Abstract
To address the synthesis of increasingly structurally diverse small-molecule drugs, methods for the generation of efficient and selective biological catalysts are becoming increasingly important. ‘Directed evolution’ is an umbrella term referring to a variety of methods for improving or altering the function of enzymes using a nature-inspired twofold strategy of mutagenesis followed by selection. This article provides an objective assessment of the effectiveness of directed evolution campaigns in generating enzymes with improved catalytic parameters for new substrates from the last decade, excluding studies that aimed to select for only improved physical properties and those that lack kinetic characterization. An analysis of the trends of methodologies and their success rates from 81 qualifying examples in the literature reveals the average fold improvement for kcat (or Vmax), Km and kcat/Km to be 366-, 12- and 2548-fold, respectively, whereas the median fold improvements are 5.4, 3 and 15.6. Further analysis by enzyme class, library-generation methodology and screening methodology explores relationships between successful campaigns and the methodologies employed.
In the ongoing race to identify new therapeutics, there has been an increase in recent years in the both the complexity of clinically targeted systems and the regulatory hurdles entailed in the development of lead compounds. Developing small molecules for complex targets that are often components of larger highly interconnected signaling pathways increasingly requires the synthesis of bioeffectors (i.e., biologically active small molecules, peptides and antibodies) that selectively act on the target, without interacting with unwanted off-target sites in related or unrelated families. This necessity often demands a correspondingly high degree of chemical complexity, including multiple functional groups and, increasingly, defined stereochemistry. Once molecules have been demonstrated to traverse the selectivity hurdle, they are required to pass through numerous drug-development barriers in pharmacology, toxicology and ultimately human efficacy. To fulfill requirements in these categories, additional chemical modifications are normally needed, often increasing the structural complexity of drug candidates further. As the complexity of lead compounds in clinical trials has increased, the costs related to their chemical synthesis have increased as well. This trend has lead to the genesis of a new term in drug development that was recently highlighted in a New York Times article reporting on trends in anticancer drug development: “Some experts now talk about ‘financial toxicity’ as a side effect of cancer drug treatment, along with nausea and hair loss [1].”
While the greatest manufacturing costs are likely incurred by biologics, there is an ongoing need for the development of catalysts to address the synthesis of these small-molecule drugs [2]. For example, in the case of the class of drugs known as nucleoside analogs, up to 99% of the final drug cost can be attributed to manufacturing [3]. In addition to a need for synthetic catalysts with greater efficiency and selectivity, biological catalysts are of increasing importance. Some beneficial properties of enzymes include significant rate enhancements, a high degree of inherent stereoselectivity, the ability to function with or without activity in organic solvents and, potentially, a lower environmental impact.
Engineering enzymes to catalyze new biosynthetic reactions
Enzymes have indeed found significant applications in synthetic processes but the full realization of their potential has been limited because they are often unstable, rarely accommodate alternate substrates, function under a limited range of reaction conditions and frequently require expensive cofactors. The drawbacks an enzyme exhibits are determined by the architecture and dynamics of the active site and their encompassing protein scaffold. To address these issues, structure-based redesign strategies are often undertaken that endeavor to identify mutations, usually based on analysis of enzyme active sites, to improve binding and catalysis for a desired substrate or reaction context. Many successes have been reported, but it is likely that for every success there are at least as many unreported failures. This is perhaps unsurprising, since there are inherent limits to the mutability of active sites for very divergent substrates and because enzyme-engineering hypotheses are often based on structural studies that are incomplete, both in terms of knowledge of substrate, transition state, product-binding modes and in understanding the dynamics of active sites and their ancillary-protein scaffolding.
Directed evolution methodologies have been developed, in part, to bypass this knowledge gap [4,5]. Inspired by natural selection, these methods endeavor to identify productive protein-sequence substitutions and their useful combinations by generating libraries of enzyme variants. For instance, random mutagenesis of an encoding gene and then selecting for improved characteristics via desired function- or property-screening strategies. In principle, by emulating the search algorithm employed by nature (mutation followed by selection), directed evolution is capable of identifying solutions to problems in the generation of new biocatalysts from progenitor enzymes lacking complete characterization, without knowledge-based biases. In practice, however, structural and biochemical data are often used to guide directed evolution experimental designs for greater success.
For over two decades, directed evolution methods have been applied throughout numerous studies, for the generation of optimized biocatalysts, in some cases with impressive results. Rate enhancements in excess of five orders of magnitude have been reported [6], in addition to the generation of biocatalysts for previously unknown biochemical reactions. While these successes have been reviewed in other recent publications [2,5,7,8], we will highlight a few here to illustrate the power and scope of these methods.
P450 monooxygenases have been developed by Arnold and co-workers for the biochemical fermentation of alcohols from straight chain alkanes. Using a medium-chain fatty acid oxidase as a progenitor enzyme, directed evolution methodologies introduced catalytic competence for the oxidation of successively shorter and less oxidized alkyl chain precursors, first octane [9] and later to propane [10]. Step-wise improvements via mutagenesis throughout the individual domains of the monooxygenase variants ultimately resulted in an efficient P450, propane monooxygenase [11]. Furthermore, one of the variants resulting from these campaigns was evolved to convert ethane to ethanol, currently an important biofuel for use in automobiles [12]. Zhao and co-workers have demonstrated how the cofactor-regeneration problem may be addressed using directed evolution of phosphite dehydrogenase. Through a combination of multiple-mutagenesis methods over several generations, the t1/2 of phosphite dehydrogenase at 45°C was improved greater than 23,000-fold from the parent enzyme without sacrificing catalytic efficiency, providing a useful biochemical method for NADH cofactor regeneration [13,14]. Impressive results have also been reported in the improvement of enzyme enantioselectivity for new substrates through the process of iterative saturation mutagenesis and Combined Active site Saturation Testing (CASTing) [15]. To demonstrate the utility of these methods, Reetz applied iterative saturation mutagenesis to Pseudomonas aeuriginosa lipase, a well studied enzyme, to increase the enantioselectivity for a selected chiral ester, succeeding in generating a mutant with an enantiomeric ratio (E-value) 594-fold improved over the original enzyme. In comparison, previous efforts using conventional mutagenesis protocols (error prone polymerase chain reaction [epPCR], saturation mutagenesis of hot spots, gene shuffling and recombination) identified a lipase possessing a comparatively lower E-value of 51 [16].
While there are several similar impressive success stories, a cursory survey of the literature also reveals a substantial number of studies in which the enhancements are more modest. Further complicating the task of quantifying the successes of many studies is that many articles report improvement in properties that do not refer directly to kinetic parameters (e.g., total yield of reaction). Presumably, this is not an intentional obfuscation. Rather, many directed evolution studies are simply goal oriented and the goal is a desired phenotype. Desired phenotypes may be ‘an organism survives’, ‘a colony changes color’ or that ‘a percent conversion is achieved’. Indeed, almost all reported studies to date appear to succeed in generating their desired phenotype or chemophenotype and by these criteria, can be viewed as bona fide successes in directed evolution.
In the context of combating the problem of ‘financial toxicity’, two recent studies demonstrate the potential of directed evolution methods for reducing costs. A synthetic-biology campaign for the economical production of artemisinin has used directed evolution methodology as a tool in optimizing production in heterologous expression of this antimalarial metabolite [17–19]. In a more recent example, the Sitagliptin synthetic pathway was streamlined via the directed evolution of an enantioselective transaminase, which obviates the need for a costly resolution step in the synthesis of this clinically prescribed antihyperglycemic compound. Given the annual market for sitagliptin (tradename Januvia/Janumet) of US$2.8 billion, these improvements have a large potential economic and health impact [20].
However, given the substantial body of literature describing successes in directed evolution, the diversity of the methods employed and the variety of enzyme classes improved, we became interested in performing an objective assessment of success of the methods in directed evolution for the creation or optimization of discreet biosynthetic enzymes. Correspondingly, for this study, we selected articles from the last ten years using objective selection criteria. Using the GoPubMed.com semantic server [21], we identified the top 20 research journals publishing articles with the search phrases ‘directed evolution’ or ‘directed molecular evolution’. Additionally, all citations comprised studies that reported improvements in a small molecule biotransformation enzyme, reported kinetic parameters, including kcat/Km and/or kcat (or Vmax) and/or Km, for both the progenitor enzyme and the evolved enzyme, are target-based studies; in other words, the reported improved turnover was for the substrate used in the screen and more than one site was mutated, reflecting a departure from single site saturation mutagenesis.
Before presenting the results of our literature analysis we review the most common methods used in typical directed evolution campaigns, to investigate the current state of the art and to define the classifications used in our subsequent analysis.
