Abstract
Post-translational methylation of arginine residues profoundly affects the structure and functions of protein and, hence, implicated in a myriad of essential cellular processes such as signal transduction, mRNA splicing and transcriptional regulation. Protein arginine methyltransferases (PRMTs), the enzymes catalyzing arginine methylation have been extensively studied in animals, yeast and, to some extent, in model plant Arabidopsis thaliana. Eight genes coding for the PRMTs were identified in Oryza sativa, previously. Here, we report that these genes show distinct expression patterns in various parts of the plant. In vivo targeting experiment demonstrated that GFP-tagged OsPRMT1, OsPRMT5 and OsPRMT10 were localized to both the cytoplasm and nucleus, whereas OsPRMT6a and OsPRMT6b were predominantly localized to the nucleus. OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b and OsPRMT10 exhibited in vitro arginine methyltransferase activity against myelin basic protein, glycine-arginine-rich domain of fibrillarin and calf thymus core histones. Furthermore, they depicted specificities for the arginine residues in histones H3 and H4 and were classified into type I and Type II PRMTs, based on the formation of type of dimethylarginine in the substrate proteins. The two homologs of OsPRMT6 showed direct interaction in vitro and further titrating different amounts of these proteins in the methyltransferase assay revealed that OsPRMT6a inhibits the methyltransferase activity of OsPRMT6b, probably, by the formation of heterodimer. The identification and characterization of PRMTs in rice suggests the conservation of arginine methylation in monocots and hold promise for gaining further insight into regulation of plant development.
Introduction
Post-translational methylation at arginine significantly influences the structure and functions of affected protein, by changing the bulkiness and hydrophobicity of modified residues, and hence modulates a myriad of essential biological processes, including transcriptional regulation, RNA metabolism, DNA repair, signal transduction, protein sorting, and apoptosis etc (reviewed in [1]–[6]). During this reaction methyl group is transferred from S-adenosyl-L-methionine (SAM), a universal methyl donor, to the arginines in the substrate proteins. The family of enzymes catalyzing the methylation at arginine residues is called protein arginine methyltransferases (PRMTs). Protein arginine methylation was reported in 1966, but, the first gene coding for the arginine methyltransferase enzyme was discovered only in 1996 (reviewed in [7]). Following their discovery, emphasis has been given to characterize PRMTs in different organisms, in order to understand their important biological functions. Hitherto, 11 PRMT members, differing in sequences and substrate specificities, have been characterized in humans [1]; however, the molecular mechanisms through which they regulate cellular processes are still largely unknown.
Arginine residue contains 3 nitrogen atoms which can replace their 5 hydrogen atoms and form different methylated arginines [3]. Based on the formation of different methylarginines, protein arginine methyltransferases can be divided into four types [8]. Type I PRMTs, including PRMT1, PRMT2, PRMT3, PRMT4/coactivator-associated arginine methyltransferase 1 (CARM1), PRMT6, PRMT8 and RMT1, catalyze the formation of ω-NG-monomethyl arginine (MMA) and asymmetric ω-NG, NG-dimethylarginine (aDMA). Type II PRMTs result in the formation of monomethyl arginine and symmetric ω-NG, N′G-dimethylarginine (sDMA), this class includes PRMT5 and Saccharomyces cerevisiae histone synthetic lethal 7 (Hsl7) [9]. Type III PRMTs form only ω-NG-monomethyl arginine; Trypanosoma brucie homolog of human PRMT7, TbPRMT7 displays this type of activity [10]. Type IV enzymes catalyze the formation of δ-NG-monomethyl arginine and PRMT2 (RMT2) from Saccharomyces cerevisiae and Candida albicans has been found to harbor this activity so far [11], [12].
PRMTs methylate a large number of essential proteins, most of them are either RNA binding proteins or involved in transcription [4], [5], [13]. PRMT1 is the main arginine methyltransferase, responsible for at least 85% of all the PRMT activity in mouse [14], which catalyses the transfer of methyl group to a wide variety of substrates, including histones as well as non-histone proteins [2]–[4], [15], [16]). PRMT1 plays a critical role in early mouse development since the growth of PRMT1-null embryo was arrested shortly after implantation [14]. Human PRMT2 methylates histone H4 [17] and heterogeneous nuclear ribonucleoprotein E1B-AP5 [18] and acts as a co-activator of the androgen and estrogen receptors [19], [20]. PRMT3 catalyzes the methylation of ribosomal protein S2 (rpS2) [21], inhibits its ubiquitination [22] and regulates ribosome biosynthesis at a stage beyond pre-rRNA processing [23]. PRMT4/CARM1 regulates many cellular processes (reviewed in [2], [3]) including gene expression and pre-mRNA splicing [24], [25] and PRMT4/CARM1-null mice were shown to be deficient in some aspects of estrogen-responsive gene expression [26]. PRMT5 plays an important role in spliceosome assembly by methylating sphingomyelin (Sm) and survival of motor neuron (SMN) [27]. It regulates interleukin 2 (IL-2) gene expression [28] and is required for myogenesis because it facilitates ATP-dependent chromatin remodeling [29]. PRMT6 mediated methylation plays role in defense against HIV1 by regulating the HIV1 gene expression [30]. PRMT7 methylates histones, myelin basic protein, Glycine-Arginine-Rich domain (GAR), and spliceosomal protein SmB. PRMT7 was reported to probably playing role in male germ line imprinted gene methylation [31]. PRMT8 remains associated with membranes due to myristoylation and has an adopter role for nuclear proteins at the cell membrane in addition to its methyltransferase activity [32]. The protein encoded by the genes 4q31and 9p13.2 have been named as human PRMT9 and PRMT10, however, they remain to be characterized.
Since they regulate many important biological processes in animals and yeast, great prominence has been given to explore the significance of PRMTs in model plant Arabidopsis thaliana, recently. Arabidopsis thaliana genome contains two homologs of human PRMT1; a major type I PRMT, AtPRMT1a and AtPRMT1b. AtPRMT1a and AtPRMT1b methylate the RNA methyltransferase, fibrillarin and histone H4 at R3 in vitro [33]. Moreover, AtPRMT1b (PRMT11) interacts with methyl-DNA-binding protein 7 (MBD7) [34], however, neither single nor double mutant of AtPRMT1a and AtPRMT1b show any visible phenotype under long day conditions (unpublished data, Yong Zhang and Xiaofeng Cao). AtPRMT4a and AtPRMT4b are the Arabidopsis homologs of human PRMT4/CARM1, which methylate histone H3 at R2, R17 and R26 in vitro and are required for methylation at H3R17 in vivo. Their double mutant shows an FLC-dependent late flowering phenotype [35]. AtPRMT5/(Shk1binding protein 1) Skb1 is a type II PRMT, resulting in the formation of symmetric dimethylarginines in the substrate proteins; and its knockout mutants exhibit pleiotropic phenotype including retarded growth, dark green and curly leaves and FLC-dependent late flowering [36], [37]. Lines of evidences show that arginine methylation mediated by PRMT5 is required for normal pre-mRNA splicing both in plants and animals [38], [39] and that the late flowering phenotype of atprmt5-1 and atprmt5-2 might be due to the splicing defects in the RNA processing-related flowering time regulators [38]. It has also been shown that the FLC expression is up-regulated by the splicing defects in FLK in the atprmt5 mutant. It was reported recently that PRMT5 is a critical determinant of circadian period in Arabidopsis and Drosophila [39], [40] and it probably links the circadian cycle to the alternative splicing [39]. Furthermore, PRMT5/Skb1 regulates transcription and pre-mRNA splicing by symmetrically dimethylating histone H4R3 and small nuclear ribonucleoprotein LSM4 and confers high salt tolerance in Arabidopsis [41]. Another plant-specific type I PRMT, AtPRMT10 was shown to asymmetrically dimethylate histone H4 at R3 and regulate the Arabidopsis flowering time in an FLC-dependent manner [42].
Oryza sativa (rice), a representative of the monocots, is one of the most important cereals, feeding more than half of the world population and has the smallest genome in the cultivated cereals. In this study, we aligned the amino acid sequences of eight OsPRMT genes and found that they contain the conserved PRMT signature motifs. We compared the expression patterns, subcellular localization, in vitro enzymatic activity, and specificity of the Oryza sativa PRMT homologs for the arginine residues in histones H3 and H4. Our study suggests that, like animals and Arabidopsis, this conserved family of PRMTs may play diverse roles in transcriptional regulation and other vital cellular processes by methylating histones and non-histone proteins in vivo; studying the knockdown and knockout mutants will further strengthen this notion.
