Abstract
Prolonged exposure to the chemical intermediate, 1,3-dinitrobenzene (1,3-DNB), produces neuropathology in the central nervous system of rodents analogous to that observed in various conditions of acute energy deprivation including thiamine deficiency and Leigh's necrotizing encephalopathy. Increased production of reactive intermediates in addition to induction of oxidative stress has been implicated in the neurotoxic mechanism of 1,3-DNB, but a clear metabolic target has not been determined. Here we propose that similar to thiamine deficiency, the effects of 1,3-DNB on metabolic status may be due to inhibition of the thiamine-dependent α-ketoacid dehydrogenase complexes. The effects of 1,3-DNB on astroglial metabolic status and α-ketoacid dehydrogenase activity were evaluated using rat C6 glioma cells. Exposure to 1,3-DNB resulted in altered morphology and biochemical dysfunction consistent with disruption of oxidative energy metabolism. Cotreatment with acetyl-carnitine or acetoacetate attenuated morphological and metabolic effects of 1,3-DNB exposure as well as increased cell viability. 1,3-DNB exposure inhibited pyruvate dehydrogenase complex (PDHc) and the inhibition correlated with the loss of lipoic acid (LA) immunoreactivity, suggesting that modification of LA is a potential mechanism of inhibition. Treatment with antioxidants and thiol-containing compounds failed to protect against loss of LA. Alternatively, inhibition of dihydrolipoamide dehydrogenase, the E3 component of the complex attenuated loss of LA. Collectively, these data suggest that 1,3-DNB impairs oxidative energy metabolism through direct inhibition of the PDHc and that this impairment is due to perturbations in the function of protein-bound LA.
Keywords: 1,3-DNB; PDHc; astrocytes
Nitroaromatic compounds are key intermediates used in the production of industrial and commercial products including industrial solvents, dyes, plastics, and explosives (Harter, 1985). Although important for the chemical industry, nitroaromatics also pose environmental issues and potential health risks as a result of their over abundant use. 1,3-Dinitrobenzene (1,3-DNB), important in azo and aniline dye synthesis, as well as a byproduct in the production of the most widely used explosive, trinitrotoluene, is a multitarget toxicant affecting both the reproductive and nervous systems (Cody et al., 1981). Prolonged exposure to 1,3-DNB produces neuropathology in the central nervous system (CNS) of rodents analogous to that observed in conditions of acute energy deprivation (AED; Cavanagh, 1988). AED syndromes represent a distinct family of metabolic disorders with diverse etiologies, ranging from nutritional deficiencies, chemical intoxication, and genetic abnormalities. These disorders are characterized by bilaterally symmetrical spongiform degeneration confined to specific brainstem and deep cerebellar nuclei involved in auditory and vestibular functions. Similar to other AED syndromes, 1,3-DNB initially affects astrocytes, with secondary neuronal dysfunction resulting from loss of astrocyte trophic and homeostatic support (Philbert et al., 1987). Brainstem astrocytes sensitive to 1,3-DNB show clear signs of disrupted metabolic function, including increased glucose utilization and elevated lactate production (Phelka et al., 2003; Philbert et al., 1987; Romero et al., 1995). Although the induced metabolic perturbation is likely resultant from disruption in multiple sites of the overall energy metabolism pathway, the specific pattern of neuropathology induced by 1,3-DNB is remarkably similar in cellular and regional topography to thiamine deficiency.
Thiamine is a required cofactor for a family of multicomponent complexes termed the α-ketoacid dehydrogenase complexes (Bâ, 2008). Reduction in thiamine availability due to nutritional deficiency results in AED through reduced activity of these complexes that include α-ketoglutarate dehydrogenase (KGDHc) and pyruvate dehydrogenase (PDHc). Furthermore, genetic mutations in PDHc such as seen in cases of Leigh's necrotizing encephalopathy, result in AED (Finsterer, 2008). These two examples of AED syndromes suggest key roles for these enzyme complexes in AED induction and points to them as potential primary targets for nitroaromatic compounds.
α-Ketoacid dehydrogrenases occupy critical sites in cellular energy metabolism with PDHc linking glycolysis to the tricarboxylic acid (TCA) cycle through oxidative decarboxylation of pyruvate (Patel and Roche, 1990) and KGDHc being a member of the TCA cycle. The catalytic core of these complexes consists of multiple copies of three enzymes. These include α-ketoacid dehydrogenase (E1), responsible for decarboxylation of α-ketoacids; dihydrolipoyltransacylase (E2), which catalyzes the transfer of α-ketoacid–derived acyl groups to coenzyme A (CoA) through protein-bound lipoic acid; dihydrolipoyl dehydrogenase (E3), which is charged with the reoxidation of lipoic acid with concomitant production of NADH (Reed, 1998; Yeaman, 1989).
Chemical modification of E2-bound lipoic acid is considered a primary mechanism for inhibition of α-ketoacid dehydrogenases. Classically, this is demonstrated by arsenical compounds, which have a high affinity for vicinal dithiols resulting in the formation of stable dithioarsinites (Samikkannu et al., 2003). Various strong electrophiles such as the ubiquinone analog, coenzyme Q0, and the reactive lipid peroxidation products, 4-hydroxynonenal (4-HNE) and acrolein, have also demonstrated a similar mechanism of action through forming stable adducts on lipoic acid (Humphries and Szweda, 1998; MacDonald et al., 2004; Pocernich and Butterfield, 2003). Furthermore, direct inactivation of PDHc by reactive oxygen and nitrogen species such as hydroxyl radical and peroxynitrite has been reported (Tabatabaie et al., 1996). During nitro reductive metabolism of 1,3-DNB, highly reactive intermediates, 3-nitro-nitrosobenzene (3-NNB) and 3-nitropheny-hydroxylamine (3-NPHA) are formed in addition to the production of superoxide through futile redox cycling (Cossum and Rickert, 1985). Both metabolites react with thiols such as glutathione (GSH), indicating a potential for enzyme inhibition through modification of protein-bound thiols by 1,3-DNB and similar nitroaromatic compounds (Ellis et al., 1992; Kazanis and McClelland, 1992).
The propensity for nitro-reductive metabolism to produce reactive intermediates that may interact with oxidizable dithiol moieties such as those present in α-ketoacid dehydrogenase complexes points to these critical metabolic enzymes as potential prime targets involved in 1,3-DNB–induced metabolic disruption. This study demonstrates the ability of 1,3-DNB to interfere with cellular energy metabolism through inhibition of the PDHc and suggests that inhibition is the result of direct adduction of lipoic acid constituents by 1,3-DNB or its metabolites.
MATERIALS AND METHODS
Cell culture.
Rat C6 glioma cells, obtained from the American Type Culture Collection, were maintained at 37°C under an atmosphere of 5% CO2/95% air (vol/vol) in Dulbecco's modified eagles medium (DMEM) supplemented with 10% fetal bovine serum and 1× antibiotics (penicillin, streptomyosin, and neomycin). Cells were subcultured using 0.25% trypsin and used 24 h after plating. Prior to treatment, cells were rinsed with Dulbecco's phosphate-buffered saline (D-PBS), and all treatments were done in serum-free DMEM.
