Abstract
The formation of biogenic materials requires the interaction of organic molecules with the mineral phase. In forming enamel, the amelogenin proteins contribute to the mineralization of hydroxyapatite (HAp). Leucine-rich amelogenin protein (LRAP) is a naturally occurring splice variant of amelogenin that comprises amelogenin's predicted HAp binding domains. We determined the partial structure of phosphorylated and non-phosphorylated LRAP variants bound to HAp using combined solid-state NMR (ssNMR) and ssNMR-biased computational structure prediction. New ssNMR measurements in the N-terminus indicate a largely extended structure for both variants, though some measurements are consistent with a partially helical N-terminal segment. The N-terminus of the phosphorylated variant is found to be consistently closer to the HAp surface than the non-phosphorylated variant. Structure prediction was biased using 21 ssNMR measurements in the N- and C-terminus at five HAp crystal faces. The predicted fold of LRAP is similar at all HAp faces studied, regardless of phosphorylation. Largely consistent with experimental observations, LRAP's predicted structure is relatively extended with a helix-turn-helix motif in the N-terminal domain and some helix in the C-terminal domain, and the N-terminal domain of the phosphorylated variant binds HAp more closely than the N-terminal domain of the non-phosphorylated variant. Predictions for both variants show some potential binding specificity for the {010} HAp crystal face, providing further support that amelogenins block crystal growth on the a and b faces to allow elongated crystals in the c-axis.
Keywords: LRAP, Amelogenin, RosettaSurface, Biomineralization, Solid-state NMR
Introduction
Proteins play an important role in the formation of hard tissues and other structural and functional materials in vivo.1,2 Some examples include the silica-based optical waveguide of the sponge spicule,3 magnetite compasses of magnetotactic bacteria,4 and hydroxyapatite (HAp)-based dental and skeletal tissues in vertebrates.5 Proteins can directly bind to crystal faces to accelerate6 or inhibit crystal growth,7,8 altering the resulting crystal morphology.9,10 Because the structure of interacting biomolecules influences function and mechanism, determining the structure of mineral-associated proteins is important.
Amelogenin and its proteolytic and genetic variants constitute over 90% of proteinaceous mass in developing tooth enamel.11 Several groups demonstrated the importance of amelogenin in healthy enamel formation in vivo using genetic knock-out,12 transcription regulation,13 and mutagenesis.14 Amelogenin also binds HAp15 and alters HAp crystal growth kinetics16,17in vitro. In vivo, in the presence of amelogenin, HAp crystallizes in highly elongated crystals that are ~3000 times longer than the HAp crystals in bone.18 The observed growth along the c-axis suggests a specific interaction with the {100} and {010} faces.
Typically, proteins involved in biomineralization have a high content of charged amino acids which interact with the charged mineral surface; however, of amelogenin's 180 residues, only 13 are charged. Most of amelogenin's charged residues are in the N- and C-terminal domains, suggesting these regions bind to the charged HAp surface. Only one of the charged amino acids is phosphorylated, phosphoserine at position 16. The phosphorylation status of S16 can vary in wild-type amelogenin, making the role of phosphorylation in contributing to HAp interaction unclear.
Biomineral-associated protein structure cannot be determined by X-ray crystallography or solution NMR; as a result, the mechanism of protein biomineralization is not well understood. Many experimental methods exist for studying proteins adsorbed to solid surfaces, but cannot resolve atomic features.19 Using dipolar recoupling techniques such as DRAWS and REDOR, solid-state NMR (ssNMR) is uniquely suited for determining the distance between a pair of spin-1/2 nuclei at the protein-surface interface, providing molecular-level structure and orientation detail.20–23 However, the requirement for isolated spin pairs limits studies to proteins that can be chemically synthesized (~60 amino acids or less), which precludes investigating full-length amelogenin. Leucine-rich amelogenin protein (LRAP) is a 59-residue naturally occurring splice variant of amelogenin, comprising amelogenin's N- and C-terminal domains,24 the domains that are thought to bind HAp. Because LRAP contains amelogenin's HAp interaction regions and can be chemically synthesized, it provides a tractable model for structural studies of the surface-bound protein using ssNMR. Using this technique, we previously determined the partial structure and orientation of LRAP's C-terminal domain adsorbed to HAp.20,23
While ssNMR is a good tool for investigating surface immobilized proteins, high-resolution ssNMR data exists for only two protein-biomineral complexes: LRAP20,21,23 and statherin.25–31 Typically, a single isolated isotopic pair gives one or two data points, such as one backbone distance measurement and one orientation measurement. This requires the preparation of many samples to investigate even a small region of any given protein, and lengthy experiment times make thorough structure analysis time consuming and incomplete.
Computational methods have the potential to reduce experiment time. For instance, methods combining solution NMR and computational structure prediction have drastically reduced the amount of NMR data required to solve high-resolution protein structures in solution.32–34 In particular, the Rosetta structure prediction method was combined with chemical shift,34 nuclear overhauser effect,35 or residual dipolar coupling NMR data.36,37 In these studies, protein structures predicted by Rosetta were at or near atomic-level accuracy relative to the corresponding crystal structures.
Recently, we developed Rosetta to predict protein structure at the biomineral interface (RosettaSurface)38–42 and added the ability to bias structure prediction using ssNMR (RosettaSurface.NMR).43 As has been seen for solution NMR, RosettaSurface.NMR can enhance the utility and scope of relatively sparse ssNMR data. In a test case, we predicted the structure of HAp-adsorbed statherin, a 43 amino acid enamel protein, and obtained similar results with or without ssNMR bias, demonstrating the power of RosettaSurface to complement experimental studies.43 Structures predicted from ssNMR-biased RosettaSurface.NMR provide a means to visualize data, extrapolate data to nearby regions of the protein, and to focus subsequent experimental studies on the most relevant regions of the protein-crystal interface.
Here we extend these computational studies to understand and complement the experimentally determined structure of LRAP bound to HAp. First we use ssNMR to determine the partial HAp-adsorbed structure and orientation of LRAP's N-terminal domain (in the context of full-length LRAP). Then, we investigate the structure and preferential adsorption of LRAP using ssNMR-biased RosettaSurface.NMR predictions at five HAp crystal faces. We also investigate the effect of phosphorylation at S16 on LRAP's structure and binding orientation to address the importance of phosphorylation on HAp interaction.
Materials and Methods
Materials
Labeled amino acids were purchased from Cambridge Isotopes and used as received. Solvents were used without further purification. FMOC-protected labeled amino acids were prepared according to standard procedures and used without further purification.44
Protein preparation, purification and characterization
Proteins were prepared using standard FMOC chemistry45 by the University of Texas, Southwestern Medical Center (Dallas, TX). Proteins were purified using prep scale reverse phase HPLC, buffer A: 0.1% trifluoroacetic acid in water, buffer B: 0.1% trifluoroacetic acid in acetonitrile. LRAP eluted at 54% B. Proteins were analyzed for molecular weight and purity using electrospray MS.
Sample preparation of the free protein
To prepare a solid state sample of the free protein, 20 mg of LRAP was dissolved into 1 mL of phosphate buffer, consisting of a solution of 0.15M NaCl and saturated with respect to hydroxyapatite (PB), and diluted to 20 mL with water. The pH was adjusted to 7.4, the solution was frozen in liquid nitrogen, lyophilized, and the entirety of the resulting powder packed into the NMR rotor.
Sample preparation of the protein bound to HAP
The protein sample was bound to HAp as described previously. HAp crystals were prepared according to published procedures46 and had the expected needle-shaped morphology with approximate dimensions of 50 × 50 × 200 nm, with the c-axis elongated. Briefly, a solution of 0.33 mg/mL LRAP at pH 7.4 was bound to 75 mg of 94 m2/g HAP (protein to HAp weight ratio of 1:3) for 1 hour. The amount of protein bound was determined by measuring the change in concentration before and after binding and for each wash using UV absorbance measurements (λ=275 nm). Amounts bound were: 12.8, 15.3, 13.1, 15.3, 14.5, 15.9 and 16.4 mg for G8-Y12(pS), G8-Y12, L15-V19(pS), L15-V19, V19-L23(pS), V19-L23 and K24-S28 respectively, to 100 mg of HAp. The sample was packed into an NMR rotor as a wet paste for the hydrated, surface bound sample.