Common methods for the generation of diverse gene libraries
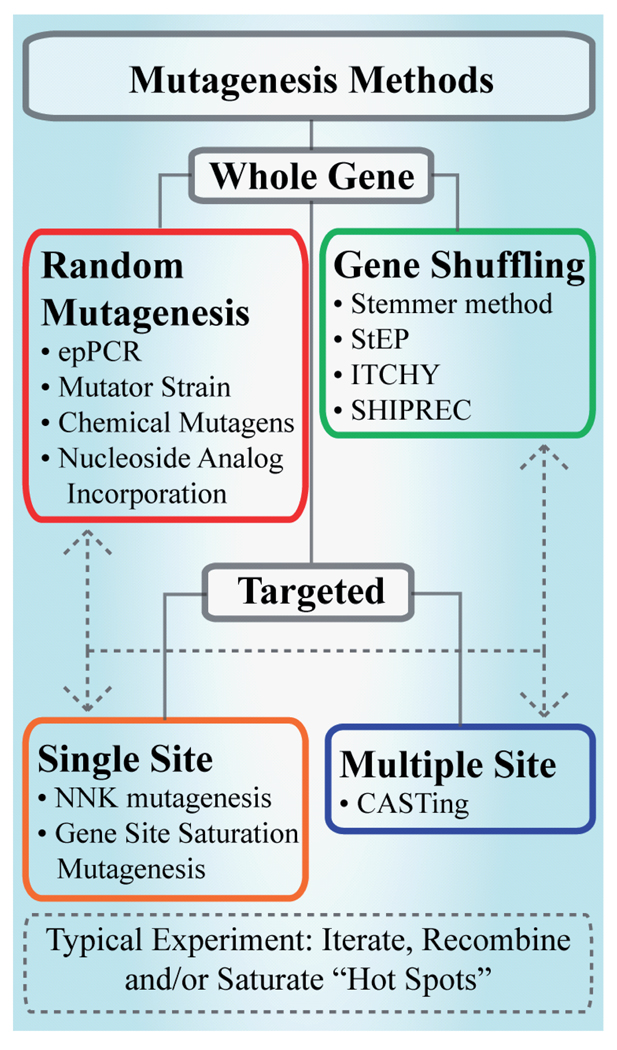
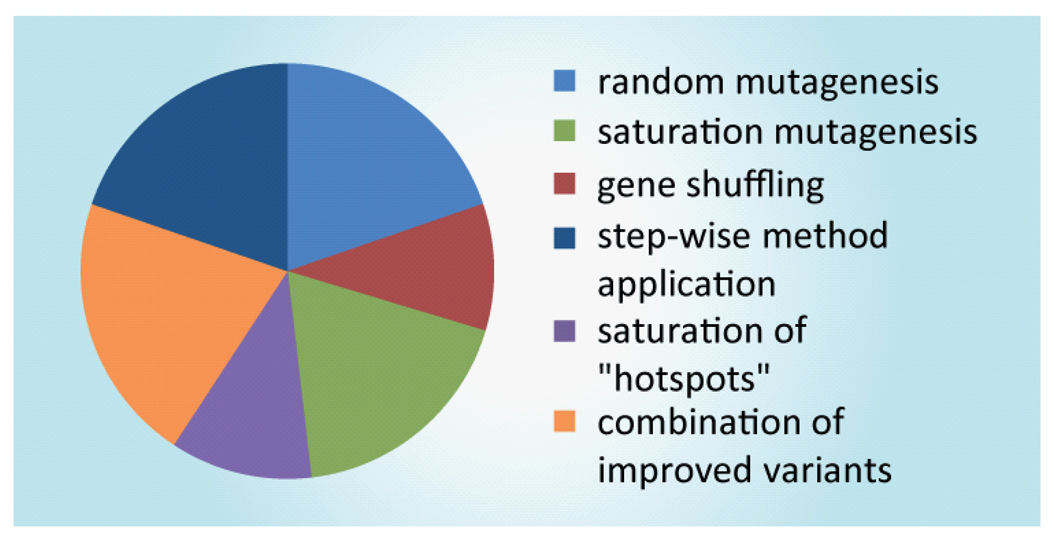
A common element shared by directed evolution experiments, as we have defined them here, is the generation of a diverse sequence library. In general, the three primary strategies for library generation are random mutagenesis, site-directed mutagenesis and genetic recombination (Figure 1).
Figure 1. Methods for introducing mutations into progenitor genes target the whole gene or particular sites are targeted rationally through structural or other functional data.
CASTing: Combinatorial active-site saturation testing; epPCR: Error prone polymerase chain reaction; ITCHY: incremental truncation of hybrid enzymes; SHIPREC: Sequence homology-independent recombination; StEP: Staggered extension process.
Random mutagenesis
Random mutagenesis methods include epPCR, chemical mutagenesis, UV irradiation and mutating bacterial strains. The most common is epPCR, (see [22]) of which there are several variations. Typical early experiments used an error prone, thermostable polymerase lacking proofreading ability (Taq) to amplify a gene of interest, incorporating random mutations along the entire length of the gene [23]. The error rate of the polymerase alone was titrated through addition of Mn2+ and by manipulation of the number of thermocycles and template and dNTP concentrations. More recently, polymerases have been engineered to possess a higher inherent mutation rate, with less bias (e.g., balancing transitions vs transversions) than the typical Taq polymerase; Mutazyme (Stratagene Inc.) is one such commercially available engineered error-prone polymerase. Alternatively, chemical mutagens may be employed to modify template DNA and foster mutation. Reactions of template DNA with nitrous acid, formic acid, hydrazine or ethyl methane sulfonate can result in chemical changes in nucleotide bases, altering their hydrogen-bonding properties and increasing the propensity for the formation of noncanonical pairings. [24,25] Another method with a similar end result involves adding unnatural nucleotide analogs capable of pairing with multiple canonical nucleotides in the PCR reaction [26]. Mutator strains and whole-cell mutation by UV irradiation have been used to generate mutations in vivo [27]. This provides the advantage of avoiding low-yield ligation steps, but this is at the expense of possibly deleteriously mutating the host genome, as well as resistance genes, copy number control sequences or promoter regions of the heterologous plamids. Some researchers have used multiple epPCR methods, balancing the bias of transitions (A→G, T→C) and transversions (A/G↔C/T) and thereby increasing the mutational diversity in the library [28–30]. Wong and co-workers have performed a statistical analysis of the mutational spectrum achieved through the application of 19 random mutagenesis experiments to three genes and developed a web application (MAP analysis) to aid researchers in choosing an optimal method for a given target gene [23].
Site-directed mutagenesis
The preceding random mutagenesis methods can facilitate the identification of advantageous mutations or tunable sites located throughout a protein sequence and can be applied fruitfully in the absence of structural data. Alternatively, site-directed mutagenesis can be used when structural data are readily available. Saturation mutagenesis protocols test all 20 amino acids at a targeted site or sites and enable a controlled and comprehensive search of mutational sequence space via degenerate oligonucleotide primers. Although saturation mutagenesis can be applied to every position in a gene, as in gene site saturation mutagenesis [31], the method is most commonly used as a targeted approach by identifying specific potentially beneficial mutation sites using structural data or through homology modeling. Saturation mutagenesis can be applied to a single site, to multiple sites in an iterative fashion or to multiple sites simultaneously. Cassette mutagenesis involves simultaneous saturation mutagenesis of multiple, proximal residues [32]. In this category, Reetz and co-workers introduced a method dubbed the combinatorial active-site saturation test or CASTing, in 2005 [15]. Most often, CASTing has been applied to alter the shape of binding sites in order to change the enantioselectivity of an enzyme [33]. Saturation mutagenesis methods have the advantage of reducing library-size requirements substantially by focusing on a subset of ‘hot spots’ either within the active site or as identified by broader sequence scans.
Gene shuffling
The term ‘gene shuffling’ refers to the in vitro recombination of homologous progenitor genes and ostensibly allows for the generation of chimeric sequences more diverse than standard epPCR and without resorting to high polymerase-dependent mutation rates [34]. The progenitor genes may comprise either naturally occurring homologs of a gene family or selected mutants of a single gene generated by other mutagenesis methods. The archetypal DNA-shuffling method introduced by Stemmer and co-workers shuffles homologous DNA sequences by generating oligonucleotide fragments from selected progenitor genes by treatment with DNAse, followed by reassembly of the mixed oligonucleotides by PCR [35]. There now exists an array of new protocols for gene recombination, many of which appear to be inspired by Stemmer’s seminal process. In staggered extension process (StEP), homologous DNA templates are mixed together with one or more primers and subjected to very short annealing and extension steps [36]. Methods for recombination of nonhomologous sequences include incremental truncation of hybrid enzymes (ITCHY), a combination of ITCHY and DNA shuffling (SCRATCHY) and sequence-homolog y independent recombination (SHIPREC) [37–39].