Microscopy.
C6 glioma cells plated in six well plates were exposed to 1,3-DNB (dissolved in DMSO) for 24 h. Plates were then washed twice with D-PBS, and phase-contrast light microscopy was performed using an Olympus CKX41 inverted microscope equipped with a Spot Insight QE digital CCD camera. Images were acquired at ×10 magnification (Olympus 10× PHP objective with 0.25 NA) using spot imaging software (Version 3.5.6, Diagnostic Instruments, Inc., Sterling Heights, MI).
Cellular reducing potential.
Establishment of a dose range for the cytotoxic effects of 1,3-DNB on C6 glioma cells was determined by monitoring the reduction of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT; Sigma, St Louis, MO) as described (Mosmann, 1983). Cells were seeded in a 96-well plate at a cell density of 104 cell per well. Cells, 24 h after plating, were exposed to increasing concentrations of 1,3-DNB for 36 h in the presence or absence of acetyl-carnitine (ALCAR) (5mM). Cells were rinsed with D-PBS, MTT assay solution (1 mg/ml in DMEM) was added to each well, and allowed to incubate for 2 h at 37°C. MTT solution was replaced with DMSO, and after 30-min incubation with vigorous mixing, the absorbance was determined at 550 nm. Mean absorbance values were compared with vehicle control, and IC50 values were determined by fitting data to a sigmoidal dose-response equation using Prism software (Version 2.0, Graph Pad Software, Inc., San Diego, CA).
Lactate dehydrogenase release.
As an indicator of cytotoxicity, extracellular release of lactate dehydrogenase (LDH) was determined using the 2-p-iodophenyl-3-nitrophenyl tetrazolium chloride (INT; Sigma)–coupled colorimetric assay. C6 glioma cells were seeded in 96-well plates at a cell density of 104 per well. After 24-h incubation, cells were rinsed with D-PBS. The cells were then incubated with varying concentrations of 1,3-DNB. After 36-h exposure to 1,3-DNB, culture medium was clarified by centrifugation, and 100 μl of supernatant was placed in a microplate containing 60 μl of assay solution consisting of 4.5mM NAD+, 4mM INT, 13.5 U/ml diaphorase, 0.03% wt/vol bovine serum albumin (BSA), and 0.2M Tris-HCl, pH 8.2. The reaction was stopped by adding 20 μl of 2mM oxamate, and absorbance was read at 490 nm. The amount of lactate dehydrogenase (LDH) released was determined through comparison of obtained values to a standard curve for LDH activity.
Determination of cellular adenosine triphosphate content.
Adenosine triphosphate (ATP) levels were determined using an isocratic reverse-phase high-performance liquid chromatography (HPLC) method described previously with slight modifications (Yang et al., 2004). Briefly, treated cells were rinsed in D-PBS and immediately snap frozen in liquid N2. ATP was acid extracted with ice-cold perchloric acid (0.3 M) containing EDTA (1mM). Protein-free extracts were neutralized with KOH, and extracts were stored at –80°C until HPLC analysis was performed. Adenine nucleotides were separated using a Waters HPLC system equipped with a dual λ-absorbance ultraviolet detector and an inline degasser. Isocratic elution was performed on a Symmetry 300 C-18 column (4.6 × 150 mm, 5 μm) at a flow rate of 0.6 ml/min and peak absorbance recorded at 206 nm. The mobile phase consisted of 0.1M ammonium dihydrogen phosphate with 1% methanol. ATP concentrations were calculated using peak height, which was determined to be directly proportional to ATP concentration when compared with ATP standards treated in the same manner as samples.
Measurement of extracellular lactate concentrations.
Extracellular lactate accumulation was determined by the lactate dehydrogenase method through monitoring the conversion of NAD to NADH at 340 nm as previously detailed with slight modification (Yang and Balcarcel, 2004). Lactate assay reagent contained 0.2M glycine buffer (pH 9.2), 0.15M hydrazine, and 10mM NAD+. The reaction was started with the addition of lactate dehydrogenase (10 U), and the rate of conversion was monitored. The rate of conversion is directly proportional to lactate concentration that was determined using lactate standards.
Purified PDHc and KGDHc activity.
A standard spectrophotometric assay was performed to determine the activities of PDHc and KGDHc as described previously (Hard et al., 2001) with minor alterations. In brief, commercially available porcine heart PDHc (15 mg/ml; Sigma) or KGDHc (10.2 mg/ml; Sigma) were employed in this assay. The reaction mixture contained 50mM potassium phosphate buffer, pH 7.4, 2.5mM NAD+, 0.2mM thiamine pyrophosphate (TPP), 1mM MgCl2, 1mM pyruvate or α-ketoglutarate, PDHc (final concentration 75 μg/ml), or KGDH (final concentration 50 μg/ml). Reaction buffer combined with the appropriate enzyme were incubated for 2 min at 30°C. Varying concentrations of inhibitor were then added and incubated for an additional 3 min. At 5 min, the reaction was started with addition of CoA (final concentration 0.2mM), and NADH absorbance was monitored at 340 nm at 30°C using a Spectramax microplate spectrophotometer (Molecular Devices, Inc., Sunnyvale, CA). Enzyme activity assays were performed at 30°C in order to reduce thermal denaturation as well as to minimize variations in room temperature.
Cytochemical determination of PDHc and KGDHc activity.
Cellular PDHc and KGDHc activity were determined by the quantitative cytochemical method as described by (Park et al., 2000), with slight modifications. Briefly, cells were rinsed with D-PBS twice, and 1 ml of reaction buffer containing 50mM Tris-HCl (pH 7.6), 1mM MgCl2, 0.1mM CaCl2, 0.05mM EDTA, 0.3mM TPP, 0.5 μg/ml rotenone (dissolved in 100% ethanol; final ethanol concentration 0.1%), 0.2% Triton X-100, 3.5% polyvinyl alcohol, 3mM pyruvate or α-ketoglutarate, 3mM NAD+, 0.75 mg/ml CoA, 0.75mM nitroblue tetrazolium (NBT), and 0.05mM phenazine methosulfate (PMS) was added to the cells. Both NBT and PMS were added immediately before the reaction was initiated. The reaction was allowed to proceed for 1 h and stopped by washing the plates twice with D-PBS. Solubilization of reduced NBT was accomplished by the method previously described (Rook et al., 1985). Unreduced NBT was rinsed away with methanol and allowed to air dry. To the dried plates, 2M KOH was added followed by the addition of DMSO. The contents of each well were mixed thoroughly, and absorbance was read at 630 nm. Wells treated with reaction mixture minus pyruvate or α-ketoglutarate were used as blanks to account for nonspecific reduction of NBT.
