Abstract
Recombination, together with mutation, is the ultimate source of genetic variation in populations. We leverage the recent mixture of people of African and European ancestry in the Americas to build a genetic map measuring the probability of crossing-over at each position in the genome, based on about 2.1 million crossovers in 30,000 unrelated African Americans. At intervals of more than three megabases it is nearly identical to a map built in Europeans. At finer scales it differs significantly, and we identify about 2,500 recombination hotspots that are active in people of West African ancestry but nearly inactive in Europeans. The probability of a crossover at these hotspots is almost fully controlled by the alleles an individual carries at PRDM9 (P<10−245). We identify a 17 base pair DNA sequence motif that is enriched in these hotspots, and is an excellent match to the predicted binding target of African-enriched alleles of PRDM9.
In humans and many other species, recombination is not evenly distributed across the genome, but instead occurs in “hotspots”: two kilobase (kb) segments where the crossover rate is far higher than in the flanking DNA sequence1,2,3. The highest resolution genetic map in contemporary humans to date, the “deCODE Map”, is based on about 500,000 crossovers identified in 15,000 Icelandic meioses4. However, a limitation of maps built in people of European descent4,5,6 is that they may not apply equally well in other populations, as suggested by comparisons of maps across ethnic groups4,7,8,9 and patterns of linkage disequilibrium (LD) breakdown which suggest that more of the genome may be recombinationally active in West Africans10. It is known that a major determinant of the positions of recombination hotspots is PRDM9, a meiosis-specific histone H3 methyltransferase whose zinc finger (ZF) domain binds DNA sequence motifs11,12,13. In Europeans, PRDM9 ZF arrays are predominantly of two similar types, “A” and “B”, both of which bind the 13-bp motif CCNCCNTNNCCNC11. In contrast, 36% of West African alleles are not of the A or B type9,13. Sperm typing of males who carry neither the A nor the B allele has shown no evidence of crossover activity at recombination hotspots associated with the 13-bp motif9.
To investigate differences in the crossover landscape across human populations, we built a genetic map in African Americans, who have an average of about 80% West African and 20% European ancestry, leading to genomes comprised of multi-megabase stretches of either West African or European ancestry14. Computational approaches, including HAPMIX15, have been developed to infer the probability of 0, 1 or 2 European or African alleles at each locus in individuals genotyped at hundreds of thousands of single nucleotide polymorphisms (SNPs)15,16,17. Positions where the inferred number of European or African alleles changes reflect crossover events that have occurred since admixture began (on average six generations ago15). The change in the probability of European ancestry between adjacent SNPs can be interpreted as the probability of such a crossover between them. We inferred crossover events in 29,589 apparently unrelated African Americans who had been genotyped on SNP arrays in genetic association studies (Methods; Figure 1A). To minimize false-positive crossovers, we restricted to crossovers that HAPMIX inferred with probability of >95%, and that were flanked by a minimum of 2 centimorgan (cM) stretches where the ancestry was inferred to be unchanging (Note S1). This produced 2,113,293 highconfidence crossovers, with a typical switch point resolved within 70kb with probability 50% (Note S1).
Figure 1.