NMR experiments
NMR experiments were performed on a 3-channel Chemagnetics Infinity console operating at 300 MHz proton frequency. A 3-channel, variable temperature Chemagnetics probe was used, employing a 6 μs 90° pulse for 1H and a 0.5–1 ms contact time for cross polarization experiments. Temperatures in the rotor were calibrated using 207Pb(NO3)2.47 Chemical shifts were referenced to glycine, 177.0 ppm.
REDOR
For Rotational Echo DOuble Resonance (REDOR) experiments, XY8 phase cycling was used on both observe and dephasing channels. For both REDOR experiments, 13C{31P} and 13C{15N}, 180° pulses of 13.0–15.0 μs were used for both the observe and dephasing nuclei and samples were spun at 4 kHz. Two Pulse Phase Modulated (TPPM) decoupling48 with a 65 kHz decoupling field was used throughout. Data for the hydrated samples were collected at −80 °C. Typically, 4096 scans were taken for shorter dephasing periods and 8192–16384 scans were taken for longer dephasing times, with a 3 s pulse delay. Data was collected at every 8 or every 16 rotor periods, out to 104 rotor periods for 13C{31P} and 13C{15N} REDOR. In all cases, the final dephasing curve represents the average of at least 3–5 repetitions. The contribution of the natural abundance background (58 backbone carbonyls and 7 sidechain carbonyls) has been removed from the 13C{31P} and 13C{15N} dephasing curves using the following equation:
where S is the signal from the dephasing experiment and S0 is the control experiment without the dephasing pulses, %NA is the percent of the signal resulting from the natural abundance background (39.6%) and %L is the percentage of the signal resulting from the isotopic label (60.4%). Simulated REDOR dephasing curves were calculated using numerical methods that incorporated chemical shift anisotropies and experimental parameters. 3-spin models assumed a 90° orientation between the C-P vectors.
The Algorithm
For this study we used our recently developed RosettaSurface.NMR protocol.43 Briefly, each execution of RosettaSurface.NMR begins by folding a protein from a fully extended conformation in solution using a united-atom model. At this low-resolution stage, protein intramolecular ssNMR constraints are applied, but protein-surface intermolecular constraints are not. Next, the high-resolution (all-atom) representation of the protein is refined in solution and adsorbed to the surface in a random orientation. The fold and orientation of the protein are refined on the surface, resulting in one candidate adsorbed-state structure. All calculations on the surface include the biasing influence of both protein intramolecular and protein-surface intermolecular ssNMR measurements. High-resolution refinement includes backbone, side-chain, and rigid-body optimization using a Monte Carlo-plus-minimization protocol.
The full-atom energy (E) used for decoy discrimination is a linear combination of attractive and repulsive Lennard-Jones interactions (Eatt and Erep), solvation (Esol), hydrogen bonding (Ehb), electrostatics (Ecoul), and a term to enforce ssNMR constraints (Econstraint):
| (1) |
where:
| (2) |
where:
| (3) |
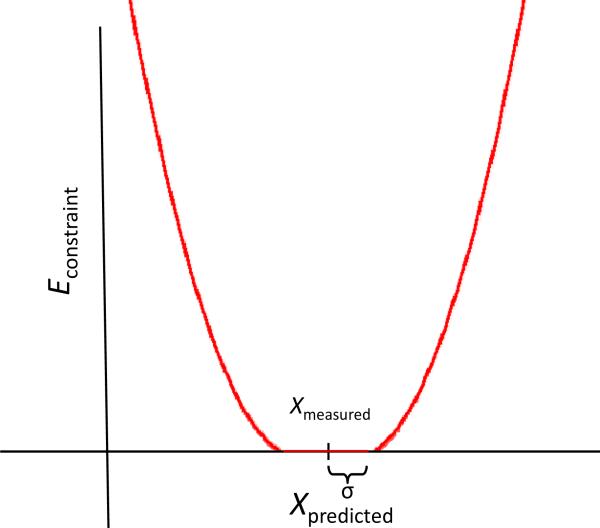
In equation 3, ximeasured and xipredicted are the ith ssNMR determined distance and RosettaSurface. NMR predicted distance, respectively; σi is the experimental error of the ith measurement; n is the number of biasing constraints used during structure prediction (equation 2). Figure 1 shows the functional form of the constraint energy. During biased structure prediction, the constraint weight w (equation 1) modulates the strength of the bias toward experimental measurements; the units of the weight are kcal/mol. Because the RosettaSurface energy function used for structure generation in this study includes a constraint energy term, a true binding energy is not calculated.
Figure 1. Functional form of Econstraint for a single measured distance from equation 3, showing the flat-bottom quadratic potential used to bias RosettaSurface. NMR structure prediction.
The energetic penalty is zero for predictions within experimental error, and increases quadratically with increasing deviation from ssNMR measurement for predictions outside the experimental error.
For all predictions, 105 candidate HAp-adsorbed LRAP structures were generated using RosettaSurface.NMR, each resulting from ~35×103 sampled conformations. Each candidate structure is assessed using the constraint energy Econstraint (for all n ssNMR measurements). Structures with the smallest constraint energy are used for further analysis and model representation.
LRAP Models
For the starting structures, we built extended molecular structures of phosphorylated and non-phosphorylated LRAP variants using PyMol.49 The atomic parameter set was previously reported in Masica et al.39
Hydroxyapatite Models
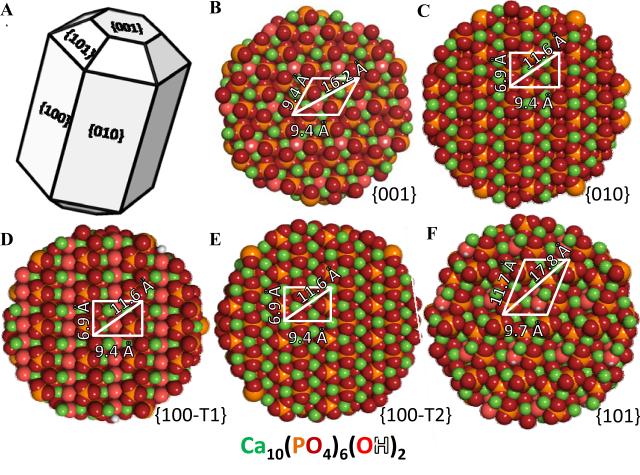
We built monoclinic hydroxyapatite crystals using CrystalMaker.50 In total, five HAp surfaces were used in this study: {001}, {010}, {101}, and two differentially terminated {100} faces (Figure 2). The atomic parameter set was previously reported in Masica et al.39
Figure 2. Hydroxyapatite model.
(A) Schematic of hexagonal HAp showing the five crystal faces (two differentially terminated surfaces at the {100} crystal plane) and the smallest periodic motif of open phosphate clusters at the (B) {001}, (C) {010}, (D) {100}-T1, (E) {100}-T2, and (F) {101} used for biased RosettaSurface. NMR structure prediction.