Materials and Methods
Amino acid sequence alignment and chromosomal distribution of the OsPRMTs
The amino acid sequences of the eight putative OsPRMT genes reported earlier were aligned in the multiple sequences alignment tool CLUSTALX1.83, with default parameters. The conserved PRMTs signature motifs were identified and edited manually with GeneDoc software.For finding the percent identity/similarity to the Arabidopsis PRMTs, the amino acid sequences of OsPRMTs were aligned in BLAST (bl2seq) at the NCBI website. Chromosomal distribution of these genes was determined by making use of the NCBI mapviewer tool on the NCBI website http://www.ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=4530.
RNA extraction, cDNA synthesis and relative gene expression patterns analysis
Total RNAs were isolated from different tissues, namely root, shoot, young leaf, mature leaf, flower and whole seedlings of the wild type Oryza sativa L. ssp. Japonica, Nipponbare plants using TRNzol reagent (TIANGEN, DP405-02) according to the manufacturer's instructions. RNAs were reverse transcribed using TransScript II reverse transcriptase (TransGen, AH301-02) to make the first strand of the complementary DNAs, as per manufacturer's instructions. These cDNAs were then used in the quantitative real time PCR (BIO-RAD, CFX96) using the gene specific primers; CX4699 (GACTCCTACTCCCACTTCGG) and CX4700 (GAGAACACTCAATCGCATAGACAT) for OsPRMT1, CX5008 (GGAACCCTCTTGGTGAATG) and CX5009 (GAAACGTAGGCGGAGAAAC) for OsPRMT4, CX4282 (GGGAAGATCAGTGAGTGGATT) and CX4283 (AGCATAGTTGGCACAAGAAGA) for OsPRMT5, CX4693 (AGTGCGCCAGAGATCCAAGAAG) and CX4694 (GATCTCTAGTAAAATATTTC) for OsPRMT6a, CX4695 (CTTCAAATCCCACAGAC) and CX4696 (CCGTGGTGCAATCTAGTTG) for OsPRMT6b, CX4697 (GCTATTATTTCATGGTGGGTACTTC) and CX4698 (GAGATGCTGGTTTGGTTATGTC) for OsPRMT7, and CX4284 (CGTCGAGGTGATACAGGG) and CX4285 (TAGCCACATCCGAGCAT) for OsPRMT10) to compare the expression of OsPRMTs in these tissues; the ACTIN gene (primers CX0909 (CCAATCGTGAGAAGATGACCCA) and CX0910 (CCATCAGGAAGCTCGTAGCTCT)) was used as internal control. Note: all the primer sequences given are in the 5′ to 3′ order.
Construction of prokaryotic (E.coli) expression vectors
Constructs for prokaryotic expression of recombinant GST-fusion OsPRMT1, OsPRMT4, OsPRMT6a, OsPRMT6b, and OsPRMT10 were made by molecular cloning methods. Coding sequences of the OsPRMT1, OsPRMT4, OsPRMT6a, OsPRMT6b, and OsPRMT10 were amplified from the cDNAs of Oryza sativa L. ssp. Japonica, Nipponbare strain using the following primer pairs: CX0494 (CTCGAATTCATGGATCAGCGCAAGGGCAGCGGCAGCGAC) and CX0495 (CTCGAATTCTCTCATATCGGCAGGCCAGTTC) for GST-OsPRMT1, CX0500 (CGCGGATCCATGGCGTCGCCGGACCTGTTCCCGAAC) and CX 0501 (CTCGAATTCTAGTGAAGGCCGCCGAGTCTTG) for OsPRMT4, CX0496 (CGCGGATCCATGTTCGCCGGCGGCGCGGATGGCGGCAA) and CX0497 (CTCGAATTCACCTATCCCACCCTTGGTCGGC) for GST-OsPRMT6a, CX3254 (GTCGGATCCATGCTGCCGTCGCACCTCAAC) and CX3255 (GTGCTCGAGTCATCGCATGGCATAATCT) for GST-OsPRMT6b and CX0696 (ATAGGATCCATGGCGTCCCTCCCCAACGG) and CX0697 (CGCGAATTCGGTAGTATTACACAATGTGGCC) for GST-OsPRMT10. The PCR products were separated on 1% agarose gel, purified and digested with EcoRI (For OsPRMT1), BamHI and EcoRI (for OsPRMT4, OsPRMT6a and OsPRMT10), and BamHI and XhoI (for OsPRMT6b). Vector pGEX-4T-2 (Amersham Biosciences) was digested simultaneously with respective restriction enzymes and the digested and purified coding sequences of the OsPRMTs were ligated into it, to construct the Escherichia coli expression vectors. The GST-fusion OsPRMT5 E. coli expression vector was generated by the ligation independent cloning (LIC) strategy [43]; primers CX2492 ( TACTTCCAATCCAATGCGATGCCGCTGGGGCAACGCGCGGGGG) and CX2493 ( TTATCCACTTCCAATGCGCTATTATAGGCCGACCCAATATGATCG) were used to amplify the coding sequence of OsPRMT5.
In order to generate the MBP (maltose binding protein)-tagged OsPRMT6a, OsPRMT6a fragment amplified with the above stated primers and vector pMal-2×His-TE were digested with the BamHI and EcoRI and ligated with T4 DNA ligase (New England BioLabs, M0202S). Note: all the primers sequences given are in the 5′ to 3′ order.
Construction of eukaryotic (plant) expression vectors
Binary vectors for the expression of GFP-fusion OsPRMT1, OsPRMT5, OsPRMT6a, OsPRMT6b, and OsPRMT10 in plant were made by following the conventional molecular cloning methods. Details of the primers are as: CX4334 (GGAAGATCTATGGATCAGCGCAAGGG) and CX4335 (GCGTCTAGAATCACGGGAACAAGCAC) for GFP-OsPRMT1, CX0500 and CX4336 (GCGTCTAGACTATCTAGTGAAGGCCGCC) for GFP-OsPRMT4, CX4337 (GTGCCATGGAGATGCCGCTGGGGCAACGCGC) and CX4338 (GTGCCATGGATAGGCCGACCCAATATGATC) for GFP-OsPRMT5, CX0496 and CX0497 for GFP-OsPRMT6a, CX3254 and CX4333 (GTGTCTAGATCATCGCATGGCATAATCT) for GFP-OsPRMT6b and CX0696 and CX4339 (GCGTCTAGACAAGCAAGGTTCAGACAT) for GFP-OsPRMT10. The purified PCR products and vector pCAMBIA1300 (CAMBIA, Canberra, Australia)-GFP were simultaneously digested with BglII and XbaI for GFP-OsPRMT1, BamHI and XbaI for GFP-OsPRMT4, GFP-OsPRMT6b and GFP-OsPRMT10, NcoI for GFP-OsPRMT5, and BamHI and EcoRI for GFP-OsPRMT6a. The digested OsPRMTs' fragments and vectors were ligated using T4 DNA ligase. Note: all the primers sequences given are in the 5′ to 3′ order.
Tobacco (Nicotiana benthamiana) infiltration and fluorescent microscopy
The Agrobacterial strain (EHA105) transformed with binary vectors for the expression of GFP-fused OsPRMTs and P19 protein (suppressor of the RNAi silencing) were grown in the LB medium supplemented with selection antibiotics, acetosyringone (0.4 µM) and MES (10 mM). The cultures were centrifuged to pellet the Agrobacterial cells, resuspended in 10 mM magnesium chloride (MgCl2) and 0.2 mM acetosyringone and left to stand for 3 hours. The resuspended cultures harboring GFP-OsPRMTs and P19 construct were mixed in 1∶1 ratio and infiltrated into the 3-week-old healthy tobacco leaves by using syringes. The leaves were sectioned after 3 days of infiltration and the GFP signal was captured using fluorescence microscope (Olympus, Japan).
Expression, purification and in vitro methyltransferase activity assay
The GST-fused OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b, and OsPRMT10 were expressed in E. coli expression strain, BL21, affinity purified by binding to the glutathione-Sepharose (GST) 4B beads (Amersham Biosciences, 17-0756-01). In vitro methyltransferase assays were performed as described previously [44]. The incubated proteins were separated by 15% SDS-PAGE, stained with Coomassie blue and visualized by fluorography.