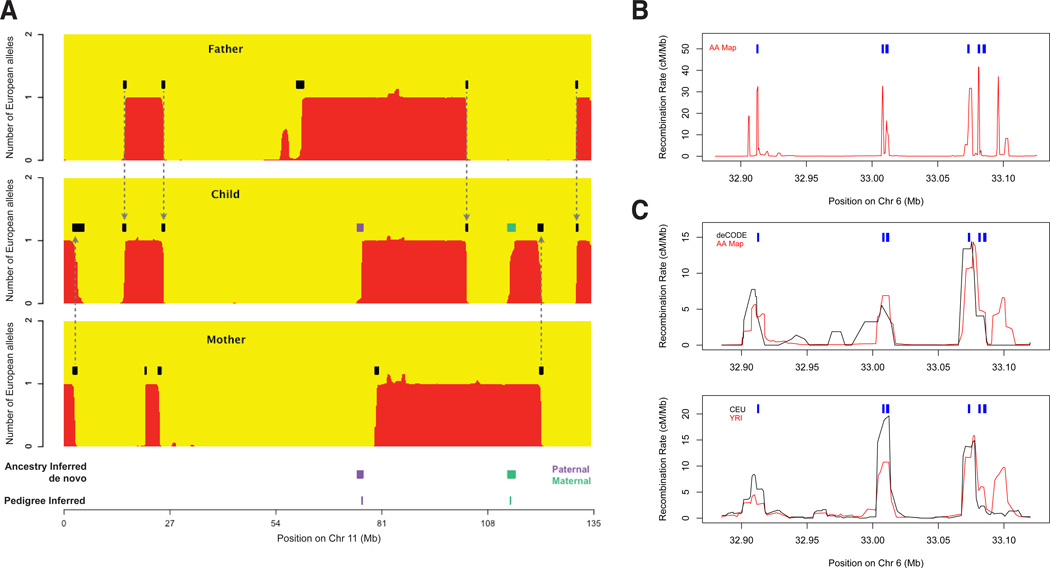
Building an African American genetic map. (A) HAPMIX detection of crossovers between segments of inferred ancestry is illustrated in a father-mother-child trio. Black segments show inferred crossovers; arrows showtransmission of ancestral crossovers from parent to child, Purple/green segments show de novo events (paternal/maternal origin respectively), corresponding to events identified directly using two additional children (bottom,“Pedigree inferred”).(B) The AA Map localizes five hotspots in a region of the MHC whose positions (blue) were previously mapped by sperm typing1. (C) Comparison of maps shows a hotspot at 33.1Mb in the African-derived AA and YRI maps, but not the deCODE and CEU maps (all maps smoothed to 10kb).
To build a high resolution African American genetic map (AA Map), we leveraged the fact that most crossovers occur in hotspots shared across individuals1 (Methods). Intuitively, while any crossover can only be roughly localized, inter-SNP intervals that are inferred to have an appreciable probability of crossover in multiple individuals are likely to contain recombination hotspots, allowing much better localization (Figure S1). To implement this idea, we modeled the recombination rate for each inter-SNP interval as shared across individuals, and used a Markov Chain Monte Carlo (MCMC) to sample rates consistent with the data (Methods). This provides well-calibrated estimates of the crossing-over rate between all pairs of markers as well as estimates of rate uncertainty (Note S1 and Figure S2). We find that the interval size at which the average recombination rate is equal to the standard error is 6 kb, which is the same accuracy that would be expected from a map based on 500,000 crossovers whose boundaries were precisely resolved (Note S1). Despite this high resolution, there are also some limitations. First, the AA Map does not separately infer male and female recombination rates (it is a sex-averaged map) and requires normalization by the total map length (like LD maps3,18). Second, the map has less resolution and may miss a higher fraction of true crossovers at loci where it is more difficult to detect and resolve crossovers due to low SNP density or low differentiation between West Africans and Europeans. Third, the map may be biased where ancestry deviates from the average, for example at 8q24, where the 10% of the people in this study who have prostate cancer have an elevated proportion of African ancestry19. Fourth, the map assumes that all individuals are unrelated, whereas in fact there is likely some shared ancestry, resulting in multiple counting of some crossovers and an overestimate of map precision.
To assess the accuracy of the AA Map, we generated an independent African American pedigree map by analyzing 222 nuclear families that included 1,056 meioses in which we could directly detect crossovers between parent and child (Methods; Figure 1A). Examination of the AA Map rate around directly detected crossovers confirms the high resolution: the rate around such crossovers shows at least as strong a peak as that observed in maps based on LD2,3,18 (Figure S3). We next computed correlation coefficients for both the AA Map and the deCODE Map4 to maps derived from the breakdown of LD in Europeans (CEU) and West Africans (YRI)18. At broad scales (>3 Mb) they are almost identical (ρ>0.97; Table 1) At fine scales, the AA Map is more accurate (Table 1 and Table S1), as reflected in a modest improvement in correlation to the CEU Map at a 3kb scale (ρAA,CEU(3kb) = 0.66 vs. ρdeCODE,CEU(3kb) = 0.58), and a major improvement for the YRI Map (ρAA,YRI(3kb) = 0.71 vs. ρdeCODE,YRI(3kb) = 0.53). The deCODE Map is more correlated to the CEU Map than to the YRI Map at scales <1 Mb, suggesting that this map, built in Icelanders, reflects more European recombination rates. The AA Map shows the opposite pattern, suggesting that it reflects more West African recombination patterns.