SDS-PAGE and Western Blotting
Total cellular protein was extracted with ice-cold cell lysis buffer consisting of Tris-HCl (50mM, pH 7.4), NaCl (150mM), Triton X-100 (1% vol/vol), and protease and phosphatase inhibitor cocktails (0.1% vol/vol; Sigma). Cell lysates were snap frozen in liquid N2 and stored at –80°C until protein quantification and SDS-PAGE. Protein concentration was determined by the BCA method (Smith et al., 1985) using a commercial kit (Thermo Scientific, Rockford, IL) as described by the manufacturer. The proteins was resolved on 4–20% Tris-glycine gels and electroblotted to PVDF membrane. Membranes were blocked with nonfat milk (5% wt/vol) and probed with antilipoic acid (1:2000; Calbiochem), anticomplex II (1:2000; Invitrogen), or anti-E2 (1:1000; Abcam) overnight at 4°C. The membranes were washed with TBS-T followed by probing with appropriate alkaline phosphatase–conjugated secondary antibodies (1:10000; Amersham Biosciences). The membranes were developed using an enhanced chemifluorescence detection system (Amersham Biosciences) and visualized on a FLA-5000 imaging system (Fujifilm Medical Systems USA, Inc, Stamford, CT). Densitometry analysis was performed using Multigauge software (Version 2.2, Fujifilm Medical Systems USA).
Oxyblot analysis.
Oxidative modification of mitochondrial protein was determined using the Oxyblot detection kit (Millipore, Temecula, CA). Mitochondrial protein was isolated from treated C6 glioma cells following the protocol MITOISO1 from Sigma with slight modification. Briefly, cells from 8 T-75 flasks were homogenized in 10 volumes of BSA (2 mg/ml) containing extraction buffer (220mM mannitol, 70mM sucrose, 0.5mM EGTA, and 2mM HEPES at a final pH of 7.4) with a dounce glass homogenizer and Teflon pestle. Intact mitochondria were pelleted by centrifugation (11,000g, 4°C, 10 min), and the pellet was washed twice with extraction buffer. The final mitochondrial pellet was then resuspended in storage buffer (10mM HEPES, 250mM sucrose, 1mM ATP, 0.08mM ADP, 5mM sodium succinate, 2mM K2HPO4, and 1mM dithiothreitol and protease inhibitor cocktail). Mitochondria were lysed by repeated freeze thaw cycles, and total mitochondrial protein was quantified by BCA as described earlier. Mitochondrial protein (20 μg) was denatured with 12% SDS and derivatized with 1× solution of dinitrophenylhydrazine. The derivatized samples were Western blotted with anti-DNP (1:150) as described earlier.
Statistical analysis.
All results are presented as means ± SEM with n representing the number of biological replicates for each experiment. Comparative analysis of treatments were made by one-way or two-way ANOVA where appropriate followed by Bonferroni post hoc analysis. All statistical analyses were conducted using Prism software (Version 2.0, Graph Pad Software, Inc.).
RESULTS
Cytotoxicity
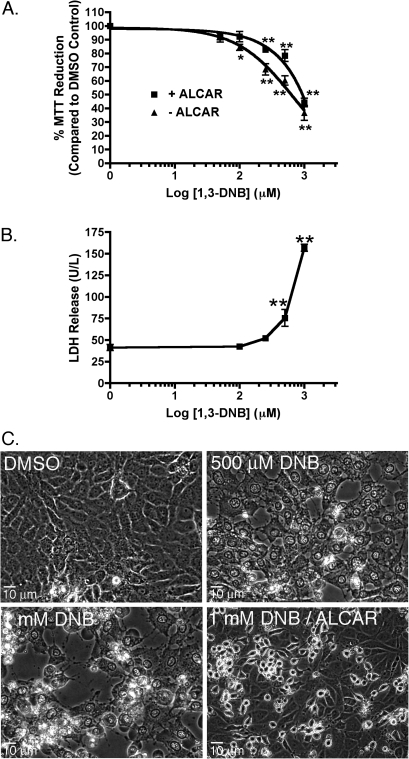
Exposure of C6 glioma cells, in culture, to increasing concentrations of 1,3-DNB (10μM to 1mM) significantly diminished overall cellular reducing potential at 100μM and above with a calculated IC50 of 629.5μM as measured by the MTT reduction assay (Fig. 1A). Cotreatment with the alternative energy source, ALCAR, significantly reduced the cytotoxic effects of 1,3-DNB as demonstrated by an increased IC50 of 933.3μM. Similarly, extracellular LDH activity, a marker of cell membrane integrity loss, increased significantly at both 500μM and 1mM 1,3-DNB. In addition to overall cytotoxic response to 1,3-DNB, extensive morphological changes occurred during exposure to 1,3-DNB. These changes included extensive vacuolation and nuclear and cytoplasmic swelling that progressed to wide spread cell death which are consistent with the significant increase in LDH release observed at these concentrations of 1,3-DNB (Fig. 1C). ALCAR supplementation was also able to significantly reduce the morphological changes. No morphological changes were seen in controls treated with DMSO alone.
FIG. 1.
Cytotoxic responses of C6 glioma cells exposed to increasing concentrations of 1,3-DNB (50μM to 1mM) for 36 h with or without the addition of ALCAR (5mM). (A) Cellular reducing potential was determined using the MTT reduction assay. Data are expressed as percent MTT reduction compared with vehicle control (DMSO). (B) Quantification of released LDH was determined using the INT-coupled enzymatic assay. Data are expressed as mean ± SEM (n = 4). Significant differences from vehicle control (DMSO) are denoted as follows: *p < 0.05, **p < 0.01. (C) Representative photomicrographs depicting 1,3-DNB (500μM and 1mM)–induced morphological changes after 36-h exposure in the presence or absence of ALCAR (5mM).
Metabolic Status
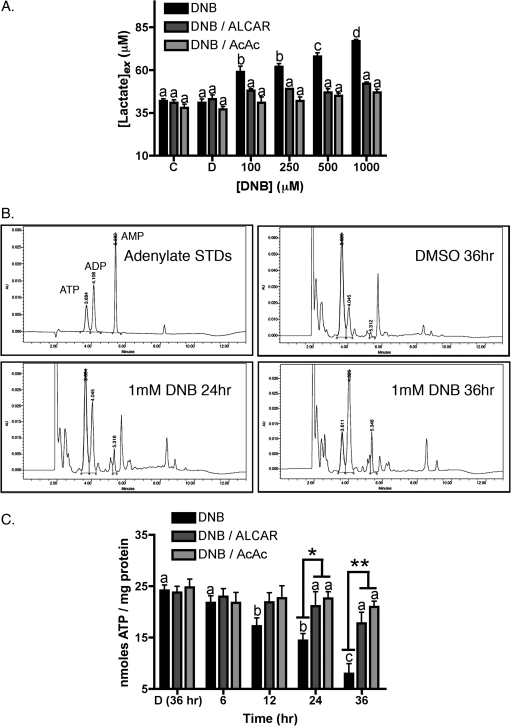
Significant accumulation of extracellular lactate was recorded at 36 h of exposure, beginning at the lowest dose employed of 100μM 1,3-DNB and increased in a dose-dependent manner with nearly a twofold increase at the highest concentration of 1mM (Fig. 2A). Cotreatment of cultures with either acetoacetate or ALCAR attenuated lactate accumulation in culture medium at all concentrations of 1,3-DNB. By contrast, equimolar levels of the nonacetylated analogue L-carnitine or free acetate did not affect lactate accumulation (data not shown) suggesting protective effects were attributable to the availability of the acetyl moiety. In addition to increased extracellular lactate concentrations, cellular ATP levels were significantly diminished by exposure to 1,3-DNB. Representative chromatographs of adenine nucleotides clearly show significant reduction in ATP levels with corresponding increases in both ADP and AMP concentrations by 12 h of exposure to 1mM 1,3-DNB and drastic depletion by 36 h compared with vehicle control (Fig. 2B). Similar to lactate accumulation, cotreatment with both acetoacetate and ALCAR preserved cellular ATP content.