Western blot analysis
GST and GST-fused OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6b, and OsPRMT10 were incubated with the purified calf thymus core histones (Roche, 10223565001) in the presence of non-radioactive SAM at 30°C for 3 hours. The reaction mixtures were separated by the 15% SDS-PAGE, proteins were transferred to the PVDF membrane (Millipore, ISEQ00010) at 4°C and probed with antibodies; anti-H3R2 (Millipore, 05-808, rabbit monoclonal, 1∶3000) anti-H3R17me2a (Abcam, 8284, rabbit polyclonal, 1∶1500), anti-H4R3me2a (Millipore, 07-213, rabbit polyclonal, 1∶1000), anti-H3R4me2s (Abcam, ab5823, rabbit polyclonal, 1∶3000), and ASYM24 (Millipore, 07-414, rabbit polyclonal, 1∶1000) in TBST (0.1% Tween20, 150 mM NaCl and 50 mM Tris/HCl, pH 7.5)]. An equal amount of the reaction mixtures were separated by the 15% SDS-PAGE, followed by western blot analysis using anti-H3 (Millipore, 05-499, mouse monoclonal, 1∶2000) and anti-H4 (Millipore, 07-108, rabbit polyclonal, 1∶3000) antibodies in TBST buffer, for equal loading. Secondary antibodies used were mouse IgG HRP conjugate (Amersham Biosciences, NA931, 1∶40000) and rabbit IgG HRP conjugate (Amersham Biosciences, NA934, 1∶40000). The signal was detected on PVDF membrane with SuperSignal substrate (Thermo Scientific, 34076).
In vitro interaction assay
Briefly; MBP-hexahistidine (HIS), GST, MBP-HIS-OsPRMT6a and GST-OsPRMT6b were expressed in E. coli and purified by affinity chromatography using GST beads (Amersham Biosciences, 17-0756-01) and HIS beads (Ni-charged resin, BIO-RAD, 156-0135). Bound proteins were eluted; about 2 µg was rebound to glutathione-Sepharose and HIS resins and used as affinity matrices. Roughly 2 µg of the resin bound proteins, GST and GST-OsPRMT6b, were used as bait to pull-down the same amount of eluted prey proteins, MBP-HIS and MBP-HIS-OsPRMT6a. The proteins mixtures were spun at 10,000 g for 1 minute, the resin was extensively washed with modified RIPA buffer (20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P40 and 0.5% sodium deoxycholate, with 1 mM PMSF and 0.5 mM dithiothreitol (DTT), added immediately before use). The bound proteins were resuspended in 2× SDS loading buffer, boiled and separated by 10% SDS-PAGE. Proteins were transferred on to PVDF membrane (Millipore, ISEQ00010) and were probed with MBP antiserum (mouse monoclonal antiserum prepared in our lab, 1∶5000). The reciprocal pull-down experiment was performed similarly by using resin bound MBP-HIS and MBP-HIS-OsPRMT6a as bait to pull-down the eluted prey proteins GST and GST-OsPRMT6b and detected with the GST antiserum (mouse monoclonal antiserum prepared in our lab, 1∶30,000).
Results
Oryza sativa genome encodes eight homologs of the PRMTs
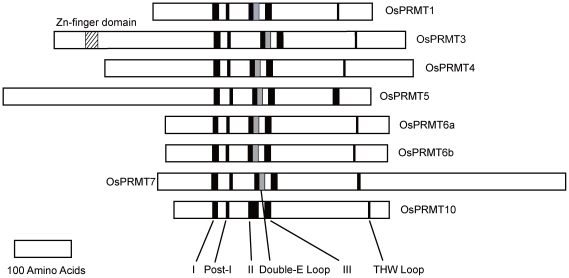
Eight loci, namely (according to TIGR/MSU database) Os09g19560 (NCBI accession NP_001062975.1), Os07g44640 (NCBI accession NP_001060419), Os07g47500 (NCBI accession NP_001060600), Os02g04660 (NCBI accession NP_00104584), Os10g34740 (NCBI accession NP_001064916), Os04g58060 (NCBI accession NP_001174145), Os06g01640 (NCBI accession NP_001056556), and Os06g05090 (NCBI accession NP_001056772) were found to encode putative OsPRMTs, previously, and were designated as OsPRMT1, OsPRMT3, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b, OsPRMT7 and OsPRMT10, respectively [42]. These eight genes coding for the OsPRMTs are distributed on six chromosomes i.e. 2, 4, 6, 7, 9, and10 (Figure S1). Here we aligned the amino acid sequences of these OsPRMTs to their Arabidopsis counterparts and found that OsPRMT1 shares 83% similarity to AtPRMT1a (and 86% to AtPRMT1b), OsPRMT3 shares 66% similarity to AtPRMT3, OsPRMT4 shares 85% similarity to AtPRMT4a (and 82% to AtPRMT4b), OsPRMT5 shares 82% similarity to AtPRMT5, OsPRMT6a shares 82% similarity to AtPRMT6, OsRMT6b share 79% similarity to AtPRMT6, OsPRMT7 shares 69% similarity to AtPRMT7 and OsPRMT10 shares 79% similarity to AtPRMT10 (Table S1). Multiple sequence alignment of the putative proteins expressed by these genes revealed a high degree of primary structure conservation in the PRMT signature motifs (I, post I, double E loop, post II, post III and THW (Thr-His-Trp)) loop (Figure 1), suggesting arginine methyltransferase activity for these proteins, as well as some additional sequences on both N- (OsPRMT3, OsPRMT4 and OsPRMT5) and C- (OsPRMT5 and OsPRMT7) terminals (Figures 1 and S2), which may be required for substrate specificity or some other still undetermined functions. The OsPRMT5 and OsPRMT7 sequences neither matched well with other OsPRMTs nor with each other (Figure S2) but when they were aligned to their Arabidopsis and human counterparts separately, they showed significant similarity in amino acid sequences (data not shown). In addition, PRMT6 has two homologs sharing 58% identity and 72% similarity at amino acid level. The molecular weights of the putative expressed proteins are in the range of 42.72 to 89.72 kD (Table 1).
Figure 1. Members of the protein arginine methyltransferase family of proteins in Oryza sativa.
Eight members of the Oryza sativa PRMT family; sharing the conserved PRMT signature motifs I, post-I, -II, -III, THW loop (Black bars) and double E loop (grey bars). PRMT3 harbors an additional Zn Finger domain (lined bar).
Table 1. Summary table.
| Tigr/IRGSP | RefSeq | Length | Molecular | Identity/ | Subcellular | Tested | Methylation | Type of | |
| Locus | Accession | (aa) | Weight (D) | Similarity to | Localization | Substrates | Sites on | PRMT | |
| AtPRMT (%) | H3 and H4 | ||||||||
| OsPRMT1 | Os09g19560/ | NP_001062975 | 387 | 43013.9 | 73/83, 77/86 | N and C | GAR, Myelin | H3R17and | I |
| Os09g0359800 | Basic Protein, | H4R3 | |||||||
| H4, H2A, H3 | |||||||||
| OsPRMT3 | Os07g44640/ | NP_001060419 | 620 | 67942.5 | 51/66 | ND | ND | ND | ND |
| Os07g0640000 | |||||||||
| OsPRMT4 | Os07g47500/ | NP_001060600 | 545 | 60108.9 | 73/85, 70/82 | ND | Myelin Basic | H3R17 | I |
| Os07g0671700 | Protein and H3 | ||||||||
| OsPRMT5 | Os02g04660/ | NP_001045843 | 649 | 72673.1 | 71/82 | N and C | Myelin Basic | H4R3 | II |
| Os02g0139200 | Protein, GAR, | ||||||||
| H4, H2A | |||||||||
| OsPRMT6a | Os10g34740/ | NP_001064916 | 395 | 43847.4 | 64/82 | Mainly N | GAR | ND | ND |
| Os10g0489100 | |||||||||
| OsPRMT6b | Os04g58060/ | Q7XKCO.2* | 392 | 43518.6 | 62/79 | Mainly N | Myelin Basic | H3R2 and | I |
| Os04g0677066 | Protein, GAR, | R17 | |||||||
| H3, H4, H2A | |||||||||
| OsPRMT7 | Os06g01640/ | NP_001056556 | 721 | 80600.8 | 54/69 | ND | ND | ND | ND |
| Os06g0105500 | |||||||||
| OsPRMT10 | Os06g05090/ | NP_001056772 | 381 | 42717.5 | 64/79 | N and C | Myelin Basic | H3R2 and | I |
| Os06g0142800 | Protein, GAR, | H4R3 | |||||||
| H4, H2A, H3 |
Abbreviations: ND, Not Determined; N, Nucleus; C, Cytoplasm; H3, histone H3; H4; histone H4; H3R2, histone H3 arginine 2; H3R17, histone H3 arginine 17; H4R3, histone H4 arginine 3; rest of the abbreviations are expanded elsewhere in the paper. TIGR/MSU loci were taken from http://rice.plantbiology.msu.edu/ and IRGSP (Japonica group) loci were taken from NCBI website http://www.ncbi.nlm.nih.gov/.