Results
Experimental data
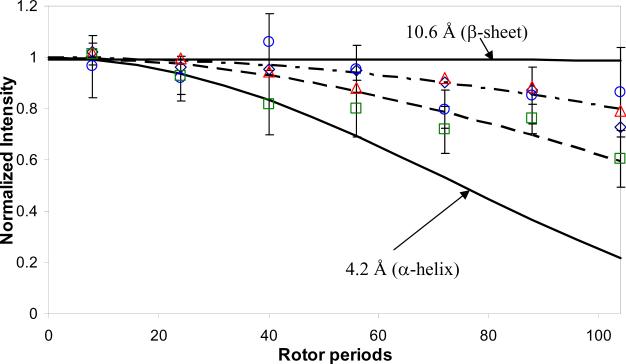
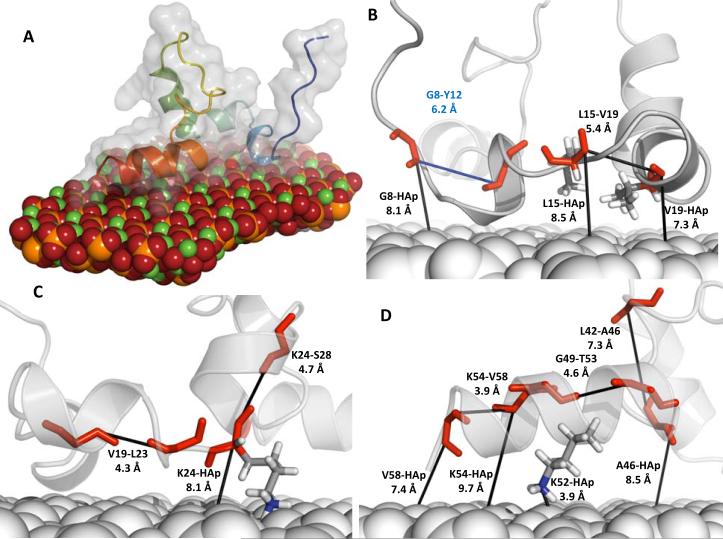
Previously, we determined the partial structure of LRAP's C-terminal domain adsorbed to HAp using ssNMR. Here, we chemically synthesized seven new constructs labeled at unique positions for acquisition of ssNMR derived distance measurements and subsequent ssNMR-biased computational structure prediction. The labels were incorporated at atomic pairs of amino acids in the N-terminal and middle domain of full-length LRAP, and include: G8-Y12, G8-Y12(pS), L15-V19, L15-V19(pS), V19-L23, V19-L23(pS), and K24-S28, where pS indicates that the labeled pair was incorporated into the LRAP variant phosphorylated at residue S16. In each labeled pair, the ito i + 4 residues were labeled at the backbone carbonyl carbon (13C′) and backbone amide nitrogen (15N) atoms respectively. This labeling scheme allowed us to probe for helix, extended, and β-sheet secondary structure in the measured regions, which have distinctly different REDOR dephasing curves as shown in Figure 3. In addition, all labeled constructs were subjected to measurements between the 13C′ isotopes and HAp-31P phosphorus atoms to determine proximity to the HAp surface; in total, 14 new ssNMR measurements were acquired for this study. Table 1 shows results for the new set of ssNMR distance measurements and all previously published measurements.
Figure 3. Representative REDOR 13C-15N dephasing curves.
Because of the similarity of the data, only the non-phosphorylated data are shown. G8-Y12 (open navy diamonds), L15-V19 (open green squares), V19-L23 (open red triangles) and K24-S28 (open blue circles). Error bars are shown only for L15-V19 and K24-S28 for clarity, but are typical for all of the data. The dashed lines show fits to the data (4.9 Å, dash), (5.5 Å, 5.6 Å, 5.7 Å, dash-dot). The solids lines are 4.2 Å (α-helix) and 10.6 Å (β-sheet).
Table 1.
Comparison of all ssNMR measurements, ensemble statistics at the {010}-LRAP(pS) and {010}-LRAP interfaces (Figure 7), and a representative structure from biased structure prediction at the {010}-LRAP(pS) interface.
| Measurement | Label | Distance off (Å) | Distance on (Å) | {010}-LRAP(pS) | {010}-LRAP | Rep. Model | Ref. |
|---|---|---|---|---|---|---|---|
| G8-Y12 (pS) | 13C′-15N | 5.4 ± 0.5 | 6.1±0.5 | 7.1±1.4 Å (21) | 6.2 Å | New | |
| G8-Y12 | 13C′-15N | 6.1 ± 0.5 | 5.5±0.5 | 6.5±1.1 Å (15) | New | ||
| L15-V19 (pS) | 13C′-15N | 5.9 ± 0.5 | 5.0±0.5 | 5.2±0.9 Å (37) | 5.4 Å | New | |
| L15-V19 | 13C′-15N | 5.0 ± 0.5 | 4.9±0.5 | 5.4±0.9 Å (26) | New | ||
| V19-L23 (pS) | 13C′-15N | 6.1 ± 0.5 | 5.6±0.5 | 5.5±1.3 Å (20) | 4.3 Å | New | |
| V19-L23 | 13C′-15N | 6.8 ± 0.5 | 5.6±0.5 | 5.3±1.0 Å (23) | New | ||
| K24-S28 | 13C′-15N | 4.5 ± 0.5 | 5.7±0.5 | 4.7±1.0 Å (6) | 4.9±0.9 Å (13) | 4.7 Å | New |
| L42-A46 | 13C′-15N | 5.3 ± 0.5 | 5.6±0.5 | 6.1±1.0 Å (30) | 6.2±1.0 Å (31) | 7.3Å | 20 |
| A49-T53 | 13C′-15N | 5.9 ± 0.5 | 6.9±1.0 | 5.8±1.6 Å (31) | 6.1±1.7 Å (39) | 4.6 Å | 20 |
| K54-V58 | 13C′-15N | 5.5 ± 0.5 | 5.5±0.5 | 5.1±1.4 Å (15) | 5.0±1.1 Å (16) | 3.9 Å | 21 |
| G8-HAp (pS) | 13C′-31p | 9.0 ± 0.5 | 6.5±0.5 | 8.6±1.6 Å (10) | 8.1 Å | New | |
| G8-HAp | 13C′-31p | >12± 1.0 | 8.0±0.5 | 9.3±1.5 Å (15) | New | ||
| L15-HAp (pS) | 13C′-31p | 6.0 ± 0.5 | 5.3±0.5 | 7.7±1.3 Å (6) | 8.5 Å | New | |
| L15-HAp | 13C′-31p | >12 ± 1.0 | 9.0±1.0 | 10.1±2.0 Å (36) | New | ||
| V19-HAp (pS) | 13C′-31p | 9.0 ± 0.5 | 7.0±0.5 | 8.9±1.3 Å (10) | 7.3 Å | New | |
| V19-HAp | 13C′-31p | >10± 1.0 | 9.0±1.0 | 10.6±2.0 Å (28) | New | ||
| K24-HAp | 13C′-31p | >12± 1.0 | 9.0±1.0 | 12.3±2.5 Å (12) | 12.4±2.6 Å (17) | 8.1 Å | New |
| A46-HAp | 13Cβ-31p | >12 ± 1.0 | 8.0±0.5 | 9.5±1.8 Å (17) | 9.2±1.9 Å (17) | 8.5Å | 20 |
| K52-HAp | 15Nζ-31p | >10 ± 1.0 | 4.0±1.0 | 6.6±3.0 Å (41) | 6.4±2.8Å (45) | 3.9 Å | 20 |
| K54-HAp | 13C′-31p | >12 ± 1.0 | 6.5±0.5 | 8.7±1.2 Å (8) | 8.6±0.1 Å (5) | 9.7 Å | 21 |
| V58-HAp | 13C′-31p | >12 ± 1.0 | 5.8±0.5 | 7.9±1.3 Å (10) | 7.5±1.3 Å (13) | 7.4 Å | 21 |
The residue for which the Measurement was acquired, the placement of the isotopic Label, the experimentally determined Distance off (Å)(distance to nearest 31P for desorbed LRAP) and Distance on (Å) (distance to nearest 31P for adsorbed LRAP), ensemble statistics at the {010}-LRAP(pS) and {010}-LRAP interfaces from biased predictions with a weight w of 1 kcal/mol (values are mean distances from the 100 structures with lowest Econstraint, errors are standard deviation, values in parenthesis indicate number of structures from the 100 structures with lowest Econstraint that met the measurement within experimental error), and corresponding prediction from a representative model (Rep. Model) at the {010}-LRAP(pS) interface. Measurements made for desorbed HAp (Distance off) are to ensure that signal in the adsorbed state (Distance on) arise from HAp-incorporated 31P. The primary structure of LRAP(pS) is: MPLPPHPGSPGYINLpSYEVLTPLKWYQSMIRQP PLSPILPELPLEAWPATDKTKREEVD
The structure of the N-terminus in both the phosphorylated and the non-phosphorylated proteins bound to HAp is consistent with a largely extended structure (5.5 Å and longer), with the L15-V19 region showing a loosely helical structure (4.9 Å–5.4 Å). The shorter measured distance for L15V19 could be consistent with either a less coiled helix resulting in weaker i to i+4 contacts, or multiple structures, one population which is helical and one which is not helical. These are indistinguishable based on the REDOR experiment as has been discussed previously.20 The non-phosphorylated REDOR dephasing curves are shown in Figure 3 as symbols and the fits are shown as dashed lines. Solid lines shown for an α-helix (4.2 Å) and a β-sheet (10.4 Å) for comparison, as indicated, demonstrate the ease of distinguishing between an α-helix and a β-sheet using the designed labeling scheme.