Frequently, beneficial mutations generated by one of the aforementioned methods are subsequently combined into a single construct to further optimize enzyme function. By generating these combinations, unproductive and/or neutral mutations may also be identified and removed. In some cases, this is accomplished by gene shuffling, which has long been used for this purpose [35], but stepwise site-directed mutagenesis is also typically employed.
In our review of the last decade, it became apparent that the most successful directed evolution campaigns utilize multiple forms of mutagenesis, both iteratively and sometimes concurrently in a given study, with some exceptional studies performing in excess of 20 rounds using a broad array of methods [40].
Typical screening & selection paradigms
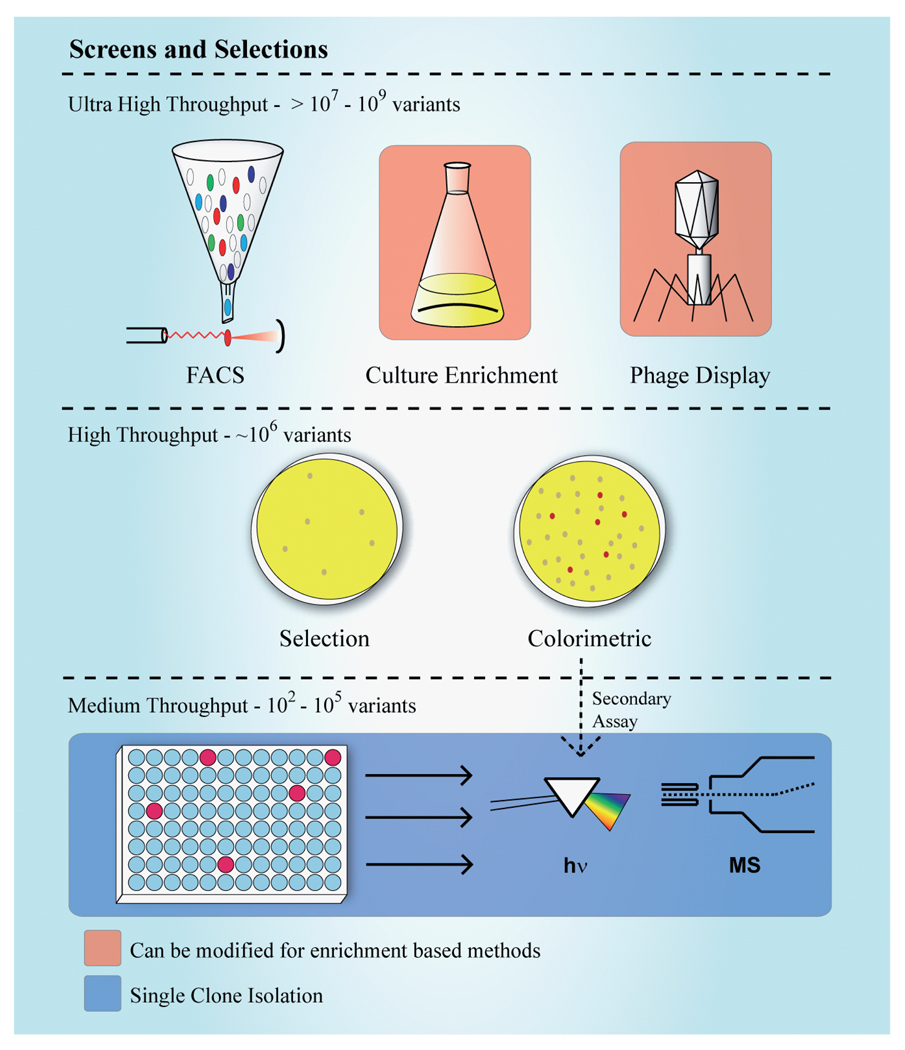
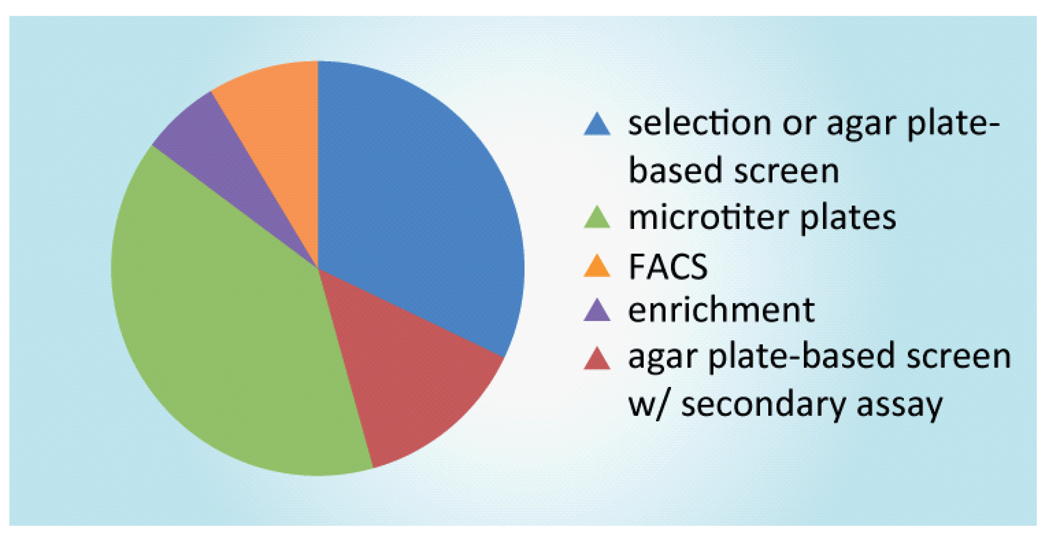
The second shared element of all directed evolution studies is the assessment paradigm. Ultimately, the size of the mutant library to be assayed is determined by the method utilized to indentify improved mutants (Figure 2). Screening and selection methods in directed evolution have been reviewed previously [41–46]. For our analysis we classify these methods into two categories: selection-based methods and biochemical screening-based approaches.
Figure 2. Representative methods for identification of improved enzyme variants are classified by throughput capacity.
Examples of methodologies capable of selecting or screening ultra-large libraries include fluorescence-activated cell sorting, enrichment and phage display. High-throughput methods include selection or screening on agar plates. Many biochemical-assay methods can be modified to 96-well format, but require colonies to be picked into individual wells prior to screening. Some plate-based colorimetric screens require secondary analysis in this format.
Selection-based methods
In selection-based methods, all mutants generated are assayed en masse for a desired biochemical activity. In vivo selections link a desired activity (e.g., β-lactamase) to host cell growth (e.g., growth in a β-lactam-containing medium). Other typical examples include the synthesis of an essential amino acid [47–49], nucleotide [38,39] or degradation of an environmental toxin [35,50–56]. Positive-selection methods can be performed on agar plates or rely on enrichment of cells harboring enzymes of the desired activity in a liquid culture [57]. Negative-selection methods, in which the desired activity results in death, often entail that cells must be sorted and replicated into both reference library and assay microtiter plates to facilitate backtracking and resurrection of the desired library cultures, thereby making them lower throughput [58,59]. A notable exception is the evolution of tRNA synthetases activity for the incorporation of unnatural amino acids, in which negative selection was performed to dial out undesired activities [60,61].
Clearly an ideal selection scheme involves engineering an organism to be dependent on reproduction via the presence of an endogenously biosynthesized small molecule in a concentration-dependent manner. A potential solution to this challenging problem was described by Schwimmer and co-workers using a yeast two-hybrid system to evolve a receptor, capable of transcriptional activation of an essential gene, to bind a new small-molecule ligand [62]. Specifically, a nuclear receptor retinoid X receptor that naturally binds 9-cis-retinoic acid was engineered to bind a synthetic retinoid-like compound, LG335, with an improvement in the EC50 from 10 µM to 40 nM. In an alternative approach, ligand-binding protein switches have been generated by engineering protein hybrids with resistance genes [63].
In addition to the molecular recognition engineering problem, challenges to engineered positive-selection schemes are numerous. The nature of the engineered response to endogenous ligand concentration is critical. For instance, it is less useful if an engineered receptor responds to the molecule as a switch and more useful that it respond as a rheostat. Once a ligand reaches a concentration sufficient for organism survival, selective pressure will diminish. Moreover, ligands need to be constrained to the intracellular environment to be of maximal utility and must not be toxic to the host organism.