FIG. 2.
1,3-DNB–induced metabolic impairment. (A) Determination of extracellular lactate accumulation in culture medium of C6 glioma cells incubated with no treatment (C), the vehicle DMSO (D), or with increasing concentrations of 1,3-DNB in the presence and absence of ALCAR or acetoacetate (AcAc, 5mM) for 36 h. (B) Representative chromatograms depicting time-dependent loss of intracellular ATP with concurrent increases in AMP. The HPLC retention times corresponding to the peaks of ATP, ADP, and AMP were 3.684, 4.108, and 5.382, respectively. (C) ATP depletion in C6 glioma cells treated with either 1mM 1,3-DNB alone or cotreated with 1,3-DNB and ALCAR for up to 36 h. Significant reduction in ATP was first observed at 12 h and continued to decline in a time-dependent manner. Results are expressed as means ± SE (n = 3). Significant differences (p < 0.05) between groups are denoted with different letters and significant differences between 1,3-DNB alone and 1,3-DNB with ALCAR or AcAc are denoted as follows *p < 0.05, **p < 0.01.
PDHc and KGDHc Activity
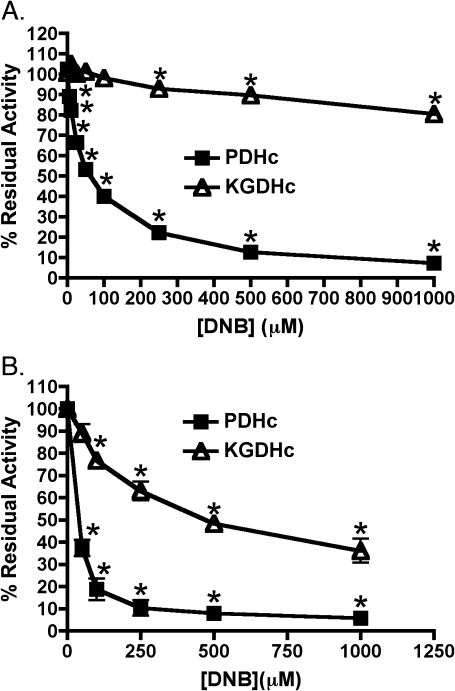
Incubation of purified porcine heart PDHc with 1,3-DNB resulted in rapid inhibition as determined by a standard spectrophotometric assay (Fig. 3A). The degree of impairment responded in a dose-dependent manner, with approximately 12% at the lowest dose of 5μM and greater than 90% inhibition at 1mM 1,3-DNB. The IC50 for 1,3-DNB impairment of PDHc was approximately 62μM. In contrast to PDHc, KGDHc showed dramatically less inhibition when incubated with 1,3-DNB with significant inhibition being observed only at 1,3-DNB concentrations above 500μM. IC50 for 1,3-DNB–mediated inhibition of KGDHc could not be accurately determined. In addition to inhibition of purified enzymes, exposure of C6 glioma cells to 1,3-DNB produced a concentration-dependent reduction in residual PDHc and KGDHc activities (Fig. 3B). At 50μM, the lowest concentration employed, there was greater then 60% reduction in PDHc activity and approximately 95% inhibition at 1mM compared with vehicle control. Similar to the results obtained with purified enzyme preparations, cellular KGDHc activity was less sensitive to the inhibitory affects of 1,3-DNB then PDHc with IC50 concentrations for PDHc and KGDHc being approximately 45 and 250μM, respectively.
FIG. 3.
Effects of 1,3-DNB on the activity of the α-ketoacid dehydrogenase complexes, PDHc and KGDHc. (A) Effects of 1,3-DNB on purified porcine heart enzyme complexes were determine by standard NADH reduction assays as described in the Materials and Methods section. Enzyme complexes were incubated for 3 min with various concentrations of 1,3-DNB, and the reaction was monitored upon addition of the substrate CoA. (B) Effects of 1,3-DNB on the α-ketoacid dehydrogenase complexes in intact C6 glioma cells were determined by a cytochemical staining method as described earlier. Data expressed as mean % residual activity. Significant differences from control are denoted as follows: *p < 0.05.
Lipoic Acid Immunoreactivity
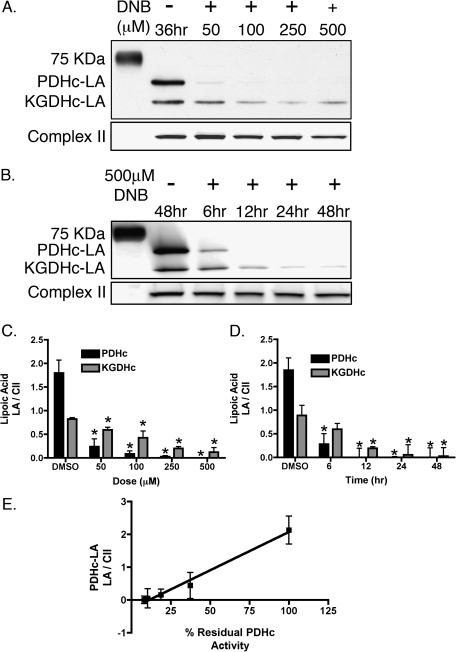
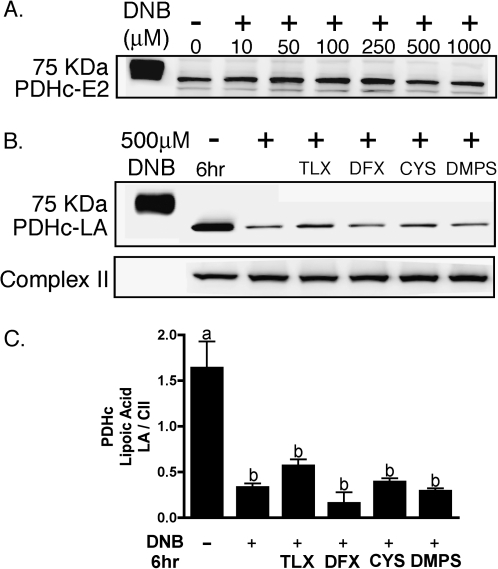
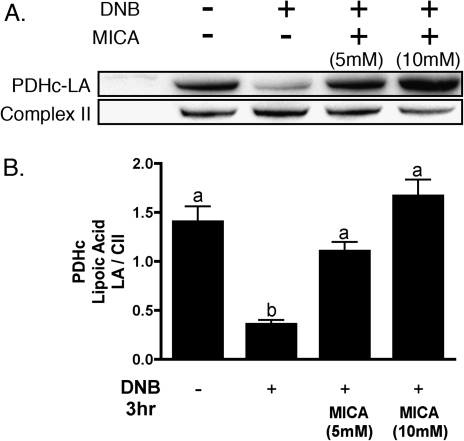
Lipoic acid (LA) immunoreactivity was determined by Western blot analysis of total cellular protein, utilizing an antibody against unmodified lipoic acid, which stains two distinct bands at approximately 70 and 55 kDa, corresponding to the dihydrolipoyltransacylase components of the PDHc and KGDHc, respectively. Exposure of C6 glioma cells to 1,3-DNB resulted in loss of LA immunoreactivity of PDHc and KGDHc in both a time- and dose-dependent manner (Fig. 4A). At 500μM 1,3-DNB, significant reduction in PDHc-LA by 6 h and complete loss by 12 h was observed. Additionally, substantial reduction in LA immunoreactivity was observed after 36 h of exposure at all concentrations of 1,3-DNB with almost complete loss apparent at the dose of 50μM 1,3-DNB. Loss of PDHc-LA immunoreactivity significantly correlated with loss of PDHc activity with a calculated correlation coefficient of 0.99. Similar to PDHc, 1,3-DNB also affects KGDHc-LA immunoreactivity with significant loss at 6 h of