Abstract
The feasibility of using solid-state MAS NMR for in situ structural characterization of the LR11 (sorLA) transmembrane domain in native Escherichia coli (E. coli) membranes is presented. LR11 interacts with the human amyloid precursor protein (APP), a central player in the pathology of Alzheimer's disease. The background signals from E. coli lipids and membrane proteins had only minor effects on LR11 TM resonances. Approximately 50% of the LR11 TM residues were assigned by using 13C PARIS data. These assignments allow comparisons of the secondary structure of LR11 TM in native membrane environments and in commonly used membrane mimics (e.g. micelles). In situ spectroscopy bypasses several obstacles in the preparation of membrane proteins for structural analysis, and offers an opportunity to investigate the consequences of membrane heterogeneity, bilayer asymmetry, chemical gradients, and macromolecular crowding on the protein structure.
Integral membrane proteins reside in a complex lipid environment. The complexity of cellular membranes is reflected in their diverse lipid composition (>1,000 different lipid species), lateral heterogeneity (e.g. lipid rafts, lipid microdomains), transbilayer asymmetry, chemical and electrical gradients, dynamics, and shapes.1–5 Membranes are no longer viewed as simple passive barriers that separate cells from their environments, but rather as active participants in important biological processes such as intracellular signal transduction, protein localization and protein trafficking.6–8 Biological membranes are crowded and contain as much protein as they do lipid.9,10 The implications of this intramolecular crowding are increasingly recognized.11–14 Although the unique lipid environment is a major determinant of membrane protein conformation and function, this environment is incompatible with the conventional methods of X-ray crystallography and solution NMR. Consequently, our structural knowledge of membrane proteins lags far behind that of soluble proteins, despite the fact that membrane proteins account for approximately 30% of all proteins in the human genome, including biologically crucial molecules such as ion channels and G-protein coupled receptors. As of April 2011, there are only ~280 unique membrane protein structures in the Protein Data Bank, mostly from prokaryotes.
The importance of membrane mimetic environments in supporting the native structure, dynamics and function of membrane proteins has been highlighted recently.23–26 So far, most structural analyses were carried out in detergent preparations and only a few in synthetic lipid bilayers. Information about protein structure in biological environments is scarce.15–18 Bacteriorhodopsin is the only protein that has been subjected to in situ detailed NMR structural characterizations in native purple membranes,19–22 thanks to its natural abundance. Recent developments in the condensed Single-Protein-Production (cSPP) system have allowed the detection of membrane proteins without purification.27–29 Here, we demonstrate the feasibility of characterizing the transmembrane domain (TM) of a human protein, LR11/SorLA, in situ in E. coli membranes by using solid-state magic-angle spinning (MAS) NMR.
LR11 is a recently identified type I transmembrane protein involved in the development of Alzheimer's disease (AD). AD causes a gradual loss of memory and general cognitive decline. It is the most common form of dementia in the elderly and currently affects more than 5.4 million Americans (http://www.alz.org). The “amyloid hypothesis” suggests that the accumulation of Aβ peptides, proteolytic products of amyloid precursor protein (APP), is the primary cause of AD.30,31 APP is a type I transmembrane protein and is continuously sorted through multiple subcellular organelles (e.g. trans-Golgi network, plasma membrane and endosome). Its aberrant intracellular trafficking is linked to the development of AD.
LR11 has emerged as a critical regulator for APP transport and processing.32–36 LR11 interacts directly with APP, regulating its subcellular localization. Variants of LR11 are associated with AD, and the expression of LR11 is dramatically decreased in the brains of patients suffering from sporadic AD.32,37,38 The transmembrane domains of LR11s from mammals are highly conserved and share >95% sequence identity, pointing to their potential functional significance. Using a new MBP-fusion expression vector, we produced human LR11 TM (residues 2132 to 2161, Figure 1a) in E. coli.39 The recombinant protein is expressed in the membranes at a much higher level relative to the background of E. coli membrane proteins, as shown in Figure 1b. We have developed a protocol to cleave MBP at the native membrane surface and have obtained LR11 TM in E. coli membranes through ultracentrifugation and buffer washes. The SDS-PAGE result for the prepared sample is also shown in Figure 1b. LR11 TM consists of 70~80% of total labeled proteins.
Figure 1.