In phage-display selection, an in vitro-selection methodology, the DNA of a gene library is fused to a viral coat protein gene and combined in a phagemid (a vector containing both plasmid and phage origins of replication). The phagemid DNA then becomes packaged in the phage capsid that displays the protein. The phage are subjected to chromatographic separation (affinity binding) to select for desired activity [64]. Very rarely is phage display connected directly to enzyme activity, although an increase in a biosynthetic enzyme activity can be identified through tighter binding to a tethered transition state analog [65]. Other display methods, such as cell-surface display, provide the advantage of a large library size in addition to facile subsequent screening capability [66].
Screening-based methods
In screening-based methods, the rate of substrate turnover is determined via biochemical assay, often via colorimetric or fluorometric measurements. In vitro (cell-free)-activity assays generally require the isolation of individual colonies into 96- or 384-well plates, followed by lysis and multiple reagent transfers. Correspondingly they are limited by cost and time to less than 106 mutants (103–104 is typical). In vivo screening-based methodologies can be applied to larger mutant libraries and it is the linking of the desired biochemical activity to a phenotype that limits the realistic library size. Examples of in vivo-activity assay techniques include agar plate colony screening [30,67–76], flow cytometric cell sorting [77–82] and phage display [83,84]. They result in larger libraries on the order of 107–1010 mutant constructs. The throughput of these methods is often limited by ligation and transformation efficiency.
In some cases, colorimetric screens can be modified and applied to agar plate colony screening. In these methods, colonies displaying a desired color within a certain time window are selected as active. It is common practice that the colorimetric agar plate assay is used as a primary screen to identify enzyme variants with functional turnover. Subsequently, a secondary- and sometimes tertiary-activity assay is generally performed in a 96-well microtiter plate biochemical assay to confirm and identify the most active clones [85–87]. These cases benefit from the larger library sizes allowable via the initial agar plate screen before encountering the time and cost limitations of single colony-isolation techniques.
Flow cytometry and phage-display methods permit the assay of larger mutant libraries. Fluorescence-activated cell sorting (FACS) is a type of flow cytometry in which a mixture of cells is sorted, one cell at a time, based upon the specific light scattering and fluorescent characteristics of each cell. Systems amenable to FACS must operate intracellularly, which often requires substrates of the screen to be cell permeable and the products impermeable. To address this requirement, FACS methods utilizing in vitro compartmentalization allow the compartmentalization of DNA, transcriptase, ribosomes and fluorescent substrates/products in artificial water–oil emulsions. This, in essence, enables the direct linking of product formation, enzyme and the enzyme-encoding gene and allows for qualitative sorting using a fluorescent marker [82].
As it is challenging to link enzymatic reactions to cell viability it is a common practice to contrive reporter systems that induce a change in color or fluorescence in vitro or in compartments. Such systems often require a creative enzymatic assay for detection of activity in cell lysates from colonies isolated in microtiter plates. Although these assays require the growth of individual cultures for assay, there is no limit to the varieties of instrumental analysis that can be performed and most assays can be implemented in a 96-well format. Examples of these methods include analytical HPLC [88], MS [50], TLC [89,90], radioactivity [91] and cofactor or substrate consumption (e.g., NADH) [92–94].
Meta-analysis of directed evolution studies for biosynthetic enzymes
Selection criteria for qualifying directed evolution studies
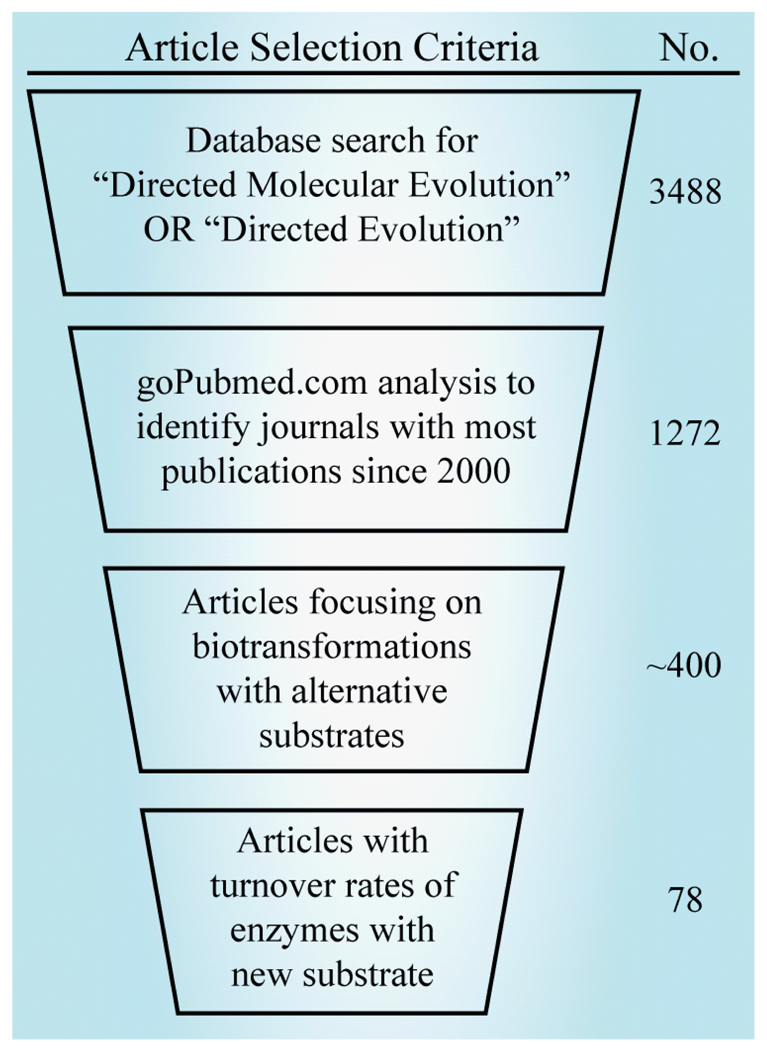
Database searches for the terms ‘directed evolution’ and ‘directed molecular evolution’ were performed in the PubMed and ISI Web of Knowledge databases resulting in the accumulation of approximately 3500 unique articles. Patents, conference proceedings and non-English language listings were removed. To focus our results, a search was performed with the GoPubMed.com semantic analysis web-server [21] to identify the top 20 journals publishing articles on ‘directed evolution’ or ‘directed molecular evolution’ during the last 10 years (2000–present). Journals that primarily published review articles (Current Opinion in Biotechnology, Current Opinion in Chemical Biology and Trends in Biotechnology) or methods (Methods in Molecular Biology) were excluded from the top 20 list. These 20 research journals have published approximately 1275 articles (including commentaries and review articles), since the year 2000. The journals included in our study were: Protein Engineering, Design and Selection, Journal of Molecular Biology, Proceedings of the National Academy of the Science, ChembioChem, Nucleic Acids Research, Angewandte Chemie International Edition in English, Applied Environmental Microbiology, Applied Microbiolog y and Biotechnology, Biochemistry, Journal of Biotechnology, Nature Biotechnology, Journal of Biological Chemistry, Journal of the American Chemical Society, Biotechnology and Bioengineering, Chemistry & Biology, Protein Science, FEBS Journal, Nature, Science, and Biochemical and Biophysical Research Communications.
In this article, we have collected examples of improvements in biocatalytic enzyme activity for non-native substrates. Correspondingly, we required that improved turnovers were not due to protein stabilization or improved production levels. Only articles reporting velocities normalized to enzyme concentration were included in our analysis; normalization to total protein content of cell-free extract was also accepted. Furthermore, we only collected articles reporting improvements in biotransformation reactions. Articles with the goal of improving protein stabilization under non-native reaction conditions (e.g., thermostablization) were not included. Studies reporting on the optimization of restriction enzymes, DNA/RNAzymes, catalytic antibodies or protein–protein interactions were excluded as they did not generally report on the improvement of a small-molecule biosynthetic enzyme. Reported improvements in activity for the natural substrate (or other common substrate, as is the case for guaiacol and cytochrome c peroxidases) were considered improvements in native reaction-condition parameters and therefore were not included. For this study, the directed evolution process was defined as the randomized mutation of a gene and screening for a desired new activity. As such, we required more than one site to be mutated, whether it be simultaneously (e.g., epPCR or CASTing) or through an iterative process (iterative saturation mutagenesis). We also required a selection scheme or activity screen to be employed as the primary method to identify improved mutants and that the compound used during screening was also the compound used for catalytic characterization (determination of Vmax, kcat, kcat/Km); however, substrate derivatives, which enabled high-throughput screening and/or selection (i.e., biotinylation [84] or fluorescent tagging [77]) were included.