Table 1.
Genetic map assessments at different size scales
| Scale (interval size) |
ρ - Pearson correlation of the AA map (deCODE Map) to the specified LD map |
Est. correlation of AA Map to the true map (inferred by MCMC)* |
Est. coefficient of variation of AA Map (std. err. divided by crossover rate expected for interval size)* |
||
|---|---|---|---|---|---|
| Combined LD§ | CEU | YRI | |||
| 3 kb | 0.75 (0.63) | 0.66 (0.58) | 0.71 (0.53) | 0.93 | 1.41 |
| 10 kb | 0.82 (0.74) | 0.73 (0.70) | 0.78 (0.65) | 0.96 | 0.73 |
| 30 kb | 0.86 (0.83) | 0.78 (0.78) | 0.83 (0.74) | 0.98 | 0.36 |
| 100 kb | 0.91 (0.89) | 0.84 (0.85) | 0.87 (0.81) | 0.99 | 0.17 |
| 300 kb | 0.94 (0.93) | 0.89 (0.90) | 0.92 (0.88) | 1.00 | 0.08 |
| 1 Mb | 0.97 (0.96) | 0.94 (0.94) | 0.95 (0.95) | 1.00 | 0.04 |
| 3 Mb | 0.98 (0.98) | 0.97 (0.97) | 0.98 (0.97) | 1.00 | 0.02 |
Note: The numbers in this table are restricted to the autosomes and genomic segments more than 5 Mb from the telomere.
The Combined map is the HapMap2 population-averaged LD-based map1818.
The standard error of the map at each size scale is determined by the posterior probability distribution of the MCMC.
We compared the rate estimates for all four maps (AA, deCODE, CEU and YRI) over a 200 kb region within the MHC locus where recombination rates in European males have been characterized through sperm typing1 (Figure 1B). The AA Map detects five of six known hotspots, and localizes them to within 1 kb (the sixth hotspot is weak, with a peak male rate below the genome average1). Strikingly, the two maps based on samples with African ancestry (AA and YRI) found a hotspot not present in either map based on samples of European ancestry (deCODE and CEU) (Figure 1C; Figure S4 gives a second example). We confirmed that such “African-enriched” hotspots also occur genome-wide, by examining 2,375 loci with recombination rate peaks in the YRI Map (>5 cM/Mb) but not the CEU Map (<1 cM/Mb), and finding a rate rise in the independently generated AA Map, but not in the deCODE Map (Figure S5A). In the reciprocal experiment searching for European-specific hotspots, we find no such evidence for genuine ancestry specificity; at loci with recombination rate peaks in the CEU Map but not the YRI Map, there are weak peaks in both the deCODE and AA maps (Methods; Figure S5B). Thus, hotspots active in Europeans are consistently “shared” with YRI and African Americans, while populations with African ancestry harbor additional, non-shared hotspots we call “African-enriched”.
To understand the features of recombination in West Africans that differ from Europeans, we estimated the degree to which each African American person’s crossovers occur in “African-enriched” hotspots, compared with “Shared” hotpots, a phenotype we refer to as their “African-enrichment” (AE). We view each individual’s crossovers as sampled from a mixture of two genetic maps—an “S Map” of shared hotspots based on the deCODE Map, and an “AE Map” of African-enriched hotspots that is learned from comparing the deCODE and AA Maps—so that the proportion of crossovers assigned to the AE Map is a person’s AE phenotype (Note S4). We tested approximately 3 million SNPs (genotyped and imputed) for association with three phenotypes: AE, usage of LD-based hotspots known to be enriched for the 13-bp motif CCNCCNTNNCCNC20, and genome-wide crossover rate (in pedigrees) (Methods and Note S4). In crossovers detected in unrelated African Americans, the alleles a person carries are only sometimes descended from the ancestor in whom the crossover occurred, thus adding noise to the association signal (nevertheless there is useful signal given the large sample size; Note S4). In the Pedigree Map, association between alleles and AE can be tested directly because we have genotypes in the parents.