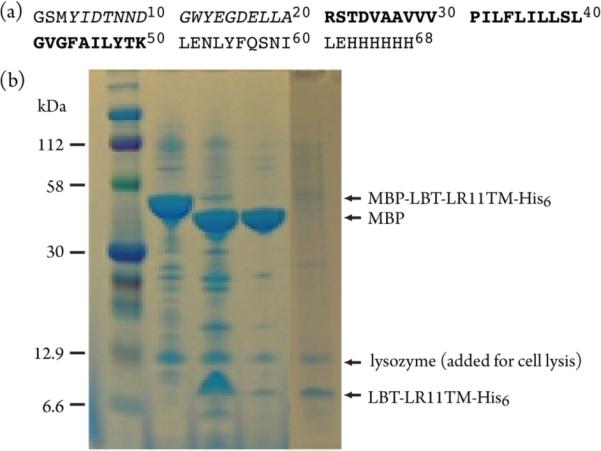
(a) Primary structure of the LBT-LR11TM-His6. The LR11 fragment is shown in bold, corresponding to residues 2132 to 2161 of the full-length protein. The LBT (lanthanide binding tag) is shown in italics. (b) SDS-PAGE results for the preparation of LR11 TM in native E. coli membranes. Lanes: 1, protein marker; 2, isolated E. coli membrane fraction; 3, thrombin cleavage of the sample in lane 2; 4, buffer washes of the sample in lane 3; 5, prepared membrane fraction for NMR experiments.
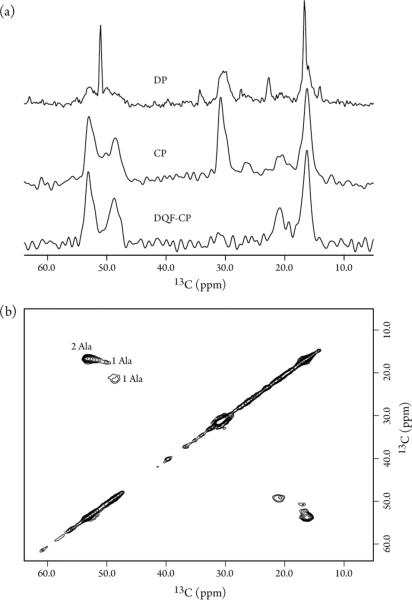
To examine sample homogeneity, spectral sensitivity and resolution, and the interference of background signals from E. coli proteins and lipids, 13C MAS NMR spectra were acquired on a 13Cα,β-Alanine enriched LR11 TM in isolated membranes. The phospholipid composition of E. coli from exponentially grown cultures is simple, mainly phosphatidylethanolamine, phosphatidylglycerol and cardiolipin.40 Despite the fact that lipids are not labeled in this preparation, their naturally abundant signals overwhelm the 1D spectrum collected with a single 90°-pulse direct polarization (DP) experiment (Figure 2a, top). The resonances at ~17 and 52 ppm are relatively sharp and likely come from the flexible lipid headgroups and methyl carbons, while the resonance at ~31 ppm comes from the lipid methylene groups. The narrow resonances are effectively suppressed in the 1H-13C cross polarization (CP) experiment, while the resonances from more rigid regions are greatly enhanced (Figure 2a, middle). The lipid 13C signals can be further suppressed in the double-quantum filter (CP-DQF)41 experiment and thus resonances solely from 13Cα,β-Alanine enriched proteins are detected (Figure 2a, bottom). These DP, CP and CPDQF spectra were collected in ~45, 17 and 70 minutes, respectively, on a 600 MHz spectrometer, suggesting that the sensitivity is sufficient for multidimensional NMR experiments to improve spectral resolution.
Figure 2.
13C MAS spectra of 13Cα,β-Ala enriched LR11 TM in native E. coli membranes recorded at 305 K on a Bruker 600 MHz spectrometer using a home-built low-E 3.2 mm probe. The spinning rate was 10 kHz. (a) 1D spectra recorded using DP, CP and CP-DQF polarization schemes with 512, 512 and 2048 scans, 5, 2 and 2s recycle delays, respectively. (b) A 2D 13C-13C PARIS spectrum collected with 20 ms mixing time, 9.2 and 7.0 ms acquisition time for direct and indirect dimensions, 1.5 s recycle delay and 512 scans per t1 point.
A 2D 13C-13C PARIS42 spectrum is shown in Figure 2b. The resonance line-width is ~1.0 ppm which is typical for non-crystalline samples and indicates good homogeneity of the preparation. Two well-resolved and two partially overlapped cross peaks are observed in the Ala Cα-Cβ chemical shift region as expected from the four Ala residues in the protein sequence, consistent with the resonances arising from LR11 TM. One cross peak at 50.1 and 17.6 ppm is much weaker and has a slow PARIS buildup. This can be tentatively attributed to the relatively flexible residue of Ala20 at the N-terminus of the TM domain.