While there is little difference in structure as a function of phosphorylation, there are significant structural changes in the N-terminus as a function of binding the protein to HAp, which are different for each construct. For the phosphorylated protein, the G8-Y12 region becomes more extended on the surface, while the region from L15 to L23 becomes more helical. For the non-phosphorylated construct, the most remarkable change is in the K24-S28 region, where this region of the protein changes from nearly a perfect helix to an extended structure on the surface. Two of the other three regions investigated for the non-phosphorylated protein also exhibit a structural change upon binding to HAP: the regions at G8-Y12 and V19-L23 become more helical while L15-V19 has little change from its loose helical character.
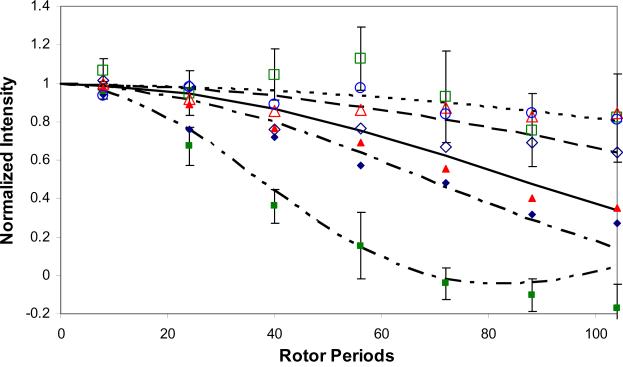
Orientation measurements from the 13C labels in the protein backbone to the 31P atoms in the surface show that in all cases the backbone of the N-terminus was oriented next to the surface. The backbone was consistently significantly closer to the surface for the phosphorylated protein than for the non-phosphorylated (Figure 4). To confirm that the presence of the side-chain phosphorous (pS16) was not resulting in a falsely low protein-surface distance, the contribution of the side-chain phosphorous to the dephasing curves was accounted for. The intraprotein 13C-31P distance was determined from the off the surface sample. Three site models were then generated for the surface-bound sample to account for the non-HAp 31P contribution. These models assumed a 90 degree orientation between the two C-P vectors. In all cases, if the residue carbonyl-pS16 distance was assumed to remain the same upon binding, the intraprotein contribution did not change the best fit distance measured between the protein and the surface. This was found to be true as long as the protein-surface distance was the shorter of the two distances, as was always the case. This has been observed before for longer dipolar recoupling measurements.20 However, the RosettaSurface.NMR distances for the bound protein do indicate a change in the intraprotein distance based on the 100 lowest Econstraint structures. In the {010}-LRAP(pS) low-energy ensemble, the L15-pS16 and pS16-V19 distances are 5.3 ± 0.6 Å and 8.2 ± 1.2 Å, respectively. If these values are used for the intraprotein distances instead of the experimentally measured values determined for the unbound protein, the dephasing data is equally well fit with a 3-spin model of 5.3 Å for the L15- pS16 and 7.0 Å L15-HAp distance (Figure 5). Similarly, the 13C-31P dephasing data for V19 is equally well fit by a 3-spin model of 8.2Å pS16-V19 and a 7.5 Å L15-HAp distance.
Figure 4. REDOR 13C-31P dephasing curves.
G8 (open navy diamonds), G8–pS (closed navy diamonds), L15 (open green squares), L15–pS (closed green squares), V19 (open red triangles) V19–pS (closed red triangles), K24 (open blue circles) and K24–pS (closed blue circles). Error bars are shown only for L15 and L15-pS for clarity, but are typical for all of the data. Lines show fits to the data (5.3 Å, dash-dot-dot; 6.5 Å, dash-dot; 7.0 Å, solid; 8.0 Å, dash; 9.0 Å, dot).
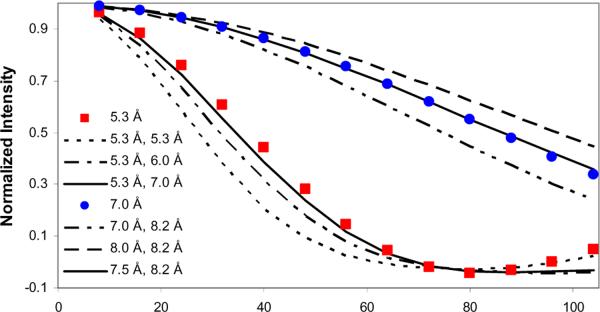
Figure 5. Modeled 13C-31P dephasing curves.
In the phosphorylated protein, 13C-31P measured distances could be a combination of a measurement to the sidechain phosphoserine, as well as to the surface phosphate groups. All simulations assumed the 3 spins were oriented at 90 degrees. For the L15-V19 distance, 5.3 Å and combinations of (5.3 Å and 5.3 Å), (5.3 Å and 6.0 Å) and (5.3 Å and 7.0 Å) are shown where the combination of (5.3 Å and 7.0 Å) is indistinguishable from a single distance of 5.3 Å within experimental error. For V19-L23, 7.0 Å and combinations of (7.0 Å and 8.2 Å), (7.5 Å and 8.2 Å) and (8.0 Å and 8.2 Å) are shown and the combination of (7.5 Å and 8.2 Å) is indistinguishable from a single distance of 7.0 Å.
RosettaSurface data
Structure prediction was biased using all new and previously published ssNMR measurements (Table 1). We performed ssNMR-biased structure prediction at five HAp crystal faces: {001}, {010}, two differentially terminated {100} faces ({100}-T1 and {100}-T2), and {101}. The {100}-T2 surface terminates with a higher density of calcium ions than the {100}-T1 surface. All other HAp surfaces were cut to expose approximately neutral (mixed-charge) surfaces. These faces were chosen because they are expressed in stable HAp crystals and have a range of surface geometries.51
For each surface, predictions were divided into 11 runs of ~9,100 candidate structures. For each of the 11 runs at each surface, the weight w (see Materials and Methods) was set to a number between 0 and 1 kcal/mol evenly divisible by 0.1 (i.e., 0, 0.1, 0.2 …. 1). This resulted in a total of 105 candidate adsorbed-state structures at each of five surfaces for phosphorylated and non-phosphorylated variants of LRAP; i.e., 106 total structures generated from ~35×109 sampled conformations (see Materials and Methods). We employed this approach to test whether the set of constraints is easier to match at particular LRAP-HAp interfaces. This result would manifest as predictions at one or more HAp surfaces exhibiting smaller Econstraint versus weight relative to predictions at the other HAp surfaces. This approach has the potential to probe surface specificity at protein-surface interfaces while determining the experimentally biased structure of the adsorbed protein.
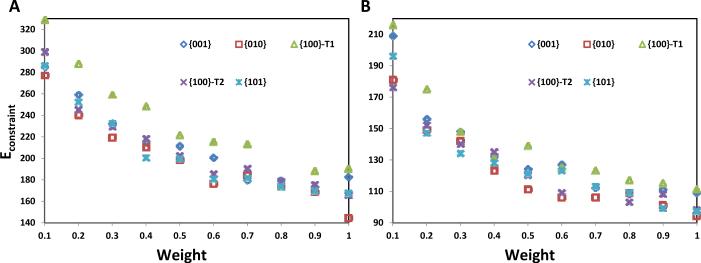
Figure 6 shows Econstraint versus weight for structures resulting from ssNMR-biased predictions. As expected, Econstraint decreases with increasing weight at all five HAp surfaces for both LRAP variants. RosettaSurface. NMR had greater difficulty meeting experimental constraints for the phosphorylated variant (Figure 6A), relative to the non-phosphorylated variant (Figure 6B), resulting in a higher average constraint weight for the phosphorylated variant. On average, Econstraint is smallest at the HAp {010} surface for phosphorylated LRAP (Figure 6A) and non-phosphorylated LRAP (Figure 6B). That is, it is easiest for RosettaSurface to create structures matching the experimental constraints when LRAP variants bind the {010} surface, suggesting the {010} surface is more likely bound in the ssNMR experiments. Convergence between prediction and experiment was slowest at the {100}-T1 surface for both LRAP variants. Structures were similar for both LRAP variants at all five HAp crystal surfaces; here we focus on structures predicted at the HAp {010} surface for both the phosphorylated and non-phosphorylated LRAP variants.