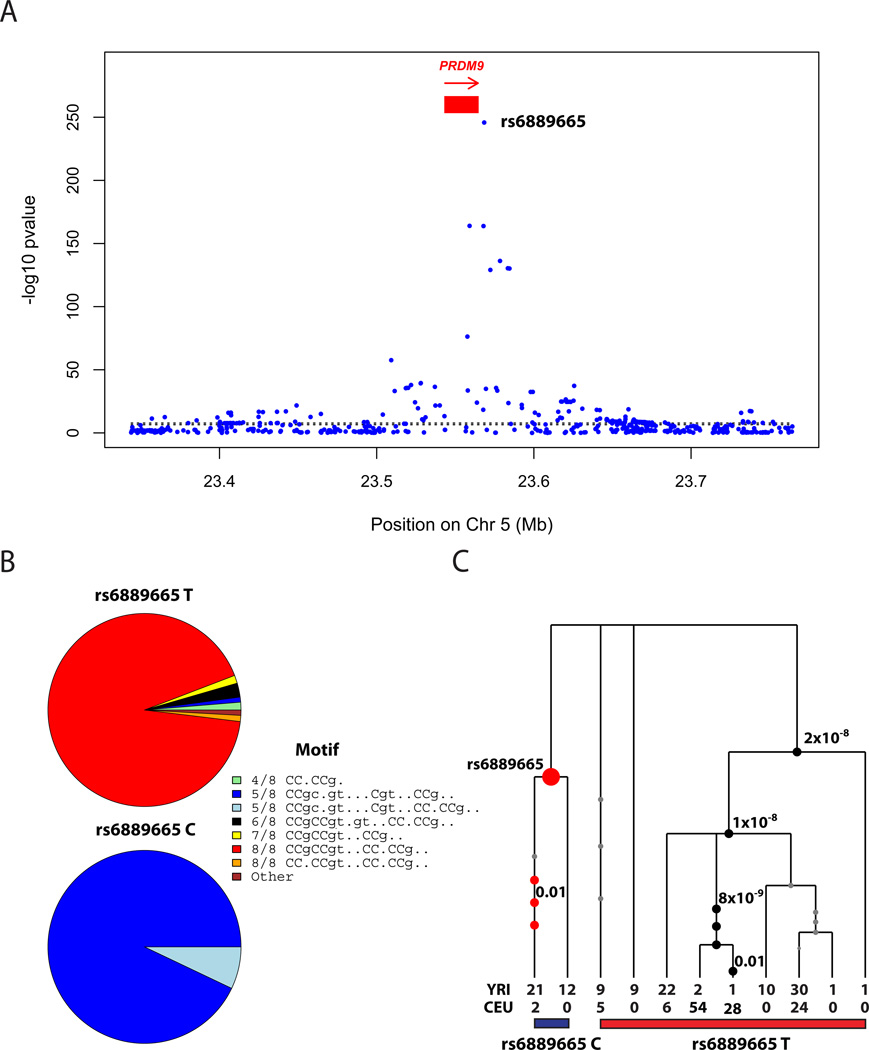
The SNP showing the strongest association with AE is rs6889665 (P=1.5×10−246; Figure 2A, Figure S6), which has a derived allele frequency of 29% in YRI and 2% in CEU, and is within 4 kb of the ZF array of PRDM94,9,11,12,13. This SNP is associated with AE in both the pedigree individuals and the unrelated individuals (Note S4), and is also the SNP most strongly associated with usage of LD-based hotspots (P=1.8×10−52) (Table S2). No locus outside PRDM9 is significant (P<0.01 after Bonferroni correction; Table S3). To better understand the association at rs6889665, we inferred the alleles in the PRDM9 ZF array carried by 139 individuals based on sequencing data from the 1000 Genomes Project21, using the reads to infer each individual’s PRDM9 alleles among 29 alleles whose full sequences were previously determined9 (Note S5). Grouping PRDM9 alleles based on how closely their binding target predictions match the 13-bp motif, following Berg et al.9, we find that the ancestral “T” variant at rs6889665 is strongly correlated to “8/8 matches” to the 13-bp motif (including the “A” and “B” alleles ), while the derived “C” variant is almost perfectly correlated to a group of “5/8 match” alleles, all predicted to bind a common, different, 17-bp motif “CCgCNgtNNNCgtNNCC”9. This implies a common historical origin for alleles matching this 17-bp motif (Figure 2B; Figure S7; Note S5). We also experimentally measured the number of zinc fingers in PRDM9 in 354 individuals including 166 African Americans from the pedigree study (Methods). This showed, again, that rs6889665 differentiates PRDM9 alleles into two different classes, with 96% of haplotypes carrying the ancestral allele having <14 zinc fingers, and 93% of haplotypes carrying the derived allele having ≥14 zinc fingers (Figure S7). After conditioning on rs6889665, there is no evidence that ZF length is associated with the AE phenotype. Several SNPs near the PRDM9 ZF array show a conditional association signal that is much weaker than rs6889665, but still significant (Figure 2C; Figure S6; Note S4), with the strongest at rs10043097 (P= 8.3×10−14), upstream of the PRDM9 transcription start site. These SNPs may tag additional variation in PRDM9 ZF array, or potentially expression levels.
Figure 2.