Studies reportedly aimed at broadening substrate specificity were excluded by the requirement that the substrate in the employed selection is the most faithful reporter of improvements in activity. Many studies report mutants functional on multiple substrates and it was often unclear how a given mutant was identified through screening with a particular substrate. In addition, studies that reported on the generation of enantioselective enzymes were typically not included on the grounds that the desired function was not strictly increased turnover of a desired substrate but a ratio of turnovers for two enantiomers. Studies that reported an improvement based on screening for improved activity for a single enantiomer (as opposed to a ratio improvement) and also reported kinetic parameters for comparison of progenitor enzymes with evolved enzymes were included [77,83].
Using these criteria we scanned the approximately 1275 articles in the GoPubMed database, first by title, then abstract and finally the full text and extracted information on enzyme class, screening method, mutagenesis protocol and enzyme-turnover parameters (Figure 3). Initially, we limited our database to articles reporting kcat and Km; but since only 32 articles were found, we expanded our criteria to articles reporting Vmax, kcat, Km or kcat/Km. The final database contains 81 improved enzymes from 77 papers published in 18 journals and is listed in Supplementary Table 1, with references contained therein.
Figure 3. An objective set of criteria was applied for collection and selection of directed evolution studies from the last decade that report turnover rates for biocatalytic enzymes utilizing alternative substrates.
Our initial goal was to identify articles that reported a fold improvement in kinetic parameter for wild-type enzyme and evolved enzyme. As the preceding discussion demonstrates this ultimately was not always a clear-cut process. Thus an important caveat to be mentioned here is that while we made every effort to identify articles within the 1275 pool that met our stringent selection criteria, it is doubtless the case that some qualifying articles were overlooked.
Analysis by enzyme class
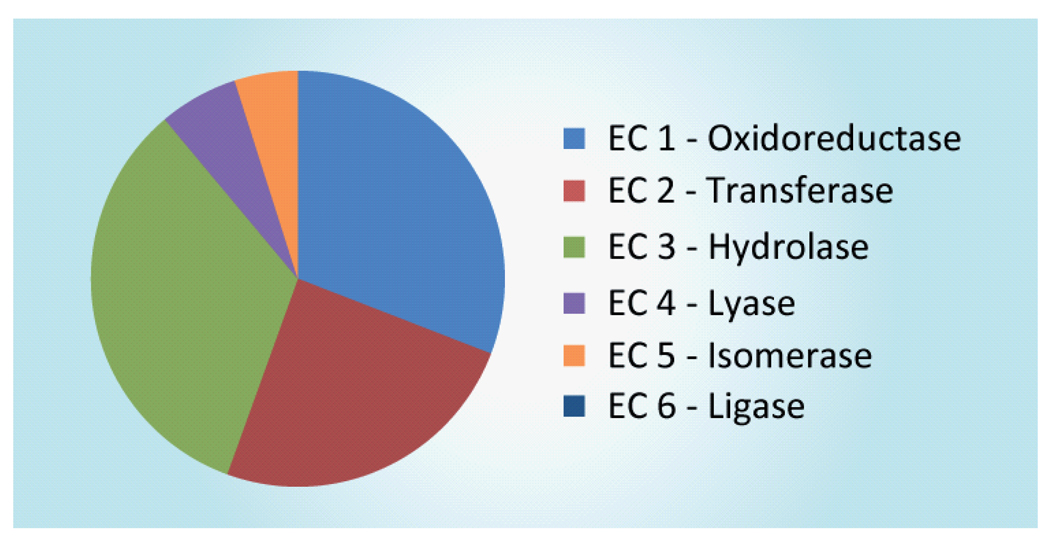
While all enzyme classes are reported to have been optimized by directed evolution methods, the majority of the 77 articles meeting our selection criteria fell into three enzyme classification (EC) classes: 23 oxidoreductases, 22 transferases and 28 hydrolases (Figure 4). Within the oxidoreductase category six enzymes were P450 or other heme-based oxidases and only one was a cofactor-dependent reductase. The prevalence of hydroxylation enzymes in recent directed evolution campaigns (seven cases) may be reflective of the importance of hydroxylation in the alternative energy industry and the difficult nature of C-H bond-activation reactions in the context of preparative organic synthesis. In addition, formal oxidation of an amino or hydroxyl group to a keto group was well represented as 6 instances of class 1. The transferase category is the most functionally diverse and included members with such assorted activities as aminotransferase, kinase and glycosyltransferase. Within the dominant class, the hydrolase category, the leading representatives were lipases, glycosyl hydrolases and esterases representing three, six and nine experiments, respectively. Ligase enzymes (e.g., synthetases) were the subject of directed evolution campaigns less frequently. The low population of this class may be a result of the inherent difficulty in evolving a multiple substrate enzymes, the lack of identification of interesting targets for evolution within this enzyme class or absence of reported kinetic parameters meeting the criteria of our analysis.
Figure 4. Distribution of experiments by enzyme class.
EC: Enzyme classification.
Analysis by library-generation protocol
As previously discussed, there is a great variety of methods for introducing mutations into progenitor genes. In practice, most studies employed a combination of methodologies over several generations to achieve an optimized enzyme (Figure 5). The workhorse of directed evolution appears to be error-prone PCR (epPCR) and it was this methodology that was responsible for the origination of many mutations and/or the determination of hot spots within a progenitor gene. Of the cases in this study, 51 used epPCR at some point in the evolutionary process. Saturation mutagenesis comprised the second largest category, being used 36 times, revealing the importance of using structural and/or experimental prescience of active site residues to guide the directed evolution experiment. Gene shuffling was the least employed as a stand-alone method for introducing diversity. However, it was most frequently applied as the last step to combine beneficial mutations and to remove deleterious or neutral changes.
Figure 5. Distribution of selected directed evolution studies by mutagenesis method.
Random mutagenesis: experiments that mutate the whole gene in the absence of guiding criteria; Saturation mutagenesis: iterative and/or simultaneous application of saturation mutagenesis; Gene shuffling: use of recombinatorial methods to generate diversity; Step-wise method application: cases where random mutagenesis, saturation mutagenesis or gene shuffling were used iteratively; Saturation mutagenesis of ‘hot spots’: probing of positions known to affect function, identified through stochastic methods; Combination of improved variants: recombination of variants through site-directed mutagenesis or gene shuffling.
Typical selection paradigms
A popular aphorism of directed evolution is ‘you get what you screen for’ [44]. To those practicing in the field, this may be prefaced with the qualification, ‘if you get anything’. Indeed, we were unable to determine the statistics for failed directed evolution experiments, although many campaigns reported null or minimal improvements in some rounds of mutagenesis and screening. In these cases, alternative mutagenesis strategies were devised to generate mutants with the desired improvement [95,96]. With regards to the successful experiments meeting our selection criteria, we identified microtiter well-based assays as the most common method of activity screening, although agar plate-based selection and screening was also frequently employed (Figure 6). Liquid culture enrichment and FACS were each used in only a small fraction of the selected articles. This distribution of methods may be a reflection of the limited range of enzymatic activities that can be tied to cell survival or a fluorescent signal in whole-cell imaging. Microtiter well-based assays in clarified cell lysate allow analysis via tracking changes in absorbance in the UV or visible light range and permit the use of tandem enzyme assays that may be required to link the desired reaction to a measurable parameter. In addition, assays designed for microtiter plates can be subsequently analyzed by using autosamplers and HPLC, MS or CE for direct product quantitation, making these approaches the most broadly applicable methods for the analysis of a wide variety of enzymatic reactions, albeit with substantially lower throughput.
Figure 6. Distribution of experiments sorted by the screening methods applied.
Blue: selection or plate-based screens use an agar plate to enable high throughput screening of enzyme variants; Green: assays that require growth of individual variants in liquid cultures; Orange: experiments utilizing fluorescence activated cell sorting; Purple: enrichment includes liquid-phase selection of improved variants and enrichment by phage display; Red: colorimetric agar plate-based screens that require a secondary or tertiary assay to distinguish the most improved variants.