Figure 6.
Econstraint versus weight for phosphorylated (A) and non-phosphorylated (B) LRAP at five different HAp surfaces.
Figure 2 shows the dimensions of the smallest periodic motif of phosphate clusters on each HAp surface used for these predictions ({100-T2} surface phosphate clusters are occupied by calcium atoms). Periodic motifs of phosphates on the {100}-T1 and {010} surfaces are similar ({010} and {100} crystal lattices are identical aside from the direction of hydroxyl groups); however, terminations chosen here have different calcium positions. The {100}-T2 and {010} surfaces are identical except for an additional calcium atom deposited into each phosphate cluster on the {100}-T2 surface. The {101} surface has a unique geometry, whose dimensions are larger than other surfaces used for these predictions. The complementarity of these motifs to the motif of LRAP's binding domain (and hence the relative positions of binding residues) plays an important role where convergence between ssNMR and structure prediction are concerned (Figure 6).
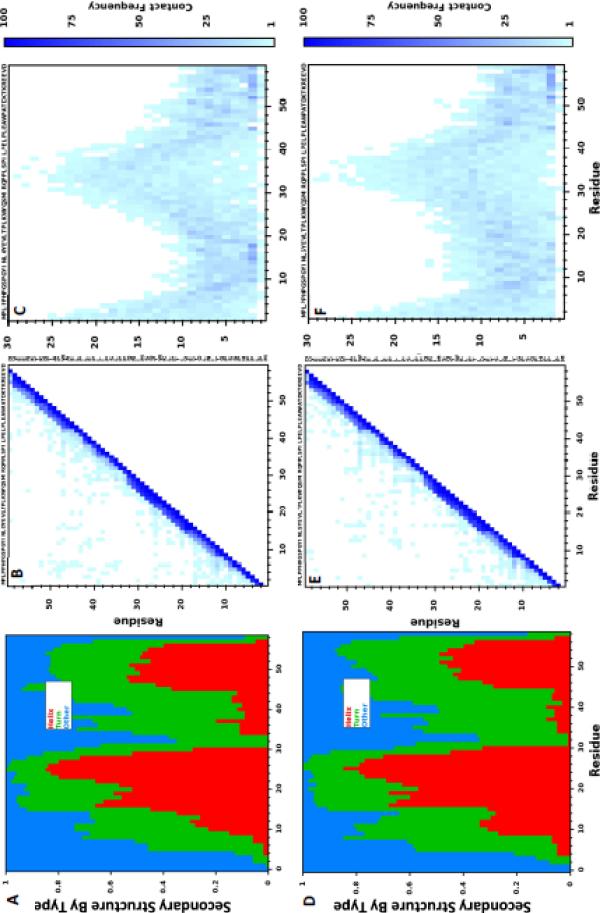
Figure 7 shows ensemble structural statistics for the 100 structures with the smallest Econstraint for HAp {010}-LRAP(pS) (Figures 7A–C) and HAp {010}-LRAP (Figures7D–F). Figures 7A and 7D show similar secondary structure for the HAp {010}-LRAP(pS) and HAp {010}-LRAP complexes respectively. For both complexes, there is increasing locally hydrogen-bonded turn-like secondary structure from residues ~3–11, with some propensity for helix formation from residues ~10–20. Residues ~18–32 form a helix-turn-helix motif at both interfaces with P22 being a common turn initiator (see also, Figure 8A). Residues ~30–48 are mostly extended or locally hydrogen-bonded turn-like secondary structure and the ~10 most C-terminal residues have significant propensity to form turn-like secondary structure with some helical structure from residue 48 to 56, at both interfaces.
Figure 7. Ensemble structure of LRAP adsorbed to HAp for the 100 structures with smallest Econstraint.
Distributions of three basic secondary structure motifs at the (A) {010}-LRAP(pS) and (D) {010}-LRAP interfaces. Pair-wise LRAP intramolecular residue-residue contacts at the (B) {010}-LRAP(pS) and (E) {010}-LRAP interfaces. Pair-wise residue-surface distances at the (C) {010}-LRAP(pS) and (F) {010}-LRAP interfaces. The structural designations “Helix” and “Turn” were assigned using the DSSP definitions49; classification relied on Rosetta's hydrogen-bond function rather than the generalized hydrogen-bond function implemented by the DSSP package. An intramolecular residue-residue contact is declared if two residues have an inter-residue atomic pair within 4 Å. A residue-surface distance reflects the closest atomic contact for that residue and the closest surface atom. Note for comparing contacts with ssNMR measurements: the intra or intermolecular atomic pairs that constitute a contact are not necessarily the same atoms that were isotopically labeled for ssNMR measurements.
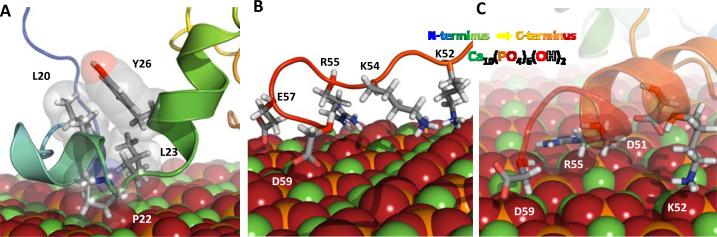
Figure 8. Phosphorylated LRAP bound to HAp {010} crystal face.
(A) Helix-turn-helix motif in LRAP's N-terminal domain is partially stabilized by a hydrophobic interaction between residues L20, L23 and Y26. Conformation-dependent differential HAp binding of charged residues in LRAP's C-terminal domain shown using two representative models, (B) and (C). In (B), a partially unfolded structure facilitating simultaneous binding of K52, K54, R55, E57, and D59. In (C), a helical secondary structure facilitating simultaneous binding of D51, K52, R55, and D59.
Figures 7B and 7E show the computationally determined protein intramolecular contacts for the HAp {010}-LRAP(pS) and HAp {101}-LRAP complexes respectively. The dense regions of i to i + 4 contacts from ~18–32 and the ~10 most C-terminal residues suggest significant helical content (in agreement with Figures 7A and 7C). There are few contacts made between residues that are greater than five residues apart in primary sequence, suggesting little tertiary structure. However, there are significant off-diagonal contacts between residues ~30–48 owing to an abundance of hydrophobic residues in that region, particularly at the HAp {010}-LRAP interface. Additionally, there are significant off-diagonal contacts from residues ~20–30, particularly at the HAp {010}-LRAP(pS) interface, owing to an abundance of hydrophobic residues in that region. Those hydrophobic contacts help facilitate the helix-turn-helix motif in that region (see Figure 8A).
Residues ~10–20 of the phosphorylated LRAP variant bind HAp more closely than the same region of the non-phosphorylated LRAP variant (Figures 7C and 7F respectively). Protein-surface distances in that region are almost exclusively within 10 Å at the HAp {010}-LRAP(pS) interface, whereas the {010}-LRAP interface shows an approximately even distribution of contacts within 15 Å from residues ~10–20. Furthermore, most residues in that region are predominately located within 3 Å of the HAp {010}-LRAP(pS) interface; these high-frequency direct contacts are mostly absent at the {010}-LRAP interface.
The C-terminal domain is tightly bound at both the HAp {010}-LRAP(pS) and HAp {010}-LRAP interfaces. Strong binding via the C-terminal domain is predominately mediated by the charged residues in that region, i.e., E45, D51, K52, K54, R55, E56, E57, and D59. Which of those residues contact the surface is dictated by the conformation of the C-terminal domain, and varies within the individual structures of the ensemble (Figures 8B and 8C).