Association of PRDM9 genetic variation with hotspot activity. (A) A GWAS measuring association of the “African-enrichment” (AE) phenotype shows a single genome-wide significant peak at PRDM9, with rs6889665 the best associated SNP. (B) Relationship between alleles at the rs6889665 and predicted binding target of the PRDM9 zinc finger array9 for West African and European samples. The alleles are grouped into 8 clusters according to their best-matching region to the 13-bp motif, and annotated by the number of bases matching the motif. The African-enriched rs6889665 “C” allele always co-occurs with motifs with a poor (5/8) match to the 13-mer. (C) Gene tree32 of the LD block containing the PRDM9 ZF array (Methods); numbered circles show SNPs and significant P-values for association, after conditioning on rs6889665.
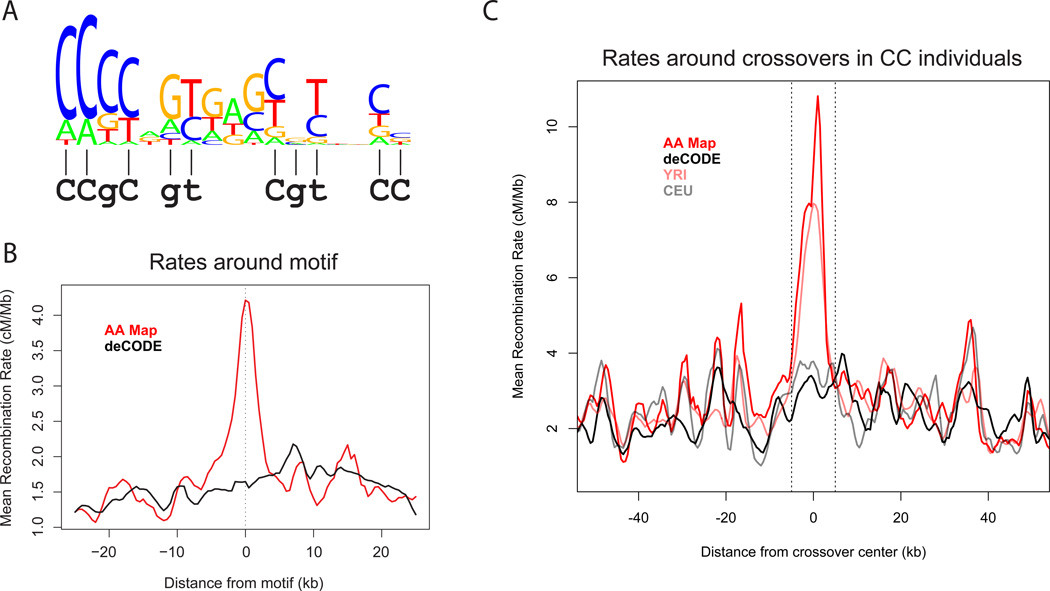
To directly identify candidate African-enriched hotspot motifs, we selected 2,454 loci with a high crossover rate in the AE Map and YRI Map (>2cM/Mb over 2kb), and no more than half this rate in the S Map and CEU Map (this set is more powerfully enriched for higher recombination in people of African ancestry than the 2,375 above, as it includes information from the contemporary maps). We compared these to a “control set” of 7,328 candidate hotspots more active in the European than the African derived maps (Methods; Note S6). To identify sequence motifs associated with the African-enriched hotspots3,22, we identified short motifs that occurred at increased frequency in the African-enriched hotspot set (Note S6). Testing all motifs of length 5–9 bases revealed a 9-mer “CCCCAGTGA” (OR=1.79, P=2.24×10−8, Bonferroni corrected P=0.004) which exhibited a kilobase-scale rate peak near occurrences of this motif in African derived maps, but in neither of the European derived maps (Figure S8). Further analysis revealed a strong influence of downstream flanking bases (Figure S9), and degeneracy, yielding a 17-bp consensus sequence “CCCCaGTGAGCGTtgCc” (Figure 3A; more strongly signaled bases are uppercase) with the same consensus obtained when we considered flanking sequence for only odd or even chromosomes, and whether we based the analysis on AE-S or YRI-CEU map comparisons (Note S6). The 500 best matches to this motif have a ~3-fold increase in average rate in the AA and YRI relative to the deCODE and CEU maps (Figure 3B, Figure S8). Hotspots associated with the motif occur in both unique and repetitive DNA (e.g. L1PA10/13 LINE elements; Figure S10) (Note S6). We also compared the 17-bp consensus to the binding motif predicted for “5/8 match” alleles, and found that they match almost precisely (Figure 3A; 10 of 11 bases, P=8.1×10−6).