Analysis by fold improvement
As previously discussed, the success of directed evolution studies can be measured in a variety of fashions, most typically by the attainment of the desired phenotype or the desired reaction yield. While providing valid criteria for success, these results form a difficult basis for evaluating these successes in the context of the degree of improvement (i.e., almost all publications report success in the attainment of a desired chemophenotype). Correspondingly, we evaluated selected papers in the context of kinetic parameters, specifically kcat, Km and/or kcat/Km, the last parameter being the most frequently cited parameter. Interestingly, while the average-fold improvement for kcat, Km and kcat/Km were 366-, 12- and 2498-fold, respectively, the median-fold improvements were significantly lower at 5.4-, 3- and 15.6-fold, respectively. This disparity is a result of the small number of extremely large-fold rate enhancements (see Supplementary Table 1 and discussion below). In both statistics, the improvements in Km were relatively low. This may be reflective of the lower dynamic range for Km values (often 10 µM–10 mM) and the difficulty in selecting for improved binding during catalysis using common activity screening.
Is there a relationship between classifications & success?
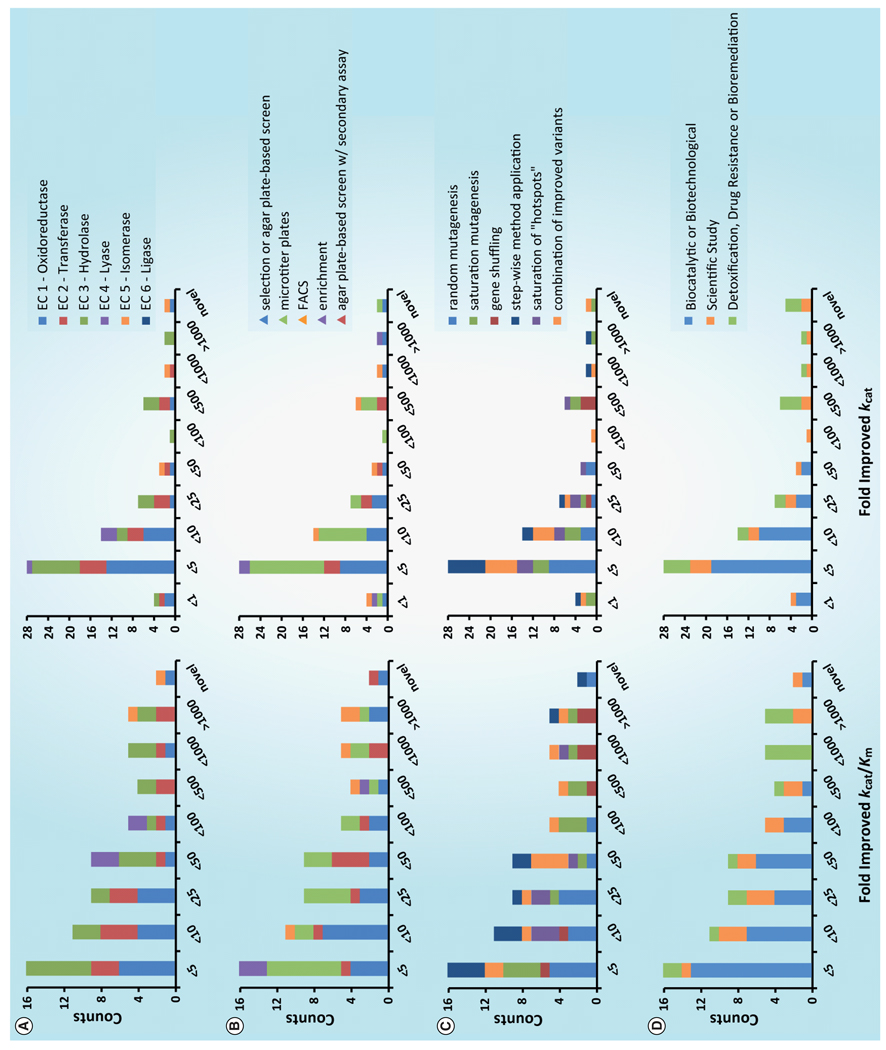
The prevalence of a given classification or method may not predict its ultimate success. Figure 7 illustrates the relationship between the parameter kcat/Km and the classification schemes presented herein. We did not identify many significant trends between the enzyme class, mutagenesis method or screening paradigm and the fold-improvement. Enzymes with the greatest fold-improvement (>100) were represented in every enzyme class, except lyases, using each mutagenesis method and from every screening method.
Figure 7. Fold improvement of kcat/Km and kcat and the relationship to various criteria.
(A) Enzyme class classification. (B) Screening method. (C) Mutagenesis method. Color schemes and classifications of A–C match those of Figures 4–6. (D) Intent of study. Blue: studies that enable small-molecule synthesis; orange: evolutionary or other scientific studies; green: generation of enzymes for detoxification of harmful agents.
Of note, seven out of the 14 cases we identified reporting more than a 100-fold improvement for a new substrate employed a form of saturation mutagenesis in their directed evolution strategy. This may be expected as it has been demonstrated that mutations closer to the binding pocket have a greater impact on enzymatic activity [97]. There are numerous examples of studies comparing saturation mutagenesis of residues in the binding site to whole-gene random mutagenesis. In each case, targeted mutagenesis strategies allow for smaller library sizes and equal or better, levels of improvement. While this has been thoroughly documented [16,98,99], our data suggest that saturation mutagenesis is not the only method resulting in large-fold improvements and it also does not guarantee a high level of success.
An assumption of directed evolution is that a more complete search of the mutational space may allow identification of more improved variants. While large library screens have been reported (e.g., >107 by FACS), the fold-improvements did not scale with the size of the library screened. For instance, screening of only 300 clones in each of two rounds identified a carboxylesterase with 840-fold improved catalytic efficiency [89], whereas a similar FACS-based study resulted in a modest 5.5-fold improvement [81].
Finally, in the context of the generation of new biocatalytic enzymes we examined the intent of each directed evolution study and sorted each case into three types: biocatalytic or biotechnological application studies; studies that aimed to answer a scientific question, such as the evolutionary process; and enzymes that were evolved to remove toxins for survival, for bioremediation or to study drug-resistance mechanisms (Figure 7). Despite making up >50% of the cases, only one study out of 14 with more than 100-fold improvement in enzyme function has biocatalytic application as the ultimate goal. Overall 41 studies of 81 were enzymes with potential biocatalytic application.
The magnitude of rate enhancements may be perceived as modest from a biophysical perspective, with a median value of 5.4-fold for kcat. However, this improvement and the improvements frequently observed as a result of improved protein-production levels, may be deemed highly significant from a biotechnological perspective as the time and/or cost savings scale with increased turnover.
Future perspective
Best practices
Directed evolution studies containing comparable kinetic data over the last 10 years were surprisingly sparse. However, despite a relatively small sample from which to draw conclusions, the summary provided by this article hints at some putative best practices to consider when implementing contemporary directed evolution methodologies:
-
▪
Identify a progenitor enzyme acting on a substrate as similar as possible to the new substrate. This point is somewhat facile, but most of the new substrates identified by our retrospective analysis in Supplementary Table 1 are closely structurally related to the parent enzyme’s natural substrate. In many cases it may not be possible to identify an existing enzyme capable of performing a reaction similar to a target reaction. In such cases, the next best option is to select a progenitor enzyme performing a desired functional-group transformation.
-
▪
As previously noted, saturation mutagenesis methods are overrepresented in the high-fold improvement bins, pointing to a greater potential in targeting particular sites. When possible, it appears that structure- or homology-based site-directed mutagenesis methods should be attempted first. When structural data for progenitor enzymes are unavailable, error prone PCR and gene shuffling are the next alternative.
-
▪
Expect to use combinations of mutagenesis methods in a single campaign. While it is clear that mutations of active site residues provides a clear benefit, it is also clear that distal mutations serve to improve the activity (and/or stability) of enzymes.
-
▪
Recombine identified mutations using site-directed mutagenesis and gene shuffling [75,100]. While individual mutations are rarely 100% additive, recombination frequently provides additional improvement in activity. Furthermore, recombination of the top mutants enables further mining of already-screened libraries.
-
▪
The steep triage from over 1200 articles to only 81 qualifying cases is primarily reflective of a lack of uniformity in reporting standards for enzymes improved by directed evolution studies. To provide clearer data to guide improvements, it would also be helpful for practitioners of directed evolution studies to report complete biochemical kinetic data for progenitor enzymes and throughout each selection exercise.