Figure 9 shows a representative structural model from the 100 structures with smallest Econstraint from ssNMR-biased structure prediction at the {010}-LRAP(pS) interface. This structure was chosen because it represents a dominant ensemble conformation (Figures 7A–C) of the ssNMR-biased structure (Table 1). Figure 9A shows a global view of the HAp {010}-LRAP(pS) complex; for that model, figures 9B–D show predicted distances for which biasing was applied. These figures directly show the extent of biasing at specific regions of the complex and the structure that was predicted in those regions. Table 1 shows all experimental measurements and the corresponding predictions from the representative model (Figure 9).
Figure 9. The molecular structure of phosphorylated LRAP adsorbed to HAp {010} surface.
(A) Representative structure determined with a weight w of 1 kcal/mol. Opacity represents LRAP's molecular shape, cartoons represent secondary structure. Predicted distance measurements at constrained atoms for the (B) N-terminal, (C) middle, and (D) C-terminal domains. For comparison with ssNMR measurements see Table 1.
Figure 10 shows the local RMSD in a representative structural model (Figure 9) relative to the 100 structures with the smallest Econstraint, and is useful to assess local dispersion among models. Complementary to the ensemble statistics shown in Figure 7, Figure 10 shows that the N- and C-terminal segments have more regular structure, compared with the middle segment, among the 100 structures with smallest Econstraint. Because LRAP's predicted structure includes regions of extended structure, the relative global RMSD among structures is generally large. The method used here, of comparing all overlapping fragments (see Figure 10 caption), is useful for showing residue-specific, local structural variation while adsorbed to the surface.
Figure 10. Local RMSD of a representative structure (Figure 9) relative to the 100 structures with lowest constraint energy.
To show the dispersion among structures, we superimposed the Cα atoms of every overlapping 3mer in the representative structure and each of the 100 structures with lowest Econstraint, and calculated the RMSD of the central Cα atom in the 3mer. The color bar is in angstroms and corresponds to the opaque profile defining LRAP's shape. LRAP pictured from C-terminus (left) to N-terminus (right).
Discussion
Structure
Our ssNMR and computational results show that LRAP is largely extended on the HAp surface, with regions of canonical secondary structure. This is observed regardless of the phosphorylation state or crystal face investigated and suggests that LRAP does not have a specific binding motif, but has a flexible structure to maximize surface coverage. Consistent with this observation, solution NMR studies show that both LRAP and amelogenin are extended in solution in their monomer form.52–56 The elongated HAp crystals found in enamel have led to the suggestion that one of amelogenin's roles is to block growth of the {100} and {010} faces. The less globular structure may facilitate this function by allowing the unfolded protein to cover more surface nucleation sites.
The new ssNMR measurements from residues 8–28 of LRAP bound to HAp reveal a largely extended structure, with L15-V19 exhibiting some loosely helical regions in both variants based on the shorter measured distances. An i to i+4 distance of 4.2 Å would be expected for a perfectly helical secondary structure, while distances out to 5.4 Å are interpreted as helical or loosely helical and a well-defined β-sheet structure would have a measured distance of 10.6 Å. A largely extended structure is consistent with i to i+4 distances longer than 5.5 Å, based on the distance of 5.8 Å resulting from a linear combination of the torsion angles from 4.2 to 10.6 Å.20,21,28 While these designations are valuable, it is important to note that the ssNMR measurement represents an average of the ensemble of structures present and could represent a single structure with small variations around it, a combination of two or three structures with small variations in those structures or an ensemble of structures with a large structural deviation.28 This distinction is difficult to make with ssNMR, and an added challenge is that tertiary structure, such as turns, is not included in these definitions and could modulate any of the expected distances depending on turn location. RosettaSurface.NMR has the potential to distinguish between these possibilities and given the prediction of turns, has the potential to significantly impact our interpretation of the measured distances.
In reasonable agreement with our ssNMR data, our ssNMR biased computational analysis found LRAP's HAp-adsorbed fold includes extended (non-locally hydrogen bonded) and turn-like (locally hydrogen bonded) secondary structure in the first ~18 residues. From residues 18–21 there is increasing helical structure, which is moderately perturbed by the presence of a proline at position 22, followed by a stable helix from residues ~24–30; proline is known to break and initiate helix formation owing to the lack of a backbone amide hydrogen atom and the presence of a constrained θ angle.57 These helical segments interact with one another via several hydrophobic residues, forming a helix-turn-helix motif from residues 18–30. The C-terminal half of LRAP comprises extended and turn-like secondary structure, with some propensity for helix formation in the predicted, low-energy ensemble. LRAP's observed fold appears to have little dependence on phosphorylation at S16.
While no structural change was observed as a function of phosphorylation, ssNMR measurements indicate surface induced structural changes for both variants. Of particular note is the non-phosphorylated variant, where the region of K24-S28 exists as a nearly perfect helix off the surface (4.5 Å), becoming largely extended upon binding (5.7 Å). This may suggest an important turn region or interaction region of the non-phosphorylated protein in the K24-S28 region. The RosettaSurface.NMR predictions indicate a turn away from the surface at K24 (Figure 9C), which could be the cause of the structural change. To increase sampling of relevant adsorbed-state structures, ssNMR-measured off-surface distances were not used to bias RosettaSurface.NMR structure prediction; therefore, the change in K24-S28 distance upon binding is not present in the predicted ensembles. Investigating surface-induced structural changes computationally is an area of future development.
The last nine residues of LRAP contain seven charged residues, which is an unusually high charge density for structured proteins.58 Several residues have high contact frequency at multiple distances from the HAp surface in the C-terminal region (Figures 7C and 7F). For instance, E56 locates at either 2 Å or 7 Å; R55 has many short-range contacts at 2 Å from the HAp surface with a significant distribution centered at 6 Å; K54 has a distribution centered at 7 Å in addition to a significant number of short-range contacts at 2 Å. Furthermore, LRAP's C-terminal region is predicted to adopt both helical and turn-like secondary structures at the HAp interface (Figures 7A and 7D); these observations suggest the possibility of surface-induced conformational preferences in the C-terminal segment of both LRAP variants. Figures 8B and 8C show LRAP charged residues in the C-terminal region binding HAp differentially in a conformation-dependent manner. Because the ssNMR distances are averages over the entire ensemble, this example highlights how RosettaSurface.NMR has the ability to provide additional insight into the structural conformations that these distances could represent.
Biased RosettaSurface.NMR recovers the general structural trends of the ssNMR data reasonably well, with the exception of the structure at K24-S28 and K54-V58. Experimentally, these were found to be largely extended, while the computational model predicted that these regions were largely helical. These disparities are likely due to the sparse ssNMR constraints and the larger size of the protein in this study relative to previous studies.43 It highlights the challenges of modeling surface-adsorbed proteins and emphasizes the importance of benchmarking these models with known experimental data to provide further development.
The general difficulties in protein structure prediction, such as developing an adequate energy function and sampling strategy, are expected to be a limitation of the RosettaSurface protocol. The RosettaSurface protocol as implemented in this study can only sample a fraction of the conformation space associated with folding multiple protein variants at the interface of five crystalline solids. We used RosettaSurface.NMR under the hypothesis that the addition of experimental bias will help compensate for some energy function limitations and focus sampling to relevant conformation space; however, the space is likely still under-sampled, especially with respect to compact LRAP structures. Furthermore, in the simulation constructs, HAp atomic coordinates were held fixed and not allowed to deviate from their lattice positions. All of these difficulties contribute to uncertainty in the models with respect to structure, orientation, and surface specificity.
Orientation
While LRAP's structure was similar for both variants when bound to HAp, the bound orientation in the N-terminus differed. Phosphorylation of the serine side chain at residue 16 decreased the average ssNMR measured distances from the backbone to the surface at G8, L15, and V19, and resulted in a more proximal segment from residues ~10–20 in the ssNMR-biased RosettaSurface.NMR low-energy ensemble. This result was expected due to the increased affinity of the additional charged residue for the charged HAp surface. Residues ~25–44 are predicted not to make high-affinity contacts with HAp (Figure 7C). The negatively charged segment from residues ~45–59 locate at the highly charged surface, regardless of the phosphorylation status of the distal S16 residue (Figures 7C and 7F). Despite the closer association with HAp upon binding, we observed 1–3 mg more protein binding in the non-phosphorylated protein. This result was unexpected since there is no corresponding structural change. Combined, this data suggests that phosphorylation at S16 is not required for binding, but does alter the binding interaction.