Figure 3.
A sequence motif specifying the positions of African-enriched hotspots. (A) Logo plot showing a degenerate 17-bp hotspot motif, with stack height proportional to -log P-value, and relative letter height proportional to the mean crossover rate increase given each base. Below is the bioinformatic PRDM9 binding prediction for the rs6889665 AE associated alleles (from Figure 2B), matching the motif at 10/11 bases (lines). (B) Average crossover rate (in 2 kb sliding windows) in the AA (red line) and deCODE (black line) maps surrounding the 500 strongest motif matches. (C) In seven rs6889665 “CC” individuals from the pedigree study, we localized 82 crossovers to within 10 kb, and plot average AA, YRI, deCODE and CEU map rates. There is no strong peak above local background in the deCODE or CEU maps.
How much of the African-enriched recombination pattern can be explained by PRDM9? We estimated the fraction of variation in the AE phenotype explained by rs6889665 in our pedigree data after accounting for noise in the phenotype estimation (Note S4). Over 82% of map usage variability is explained by rs6889665 genotype alone. Given there are further influential PRDM9 variants (Figure 3C), this gene may thus explain almost all differences in local rate between the West African and European populations. We next examined rates around 82 narrowly defined (<10kb) crossover sites in 7 individuals homozygous for the derived allele at rs6889665. There is no evidence of hotspots at these loci in either the deCODE or CEU Maps (Figure 3C), in contrast to events in individuals carrying the ancestral allele at rs6889665 (Figure S11). Thus, crossover positions in individuals who are homozygous for the derived allele at rs6889665 are consistent with an entirely different recombination hotspot landscape, which would imply PRDM9 control of all hotspots9. Despite the strong correlation between maps at megabase scales, there is mounting evidence that PRDM9’s influence on crossing-over may not be limited to fine scales4,11: we observe a weakly significant association of rs6889665 with the total number of crossovers genome-wide in pedigrees (P=0.04), corresponding to an average 1.3 crossovers more per meiosis per derived allele, exceeding the strongest previously known association23 at RNF212.
