uniquely methylated in the 5’ upstream region and harbors a putative sex-specific element.
-
▪
A tissue-specific differentially methylated region was identified within the gene body of Isyna1. In contrast to the prevailing view, increased gene body methylation did not correlate with increased gene expression.
-
▪
The study lays the groundwork for future studies to examine the potential role of aberrant Isyna1 methylation in contributing to altered Myo-inositol levels observed in various brain disorders.
Acknowledgments
This work was made possible by grant #5P20RR017702 from the COBRE program of the NCRR, a component of the NIH, to Robert M Greene, grant #MH69991 from the NIH to Manuel F Casanova and by the ORD, MRS, DVA, Washington DC, USA, to Ranga N Parthasarathy.
Footnotes
Author disclosure
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of National Center for Research Resources (NCRR), NIH or Office of Research and Development (ORD)/Medical Research Service (MRS) Department of Veterans Affairs (DVA).
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1. Maeda T, Eisenberg F., Jr Purification, structure, and catalytic properties of l-myo- inositol 1-phosphate synthase from rat testis. J. Biol. Chem. 1980;255:8458–8464. ▪ Pioneering study that describes the purification and enzymatic properties of myo-inositol 3-phosphate synthase.
- 2.Seelan RS, Lakshmanan J, Casanova MF, Parthasarathy RN. Identification of myo-inositol-3-phosphate synthase isoforms: characterization, expression, and putative role of a 16-kDa γc isoform. J. Biol. Chem. 2009;284:9443–9457. doi: 10.1074/jbc.M900206200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shamir A, Shaltiel G, Mark S, Bersudsky Y, Belmaker RH, Agam G. Human MIP synthase splice variants in bipolar disorder. Bipolar Disord. 2007;9:766–771. doi: 10.1111/j.1399-5618.2007.00440.x. [DOI] [PubMed] [Google Scholar]
- 4. Harwood AJ. Lithium and bipolar mood disorder: the inositol-depletion hypothesis revisited. Mol. Psychiatry. 2005;10:117–126. doi: 10.1038/sj.mp.4001618. ▪ Excellent review that highlights mechanisms that may contribute to the therapeutic effect of inositol in the treatment of bipolar disorder.
- 5.Delmas P, Coste B, Gamper N, Shapiro MS. Phosphoinositide lipid second messengers: new paradigms for calcium channel modulation. Neuron. 2005;47:179–182. doi: 10.1016/j.neuron.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Andreassi C, Zimmermann C, Mitter R, et al. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- 7.Cosker KE, Segal RA. The longer U(T)R, the further you go. Nat. Neurosci. 2010;13:273–275. doi: 10.1038/nn0310-273. [DOI] [PubMed] [Google Scholar]
- 8.Chen SQ, Wang PJ, Ten GJ, Zhan W, Li MH, Zang FC. Role of myo-inositol by magnetic resonance spectroscopy in early diagnosis of Alzheimer’s disease in APP/PS1 transgenic mice. Dement. Geriatr. Cogn. Disord. 2009;28:558–566. doi: 10.1159/000261646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yücel M, Wood SJ, Wellard RM, et al. Anterior cingulate glutamate-glutamine levels predict symptom severity in women with obsessive–compulsive disorder. Aust. NZ J. Psychiatry. 2008;42:467–477. doi: 10.1080/00048670802050546. [DOI] [PubMed] [Google Scholar]
- 10.Whiteside SP, Port JD, Deacon BJ, Abramowitz JS. A magnetic resonance spectroscopy investigation of obsessive-compulsive disorder and anxiety. Psychiatry Res. 2006;146:137–147. doi: 10.1016/j.pscychresns.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Friedman SD, Shaw DW, Artru AA, Dawson G, Petropoulos H, Dager SR. Gray and white matter brain chemistry in young children with autism. Arch. Gen. Psychiatry. 2006;63:786–794. doi: 10.1001/archpsyc.63.7.786. [DOI] [PubMed] [Google Scholar]