Emergent paradigms
A current limitation of directed evolution methods is the requirement of the identification of an existing progenitor enzyme with an active site mutable towards a new substrate. This requirement limits the scope of directed evolution. First, it limits the kinds of reactions that can be developed using directed evolution to reactions analogous to those found in nature. Comparison of Laroc’s Comprehensive Organic Transformations [101] with the enzymes identified within EC system illustrates the divide between the possible and the available in the context of developing catalysts using directed evolution. Second, even if a functional progenitor is identified, there are limitations to its adaptability towards new substrates. These limitations are a function of the geometric constraints of the active site, the positioning of side chains essential during a dynamic reaction coordinate and the geometry of the overall protein scaffold containing the active site. For example, in a recent study from our laboratory involving the modification of human purine nucleoside phosphorylase for the condensation of hypoxanthine with dideoxyribose-1-phosphate, only one residue in the active site was identified as being capable of imparting a selectivity advantage. Active site residues favoring substrates with substitution at other positions, for instance the 2´ position, were difficult to identify due to the unavailability of amino acids at this position that did not interfere with catalytically essential side chains [102].
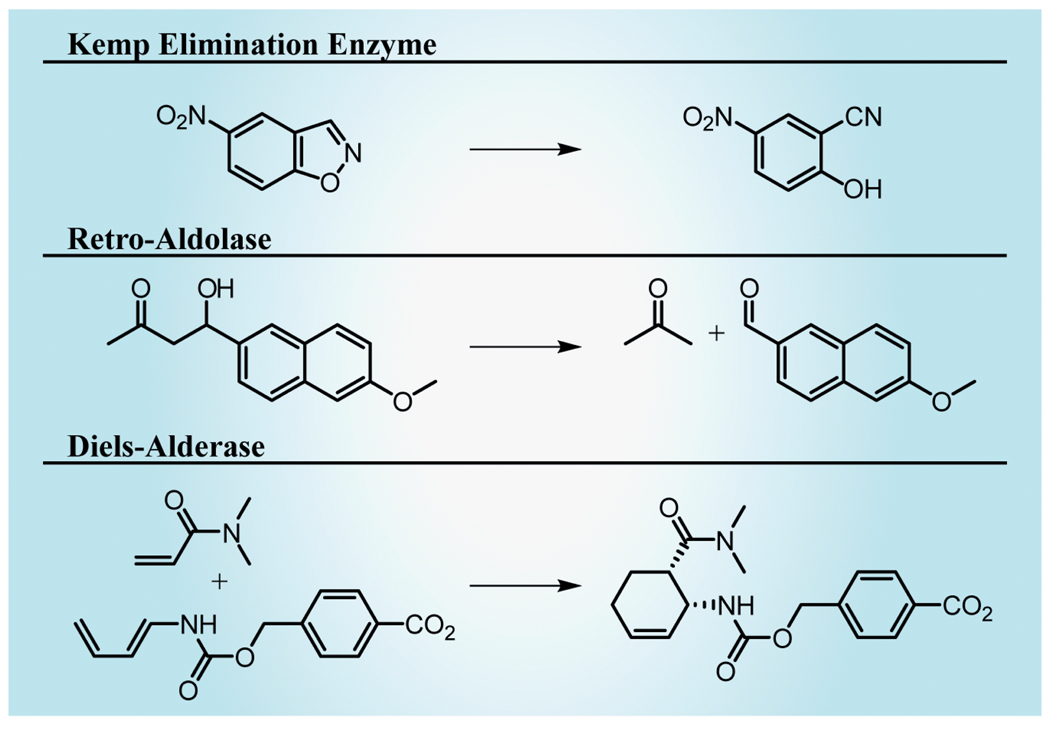
Recently, an approach has been outlined and demonstrated that has potential for addressing the limitation of identifying natural progenitors. In this approach, a transition state model for a reaction of interest is first generated using quantum mechanical calculations. Second, a Rosetta-based algorithm called RosettaMatch [103] is used to identify scaffolds in the structural databases capable of placing functional groups in idealized positions for stabilizing transition states. Third, residues within the scaffold are substituted in silico, scanning all possible combinations and ranking the results by the design’s maintenance of idealized geometries and the binding energy as calculated using a knowledge based energy scoring function. Finally, the theoretical enzymes are synthesized by gene synthesis and tested. This approach has been demonstrated with three case studies (Figure 8).
Figure 8. Computationally designed enzymes catalyze a Kemp elimination, retro-aldol and Diels–Alder reaction.
The enzymes were designed with the Rosetta algorithms [103].
A Kemp elimination catalyzing enzyme was developed using this approach [4]. Subsequent to transition state modeling, RosettaMatch was used to identify 100,000 scaffolds with potential for Kemp elimination catalysis. In the ‘design’ phase (scoring combinatorial substitutions in the putative active site), 59 designs were identified in 17 different scaffolds and were synthesized and tested. Eight of these demonstrated detectable activity with kcat/Km values of 6–160 M−1s−1. One of these candidates crystallized readily and was used to direct seven rounds of directed evolution including site-directed mutagenesis, random mutagenesis and gene shuffling. These directed evolution studies resulted in a 200-fold improvement in catalytic efficiency, from 12.2 to 2,590 M−1s−1.
In a comparable approach, a retro-aldol catalyzing enzyme using a Schiff base mechanism was developed [104]. From 181,555 RosettaMatch solutions, 72 designs in ten different scaffolds were selected for synthesis and testing. Active designs were produced with TIM-barrel and jelly-roll folds with the best design catalyzing a 0.74 M−1s−1 turnover.
Similarly, a Diels–Alderase has been developed in which some 106 matching scaffolds were identified by RosettaMatch; 84 designs were synthesized and 50 soluble enzymes were ultimately characterized [105]. From these, two active enzymes (a six-bladed β-propeller and ketosteroid isomerase scaffold) were identified. Iterative saturation mutagenesis of residues adjacent to activating residues (‘catalytic residues’) optimized 100- and 20-fold each design, respectively. Of note, this study designed stereoselectivity into the reaction.
In addition to these knowledge-based natural scaffold-selection processes, Koder and co-workers have designed completely artificial proteins based on four helix bundles to bind unnatural heme cofactors. Moreover, these artificial hemoproteins are fully competent in O2 binding [106]. It will be impressive if these methodologies can be extended to tune the redox properties of such synthetic cofactors to match the oxidation potential of organic molecules and then to orient them to facilitate asymmetric oxidation reactions.
Whether these transition state binding inspired approaches will ultimately provide solutions to the development of useful biocatalysts for unnatural reactions remains to be determined. While rate enhancements are evident, to date the absolute turnovers generated by these methods are more commonly on the order of catalytic antibodies, which have yet to be developed for preparative biocatalysis. However, there is room for optimism since the designed Diels– Alderase performs 20-fold faster than catalytic antibodies elicited for the same reaction and consumes more than 80% of the reactant at high enzyme concentrations. Regardless, these successes are landmarks in protein engineering and outline a general approach that, if successfully developed, will greatly expand potential application for engineered biocatalysts and ultimately for engineered pathways for small-molecule biosynthesis.
Conclusion
Where biocatalysts for a particular reaction system are not available in the natural repertoire, directed evolution methodologies allow the generation of new enzymes with non-native reaction characteristics such as increased rates of catalysis or altered substrate and/or product profiles, while functioning in a wide range of reaction conditions. As drug development continues to rely on the generation of chemically complex small molecules, the development of biocatalysts for regio- and stereo-selective production of these complex entities will become an important tool for the medicinal and process chemist.
The retrospective assessment provided by this article aims to assist researchers interested in applying existing directed evolution methods for the development of new biocatalysts for transformation of non-native substrates. It is evident that directed evolution methodologies continue to contribute to significant improvements in enzyme catalysis, although a single and generalized route towards the most improved enzyme is frequently unclear. The numeric assessments provided here may be useful in expectations management for both investigators and reviewers of directed evolution studies. The disparity between the average- and median-fold improvements in parameters is striking, suggesting that the median value may represent a more equitable standard for performance improvements. However, numeric assessments fail to fully capture the successes and failures of directed evolution methodologies. We lack comprehensive knowledge of the failures, which may or may not outnumber the reported successes, but regardless would reduce the median-fold improved values further. However, the success from a biocatalytic perspective must ultimately not be defined by fold improvements in biochemical parameters but by the attainment of goals for a given study. Based on our broad survey, it was clear that these criteria were met by the majority of campaigns reported in recent literature, further underlining the ongoing contribution of directed evolution methodologies to engineering of biocatalysts for new and existing substrates.
Executive summary.
-
▪
Direct costs of drug manufacturing are becoming an increasingly important component of therapeutic prices. This has led to the concept of ‘financial toxicity’, which can be defined as the financial impact to a patient as a result of the cost of a therapeutic exceeding a therapeutic benefit.