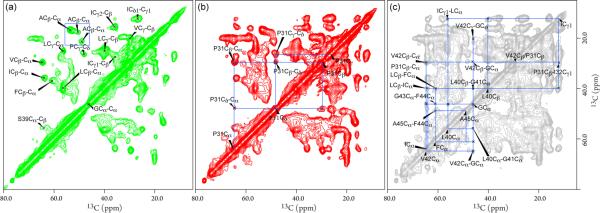
To pursue resonance assignments and validate the secondary structure of the TM domain, 2D 13C PARIS spectra with various mixing times were collected on a uniformly 13C, 15N enriched sample. The 13C-13C correlations are generated by through-spaced dipolar couplings, thus at a short mixing time of 5 ms most of the cross peaks result from directly bonded 13C sites (Figure 3a). Despite having lipids that are also 13C enriched, lipid resonances do not interfere with the protein resonances because CP and dipolar-coupling mediated magnetization transfer select against relatively flexible lipid resonances. The Ala Cα-Cβ cross peak region, highlighted in the blue box, is identical to the above spectrum from the 13Cα,β-Ala labeled sample. Based on their characteristic chemical shifts and spin systems, the resonances of Ile, Ser, Val, Leu, and Gly are easily identified. The cross peak at 37.1 and 61.1 ppm was assigned to Cβ-Cα of Phe, with the aid of its connectivity to resonances in the aromatic region (data not shown). The cross peak at 26.4 and 47.8 ppm was attributed to Cγ-Cδ of Pro based on its unique chemical shifts and connectivity at a longer mixing time (see below). Thus, all amino acid residue types of the LR 11 TM domain were readily identified. The Cα and Cβ chemical shifts of Ile at 63.9 and 35.8 ppm, Leu at 56.2 and 40.0 ppm, Phe at 60.3 and 37.1 ppm, and Val at 64.3 and 29.7 ppm are indicative of an α-helical backbone conformation.43 For helical membrane proteins, the overlapping cross peaks of the same amino acid type are common. Resonances from some of the tag residues are also detected (e.g. the cross peak at 53.6 and 40 ppm is likely from Asp and/or Asn), but they generally show a signature of chemical exchange broadening due to the relative flexibility of the tags.
Figure 3.
2D 13C-13C PARIS spectra of 13C, 15N uniformly enriched LR11 TM in native E. coli membranes recorded for resonance assignment with (a) 5 ms mixing time and 96 scans per t1 point; (b) 20 ms mixing time and 112 scans per t1 point; (c) 100 ms mixing time and 128 scans per t1 point. The acquisition times for direct and indirect dimensions were 10.3 and 4.8 ms, and the recycle delay was 1.5 s.
Figures 3b and 3c show 2D 13C-13C PARIS spectra acquired with mixing times of 20 and 100 ms, respectively. The long mixing times permit magnetization transfers between 13C spins, separated by multiple bonds or from different residues, and provide connectivity for resonance assignment. Starting from the cross peak of Pro Cγ-Cδ, the cross peaks of Cβ-Cδ and Cδ-Cα were identified in Figure 3b. Since there is only one Pro residue in the LR11 TM sequence, the Cα and Cβ chemical shifts for residue Pro31 were obtained. Ile32 was subsequently assigned based on its Cγ connectivity to the Cβ of Pro31 in Figure 3c. Six PARIS cross peaks at 64.6, 60.4, 56.2, 40.7, 29.6, and 21.9 ppm were observed for Gly at 46.1 ppm with 100 ms mixing time, and they were assigned to Cα of Val, Phe, and Leu, and Cβ of Leu, Val, and Ala, respectively, based on the amino acid type information in Figure 3a. Furthermore, a cross peak between Phe Cα at 61.1 ppm and Ala Cα at 48.4 ppm was observed. These connectivities were mapped to the LGVGFA fragment in the TM sequence. In addition, several cross peaks between Leu and Ile and between Phe and Leu were identified, but could not be unambiguously assigned to specific sites without additional data. Most of the unassigned peaks in Figure 3a come from residues of LBT and His tags, and a few of them might be E. coli background signals. From 13C-13C PARIS data, we have readily assigned 12 out of 23 residues of the LR11 TM and their chemical shifts are listed in Table S1. All assigned residues show characteristic secondary shifts of an α-helix and are in agreement with the secondary shifts44 of LR11 TM in DPC micelles (listed in Table S1) except for residue Ala45. This residue is near the C-terminus of the predicted TM domain and resides in the membrane-solution interface region, where there are substantial differences between bilayers and micelles and where structural discrepancy likely occurs.