exposure. Residual immunoreactivity for KGDHc-LA was observed at all time points following exposure to 1,3-DNB. Utilizing an antibody to the E2 subunit of PDHc, no significant loss in protein expression was observed (Fig. 5A). Cotreatment with various antioxidants (Trolox, deferoximine; 1mM) as well as thiol-containing compounds shown to be strongly protective against both phenylarsine oxide and 4-HNE–mediated inhibition of PDHc (DL-ipoic acid and 2,3-dimercapto-1-propanesulfonic acid; 3mM; Korotchkina et al., 2001;Samikkannu et al., 2003) showed no significant attenuation of LA loss at 250μM 1,3-DNB following 3 h of exposure (Fig. 5B). Alternatively, inhibition of dihydrolipoamide dehydrogenase, the E3 component of the enzyme complex by the competitive inhibitor 5-methoxyindole carboxylic acid (MICA; 5mM), resulted in attenuation of LA loss (Fig. 6).
FIG. 4.
Modification of LA by exposure to 1,3-DNB in C6 glioma cells. (A) Representative immunoblot illustrating the dose-dependent effects on LA immunoreactivity by 36-h exposure to 1,3-DNB and (B) time-dependent effects on LA immunoreactivity by exposure to 500μM 1,3-DNB for 6, 12, 24, and 48 h. Quantification of (C) dose-dependent and (D) time-dependent LA immunoreactivity loss by densitometric analysis. (E) Correlation analysis between loss of LA immunoreactivity and PDHc activity. Data are presented as mean pixel intensity of LA normalized to complex II expression. Significant differences from control are denoted as follows: *p < 0.05.
FIG. 5.
Antioxidants and thiol-containing compounds inability to protect loss of LA by 1,3-DNB in C6 glioma cells. (A) Representative immunoblot depicting the effect of increasing concentrations of 1,3-DNB on the expression of the E2 component of the PDHc. (B) Representative immunoblot depicting the effects of various antioxidants and thiol-containing compounds on 1,3-DNB–induced loss of LA immunoreactivity. C6 glioma cells were pretreated with the following compounds for 1 h followed by a cotreatment with 1,3-DNB (250μM) for 3 h (Trolox [TLX]; 1mM), deferoximine (DFX; 1mM), L-cysteine (l-CYS; 3mM), and 2,3-dimercapto-l-propane sulfonic acid (DMPS; 3mM). (C) Quantification of LA immunoreactivity loss by 1,3-DNB exposure in the presence of antioxidants and thiol-containing compounds. Data are presented as mean pixel intensity of LA normalized to complex II expression. Significant difference (p < 0.05) between groups are denoted with different letters.
FIG. 6.
Protection by 5-MICA against 1,3-DNB–induced loss of PDHc LA. (A) Representative immunoblot demonstrating the effects of cotreatment with MICA on 1,3-DNB–induced loss of LA immunoreactivity. C6 glioma cells were treated with 1,3-DNB (250μM) for 3 h in the presence or absence of MICA (5 or 10mM). (B) Quantification of LA immunoreactivity loss by 1,3-DNB in the presence of MICA. Data are presented as mean pixel intensity of LA normalized to complex II expression. Significant difference (p < 0.05) between groups are denoted with different letters.
Protein Oxidation
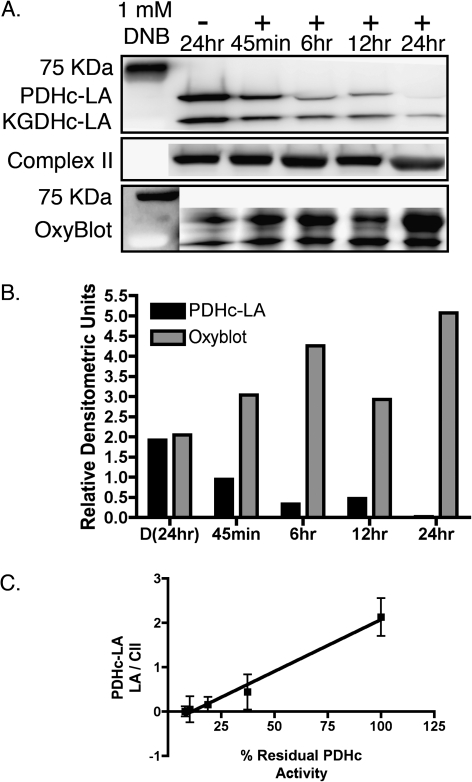
To determine if loss of LA immunoreactivity was potentially a consequence of increased protein oxidation, OxyBlot analysis of mitochondrial protein samples was performed (Fig. 7). Exposure to 1mM DNB for increasing time resulted in increased carbonylation of proteins corresponding to the molecular weights of the dihydrolipoyltransacylase component of PDHc and KGDHc with increases apparent by 45 min. Time-dependent increases in protein carbonylation correlated with the time-dependent reduction in LA immunoreactivity, with a calculated correlation coefficient of 0.92.
FIG. 7.
Loss of LA immunoreactivity mirrors the increase in mitochondria protein oxidation upon exposure to 1,3-DNB in C6 glioma cells. (A) Representative immuoblots illustrating the time-dependent loss of PDHc-LA immunoreactivity in association with increasing mitochondrial protein carbonylation after exposure to 1,3-DNB (1mM). (B) Quantitation of PDHc-LA loss and the increase in mitochondrial protein carbonylation. (C) Correlation analysis between protein oxidation and loss of LA immunoreactivity.