One confounding factor in ssNMR data acquisition in the phosphorylated variant is the uncertainty in signal origin (i.e., HAp phosphates and phosphoserine will produce similar dephasing if they are both close to the 13C being investigated). Indeed, RosettaSurface.NMR predictions suggest that L15-pS16 and pS16-V19 distances become shorter for the bound protein compared to the protein off the surface (Table 1), to a degree that they become similar to the experimentally measured distance. This is a measurement that cannot be made independently by experiment and this computational measurement allows reevaluation of the experimental data. Using the computationally derived intraprotein13C-31P distances results in longer backbone-HAp distances of 7.0 Å for L15-HAp and 7.5 Å for V19-HAp (Figure 5), in much better agreement with the computationally derived data. It is important to note that while these distances are longer then a single spin model suggests, they are still shorter than the non-phosphorylated variant, indicating an increased interaction when the phosphoserine is present.
While the orientation of the representative model was largely consistent with the experimental data, the ensemble distances were consistently longer, with 5 of the 10 distances being on average 2 Å longer than the experimentally determined values. It is difficult to determine exactly what the source of this difference might be. Differences in the surface could be one source of error. In the experimental studies, we cannot distinguish which face of HAp we are binding to, due to its small size, and even though we expect a preference for {100} and {010}, it is likely that we are observing the average of protein bound to all exposed faces. It is also possible that there is some dissolution of the surface in solution that could alter the surface morphology in an unpredictable way. Another possibility is that LRAP preferentially binds a step-edge, a surface that we did not evaluate computationally and one that would be difficult to validate experimentally. Importantly, the overall features of the protein-surface interaction have been captured with the model. Further refinements and benchmarking of this method will be aided by further experimental constraints.
Surface specificity
Amelogenins are thought to bind the {010} and {100} faces of mature HAp crystals.59 Our investigation did reveal some {010}-HAp crystal face specificity for both LRAP variants. However, the method we developed for probing specificity is novel, and it is unclear how significant our findings are. In a previous study, using ssNMR-biased RosettaSurface.NMR, we predicted that statherin could significantly discriminate some HAp surfaces. Furthermore, our method predicts that LRAP and statherin prefer different HAp surfaces.43 These predicted differences for statherin and LRAP indicate the algorithm has no intrinsic bias for a particular face.
One factor that might contribute to the increased specificity in the case of statherin is the existence of a single, well-defined binding interface. Statherin's binding domain consists of a single, stable helix. In the case of LRAP there are two predicted binding interfaces from residues ~10–20 and residues ~45–59. The predicted flexibility from residues ~30–45 and the lack of predicted global tertiary structure results in an LRAP fold where the relative rigid-body positions of the binding interfaces vary in the low-energy ensembles. Because the relative rigid-body positions of the binding interfaces are not fixed, LRAP can adopt multiple, energetically equivalent orientations on the HAp surfaces. Furthermore, our results predict that the C-terminal residues of LRAP bind differentially, in a conformation-dependent manner. This ability to adopt diverse conformations in the highly charged C-terminal binding domain could facilitate the binding of diverse surface geometries, resulting in decreased specificity and increased affinity.
Future direction
While this study represents the ambitious effort of determining the adsorbed-state structure in a relatively large protein, the full structure remains underdetermined. One way to increase structural resolution and certainty would be to identify residues that are distant in primary structure, but proximal in tertiary structure, if they exist. For instance, our structures show some hydrophobic packing in the segments intervening residues ~20–30 and residues ~30–48. Acquiring distance measurements in these segments would reveal the compactness of LRAP in the HAp-adsorbed state. Because the compactness of the molecule affects the relative orientation of the binding interfaces, long-range contacts may also help confirm or refute the existence of surface specificity. Additionally, a greater number of LRAP-HAp distance measurements might help address specificity.
In addition to a greater number of experimental measurements, development of RosettaSurface could contribute to further refinement for the HAp-adsorbed LRAP structure. Energy function calibration requires benchmarking on experimental data, which are scarce. One obvious approach would be to increase the constraint weight; however, it is unclear to what extent the constraint weight should dominate relative to other terms in the physical potential. Further algorithmic development coupled with an increased number of experimental measurements is expected to further increase the scope and utility of this novel and rigorous approach.
Conclusions
New ssNMR data reveal that the N-terminus of the amelogenin LRAP is largely extended on the surface of HAp. Computational data combining all of the N- and C-terminal experimental measurements to date reproduce the experimental data reasonably well, and suggest a very dynamic region of LRAP in the middle of the protein. Phosphorylation of the protein did not alter the conformation of LRAP on the surface, however, the interaction with HAp was modulated with the addition of the charged residue. The conformation was also not sensitive to any of the surfaces studied, but predictions for both variants showed some preference for the {010} face of HAp. Collectively, these observations are consistent with a protein that is extended and interacting closely with HAp to maximize surface coverage. The predicted preferential binding is consistent with blocking growth on the a and b faces, allowing maximal growth along the c-axis to result in elongated crystals.
Acknowledgements
This research was supported by NIH-NIDCR Grant DE-015347 (WJS) and NSF CAREER award CBET 0846324 (JJG). The research was performed at the Pacific Northwest National Laboratory (PNNL), a facility operated by Battelle for the U.S. Department of Energy, and at the W.R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by the U.S. DOE Biological and Environmental Research program.
References
- (1).Mann S. Biomineralization. Principles and Concepts in Bioinorganic Materials Chemistry. Oxford University Press; Oxford, England: 2001. [Google Scholar]
- (2).Dove PM, J. J. D. Y. S. Weiner. Biomineralization. Vol. 54. Mineralogical Society of America, Geochemical Society; Washington, DC: 2003. [Google Scholar]
- (3).Sundar VC, Yablon AD, Grazul JL, Ilan M, Aizenberg J. Nature. 2003;424:899–900. doi: 10.1038/424899a. [DOI] [PubMed] [Google Scholar]
- (4).Faivre D, Schüler D. Chem. Rev. 2008;108:4875–4898. doi: 10.1021/cr078258w. [DOI] [PubMed] [Google Scholar]
- (5).Ozawa H, Hoshi K, Amizuka N. Journal of Oral Biosciences. 2008;50:1–14. [Google Scholar]