*is the Swiss-prot identifier for OsPRMT6b protein. The no of amino acids and molecular weights of the proteins are taken from NCBI http://www.ncbi.nlm.nih.gov.
OsPRMT genes reveal distinct expression patterns
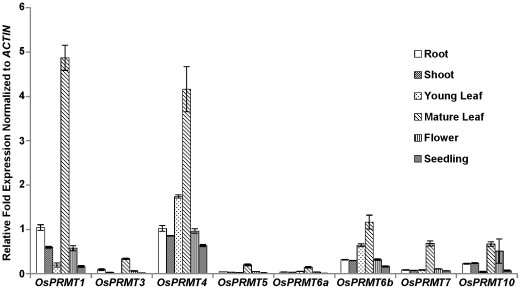
In order to illustrate the spatial expression patterns of the OsPRMTs, total RNAs were isolated from root, flower, shoot, young leaf, mature leaf and whole seedlings. These RNAs were reverse transcribed to cDNAs and analyzed by quantitative real time PCR to check the expression of the OsPRMTs in these tissues, using specific primer pairs designed for this purpose, as described in the experimental part. As shown in figure 2, OsPRMTs display distinct expression patterns in a tissue specific manner. Among all the OsPRMTs, OsPRMT1 and OsPRMT4 are expressed relatively higher than other homologs in all the tissue types analyzed in this study, which is consistent with their involvement in a variety of biological processes as studied in animals and yeast. Next in the relative abundance are OsPRMT6b and OsPRMT10. However, among the tissue types analyzed in this study, the mature leaf contains high abundance of all the PRMTs. OsPRMT6b and OsPRMT10 also exhibit significant expression in the root, shoot, young leaf and flower, the significance of which awaits further investigation. AtPRMT10 has been implicated in regulation of important traits in Arabidopsis such as flowering time and the PRMT6 in human has been reported to involve in transcriptional regulation and antiviral defense; however its role in plants remains to be unearthed. OsPRMT6b is expressed more than its homolog, OsPRMT6a, which is consistent with our in vitro enzymatic assay which shows that only the OsPRMT6b shows strong ability to catalyze the transfer of methyl group, whereas OsPRMT6a has very weak activity.
Figure 2. Relative expression patterns of the OsPRMTs.
Total RNAs were isolated from plant tissues; root, young leaf, mature leaf, shoot, flower and whole seedling of the Nipponbare strain and cDNAs were synthesized. The relative fold change in the expression of the OsPRMT1, OsPRMT3, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b, OsPRMT7 and OsPRMT10 was normalized to the expression of ACTIN. The error bars represent standard deviation.
OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6b and OsPRMT10 have bona fide methyltransferase activity in vitro
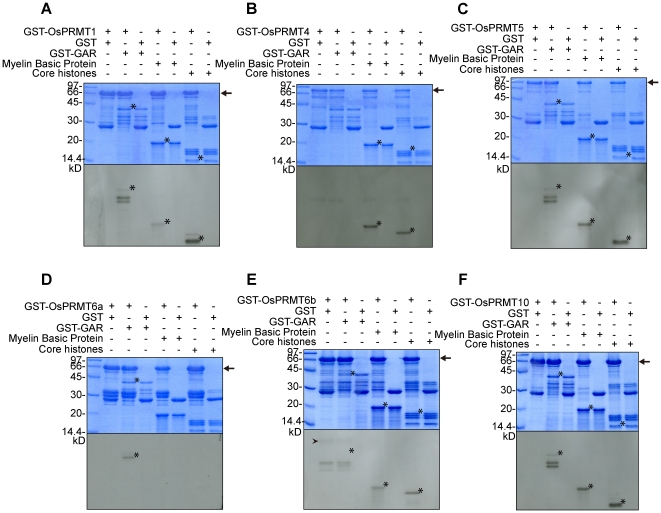
The conservation of the PRMT signature motifs in the OsPRMTs prompted us to decipher their in vitro arginine methyltransferase activity. For this purpose, GST and GST-fusion full length OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b, and OsPRMT10 were expressed in E. coli and affinity purified with the glutathione-Sepharose 4B (GST) beads. These proteins were assayed for in vitro methyltransferase activity in the presence of radioactive SAM ([3H]SAM) and the substrate proteins; GST-GAR, myelin basic protein and purified calf thymus core histones. The reaction mixtures were separated by 15% SDS-PAGE and visualized by fluorography (Figures 3 and S3). The exposure time for figure 3 and figure S3 is 48 hours and4 days, respectively. The GST-fusion OsPRMTs transferred 3H-labeled CH3-group onto the substrate proteins whereas the GST alone could not, which was used as negative control (Figures 3A–F and S3A–F). OsPRMT1 methylated GST-GAR and myelin basic protein; however, among the core histones H4 is methylated preferentially whereas histone H2A and H3 are weakly methylated, consistent with the earlier reports in human and Arabidopsis (Figures 3A and S3A). Like PRMT4/CARM1 in animals, OsPRMT4 did not methylate GST-GAR, however it methylated myelin basic protein and histone H3 (Figures 3B and S3B). Similar to the Arabidopsis counterpart [36], OsPRMT5 methylated histone H4, GST-GAR, and myelin basic protein; and to some extant H2A (Figures 3C and S3C). OsPRMT6a displayed activity for GST-GAR but none of the other substrates were methylated; showing that it either does not methylate the myelin basic protein and core histones or it has weak activity (Figures 3D and S3D). OsPRMT6b showed relatively stronger activity and methylated GST-GAR, myelin basic protein and histone H3 (Figure 3E). However, it also shows activity against H4 which is visible after longer exposure (Figure S3E). Additionally, OsPRMT6b showed automethylation like its human counterpart, as indicated by the arrow head (Figures 3E and S3E). As depicted by figure 3F, OsPRMT10 catalyzed the methylation of GST-GAR, myelin basic protein, and H4 (in accordance with the Arabidopsis PRMT10 [42]) (Figure 3F), and to a little extant H3 and H2A which is evident after four days exposure (Figure S3F). There are some non-specific bands, especially in the longer exposure films which probably show methylation of the degradation products of the enzymes and/or substrates (Figure S3). These results indicate that OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6b, and OsPRMT10 are bona fide arginine methyltransferases in vitro.
Figure 3. In vitro methyltransferase activity assay of the OsPRMTs.
Methyltransferase activity assay of OsPRMT1 (A), OsPRMT4 (B), OsPRMT5 (C), OsPRMT6a (D), OsPRMT6b (E), and OsPRMT10 (F). GST and GST-OsPRMTs were bound to the GST beads and incubated with indicated substrates; myelin basic protein, GST-GAR and calf thymus core histones in the presence of [3H]SAM for 3 hours in a final volume of 30 µl of HMT buffer (20 mM Tris/HCl, 4 mM sodium EDTA, 1 mM PMSF and 1 mM dithiothreitol). Methylated proteins were separated by 10% SDS-PAGE, stained with Coomassie blue (upper panels), destained, soaked in amplify (Amersham Biosciences, NAMP100) dried and visualized by fluorography by exposing to the x-ray film at −80°C for 48 hours (lower panels). The corresponding proteins are indicated above the upper panels. The arrow “→” points GST-Tagged full length OsPRMTs whereas the asteric “*” represents the substrate proetins i.e. GST-GAR, myelin basic protein and core histones (H3, H4, H2A and H2B). The arrow head “ ” indicates the automethylation of OsPRMT6b.