We have shown that PRDM9 alleles that bind a novel 17-bp motif and occur at greatly increased frequency in people of West African ancestry have led to a shift in the recombination landscape compared with people of non-African ancestry. The larger number of hotspots available to West Africans implies that at the population level, crossovers are more evenly distributed than in Europeans10, and thus the shorter extent of West African LD is not due to differences in demographic history alone (such as the lack of an out-of-Africa founder event)24. Our findings also have medical implications, as recombination errors leading to insertions or deletions are known to be associated with recombination hotspots9,22,25. Our results predict that the congenital abnormalities that have been associated with the recombination hotpots bound by PRDM9 “A” and “B” alleles will occur at a decreased rate in people of West African ancestry, whereas new diseases will arise due to recombination errors near African-enriched hotspots.
METHODS SUMMARY
We assembled SNP array data from 29,589 unrelated people and 222 nuclear families genotyped at 490,000–910,000 SNPs from the Candidate Gene Association Resource (CARe), studies at the Children’s Hospital of Philadelphia (CHOP), the African American Breast Cancer Consortium, the African American Prostate Cancer Consortium and the African American Lung Cancer Consortium. To build a recombination map, we used HAPMIX to localize candidate crossover positions15, and implemented a Markov Chain Monte Carlo (MCMC) that used the probability distributions for the positions of the filtered crossovers to infer recombination rates for each of 1.3 million inter-SNP intervals. We also implemented a second MCMC that models each individual’s set of crossovers as a mixture of a Shared (S) Map similar to the European deCODE Map and an African-enriched (AE) Map, and then assigns each individual an “AE phenotype” corresponding to the proportion of their newly detected crossovers assigned to the AE Map. We imputed genotypes at up to three million HapMap2 SNPs8 using MaCH26, and then tested each of these SNPs for association with the AE phenotype and other recombination-related phenotypes. We identified 2,454 candidate African-enriched hotspots with increased recombination rates in the YRI vs. CEU maps, and in the AE vs. S maps, and searched for motifs enriched at these loci, thus identifying a degenerate 17-bp motif. To study the structure of PRDM9, we measured the length of the PRDM9 zinc finger array and genotyped rs6889665 in YRI, CEU and the CARe nuclear families; we also carried out imputation based on 1000 Genomes Project short read data10 to infer the alleles individuals carry, among 29 previously characterized in a sequencing study of PRDM99.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the participants who donated DNA samples, to David Altshuler, Jerome Buard, Kasia Bryc, Joseph Kovacs, Bernard de Massy, Gil McVean, Bogdan Pasaniuc and Sriram Sankararaman for conversations and critiques, and to Adam Auton for facilitating analysis of the 1000 Genomes Project data.
Analysis was supported by the Wellcome Trust and NIH grants HL084107 and GM091332.
CARe was supported by a contract from the National Heart, Lung and Blood Institute (HHSN268200960009C) to create a phenotype and genotype database for dissemination to the biomedical research community. Eight parent studies contributed phenotypic data and DNA samples through the Broad Institute (N01-HC-65226): the ARIC, CFS, CARDIA, JHS, and MESA studies, as well as the Cardiovascular Health Study (CHS), the Framingham Heart Study (FHS), and the Sleep Heart Health Study (SHHS). Support for CARe also came from the individual research institutions, investigators, field staff and study participants. Individual funding information is available at http://public.nhlbi.nih.gov/GeneticsGenomics/home/care.aspx.
All genome-wide genotyping of the CHOP dataset was supported by an Institutional Development Award to the Center for Applied Genomics from the Children's Hospital of Philadelphia, a research award from the Landenberger Foundation and the Cotswold Foundation. We thank all study participants and the staff at the Center for Applied Genomics for performing the genotyping.