DISCUSSION
Neuropathologic changes following 1,3-DNB exposure in rats resemble those observed in both thiamine deficiency and the early phases of Leigh's necrotizing encephalopathy both of which result from a reduction in the activities of the thiamine-dependent enzymes, KGDHc and PDHc. Although significant increase in cerebral glucose utilization and extracellular lactate concentrations observed in whole animals and cultured cells treated with 1,3-DNB suggests a dysfunction in oxidative energy metabolism, the specific site affected is unknown (Phelka et al., 2003; Ray et al., 1994; Romero et al., 1991, 1995). In the absence of mitochondrial respiration, accelerated glycolytic activity occurs, which accounts for the increased glucose utilization in 1,3-DNB exposure (Ray et al., 1994; Romero et al., 1991, 1995). Consequently, NAD+ stores are exhausted and must be replenished through anaerobic glycolysis by lactate dehydrogenase resulting in increased lactate production. Acidosis-induced persistent cell swelling due to increased lactate has been shown to occur in astrocytes and C6 glioma cells likely through increased sodium uptake by the Na+/H+ exchanger (Nabekura et al., 2003). In the present study, exposure of C6 gliomas to 1,3-DNB resulted in metabolic dysfunction and significant cytotoxicity consistent with those observed in both primary astrocytes and in the CNS of 1,3-DNB–exposed rodents (Philbert et al., 1987; Romero et al., 1995). Increasing concentrations of 1,3-DNB cause decreased cell viability as indicated by loss of MTT reduction and increased LDH release. Additionally, C6 gliomas treated with 1,3-DNB underwent persistent swelling and extensive vacuolation (Fig. 1). 1,3-DNB exposure also caused an increase in extracellular lactate levels and depletion of cellular ATP (Fig. 2). Cotreatment with the energy substrates, acetoacetate and ALCAR, protected cells from 1,3-DNB–induced structural and biochemical changes and reduced overall loss in cell viability. Acetoacetate and ALCAR are alternative sources of acetyl-CoA. The ketone body, acetoacetate, contributes to cerebral metabolism during many physiologic and pathologic events including periods during postnatal development, starvation, and ischemia (Edmond, 1992; Morris, 2005). Additionally, administration of ketogenic diets has proved neuroprotective in various neurological disorders including PDHc deficiency. Similarly, ALCAR also protects in models of metabolic impairment, including ischemia/reperfusion injury (Lesnefsky et al., 2006; Rosenthal et al., 1992). The acetyl-CoA derived from both acetoacetate and ALCAR may sustain oxidative energy metabolism at the level of the TCA cycle, effectively bypassing defective PDHc.
Further evidence of a role for PDHc in 1,3-DNB–induced neurotoxicity was demonstrated by the ability of 1,3-DNB to inhibit PDHc in both purified enzymes and whole cells (Fig. 3). Dramatic loss of PDHc activity was observed at the lowest concentration of 50μM in intact cells. This dose level corresponds closely to the blood concentration of the threshold neuropathic dose of 25 mg/kg in rats that was calculated to be 43μM (Bailey et al., 1988). Additionally, this dose also represents the threshold dose for increased glucose utilization in primary astrocytes (Romero et al., 1995). α-Keto acid dehydrogenase complexes possess covalently bound LA on the E2 component. The vicinal dithiol of LA is sensitive to oxidiation and represents a key site for modification. C6 glioma cells exposed to 1,3-DNB showed dose-dependent decreases in LA immunoreactivity with no apparent changes in E2 protein expression suggesting direct affect on LA (Fig. 4). When comparing the inhibitory effects of 1,3-DNB on PDHc with loss of PDHc-LA immunoreactivity, a strong correlation between the two parameters was revealed as demonstrated by a significant correlation coefficient of 0.99 (Fig. 4).
Dysfunction of the PDHc has been reported in a number of neurological disorders including ischemia/reperfusion injury and neurodegenerative diseases (Butterworth and Besnard, 1990; Fukuchi et al., 1998; Sheu et al., 1985). In both situations, reactive oxygen species (ROS) have been implicated in disease progression. α-Ketoacid dehydrogenase complexes are sensitive to direct inactivation by hydroxyl radicals (Bogaert et al., 1994). Indirectly, ROS can result in the formation of the lipid peroxidation products, 4-hydroxy-nonenal, and 2-propen-1-al both of which adversely affect PDHc through binding of protein-bound LA (Humphries and Szweda, 1998; Pocernich and Butterfield, 2003). During nitroreduction of 1,3-DNB, reactive intermediates may be formed that interact with macromolecules such as nucleic acids, lipids, or proteins. Increased superoxide production with a subsequent reduction in total GSH levels after exposure to 1,3-DNB has been reported in primary astrocytes (Ray et al., 1994). Additionally, modification of GSH status effectively altered susceptibility to 1,3-DNB–induced neurotoxicity (Hu et al., 1999). These data suggest that oxidative stress is involved, at least in part, in the toxicity of 1,3-DNB. To investigate the involvement of oxidative stress, protein carbonylation, an indicator of protein oxidation, was determined. Time-dependent increases in carbonylation of proteins at the apparent molecular weights of the PDHc and KGDHc E2 components were observed suggesting that modification of LA by oxidation may be responsible for loss of LA immunoreactivity (Fig. 7). Analysis of the relationship between increased protein oxidation and loss of LA showed a significant correlation between the two parameters further supporting that PDHc becomes oxidized and that LA is most likely the site of oxidation. However, the addition of antioxidants showed no attenuation of LA loss implying that loss of LA may not be due directly to ROS (Fig. 5B). Additionally, no increase in 4-HNE adducts was observed indicating the absence of lipid peroxidation (data not shown).
The reactive intermediates 3-NNB and 3-NPHA, formed during 1,3-DNB biotransformation, are strong electrophiles that may react with nucleophilic centers of proteins such as primary amines, histidinyl secondary amines, and thiols. 3-NNB has been shown to react with biological thiols, specifically GSH and may potentially react with reduced thiols of LA (Ellis et al., 1992). 1,3-DNB is primarily metabolized by nitroreduction in the mitochondria in seminiferous tubules but the subcellular location of metabolism in other tissues has not been determined (Reeve et al., 2002). The E3 component (dihydrolipoamide dehydrogenase) of α-ketoacid dehydrogenases has been shown to catalyze 1,3-DNB nitroreduction (Tsai, 1987). Due to the complex structural geometry and consequential active site coupling within these enzyme systems, E3-produced reactive intermediates may potentially have direct access to E2-bound lipoic acid resulting in protein modification. This hypothesis is supported, in part, by the ability of MICA, a competitive inhibitor of the E3 component of the complex, to attenuate the loss of LA (Fig. 6). Furthermore, the inability of thiol-containing compounds to mitigate loss of LA despite the effectiveness of these compounds in blocking inhibition of PDHc by both phenylarsine oxide and 4-HNE suggests that production of reactive intermediates of 1,3-DNB occur within the complex precluding their accessibility by the protective compounds.