” indicates the automethylation of OsPRMT6b.
OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6b and OsPRMT10 methylate specific arginine residues on histone H3 and H4 and can be classified into two types of PRMTs
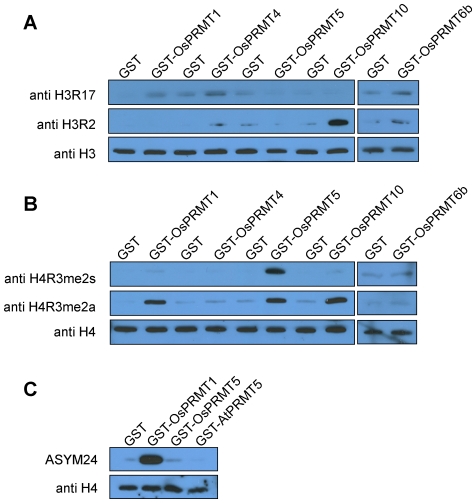
Different PRMTs have distinct specificities for the methylation sites in the histone tails which in turn contribute to the histone code (as reviewed in [45]). In order to investigate which particular arginine residues are methylated by each OsPRMT analyzed in calf thymus core histones, western blot analysis was performed after the in vitro methylation reaction using antibodies against different methylated arginine residues. Calf thymus core histones incubated with GST protein served as negative control. The methylated substrates detected by antibodies against aDMA were considered to be methylated by type I PRMT whereas those detected by antibodies against sDMA were thought to be methylated by type II PRMT. OsPRMT1 asymmetrically dimethylated H3 at R17 and H4 at R3 hence it belongs to type I PRMTs (Figures 4A and 4B, left panels). OsPRMT4 methylated H3R17 which is consistent with the earlier reports and it belongs to type I PRMT because its substrate was detected by antibody against asymmetrically dimethylated H3R17 (Figures 4A and 4B, left panels). OsPRMT5 belongs to type II PRMTs, since the substrates methylated by it were recognized by the antibody against sDMA and it symmetrically dimethylated R3 in histones H4 (Figures 4A and 4B, left panels), which is in consistence with our previous study on Arabidopsis PRMT5 [36]. However, unexpectedly the histone H4 incubated with OsPRMT5 was also detected by the antibody against aDMA at H4R3 which is probably due to the ability of the antibody to detect both the symmetric as well as the asymmetric dimethyl H4R3. To address this problem, the histone H4 incubated with the GST, GST-OsPRMT1, GST-OsPRMT5 and GST-AtPRMT5 (as control) were probed with anti-aDMA (ASYM24) antibody. The ASYM24 antibody detected the arginine methylated product from OsPRMT1 but not from the OsPRMT5 and AtPRMT5 (Figure 4C), which proves that OsPRMT5 is indeed a type II PRMT catalyzing the formation of MMA and sDMA in the substrate proteins. OsPRMT10 is a type I PRMT because it asymmetrically dimethylates H3 at R2 and H4 at R3 (Figures 4A and 4B, left panels). OsPRMT6b is a type I PRMT and it asymmetrically dimethylates H3 at R2 and 17 (Figures 4A and 4B, right panels). From the in vitro enzymatic activity assay it is evident that OsPRMT6b has relatively stronger activity at histones H3 and H4 (Figure 3E and Figure S3E), but here we did not see any activity at H4R3 which shows that it might be methylating some other arginine residue(s) in histone H4, which awaits further investigation.
Figure 4. Site specificities of the OsPRMT1, OsPRMT5, OsPRMT6b, and OsPRMT10.
(A, B and C) Purified calf thymus core histones were either incubated with negative control, GST or GST-OsPRMT1, GST-OsPRMT5, GST-OsPRMT6b, GST-OsPRMT10 and GST-AtPRMT5 and were separated by 15% SDS-PAGE, followed by western blot analysis using anti-H3R2me2a, anti-H3R17me2a, anti-H4R3me2a, anti-H4R3me2s, and ASYM24 antibodies. An equal amount of the reaction mixtures were separated by the 15% SDS-PAGE, followed by western blot analysis using anti-H3 and anti-H4 antibodies, for equal loading (lowest panels in A, B and C). The antibodies are indicated to the left of each panel, whereas the corresponding PRMTs are represented above the upper panels.
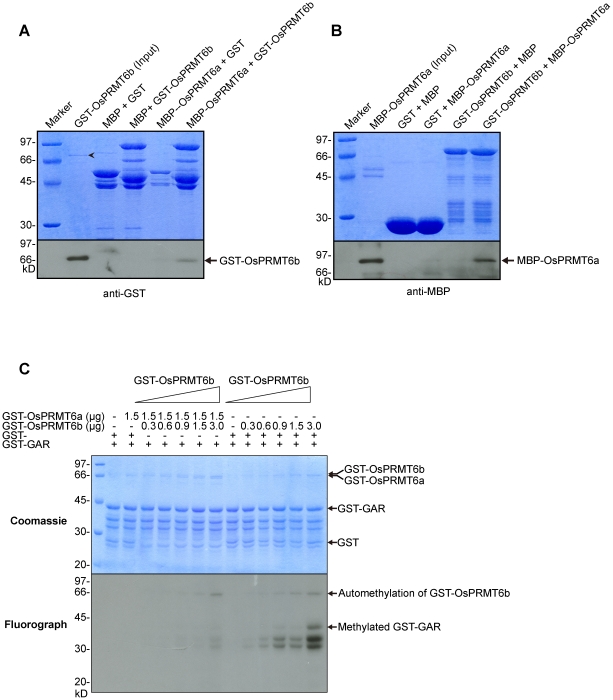
OsPRMT6a interacts with OsPRMT6b and inhibits its methyltransferase activity in vitro
Rice has two homologs of the PRMT6, OsPRMT6a and OsPRMT6b. Our in vitro enzyme activity assay showed that OsPRMT6b has strong activity but the OsPRMT6a has weak activity only against the GAR domain (Figures S3D and S3E), which indicates that OsPRMT6a has either lost its methyltransferase activity or acquired another function during the course of evolution. Since the PRMTs harbor a characteristic dimerization arm, which is considered as essential for the enzymatic activity [46], we checked if OsPRMT6a and OsPRMT6b could form a heterodimer in vitro. For this purpose we expressed the GST, MBP-HIS, GST-OsPRMT6a, MBP-HIS-OsPRMT6a, GST-OsPRMT6b, and MBP-HIS-OsPRMT6b in E. coli and performed the reciprocal pull-down experiments to check their direct interaction in vitro. In the first experiment we used the resin bound ‘bait’ proteins, MBP-HIS (as negative control) and MBP-HIS-OsPRMT6a to pull down the ‘prey’ proteins, GST (as negative control) and GST-OsPRMT6b (Figure 5A). In the reciprocal experiment the resin bound ‘bait’ proteins GST (as negative control) and GST-OsPRMT6b were used as to pull-down the ‘prey’ proteins, MBP (as negative control) and MBP-HIS-OsPRMT6a (Figure 5B). The proteins were separated on two 10% SDS-PAGE, one was stained with Coomassie blue for equal loading and other was used for western blot. The monoclonal anti-GST (Figure 5A) and anti-MBP antibodies (Figure 5B) were used to detect the GST, GST-OsPRMT6b, MBP-HIS and MBP-HIS-OsPRMT6a proteins. As expected, in the reciprocal pull-down experiment we found that OsPRMT6a and OsPRMT6b directly interact with each other in vitro but not with the GST and MBP-HIS and they may form a heterodimer in vivo. Further to explore the significance of this interaction, GST-OsPRMT6a was mixed with different amounts of GST-OsPRMT6b in an in vitro methyltransferase assay where GST-GAR was used as substrate. In control reaction the same amount of GST-OsPRMT6b was mixed with only GST protein. Interestingly, GST-OsPRMT6a but not the GST protein inhibited the in vitro methyltransferase activity of the GST-OsPRMT6b (Figure 5C). Since, dimerization is essential for the enzymatic activity of the PRMTs, this finding reveals a novel mechanism of regulation of PRMTs' activity, probably, by heterodimerization.
Figure 5. OsPRMT6a and OsPRMT6b directly interact in vitro.