Our studies have demonstrated the feasibility of in situ detection of the human LR11 TM domain in native E. coli membranes. The spectral sensitivity and resolution are adequate for a structural analysis of this small protein. Signals from lipids and membrane proteins of E. coli are minimally interfered with the detection of LR11 TM resonances. By using 13C-13C homonuclear correlation experiments, we have assigned ~50% of TM residues. Their secondary chemical shifts are consistent with expected values for an α-helix conformation. Most of the unassigned residues are Leu and Val because of their high abundance in the sequence. We expect that the spectral resolution can be improved further by using multidimensional heteronuclear correlation experiments and advanced enrichment strategies.45–51
Although the composition of E. coli membranes differs from that of human cells, in situ detection eliminates the use of detergents for extraction, purification and reconstitution of recombinant membrane proteins. Moreover, our approach offers an opportunity to validate and refine membrane protein structures in a native environment and investigate the consequences of membrane heterogeneity, bilayer asymmetry, chemical gradients, and macromolecular crowding on the protein structure, characteristics which cannot be addressed in studies using detergent micelles and synthetic lipid bilayers.
Supplementary Material
ACKNOWLEDGMENT
We are grateful for financial support from the National Institutes of Health (5R01GM081793-03 and 5DP1OD783), the NSF (MCB 1051819), the Penn State University College of Medicine and National Natural Science Foundation of China (21075134). We thank Drs. J. M. Flanagan and T. A. Cross for helpful discussions. The solid-state MAS NMR measurements were performed at the National High Magnetic Field Laboratory, which is supported by NSF Cooperative Agreement No. DMR-0654118, the State of Florida, and the U.S. Department of Energy.
Footnotes
SUPPORTING INFORMATION. Sample preparation, experimental details, chemical shifts of LR11 TM in E. coli membranes and DPC micelles, complete references 10, 34 and 38. This material is available free of charge via Internet at http://pubs.acs.org.
REFERENCE
- (1).Engelman DM. Nature. 2005;438:578–580. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- (2).Simons K, Gerl MJ. Nat. Rev. Mol. Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- (3).van Meer G, Voelker DR, Feigenson GW. Nat. Rev. Mol. Cell Bio. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Semrau S, Schmidt T. Soft Matter. 2009;5:3174–3186. [Google Scholar]
- (5).Shevchenko A, Simons K. Nat. Rev. Mol. Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- (6).Doucleff M. Cell. 2010;143:853–854. [Google Scholar]
- (7).Groves JT, Kuriyan J. Nat. Struct. Mol. Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Phillips R, Ursell T, Wiggins P, Sens P. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Dupuy AD, Engelman DM. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2848–2852. doi: 10.1073/pnas.0712379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Takamori S, et al. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- (11).Mika JT, Poolman B. Curr. Opin. Biotechnol. 2011;22:117–126. doi: 10.1016/j.copbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- (12).Zhou HX. J. Phys. Chem. B. 2009;113:7995–8005. doi: 10.1021/jp8107446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Aisenbrey C, Bechinger B, Grobner G. J. Mol. Biol. 2008;375:376–385. doi: 10.1016/j.jmb.2007.10.053. [DOI] [PubMed] [Google Scholar]
- (14).Bokvist M, Grobner G. J. Am. Chem. Soc. 2007;129:14848–+. doi: 10.1021/ja076059o. [DOI] [PubMed] [Google Scholar]
- (15).Goldbourt A, Gross BJ, Day LA, McDermott AE. J. Am. Chem. Soc. 2007;129:2338–2344. doi: 10.1021/ja066928u. [DOI] [PubMed] [Google Scholar]
- (16).Lorieau JL, Day LA, McDermott AE. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10366–10371. doi: 10.1073/pnas.0800405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Luo WB, Cady SD, Hong M. Biochemistry. 2009;48:6361–6368. doi: 10.1021/bi900716s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ieronimo M, Afonin S, Koch K, Berditsch M, Wadhwani P, Ulrich AS. J. Am. Chem. Soc. 2010;132:8822–8823. doi: 10.1021/ja101608z. [DOI] [PubMed] [Google Scholar]
- (19).Varga K, Aslimovska L, Watts A. J. Biomol. NMR. 2008;41:1–4. doi: 10.1007/s10858-008-9235-5. [DOI] [PubMed] [Google Scholar]