Although both PDHc and KGDHc are affected by 1,3-DNB, there is an apparent differential sensitivity between the two complexes, with PDHc being fivefold more sensitive to inhibition in intact cells and even greater differences encountered with purified enzymes (Fig. 3). The increased sensitivity of PDHc over KGDHc contrasts the effects seen with 4-HNE, where KGDHc is two times more sensitive to inhibition (Humphries and Szweda, 1998). This discrepancy maybe a consequence of the direct action of 4-HNE versus the apparent requirement for E3-dependent metabolism with respect to 1,3-DNB. Indeed, the two complexes possess distinct substrate specificities but catalyze practically identical reactions and share similar structures (Reed and Hackert, 1990). It is possible that slight differences in structure may confer the differential sensitivity observed with 1,3-DNB. E3BP, an additional component of PDHc, possesses a lipoyl domain with a covalently bound LA and is responsible for binding the E3 component to the E2 structural core (Brown et al., 2006). Newly proposed models of PDHc structure suggest that lipoyl domains of both E2 and E3BP may be restricted in their movements, limiting their ability to reach the various active sites of the complex. Hence, the E2-bound lipoic acid may not have direct access to the E3 component in order for reoxidation to occur. An alternate path to E2-LA reoxidation may require transfer of reducing equivalents between the lipoyl domains of E2 and E3BP (Smolle et al., 2006). This hypothesis may explain why PDHc is more sensitive to 1,3-DNB–mediated inhibition. The lack of E3BP of KGDHc potentially reduces the ability of E3-generated intermediates to access the E2-bound LA. More detailed studies on the interaction between each of the complexes and 1,3-DNB is required to confirm this hypothesis.
In conclusion, these findings support inhibition of PDHc as a likely mediator of metabolic disruption resulting from modification of LA. The correlation between loss of LA, increases in protein oxidation, and inhibition of activity all point to direct protein modification of PDHc as the most likely route for 1,3-DNB–meditated inhibition. Further work is needed to determine the specific adducting species and ultimately the role these enzymes play in the pathogenesis of 1,3-DNB–mediated encephalopathy.
FUNDING
National Institutes of Health (2R01 ES08846 to M.A.P).
Acknowledgments
This manuscript is dedicated to the memory of Dr David Ray: Professor, Scientist, and Friend.
References
- Bâ A. Metabolic and structural role of thiamine in nervous tissues. Cell. Mol. Neurobiol. 2008;28:923–931. doi: 10.1007/s10571-008-9297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey E, Peal JA, Philbert M. Determination of 1,3-dinitrobenzene and its metabolites in rat blood by capillary gas chromatography with electron-capture detection. J. Chromatogr. 1988;425:187–192. doi: 10.1016/0378-4347(88)80020-3. [DOI] [PubMed] [Google Scholar]
- Bogaert YE, Rosenthal RE, Fiskum G. Postischemic inhibition of cerebral cortex pyruvate dehydrogenase. Free Radic. Biol. Med. 1994;16:811–820. doi: 10.1016/0891-5849(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Brown RM, Head RA, Morris AAM, Raiman JAJ, Walter JH, Whitehouse WP, Brown GK. Pyruvate dehydrogenase E3 binding protein (protein X) deficiency. Dev. Med. Child Neurol. 2006;48:756–760. doi: 10.1017/S0012162206001617. [DOI] [PubMed] [Google Scholar]
- Butterworth RF, Besnard AM. Thiamine-dependent enzyme changes in temporal cortex of patients with Alzheimer's disease. Metab. Brain Dis. 1990;5:179–184. doi: 10.1007/BF00997071. [DOI] [PubMed] [Google Scholar]
- Cavanagh JB. Lesion localisation: implications for the study of functional effects and mechanisms of action. Toxicology. 1988;49:131–136. doi: 10.1016/0300-483x(88)90184-9. [DOI] [PubMed] [Google Scholar]
- Cody TE, Witherup S, Hastings L, Stemmer K, Christian RT. 1,3-Dinitrobenzene: toxic effects in vivo and in vitro. J. Toxicol. Environ. Health. 1981;7:829–847. doi: 10.1080/15287398109530024. [DOI] [PubMed] [Google Scholar]
- Cossum PA, Rickert DE. Metabolism of dinitrobenzenes by rat isolated hepatocytes. Drug Metab. Dispos. 1985;13:664–668. [PubMed] [Google Scholar]
- Edmond J. Energy metabolism in developing brain cells. Can. J. Physiol. Pharmacol. 1992;70(Suppl.):S118–S129. doi: 10.1139/y92-253. [DOI] [PubMed] [Google Scholar]
- Ellis MK, Hill S, Foster PM. Reactions of nitrosonitrobenzenes with biological thiols: identification and reactivity of glutathion-S-yl conjugates. Chem. Biol. Interact. 1992;82:151–163. doi: 10.1016/0009-2797(92)90107-v. [DOI] [PubMed] [Google Scholar]
- Finsterer J. Leigh and Leigh-like syndrome in children and adults. Pediatr. Neurol. 2008;39:223–235. doi: 10.1016/j.pediatrneurol.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Fukuchi T, Katayama Y, Kamiya T, McKee A, Kashiwagi F, Terashi A. The effect of duration of cerebral ischemia on brain pyruvate dehydrogenase activity, energy metabolites, and blood flow during reperfusion in gerbil brain. Brain Res. 1998;792:59–65. doi: 10.1016/s0006-8993(98)00121-8. [DOI] [PubMed] [Google Scholar]
- Hard ML, Raha S, Spino M, Robinson BH, Koren G. Impairment of pyruvate dehydrogenase activity by acetaldehyde. Alcohol. 2001;25:1–8. doi: 10.1016/s0741-8329(01)00156-2. [DOI] [PubMed] [Google Scholar]
- Harter, D. R. (1985). The use and importance of nitroaromatic chemicals in the chemical industry. In Toxicity of Nitroaromatic Chemicals (D. E. Ricket, Ed.), pp. 1--14. Chemical Industry Institute of Toxicology Series, New York, NY. [Google Scholar]
- Hu HL, Bennett N, Holton JL, Nolan CC, Lister T, Cavanagh JB, Ray DE. Glutathione depletion increases brain susceptibility to m-dinitrobenzene neurotoxicity. Neurotoxicology. 1999;20:83–90. [PubMed] [Google Scholar]
- Humphries KM, Szweda LI. Selective inactivation of alpha-ketoglutarate dehydrogenase and pyruvate dehydrogenase: reaction of lipoic acid with 4-hydroxy-2-nonenal. Biochemistry. 1998;37:15835–15841. doi: 10.1021/bi981512h. [DOI] [PubMed] [Google Scholar]
- Kazanis, S., and McClelland, R. (1992). Electrophilic intermediate in the reaction of glutathione and nitroso arenes. J. Am. Chem. Soc. 114, 3052–3059. [Google Scholar]