(A and B) GST, MBP, MBP-OsPRMT6a and GST-OsPRMT6b were expressed in E. coli. Reciprocal pull-down assay was performed as described in the experimental part. Equal amount of each mixture of proteins was separated on two separate 10% SDS-PAGE, one for Coomassie staining (upper panels A and B) for equal loading and the other for western blotting (lower panels A and B). GST and MBP alone were used as negative controls. The protein pairs are indicated at the top of the upper panels. MBP-OsPRMT6a served as bait and GST-OsPRMT6b as prey (A) and vice versa (B). A 10 µl cell extract from E. coli containing MBP-OsPRMT6a and GST-OsPRMT6b was used as input. Western blotting using GST and MBP monoclonal antisera revealed the interaction between OsPRMT6a and OsPRMT6b. The arrow head “???” indicates MBP-OsPRMT6a. (C) 0.3, 0.6, 0.9, 1.5 and 3.0 µg of GST-OsPRMT6b was mixed either with 1.5 µg of GST-OsPRMT6a or with GST protein alone (as control) in an in vitro methyltransferase activity assay as described in figure 3. The Coomassie blue stained and dried gel was exposed at −80°C for four days.
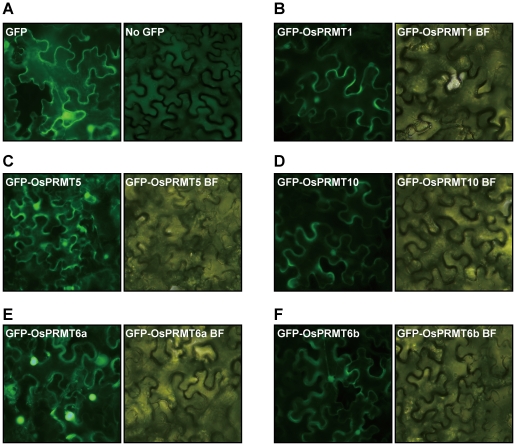
Subcellular localization of the OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b, and OsPRMT10
Since OsPRMTs methylate a variety of substrates such as histones (nuclear basic proteins), GAR domain of Arabidopsis fibrillarin (an RNA binding nucleolar protein) and myelin basic protein (a cell membrane protein); they may localize to and function in different compartments of the cell. To find out the specific subcellular localization of the OsPRMTs, the coding sequences of the OsPRMT1, OsPRMT5, OsPRMT6a, OsPRMT6b, and OsPRMT10 were fused in frame to the GFP, driven by the CMV 35S promoter and transfected into the Nicotiana benthamiana leaves by the Agrobacterial strain EHA105. After 3 days of injection, the GFP signal was checked under the fluorescence microscope. GFP alone (used as control) was localized both to the nucleus as well as to the cytoplasm (Figure 6A). OsPRMT1, OsPRMT5, and OsPRMT10 exhibited the same subcellular location both in the cell nucleus and cytoplasm (Figures 6B–6D), which is consistent with the localization of their Arabidopsis and human counterparts. However, the OsPRMT6a and OsPRMT6b were mainly localized into the nucleus (Figures 6E and 6F), which is also in agreement with the localization of human PRMT6 [47]. The localization of the OsPRMTs into both the cytoplasm and nucleus designates their general requirement in many basic cellular processes.
Figure 6. Subcellular localization of the OsPRMT1, OsPRMT5, OsPRMT6a, OsPRMT6b, and OsPRMT10.
OsPRMT1, OsPRMT5, OsPRMT6a, OsPRMT6b, and OsPRMT10 were fused in frame to GFP in binary vector, transformed to the Agrobactrial strain, EHA105 and injected into the Nicotiana benthamiana leaves. Expression of GFP and GFP-tagged OsPRMT1, OsPRMT5, OsPRMT6a, OsPRMT6b, and OsPRMT10 was observed under fluorescence microscopy and bright field microscopy (designated by BF) after 2–3 days.
Discussion
The evolutionary conservation of arginine methylation from unicellular eukaryotes to plants and humans indicates its involvement in the basic cellular machinery. Saccharomyces cerevisiae genome encodes three PRMTs (HMT1, Hsl7 and Rmt2) [12], Drosophila genome contains 9 (DART1 to DART9) [48], Arabidopsis genome bears 9 (AtPRMT1a, AtPRMT4b, AtPRMT3, AtPRMT4a, AtPRMT4b, AtPRMT5, AtPRMT6, AtPRMT7, and AtPRMT10) [42] and human genome bears 11 (hPRMT1-11) [1]; as known, so far. PRMTs have been studied extensively in animals and yeast. However, in the recent years there is a growing tendency to substantiate their functional importance in plants, especially in the model plant Arabidopsis thaliana [33], [35]–[38], [40]–[42]. PRMT1, PRMT3 and PRMT5 have been found to be strictly conserved during the evolution of eukaryotes from unicellular yeasts, molds, amoebae, and protozoa to higher plants and animals; however, the other PRMTs are less conserved which might have evolved in the multicellular organisms as requirement for tissue specific functions. Consistent with this hypothesis, PRMT2 and PRMT8 exhibit tissue specific expression [49], [50]. Additionally, no homologs of PRMT2 and PRMT8 exist in the Arabidopsis and rice genomes which further strengthen the notion that these PRMTs arose for the tissue specific functions in higher eukaryotes.
Gene duplication is common phenomenon in eukaryotes which occurs at a rate of 0.01 per gene per million years and is a source of evolutionary novelties [51]. The duplication of gene may result in nonfunctionalization (one copy of gene is silenced), neofunctionalization (one copy acquire a new function) or subfunctionalization (both copies become partially compromised). Unlike animals and yeast, there are two homologs each of PRMT1 and PRMT4 in Arabidopsis and PRMT6 in rice. It seems that in Arabidopsis, duplication of PRMT genes, AtPRMT1 and AtPRMT4, resulted in subfunctionalization, because the duplicated genes showed same expression patterns and methyltransferase activities. In addition, the double knockout mutant of AtPRMT4a and AtPRMT4b results in more severe phenotype than that of single knockout of these two copies of the gene [35]. In Oryza; although the expression pattern and cellular location of OsPRMT6a and OsPRMT6b are similar, OsPRMT6a almost lost methyltransferase activity, which suggests a nonfunctionalization outcome of OsPRMT6 duplication. Our data also suggest that OsPRMT6b may be regulated by OsPRMT6a which points to neofunctionalization; however, more solid evidence, such as the resolution of crystal structure of the heterodimer of the OsPRMT6a and OsPRMT6b is needed to support this idea.
The significance/functional importance of the PRMTs is evident from the quantitative real time PCR analysis data of the mRNAs from different tissues, revealing that they are widely expressed in many tissues, rather than limited to specific tissues/cell types. Among the tissue types studied; root, shoot, young leaf, mature leaf, flower and total seedling, mature leaf has highest pool of all the OsPRMTs (the physiological meaning of which awaits further investigation), whereas among the OsPRMTs, OsPRMT1 and OsPRMT4 are more abundant in all the tissue types. Additionally, the in vivo targeting of the OsPRMT1, OsPRMT4, OsPRMT5, and OsPRMT10 to both the nucleus and cytoplasm indicates the redundancy and more general requirement for these PRMTs; however, the predominant nuclear localization of the OsPRMT6a and OsPRMT6b reflects somewhat specificity for these enzymes. Further studying their expression pattern by the promoter-genomic-GUS analysis will hint their probable function in specific tissues and cells in more detail.