- (6).Elhadj S, De Yoreo JJ, Hoyer JR, Dove PM. Proc. Natl. Acad. Sci. USA. 2006;103:19237–19242. doi: 10.1073/pnas.0605748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Boskey AL, Maresca M, Ullrich W, Doty SB, Butler WT, Prince CW. Bone and Mineral. 1993;22:147–159. doi: 10.1016/s0169-6009(08)80225-5. [DOI] [PubMed] [Google Scholar]
- (8).Shiraga H, Min W, VanDusen WJ, Clayman MD, Miner D, Terrell CH, Sherbotie JR, Foreman JW, Przysiecki C, Neilson EG. Proc. Natl. Acad. Sci. USA. 1992;89:426–430. doi: 10.1073/pnas.89.1.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Naka K, Chujo Y. Chem. Mater. 2001;13:3245–3259. [Google Scholar]
- (10).Sollner C, Burghammer M, Busch-Nentwich E, Berger J, Schwarz H, Riekel C, Nicolson T. Science. 2003;302:282–286. doi: 10.1126/science.1088443. [DOI] [PubMed] [Google Scholar]
- (11).Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. J. Biol. Chem. 1980;255:9760–9768. [PubMed] [Google Scholar]
- (12).Gibson CW, Yuan Z-A, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. J. Biol. Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- (13).Mitsiadis TA, Tucker AS, De Bari C, Cobourne MT, Rice DPC. Dev. Biol. 2008;320:39–48. doi: 10.1016/j.ydbio.2008.04.006. [DOI] [PubMed] [Google Scholar]
- (14).Hu JCC, Chun YHP, Al Hazzazzi T, Simmer JP. Cells Tissues Organs. 2007;186:78–85. doi: 10.1159/000102683. [DOI] [PubMed] [Google Scholar]
- (15).Moradian-Oldak J, Bouropoulos N, Wang L, Gharakhanian N. Matrix Biol. 2002;21:197–205. doi: 10.1016/s0945-053x(01)00190-1. [DOI] [PubMed] [Google Scholar]
- (16).Aoba T, Fukae M, Tanabe T, Shimizu M, Moreno E. Calcif. Tissue Int. 1987;41:281–289. doi: 10.1007/BF02555230. [DOI] [PubMed] [Google Scholar]
- (17).Tarasevich BJ, Howard CJ, Larson JL, Snead ML, Simmer JP, Paine M, Shaw WJ. J. Cryst. Growth. 2007;304:407–415. doi: 10.1016/j.jcrysgro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hunter GK. Curr. Opin. Solid State Mater. Sci. 1996;1:430–435. [Google Scholar]
- (19).Gray JJ. Curr. Opin. Struct. Biol. 2004;14:110–115. doi: 10.1016/j.sbi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- (20).Shaw WJ, Ferris K, Tarasevich B, Larson JL. Biophys. J. 2008;94:3247–3257. doi: 10.1529/biophysj.107.119636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Shaw WJ, Campbell AA, Paine ML, Snead ML. J. Biol. Chem. 2004;279:40263–40266. doi: 10.1074/jbc.C400322200. [DOI] [PubMed] [Google Scholar]
- (22).Stayton PS, Drobny GP, Shaw WJ, Long JR, Gilbert M. Crit. Rev. Oral Biol. Medicine. 2003;14:370–376. doi: 10.1177/154411130301400507. [DOI] [PubMed] [Google Scholar]
- (23).Shaw WJ, Ferris K. J. Phys. Chem. B. 2008;112:16975–16981. doi: 10.1021/jp808012g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gibson C, Golub E, Herold R, Risser M, Ding W, Shimokawa H, Young M, Termine J, Rosenbloom J. Biochemistry. 1991;30:1075–1079. doi: 10.1021/bi00218a028. [DOI] [PubMed] [Google Scholar]
- (25).Long JR, Shaw WJ, Stayton PS, Drobny GP. Biochemistry. 2001;40:15451–15455. doi: 10.1021/bi010864c. [DOI] [PubMed] [Google Scholar]
- (26).Gibson JM, Popham JM, Raghunathan V, Stayton PS, Drobny GP. J. Am. Chem. Soc. 2006;128:5364–5370. doi: 10.1021/ja056731m. [DOI] [PubMed] [Google Scholar]
- (27).Gibson JM, Raghunathan V, Popham JM, Stayton PS, Drobny GP. J. Am. Chem. Soc. 2005;127:9350–9351. doi: 10.1021/ja050910m. [DOI] [PubMed] [Google Scholar]
- (28).Shaw WJ, Long JR, Dindot JL, Campbell AA, Stayton PS, Drobny GP. J. Am. Chem. Soc. 2000;122:1709–1716. [Google Scholar]
- (29).Goobes G, Goobes R, Schueler-Furman O, Baker D, Stayton PS, Drobny GP. Proc. Natl. Acad. Sci. USA. 2006;103:16083–16088. doi: 10.1073/pnas.0607193103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ndao M, Ash JT, Breen NF, Goobes G, Stayton PS, Drobny GP. Langmuir. 2009;25:12136–12143. doi: 10.1021/la901647n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ndao M, Ash JT, Stayton PS, Drobny GP. Surf. Sci. 2010;604:L39–L42. doi: 10.1016/j.susc.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Cavalli A, Salvatella X, Dobson CM, Vendruscolo M. Proc. Natl. Acad. Sci. USA. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gong H, Shen Y, Rose GD. Protein Sci. 2007;16:1515–1521. doi: 10.1110/ps.072988407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Shen Y, et al. Proc. Natl. Acad. Sci. USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bowers PM, Strauss CEM, Baker D. J. Biomol. NMR. 2000;18:311–318. doi: 10.1023/a:1026744431105. [DOI] [PubMed] [Google Scholar]
- (36).Meiler J, Baker D. Proc. Natl. Acad. Sci. USA. 2003;100:15404–15409. doi: 10.1073/pnas.2434121100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Rohl CA, Baker D. J. Am. Chem. Soc. 2002;124:2723–2729. doi: 10.1021/ja016880e. [DOI] [PubMed] [Google Scholar]
- (38).Makrodimitris K, Masica DL, Kim ET, Gray JJ. J. Am. Chem. Soc. 2007;129:13713–13722. doi: 10.1021/ja074602v. [DOI] [PubMed] [Google Scholar]
- (39).Masica DL, Gray JJ. Biophys. J. 2009;96:3082–3091. doi: 10.1016/j.bpj.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Masica DL, Schrier SB, Specht EA, Gray JJ. J. Am. Chem. Soc. 2010;132:12252–12262. doi: 10.1021/ja1001086. [DOI] [PubMed] [Google Scholar]
- (41).Addison WN, Masica DL, Gray JJ, McKee MD. J. Bone Miner. Res. 2010;25:695–705. doi: 10.1359/jbmr.090832. [DOI] [PubMed] [Google Scholar]
- (42).Chien Y-C, Masica DL, Gray JJ, Nguyen S, Vali H, McKee MD. J. Biol. Chem. 2009;284:23491–23501. doi: 10.1074/jbc.M109.021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Masica DL, Ash JT, Ndao M, Drobny GP, Gray JJ. Structure. 2010;18:1678–1687. doi: 10.1016/j.str.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Carpino LA, Han GY. J. Org. Chem. 1972;37:3404–3409. [Google Scholar]
- (45).Meienhofer J, Waki M, Heimre EP, Lambros TJ, Makofske RC, Chang C-D. Int. J. Pept. Protein Res. 1979;13:35–42. [PubMed] [Google Scholar]
- (46).Ebrahimpour A, Johnsson M, Nancollas CFRGH. J. Colloid Interface Sci. 1993;159:158–163. [Google Scholar]
- (47).Bielecki A, Burum DP. Journal of Magnetic Resonance A. 1995;116:215–220. [Google Scholar]
- (48).Bennett A, Rienstra C, Auger M, Lakshmi KV, Griffen RG. Journal of Chemical Physics. 1995;103:6951–6958. [Google Scholar]
- (49).DeLano W. The PyMOL molecular graphics system. 2002 [Google Scholar]
- (50).Palmer D. CrystalMaker: A computer program for plotting crystal structures. 2001;5:3. [Google Scholar]
- (51).Astala R, Stott MJ. Physical Review B. 2008;78:075427. [Google Scholar]
- (52).Matsushima N, Izumi Y, Aoba T. J. Biochem. 1998;123:150–156. doi: 10.1093/oxfordjournals.jbchem.a021902. [DOI] [PubMed] [Google Scholar]
- (53).Renugopalakrishnan V, Strawich ES, Horowitz PM, Glimcher MJ. Biochemistry. 1986;25:4879–4887. doi: 10.1021/bi00365a023. [DOI] [PubMed] [Google Scholar]
- (54).Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan Y, Moradian-Oldak J, Evans JS. Biochemistry. 2009;48:2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Buchko GW, Tarasevich BJ, Roberts J, Snead ML, Shaw WJ. Biochim. Biophys. Acta: Proteins Proteomics. 2010;1804:1768–1774. doi: 10.1016/j.bbapap.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Buchko G, Bekhazi J, Cort J, Valentine N, Snead M, Shaw W. Biomolecular NMR Assignments. 2008;2:89–91. doi: 10.1007/s12104-008-9092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Piela L, Némethy G, Scheraga HA. Biopolymers. 1987;26:1587–1600. doi: 10.1002/bip.360260910. [DOI] [PubMed] [Google Scholar]
- (58).Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- (59).Wallwork ML, Kirkham J, Zhang J, Smith DA, Brookes SJ, Shore RC, Wood SR, Ryu O, Robinson C. Langmuir. 2001;17:2508–2513. [Google Scholar]