AABCC was supported by a DoD Breast Cancer Research Program Era of Hope Scholar Award to CAH and the Norris Foundation, and by grants to the component studies: MEC (CA63464, CA54281); CARE (HD33175); WCHS (CA100598, DAMD 170100334, Breast Cancer Research Foundation); SFBC (CA77305, DAMD 17966071); CBCS (CA58223, ES10126), PLCO (NCI Intramural Research Program); NHBS (CA100374), WFBC (R01-CA73629) and CPS-II (the American Cancer Society).
AAPCC was supported by grants CA63464, CA54281, CA1326792, CA148085 and HG004726, and by grants to the component studies: PLCO (NCI Intramural Research Program), LAAPC (Cancer Research Fund 99-00524V-10258), both MEC and LAAPC (PC35139, DP000807); MDA (CA68578, CA140388, ES007784, DAMD W81XWH0710645); GECAP (ES011126); CaP Genes (CA88164); IPCG (W81XWH0710122); DCPC (GM08016, DAMD W81XWH0710203, DAMD W81XWH0610066); SCCS (CA092447, CA68485).
AALCC was supported by grants CA060691, CA87895, PC35145 and CA22453, CA68578, CA140388, ES007784, ES06717, CA55769, CA127219, CA1116460S1, CA1116460, CA121197, CA141716, CA121197S2, CPRIT RP100443, CA148127, DAMD W81XWH0710645, University Cancer Foundation, Duncan Family institute, Center for Community, Implementation, and Dissemination Research Core, and by grants to the component studies: PLCO and the Maryland Studies (NCI Intramural Research Program), LAAPC (Cancer Research Fund 99-00524V-10258), and both MEC and LAAPC (PC35139, DP000807).
Footnotes
AUTHOR CONTRIBUTIONS
DR and SRM conceived the study. AGH, AT, NP, YS, NR CDP, GKC, KW, SGB, DR and SRM performed analyses. NR performed the experimental work (genotyping of polymorphisms at PRDM9). AGH, NP, JNH, BEH, HAT, ALP, HH, SJC, CAH, JGW, DR and SRM coordinated the study. AGH, DR and SRM wrote the paper. NR, CDP, GKC, KW, SGB, SR, JNH, BEH, HAT, HH, CJC, CAH, JGW, DR and all the alphabetically listed authors contributed to sample collection and generation of SNP array data. All authors contributed to revision and review of the manuscript.
AUTHOR INFORMATION
Crossover rate estimates for the AA Map, Pedigree Map, AE Map and S Map can be found at http://www.well.ox.ac.uk/~anjali/AAmap/. We also provide estimates of uncertainty for each map based on samples from the Markov Chain Monte Carlo. Association testing results for each SNP are available from the authors on request.
REFERENCES
- 1.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 2001;29:217–212. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 2.McVean GA, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 3.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 4.Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 5.Kong A, et al. A high-resolution recombination map of the human genome. Nature Genetics. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 6.Matise TC, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weitkamp LR. Proceedings: Population differences in meiotic recombination frequency between loci on chromosome 1. Cytogenet Cell Genet. 1974;13:179–182. doi: 10.1159/000130267. [DOI] [PubMed] [Google Scholar]
- 8.Jorgenson E, et al. Ethnicity and human genetic linkage maps. Am J Hum Genet. 76;2005:276–290. doi: 10.1086/427926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nature Genetics. 2010;10:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers S, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith MW, et al. A high-density admixture map for disease gene discovery in African Americans. Am J Hum Genet. 2004;74:1001–1013. doi: 10.1086/420856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price AL, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. Am J Hum Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patterson N, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The International Haplotype Map Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman ML, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 21.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1128. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 23.Kong A, et al. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science. 2008;319:1398–1401. doi: 10.1126/science.1152422. [DOI] [PubMed] [Google Scholar]
- 24.Reich DE, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 25.Raedt TD, et al. Conservation of hotspots for recombination in low-copy repeats associated with the NF1 microdeletion. Nat Genet. 2006;38:1419–1423. doi: 10.1038/ng1920. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiology. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.