Our in vitro enzymatic activity assay demonstrated that OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6b, and OsPRMT10 can directly catalyze the transfer of methyl group from SAM to their substrate proteins and no additional cofactor(s), partner molecule or conformation is required for this activity. OsPRMT6a showed weak methyltransferase activity which suggests that it might have some additional requirement for its activity. Another possibility is that, this protein harbors the core PRMT motifs probably for some other unknown functions. This notion is strengthened by our in vitro titration experiment in which the addition of GST-OsPRMT6a inhibits the methyltransferase activity of GST-OsPRMT6b. However, further studies dedicated to finding the effect of making different combinations of the PRMTs on the methyltransferase activity of these PRMTs and resolving the crystal structures of heterodimers will give the conclusive evidence to this phenomenon. In the in vitro enzyme activity assay we used myelin basic protein, GST-GAR and core histones from the calf thymus as substrates for the OsPRMTs. Myelin basic protein is widely used as substrate for the enzymes responsible for post-translational modifications because it readily undergoes various posttranslational modifications such as phosphorylation, ubiquitination and methylation [52]. In vivo, the modifier enzymes affect its structure and, consequently, interactions with other membrane components in the membrane assembly which is a predominantly post-synthetic phenomenon. Arginine methylation of the myelin basic protein results in its compact structure; antagonizes the effect of citrolination by peptidylarginine deiminases (PADs), in vivo dimerization, and resistance to trypsin digestion. OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6b and OsPRMT10 displayed significant activity against the myelin basic protein in our methyltransferase assay, suggesting that these enzymes may methylate myelin basic protein in vivo and modulate the above stated as well as some yet unknown properties of this biologically important protein. GAR domains, characterized by the tri-peptide motif, RGG, are frequently found in RNA binding proteins including, nucleolar protein 1 (Nop1), Nop3, Npl3, human DNA 35 helicase II, ewing sarcoma protein, hnRNP A1, hnRNP A2, human MRE11, nucleolin, and fibrillarin. The structural integrity of GAR is required for the nucleic acid and protein binding, intracellular targeting and hence proper functioning of the proteins harboring this domain. The GAR domain of hnRNP A1 is required for cooperative binding to RNA, but it is also required for interactions with other pre-mRNA binding proteins. Nucleolin's GAR domain shows a specific interaction with ribosomal protein L3, thus strengthening a possible role in protein-protein interactions and perhaps assembly of ribosomal proteins into ribosome. AtPRMT1a and AtPRMT1b have been reported to directly interact with and methylate the fibrillarin protein in the GAR domain [33]. Here we outlined that all the tested OsPRMTs; OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b, and OsPRMT10 methylate the GAR domain in vitro. PRMT6 has two homologs in rice, namely OsPRMT6a and OsPRMT6b. OsPRMT6b has strong in vitro methyltransferase activity against myelin basic protein, GAR and histones H2A, H3 and H4; however, OsPRMT6a shows only weak activity against GAR domain, which shows that OsPRMT6b is the functional enzyme, whereas OsPRMT6a might has lost its activity during the course of evolution or it requires some additional cofactor or partner for its methyltransferase activity. Another possible explanation for the lack of enzymatic activity of OsPRMT6a is due to the requirement of hetero-dimerization with OsPRMT6b. Our in vitro pull-down experiment proved that they directly interact with each other. Taken together, our data implies that OsPRMTs may redundantly methylate the GAR domain in a wide variety of proteins, involved in a multitude of essential biological processes. Nonetheless, studying the plants with lesions in these genes and especially multiple mutants might be very handy to unveil the biological significance of arginine methylation in regulating plant development.
Histones are basic proteins and essential component of the chromatin, which carries the genetic information from one generation to other generation. Histone tails are prone to many post translational modifications (phosphorylated, acetylated, methylated, ubiquitinated, etc) at various amino acid residues such as lysine, serine, arginine, dicarboxylic acid etc; which in turn constitute the histone code. Different combinations of these modifications have distinct transcriptional outcomes. Among these modifications is the methylation of arginine in histone tails, both in animals and plants; Thus far, the major known sites liable to arginine methylation are R2, R8, R17, R26 of histone H3, and R3 of histone H4 [45]. We have recently summarized that how the epigenetic changes, especially chromatin modifications affect the plant development [45], [53]. In mammals, PRMT1 and PRMT5 asymmetrically and symmetrically dimethylate H4R3, respectively and are required for the normal development [6]. Arabidopsis PRMT1a and PRMT1b were reported to asymmetrically dimethylate H4R3 in vitro but its functional importance is yet to be discovered [33]. Furthermore, AtPRMT5/Skb1-mediated symmetric dimethylation at H4R3 regulates flowering time by repressing FLC transcription [36]. Our in vitro methyltransferase activity assay and western blot data consistently suggest that the OsPRMT1 and OsPRMT10 asymmetrically and OsPRMT5 symmetrically dimethylate H4R3. PRMT6-mediated asymmetric dimethylation at H3R2 antagonizes the H3K4 tri-methylation in mammals, which is a well-known transcriptional activation mark [54]. AtPRMT4a and AtPRMT4b, the Arabidopsis homologs of human PRMT4/CARM1, asymmetrically dimethylate R2, R17, and R26 in histone H3 in vitro and are required for the in vivo H3R17me2a and atprmt4a atprmt4b double mutant displays FLC-dependent late flowering phenotype [35]. In addition, asymmetric dimethylation at H4R3 catalyzed by AtPRMT10 regulates the Arabidopsis flowering time in an FLC-dependent manner. In vitro enzymatic activity assay combined with the immunoblotting experiment, in this study, proved that OsPRMT1, OsPRMT4 and OsPRMT6b asymmetrically dimethylated H3 at R17 whereas OsPRMT10 and OsPRMT6b methylated H3 at R2. In addition, OsPRM1 and OsPRMT10 also asymmetrically dimethylate H4R3, OsPRMT5 catalyzes symmetric dimethylation at this site. This in vitro data manifests redundancy for the methylation at these sites in H3 and H4; nevertheless, investigating the in vivo global changes in the methylation levels at these sites of histones, isolated from knockout mutants (single as well as multiple) plants, would probably uncover the specificities of OsPRMTs, which may require some proper conformation of the histones, for activity at these sites. Furthermore, unveiling the physiological importance of these conserved methylation marks also awaits further investigation of the mutant plants with lesion in the genes coding for these enzymes. In the nut-shell; this study strongly suggests the conservation of this important posttranslational modification of proteins, arginine methylation, in monocots and will surely trigger the understanding of biological significance of protein arginine methylation in the important cereal, Oryza sativa, and provide additional challenges and surprises about this evolutionarily conserved posttranslational modification event.
Supporting Information
Chromosomal distribution of OsPRMTs. Chromosomal distribution of PRMT gene family member in Oryza sativa. Eight PRMT genes are distributed throughout the Oryza genome as single genes. Gene positions on the physical map of NCBI mapviewer are indicated in megabases for each gene. Six chromosomes on which eight PRMT genes localize are indicated by numbers (2, 4, 6, 7, 9, and10).
(TIF)
Amino acid sequence alignment of the PRMT gene family members in Oryza sativa . The TIGR/MSU identifiers for OsPRMT1, OsPRMT3, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b, OsPRMT7 and OsPRMT10 are Os09g19560, Os07g44640, Os07g47500, Os02g04660, Os10g34740, Os04g58060, Os06g01640, and Os06g05090, respectively. Identical amino acids are boxed in black and similar amino acids are boxed in grey. Characteristic methyltransferase motifs, I, post-I, post-II, post-III are underlined, whereas Double E loop and THW loop are enclosed in boxes.
(TIF)
In vitro methyltransferase activity assay of the OsPRMTs (longer exposure time). S3A–S3F represent the in vitro enzyme activity assay result for the OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b and OsPRMT10, respectively. All the steps are essentially the same as in figure 3, however, the exposure time is four days.