- 12.Shimon H, Agam G, Belmaker RH, Hyde TM, Kleinman JE. Reduced frontal cortex inositol levels in postmortem brain of suicide victims and patients with bipolar disorder. Am. J. Psychiatry. 1997;154:1148–1150. doi: 10.1176/ajp.154.8.1148. [DOI] [PubMed] [Google Scholar]
- 13.Mader I, Rauer S, Gall P, Klose U. (1)H MR spectroscopy of inflammation, infection and ischemia of the brain. Eur. J. Radiol. 2008;67:250–257. doi: 10.1016/j.ejrad.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Beacher F, Simmons A, Daly E, et al. Hippocampal myo-inositol and cognitive ability in adults with Down syndrome: an in vivo proton magnetic resonance spectroscopy study. Arch. Gen. Psychiatry. 2005;62:1360–1365. doi: 10.1001/archpsyc.62.12.1360. [DOI] [PubMed] [Google Scholar]
- 15.Copp AJ, Greene ND. Genetics and development of neural tube defects. J. Pathol. 2010;220:217–230. doi: 10.1002/path.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris MJ, Juriloff DM. Mouse mutants with neural tube closure defects and their role in understanding human neural tube defects. Birth Defects Res. A Clin. Mol. Teratol. 2007;79:187–210. doi: 10.1002/bdra.20333. [DOI] [PubMed] [Google Scholar]
- 17.Palatnik A, Frolov K, Fux M, Benjamin J. Double-blind, controlled, crossover trial of inositol versus fluvoxamine for the treatment of panic disorder. J. Clin. Psychopharmacol. 2001;21:335–339. doi: 10.1097/00004714-200106000-00014. [DOI] [PubMed] [Google Scholar]
- 18.Vaden DL, Ding D, Peterson B, Greenberg ML. Lithium and valproate decrease inositol mass and increase expression of the yeast INO1 and INO2 genes for inositol biosynthesis. J. Biol. Chem. 2001;276:15466–15471. doi: 10.1074/jbc.M004179200. [DOI] [PubMed] [Google Scholar]
- 19.Levine J, Barak Y, Gonzalves M, et al. Double-blind, controlled trial of inositol treatment of depression. Am. J. Psychiatry. 1995;152:792–794. doi: 10.1176/ajp.152.5.792. [DOI] [PubMed] [Google Scholar]
- 20.Seelan RS, Parthasarathy LP, Parthasarathy RN. E2F1 regulation of the human myo-inositol 1-phosphate synthase (ISYNA1) gene promoter. Arch. Biochem. Biophys. 2004;431:95–106. doi: 10.1016/j.abb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Takaya T, Kasatani K, Noguchi S, Nikawa J. Functional analyses of immediate early gene ETR101 expressed in yeast. Biosci. Biotechnol. Biochem. 2009;73:1653–1660. doi: 10.1271/bbb.90162. [DOI] [PubMed] [Google Scholar]
- 22.Malo MS, Abedrapo M, Chen A, Mozumder M, et al. Improved eukaryotic promoter-detection vector carrying two luciferase reporter genes. Biotechniques. 2003;35:1150–1154. doi: 10.2144/03356bm05. [DOI] [PubMed] [Google Scholar]
- 23.Tusnady GE, Simon I, Varadi A, Aranyi T. BiSearch: primer-design and search tool for PCR on bisulfite-treated genomes. Nucleic Acids Res. 2005;33:e9. doi: 10.1093/nar/gni012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS ONE. 2010;5(9) doi: 10.1371/journal.pone.0013100. pii e13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:e65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcourt L, Cordier C, Couesnon T, Dodin G. Impact of C5-cytosine methylation on the solution structure of d(GAAAACGTTTTC)2: an NMR and molecular modelling investigation. Eur. J. Biochem. 1999;265:1032–1042. doi: 10.1046/j.1432-1327.1999.00819.x. [DOI] [PubMed] [Google Scholar]
- 28. Bakker J, De Mees C, Douhard Q, et al. α-fetoprotein protects the developing female mouse brain from masculinization and defeminization by estrogens. Nat. Neurosci. 2006;9:220–226. doi: 10.1038/nn1624. ▪▪ Significant study that describes the role of prenatal estrogens in determining the developmental fate of male and female brains.