- Korotchkina LG, Yang H, Tirosh O, Packer L, Patel MS. Protection by thiols of the mitochondrial complexes from 4-hydroxy-2-nonenal. Free Radic. Biol. Med. 2001;30:992–999. doi: 10.1016/s0891-5849(01)00491-9. [DOI] [PubMed] [Google Scholar]
- Lesnefsky EJ, He D, Moghaddas S, Hoppel CL. Reversal of mitochondrial defects before ischemia protects the aged heart. FASEB J. 2006;20:1543–1545. doi: 10.1096/fj.05-4535fje. [DOI] [PubMed] [Google Scholar]
- MacDonald MJ, Husain RD, Hoffmann-Benning S, Baker TR. Immunochemical identification of coenzyme Q0-dihydrolipoamide adducts in the E2 components of the alpha-ketoglutarate and pyruvate dehydrogenase complexes partially explains the cellular toxicity of coenzyme Q0. J. Biol. Chem. 2004;279:27278, 272–85. doi: 10.1074/jbc.M314148200. [DOI] [PubMed] [Google Scholar]
- Martin E, Rosenthal RE, Fiskum G. Pyruvate dehydrogenase complex: metabolic link to ischemic brain injury and target of oxidative stress. J. Neurosci. Res. 2005;79:240–247. doi: 10.1002/jnr.20293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AAM. Cerebral ketone body metabolism. J. Inherit. Metab. Dis. 2005;28:109–121. doi: 10.1007/s10545-005-5518-0. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Nabekura T, Morishima S, Cover TL, Mori S-I, Kannan H, Komune S, Okada Y. Recovery from lactacidosis-induced glial cell swelling with the aid of exogenous anion channels. Glia. 2003;41:247–259. doi: 10.1002/glia.10190. [DOI] [PubMed] [Google Scholar]
- Park LC, Calingasan NY, Sheu KF, Gibson GE. Quantitative alpha-ketoglutarate dehydrogenase activity staining in brain sections and in cultured cells. Anal. Biochem. 2000;277:86–93. doi: 10.1006/abio.1999.4359. [DOI] [PubMed] [Google Scholar]
- Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990;4:3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- Phelka AD, Beck MJ, Philbert MA. 1,3-Dinitrobenzene inhibits mitochondrial complex II in rat and mouse brainstem and cortical astrocytes. Neurotoxicology. 2003;24:403–415. doi: 10.1016/S0161-813X(03)00031-7. [DOI] [PubMed] [Google Scholar]
- Philbert MA, Nolan CC, Cremer JE, Tucker D, Brown AW. 1,3-Dinitrobenzene-induced encephalopathy in rats. Neuropathol. Appl. Neurobiol. 1987;13:371–389. doi: 10.1111/j.1365-2990.1987.tb00192.x. [DOI] [PubMed] [Google Scholar]
- Pocernich CB, Butterfield DA. Acrolein inhibits NADH-linked mitochondrial enzyme activity: implications for Alzheimer's disease. Neurotox. Res. 2003;5:515–520. doi: 10.1007/BF03033161. [DOI] [PubMed] [Google Scholar]
- Ray DE, Abbott NJ, Chan MW, Romero IA. Increased oxidative metabolism and oxidative stress in m-dinitrobenzene neurotoxicity. Biochem. Soc. Trans. 1994;22:407S. doi: 10.1042/bst022407s. [DOI] [PubMed] [Google Scholar]
- Reed LJ. From lipoic acid to multi-enzyme complexes. Protein Sci. 1998;7:220–224. doi: 10.1002/pro.5560070125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Hackert ML. Structure-function relationships in dihydrolipoamide acyltransferases. J. Biol. Chem. 1990;265:8971–8974. [PubMed] [Google Scholar]
- Reeve IT, Voss JC, Miller MG. 1,3-Dinitrobenzene metabolism and GSH depletion. Chem. Res. Toxicol. 2002;15:361–366. doi: 10.1021/tx0155552. [DOI] [PubMed] [Google Scholar]
- Robinson BH. Lactic acidemia and mitochondrial disease. Mol. Genet. Metab. 2006;89:3–13. doi: 10.1016/j.ymgme.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Romero I, Brown AW, Cavanagh JB, Nolan CC, Ray DE, Seville MP. Vascular factors in the neurotoxic damage caused by 1,3-dinitrobenzene in the rat. Neuropathol. Appl. Neurobiol. 1991;17:495–508. doi: 10.1111/j.1365-2990.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Romero IA, Lister T, Richards HK, Seville MP, Wylie SP, Ray DE. Early metabolic changes during m-dinitrobenzene neurotoxicity and the possible role of oxidative stress. Free Radic. Biol. Med. 1995;18:311–319. doi: 10.1016/0891-5849(94)e0143-7. [DOI] [PubMed] [Google Scholar]
- Rook GA, Steele J, Umar S, Dockrell HM. A simple method for the solubilisation of reduced NBT, and its use as a colorimetric assay for activation of human macrophages by gamma-interferon. J. Immunol. Methods. 1985;82:161–167. doi: 10.1016/0022-1759(85)90235-2. [DOI] [PubMed] [Google Scholar]
- Rosenthal RE, Williams R, Bogaert YE, Getson PR, Fiskum G. Prevention of postischemic canine neurological injury through potentiation of brain energy metabolism by acetyl-L-carnitine. Stroke. 1992;23:1312–1317. doi: 10.1161/01.str.23.9.1312. discussion 1317–1318. [DOI] [PubMed] [Google Scholar]
- Samikkannu T, Chen C-H, Yih L-H, Wang ASS, Lin S-Y, Chen T-C, Jan K-Y. Reactive oxygen species are involved in arsenic trioxide inhibition of pyruvate dehydrogenase activity. Chem. Res. Toxicol. 2003;16:409–414. doi: 10.1021/tx025615j. [DOI] [PubMed] [Google Scholar]
- Sheu KF, Kim YT, Blass JP, Weksler ME. An immunochemical study of the pyruvate dehydrogenase deficit in Alzheimer's disease brain. Ann. Neurol. 1985;17:444–449. doi: 10.1002/ana.410170505. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Smolle M, Prior AE, Brown AE, Cooper A, Byron O, Lindsay JG. A new level of architectural complexity in the human pyruvate dehydrogenase complex. J. Biol. Chem. 2006;281:19772–19780. doi: 10.1074/jbc.M601140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabatabaie T, Potts JD, Floyd RA. Reactive oxygen species-mediated inactivation of pyruvate dehydrogenase. Arch. Biochem. Biophys. 1996;336:290–296. doi: 10.1006/abbi.1996.0560. 210. [DOI] [PubMed] [Google Scholar]
- Tsai CS. Nitroreductase activity of heart lipoamide dehydrogenase. Biochem. J. 1987;242:447–452. doi: 10.1042/bj2420447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MS, Yu LC, Gupta RC. Analysis of changes in energy and redox states in HepG2 hepatoma and C6 glioma cells upon exposure to cadmium. Toxicology. 2004;201:105–113. doi: 10.1016/j.tox.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Yang Y, Balcarcel RR. 96-Well plate assay for sublethal metabolic activity. Assay Drug Dev. Technol. 2004;2:353–361. doi: 10.1089/adt.2004.2.353. [DOI] [PubMed] [Google Scholar]
- Yeaman SJ. The 2-oxo acid dehydrogenase complexes: recent advances. Biochem. J. 1989;257:625–632. doi: 10.1042/bj2570625. [DOI] [PMC free article] [PubMed] [Google Scholar]