(TIF)
Table representing the identities/similarities of OsPRMTs to the Arabidopsis PRMTs at amino acids level. Amino acid sequences of the OsPRMTs and AtPRMTs were aligned in the bl2seq tool on the NCBI website to find the percent identity/similarity.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The first author would like to cordially thank the Higher Education Commission of Pakistan for awarding him a scholarship for this study. This work was supported by National Basic Research Program of China (grant nos. 2011CB915400, 2009CB941500), by National Natural Science Foundation of China (grant nos. 30930048 and 30621001) to X.C. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wolf SS. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci. 2009;66:2109–2121. doi: 10.1007/s00018-009-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pahlich S, Zakaryan RP, Gehring H. Protein arginine methylation: Cellular functions and methods of analysis. Biochem Biophys Acta. 2006;1764:1890–1903. doi: 10.1016/j.bbapap.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Krause CD, Yang ZH, Kim YS, Lee JH, Cook JR, et al. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther. 2007;113:50–87. doi: 10.1016/j.pharmthera.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120:4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 5.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 6.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paik WK, Paik DC, Kim S. Historical review: the field of protein methylation. Trends Biochem Sci. 2007;32:146–152. doi: 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Bachand F. Protein arginine methyltransferases: from unicellular eukaryotes to humans. Eukaryot Cell. 2007;6:889–898. doi: 10.1128/EC.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayegh J, Clarke SG. Hsl7 is a substrate-specific type II protein arginine methyltransferase in yeast. Biochem Biophys Res Commun. 2008;372:811–815. doi: 10.1016/j.bbrc.2008.05.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisk JC, Sayegh J, Zurita-Lopez C, Menon S, Presnyak V, et al. A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei. J Biol Chem. 2009;284:11590–11600. doi: 10.1074/jbc.M807279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niewmierzycka A, Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 12.McBride AE, Zurita-Lopez C, Regis A, Blum E, Conboy A, et al. Protein arginine methylation in Candida albicans: role in nuclear transport. Eukaryot Cell. 2007;6:1119–1129. doi: 10.1128/EC.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YH, Stallcup MR. Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol. 2000;20:4859–4869. doi: 10.1128/mcb.20.13.4859-4869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kzhyshkowska J, Kremmer E, Hofmann M, Wolf H, Dobner T. Protein arginine methylation during lytic adenovirus infection. Biochem J. 2004;383:259–265. doi: 10.1042/BJ20040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 17.Lakowski TM, Frankel A. Kinetic analysis of human protein arginine N-methyltransferase 2: formation of monomethyl-and asymmetric dimethyl-arginine residues on histone H4. Biochem J. 2009;421:253–261. doi: 10.1042/BJ20090268. [DOI] [PubMed] [Google Scholar]
- 18.Kzhyshkowska J, Schütt H, Liss M, Kremmer E, Stauber R, et al. Heterogeneous nuclear ribonucleoprotein E1B-AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem J. 2001;358:305–314. doi: 10.1042/0264-6021:3580305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, et al. Identification of protein arginine methyltransferase 2 as a coactivator for estrogen receptor alpha. J Biol Chem. 2002;277:28624–28630. doi: 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- 20.Meyer R, Wolf SS, Obendorf M. PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor. J Steroid Biochem Mol Biol. 2007;107:1–14. doi: 10.1016/j.jsbmb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Swiercz R, Person MD, Bedford MT. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem J. 2005;386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi S, Jung CR, Kim JY, Im DS. PRMT3 inhibits ubiquitination of ribosomal protein S2 and together forms an active enzyme complex. Biochem Biophys Acta. 2008;1780:1062–1069. doi: 10.1016/j.bbagen.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Bachand F, Silver PA. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 2004;23:2641–2650. doi: 10.1038/sj.emboj.7600265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, et al. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF- B-dependent gene expression. EMBO J. 2004;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohkura N, Takahashi M, Yaguchi H, Nagamura Y, Tsukada T. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J Biol Chem. 2005;280:28927–28935. doi: 10.1074/jbc.M502173200. [DOI] [PubMed] [Google Scholar]
- 26.Yadav N, Lee J, Kim J, Shen J, Hu MC, et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richard S, Morel M, Cleroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5). Biochem J. 2005;388:379–386. doi: 10.1042/BJ20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dacwag CS, Bedford MT, Sif S, Imbalzano AN. Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol Cell Biol. 2009;29:1909–1921. doi: 10.1128/MCB.00742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulanger MC, Liang C, Russell RS, Lin R, Bedford MT, et al. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J Virol. 2005;79:124–131. doi: 10.1128/JVI.79.1.124-131.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, et al. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2009;94:585–594. doi: 10.1016/j.fertnstert.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 32.Pahlich S, Zakaryan RP, Gehring H. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins. 2008;72:1125–1137. doi: 10.1002/prot.22004. [DOI] [PubMed] [Google Scholar]
- 33.Yan D, Zhang Y, Niu L, Yuan Y, Cao X. Identification and characterization of two closely related histone H4 arginine 3 methyltransferases in Arabidopsis thaliana. Biochem J. 2007;408:113–121. doi: 10.1042/BJ20070786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scebba F, De Bastiani M, Bernacchia G, Andreucci A, Galli A, et al. PRMT11: a new Arabidopsis MBD7 protein partner with arginine methyltransferase activity. Plant J. 2007;52:210–222. doi: 10.1111/j.1365-313X.2007.03238.x. [DOI] [PubMed] [Google Scholar]
- 35.Niu L, Zhang Y, Pei Y, Liu C, Cao X. Redundant requirement for a pair of PROTEIN ARGININE METHYLTRANSFERASE4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol. 2008;148:490. doi: 10.1104/pp.108.124727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei Y, Niu L, Lu F, Liu C, Zhai J, et al. Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 2007;144:1913–1923. doi: 10.1104/pp.107.099531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Zhang Y, Ma Q, Zhang Z, Xue Y, et al. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 2007;26:1934–1941. doi: 10.1038/sj.emboj.7601647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng X, Gu L, Liu C, Lu T, Lu F, et al. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc Natl Acad Sci U S A. 2010;107:19114–19119. doi: 10.1073/pnas.1009669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 40.Hong S, Song HR, Lutz K, Kerstetter RA, Michael TP, et al. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc Natl Acad Sci. 2010;107:21211–21216. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z, Zhang S, Zhang Y, Wang X, Li D, et al. Arabidopsis Floral Initiator SKB1 Confers High Salt Tolerance by Regulating Transcription and Pre-mRNA Splicing through Altering Histone H4R3 and Small Nuclear Ribonucleoprotein LSM4 Methylation. Plant Cell. 2011;23:396–411. doi: 10.1105/tpc.110.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu L, Lu F, Pei Y, Liu C, Cao X. Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep. 2007;8:1190–1195. doi: 10.1038/sj.embor.7401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 44.Ding Y, Wang X, Su L, Zhai J, Cao S, et al. SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell. 2007;19:9–22. doi: 10.1105/tpc.106.048124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 46.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, et al. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 48.Boulanger MC, Miranda TB, Clarke S, Di Fruscio M, Suter B, et al. Characterization of the Drosophila protein arginine methyltransferases DART1 and DART4. Biochem J. 2004;379:283–289. doi: 10.1042/BJ20031176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Sayegh J, Daniel J, Clarke S, Bedford MT. PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J Biol Chem. 2005;280:32890–32896. doi: 10.1074/jbc.M506944200. [DOI] [PubMed] [Google Scholar]
- 50.Scott HS, Antonarakis SE, Lalioti MD, Rossier C, Silver PA, et al. Identification and characterization of two putative human arginine methyltransferases (HRMT1L1 and HRMT1L2). Genomics. 1998;48:330–340. doi: 10.1006/geno.1997.5190. [DOI] [PubMed] [Google Scholar]
- 51.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 52.Boggs J. Myelin basic protein: a multifunctional protein. Cell Mol life Sci. 2006;63:1945–1961. doi: 10.1007/s00018-006-6094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahmad A, Zhang Y, Cao XF. Decoding the epigenetic language of plant development. Mol Plant. 2010;3:719–728. doi: 10.1093/mp/ssq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, et al. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chromosomal distribution of OsPRMTs. Chromosomal distribution of PRMT gene family member in Oryza sativa. Eight PRMT genes are distributed throughout the Oryza genome as single genes. Gene positions on the physical map of NCBI mapviewer are indicated in megabases for each gene. Six chromosomes on which eight PRMT genes localize are indicated by numbers (2, 4, 6, 7, 9, and10).
(TIF)
Amino acid sequence alignment of the PRMT gene family members in Oryza sativa . The TIGR/MSU identifiers for OsPRMT1, OsPRMT3, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b, OsPRMT7 and OsPRMT10 are Os09g19560, Os07g44640, Os07g47500, Os02g04660, Os10g34740, Os04g58060, Os06g01640, and Os06g05090, respectively. Identical amino acids are boxed in black and similar amino acids are boxed in grey. Characteristic methyltransferase motifs, I, post-I, post-II, post-III are underlined, whereas Double E loop and THW loop are enclosed in boxes.
(TIF)
In vitro methyltransferase activity assay of the OsPRMTs (longer exposure time). S3A–S3F represent the in vitro enzyme activity assay result for the OsPRMT1, OsPRMT4, OsPRMT5, OsPRMT6a, OsPRMT6b and OsPRMT10, respectively. All the steps are essentially the same as in figure 3, however, the exposure time is four days.
(TIF)
Table representing the identities/similarities of OsPRMTs to the Arabidopsis PRMTs at amino acids level. Amino acid sequences of the OsPRMTs and AtPRMTs were aligned in the bl2seq tool on the NCBI website to find the percent identity/similarity.
(DOC)