- (20).Saito H, Naito A. Biochim. Biophys. Acta-Biomembr. 2007;1768:3145–3161. doi: 10.1016/j.bbamem.2007.08.026. [DOI] [PubMed] [Google Scholar]
- (21).Smith SO, Braiman MS, Myers AB, Pardoen JA, Courtin JML, Winkel C, Lugtenburg J, Mathies RA. J. Am. Chem. Soc. 1987;109:3108–3125. [Google Scholar]
- (22).Kamihira M, Vosegaard T, Mason AJ, Straus SK, Nielsen NC, Watts A. J. Struct. Biol. 2005;149:7–16. doi: 10.1016/j.jsb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- (23).Cross TA, Sharma M, Yi M, Zhou HX. Trends Biochem. Sci. 2011;36:117–125. doi: 10.1016/j.tibs.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Poget SF, Girvin ME. Biochim. Biophys. Acta. 2007;1768:3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kim HJ, Howell SC, Van Horn WD, Jeon YH, Sanders CR. Prog. NMR Spectrosc. 2009;55:335–360. doi: 10.1016/j.pnmrs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Traaseth NJ, Shi L, Verardi R, Mullen DG, Barany G, Veglia G. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10165–10170. doi: 10.1073/pnas.0904290106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mao LL, Tang YF, Vaiphei ST, Shimazu T, Kim SG, Mani R, Fakhoury E, White E, Montelione GT, Inouye M. J. Struct. Funct. Genomics. 2009;10:281–289. doi: 10.1007/s10969-009-9072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Vaiphei ST, Tang YF, Montelione GT, Inouye M. Mol. Biotechnol. 2011;47:205–210. doi: 10.1007/s12033-010-9330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Mao LL, Inoue K, Tao YS, Montelione GT, McDermott AE, Inouye M. J. Biomol. NMR. 2011;49:131–137. doi: 10.1007/s10858-011-9469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hardy JA, Higgins GA. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- (31).Tanzi RE. Nat. Neurosci. 2005;8:977–979. doi: 10.1038/nn0805-977. [DOI] [PubMed] [Google Scholar]
- (32).Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, Lah JJ. J. Neurosci. 2006;26:1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Willnow TE, Petersen CM, Nykjaer A. Nat. Rev. Neurosci. 2008;9:899–909. doi: 10.1038/nrn2516. [DOI] [PubMed] [Google Scholar]
- (34).Andersen OM, et al. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, Willnow TE. J. Biol. Chem. 2007;282:32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- (36).Nielsen MS, Gustafsen C, Madsen P, Nyengaard JR, Hermey G, Bakke O, Mari M, Schu P, Pohlmann R, Dennes A, Petersen CM. Mol. Cell. Biol. 2007;27:6842–6851. doi: 10.1128/MCB.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. J. Neuropathol. Exp. Neurol. 2006;65:866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Rogaeva E, et al. Nat. Genet. 2007;2:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wang X, R. L. G., Zhu Q, Tian F. Protein Expres. Purif. 2011;77:224–230. doi: 10.1016/j.pep.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Cronan JE. Annu. Rev. Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- (41).Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. J. Chem. Phys. 1999;110:7983–7992. [Google Scholar]
- (42).Weingarth M, Demco DE, Bodenhausen G, Tekely P. Chem. Phys. Lett. 2009;469:342–348. [Google Scholar]
- (43).Wishart DS, Sykes BD. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- (44).Luca S, Filippov DV, van Boom JH, Oschkinat H, de Groot HJM, Baldus M. J. Biomol. NMR. 2001;20:325–331. doi: 10.1023/a:1011278317489. [DOI] [PubMed] [Google Scholar]
- (45).Renault M, Cukkemane A, Baldus M. Angew. Chem. Int. Ed. 2010;49:8346–8357. doi: 10.1002/anie.201002823. [DOI] [PubMed] [Google Scholar]
- (46).Loquet A, Lv G, Giller K, Becker S, Lange A. J. Am. Chem. Soc. 2011;133:4722–4725. doi: 10.1021/ja200066s. [DOI] [PubMed] [Google Scholar]
- (47).LeMaster DM, Kushlan DM. J. Am. Chem. Soc. 1996;118:9255–9264. [Google Scholar]
- (48).Hong M. J. Magn. Reson. 1999;139:389–401. doi: 10.1006/jmre.1999.1805. [DOI] [PubMed] [Google Scholar]
- (49).Higman VA, Flinders J, Hiller M, Jehle S, Markovic S, Fiedler S, van Rossum BJ, Oschkinat H. J. Biomol. NMR. 2009;44:245–260. doi: 10.1007/s10858-009-9338-7. [DOI] [PubMed] [Google Scholar]
- (50).Sperling LJ, Berthold DA, Sasser TL, Jeisy-Scott V, Rienstra CM. J. Mol. Biol. 2010;399:268–282. doi: 10.1016/j.jmb.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).McDermott A. Ann. Rev. Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.