- 29.Guan G, Dai P, Shechter I. cDNA cloning and gene expression analysis of human myo-inositol 1-phosphate synthase. Arch. Biochem. Biophys. 2003;417:251–259. doi: 10.1016/s0003-9861(03)00388-6. [DOI] [PubMed] [Google Scholar]
- 30.Schmidl C, Klug M, Boeld TJ, et al. Lineage- specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19:1165–1174. doi: 10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurent L, Wong E, Li G, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20:320–331. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. Erratum: 27, 485. ▪▪ High-throughput methylation profiling study that shows that gene body methylation is a general feature of all highly expressed genes in human cells.
- 33. Cahill L. Why sex matters for neuroscience. Nat. Rev. Neurosci. 2006;7:477–484. doi: 10.1038/nrn1909. ▪▪ Well-written review that highlights sex differences in the brain and their contribution to a variety of brain disorders.
- 34.Mitka M. Rising autism rates still pose a mystery. JAMA. 2010;303:602. doi: 10.1001/jama.2010.113. [DOI] [PubMed] [Google Scholar]
- 35.Autism and Developmental Disabilities Monitoring Network Surveillance Year 2006 Principal Investigators; Centers for Disease Control and Prevention: Prevalence of autism spectrum disorders – autism and developmental disabilities monitoring network, United States, 2006. MMWR Surveill. Summ. 2009;58:1–20. Erratum 2010, 59: 956. [PubMed] [Google Scholar]
- 36.Hasin DS, Stinson FS, Ogburn E, Grant BF. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States. Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch. Gen. Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- 37.Raine A, Yang Y, Narr KL, Toga AW. Sex differences in orbitofrontal gray as a partial explanation for sex differences in antisocial personality. Mol. Psychiatry. 2011;16(2):227–236. doi: 10.1038/mp.2009.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sassi RB. Attention-deficit hyperactivity disorder and gender. Arch. Womens Ment. Health. 2010;13:29–31. doi: 10.1007/s00737-009-0121-2. [DOI] [PubMed] [Google Scholar]
- 39.Støving RK, Andries A, Brixen K, Bilenberg N, Hørder K. Gender differences in outcome of eating disorders: a retrospective cohort study. Psychiatry Res. 2011;186:362–366. doi: 10.1016/j.psychres.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Kessler RC, Berglund P, Demler O, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 41.Brahmachari S, Pahan K. Gender-specific expression of β1 integrin of VLA-4 in myelin basic protein-primed T cells: implications for gender bias in multiple sclerosis. J. Immunol. 2010;184:6103–6113. doi: 10.4049/jimmunol.0804356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kessler RC, Chiu WT, Jin R, Ruscio AM, Shear K, Walters EE. The epidemiology of panic attacks, panic disorder, and agoraphobia in the National Comorbidity Survey Replication. Arch. Gen. Psychiatry. 2006;63:415–424. doi: 10.1001/archpsyc.63.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lilly MM, Pole N, Best SR, Metzler T, Marmar CR. Gender and PTSD: what can we learn from female police officers? J. Anxiety Disord. 2009;23:767–774. doi: 10.1016/j.janxdis.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurian JR, Olesen KM, Auger AP. Sex differences in epigenetic regulation of the estrogen receptor-α promoter within the developing preoptic area. Endocrinology. 2010;151:2297–2305. doi: 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. ▪▪ Landmark paper that demonstrates that tissue specific regulatory regions are located outside CpG islands and promoters in regions termed ‘CpG island shores’.
Websites
- 101.Gene regulation. www.gene-regulation.com. [Google Scholar]
- 102.EMBOSS CpGPlot. www.ebi.ac.uk/Tools/emboss/cpgplot/index.html.