-
▪
The manufacturing of drugs and drug precursors can potentially be made more efficient by the application of biocatalysts and therefore improve the cost–benefit ratio for therapeutics in the clinic or in development.
-
▪
Most existing enzymes display substrate profiles and/or physical properties that are not suitable for biocatalysis of unnatural substrates.
-
▪
Directed evolution is a proven suite of methodologies used to improve the function of a progenitor enzyme for a new substrate or reaction conditions.
-
▪Using an objective set of criteria, we selected directed evolution studies reported over the last decade. Directed evolution studies were required to:
-
▪Report improvements in a small-molecule biotransformation enzyme;
-
▪Report kinetic parameters, including kcat/Km and/or kcat (or Vmax) and/or Km, for both the progenitor enzyme and the evolved enzyme;
-
▪Be target-based studies; in other words, the reported improved turnover was for the substrate used in the screen;
-
▪Report more than one site mutated, reflecting a departure from single-site saturation mutagenesis.
-
▪
-
▪
These criteria imposed a stringent triage. While some articles may have escaped our notice, from >1200 qualifying articles in the last decade, we were only able to identify 81 articles that met our criteria.
-
▪
We evaluated selected directed evolution studies in the context of enzyme class, library-generation protocol, selection paradigms and fold improvement.
-
▪
No method or enzyme class is overwhelmingly linked to great fold-improvements.
-
▪
Only small-fold improvements are necessary for biocatalytic application.
-
▪
Methodologies with the ability to address some of the key limitations of directed evolution, particularly the problem of identifying a fruitful progenitor enzyme, were reviewed.
Supplementary Material
Acknowledgments
This work was supported in part by NIH, GM077189, T32 GM065086 and the Vanderbilt Institute of Chemical Biology.
Key Terms
- Directed evolution
Iterable two-phase process for improving or altering the physical or catalytic properties of an enzyme, consisting of generating a library of a progenitor gene capable of being expressed in a host or otherwise isolable vessel and selecting for the desired properties of improved gene products via in vitro- or in vivo-selection methods.
- Selection
Identifying members of a gene-expression library with a modified survival or growth phenotype. In the case of a biosynthetic gene library, members with improved turnover for a substrate or substrates are identified via an assay linking this turnover to growth or survival.
- Random mutagenesis
Introduction of mutations throughout a gene sequence of interest, often using error-prone DNA replication strategies.
- Screening
Identifying members of a gene expression library with a modified chemophenotype. In the case of a biosynthetic gene library, members with improved turnover for a substrate or substrates are identified via biochemical assay, often chromogenic.
- Gene shuffling
Recombination, usually in vitro, of pools of related progenitor genes, resulting in libraries of descendent genes, comprised of randomly recombined long stretches of progenitor genes.
- Gene library
Collection of compartmentalized systems, where each member contains a variation of a progenitor gene or gene family. A typical example is a library of Escherichia coli cells in 96-well microtiter plates, each harboring a protein expression plasmid with a sequence variant of a progenitor biosynthetic gene.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Supplementary data
To view the Supplementary Data that accompany this paper please visit the journal website at: www.future-science.com/doi/suppl/10.4155/FMC.11.48.
Bibliography
- 1.Pollack A. Taking big risk for big payoff, industry seeks cancer drugs. The New York Times. 2009 September;1 [Google Scholar]
- 2.Tao JH, Xu JH. Biocatalysis in development of green pharmaceutical processes. Curr. Opin. Chem. Biol. 2009;13(1):43–50. doi: 10.1016/j.cbpa.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Pinheiro E, Vasan A, Kim JY, Lee E, Guimier JM, Perriens J. Examining the production costs of antiretroviral drugs. Aids. 2006;20(13):1745–1752. doi: 10.1097/01.aids.0000242821.67001.65. [DOI] [PubMed] [Google Scholar]
- 4.Rothlisberger D, Khersonsky O, Wollacott AM, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453(7192):U190–U194. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 5.Dougherty MJ, Arnold FH. Directed evolution: new parts and optimized function. Curr. Opin. Biotechnol. 2009;20(4):486–491. doi: 10.1016/j.copbio.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt DMZ, Mundorff EC, Dojka M, et al. Evolutionary potential of (β/α)(8)-barrels: functional promiscuity produced by single substitutions in the enolase superfamily. Biochemistry. 2003;42(28):8387–8393. doi: 10.1021/bi034769a. [DOI] [PubMed] [Google Scholar]
- 7.Jackel C, Kast P, Hilvert D. Protein design by directed evolution. Annu. Rev. Biophys. 2008;37:153–173. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- 8.Johannes TW, Zhao HM. Directed evolution of enzymes and biosynthetic pathways. Curr. Opin. Microbiol. 2006;9(3):261–267. doi: 10.1016/j.mib.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Glieder A, Farinas ET, Arnold FH. Laboratory evolution of a soluble, self-sufficient, highly active alkane hydroxylase. Nat. Biotechnol. 2002;20(11):1135–1139. doi: 10.1038/nbt744. [DOI] [PubMed] [Google Scholar]
- 10.Peters MW, Meinhold P, Glieder A, Arnold FH. Regio- and enantioselective alkane hydroxylation with engineered cytochromes P450 BM-3. J. Am. Chem. Soc. 2003;125(44):13442–13450. doi: 10.1021/ja0303790. [DOI] [PubMed] [Google Scholar]
- 11.Fasan R, Chen MM, Crook NC, Arnold FH. Engineered alkane-hydroxylating cytochrome P450(BM3) exhibiting nativelike catalytic properties. Angew. Chem. Int. Ed. 2007;46:8414–8418. doi: 10.1002/anie.200702616. [DOI] [PubMed] [Google Scholar]
- 12.Meinhold P, Peters MW, Chen MMY, Takahashi K, Arnold FH. Direct conversion of ethane to ethanol by engineered cytochrome P450BM3. ChemBioChem. 2005;6(10):1765–1768. doi: 10.1002/cbic.200500261. [DOI] [PubMed] [Google Scholar]
- 13.Johannes TW, Woodyer RD, Zhao H. Directed evolution of a thermostable phosphite dehydrogenase for NAD(P)H regeneration. Appl. Environ. Microbiol. 2005;71(10):5728–5734. doi: 10.1128/AEM.71.10.5728-5734.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLachlan MJ, Johannes TW, Zhao H. Further improvement of phosphite dehydrogenase thermostability by saturation mutagenesis. Biotechnol. Bioeng. 2008;99(2):268–274. doi: 10.1002/bit.21546. [DOI] [PubMed] [Google Scholar]
- 15.Reetz MT, Bocola M, Carballeira JD, Zha DX, Vogel A. Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test. Angew. Chem. Int. Ed. 2005;44(27):4192–4196. doi: 10.1002/anie.200500767. [DOI] [PubMed] [Google Scholar]
- 16.Reetz MT, Prasad S, Carballeira JD, Gumulya Y, Bocola M. Iterative saturation mutagenesis accelerates laboratory evolution of enzyme stereoselectivity: rigorous comparison with traditional methods. J. Am. Chem. Soc. 2010;132(26):9144–9152. doi: 10.1021/ja1030479. [DOI] [PubMed] [Google Scholar]
- 17.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nature Biotechnology. 2006;24(8):1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 18.Ro DK, Paradise EM, Ouellet M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 19.Yoshikuni Y, Ferrin TE, Keasling JD. Designed divergent evolution of enzyme function. Nature. 2006;440(7087):1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 20.Savile CK, Janey JM, Mundorff EC, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329(5989):305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 21.Doms A, Schroeder M. Gopubmed: exploring pubmed with the gene ontology. Nucleic Acids Res. 2005;33:W783–W786. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong TS, Zhurina D, Schwaneberg U. The diversity challenge in directed protein evolution. Comb. Chem. High Throughput Screening. 2006;9(4):271–288. doi: 10.2174/138620706776843192. [DOI] [PubMed] [Google Scholar]
- 23.Wong TS, Roccatano D, Zacharias M, Schwaneberg U. A statistical analysis of random mutagenesis methods used for directed protein evolution. J. Mol. Biol. 2006;355(4):858–871. doi: 10.1016/j.jmb.2005.10.082. [DOI] [PubMed] [Google Scholar]
- 24.Lai YP, Huang J, Wang LF, Li J, Wu ZR. A new approach to random mutagenesis in vitro. Biotechnol. Bioeng. 2004;86(6):622–627. doi: 10.1002/bit.20066. [DOI] [PubMed] [Google Scholar]
- 25.Myers RM, Lerman LS, Maniatis T. A general-method for saturation mutagenesis of cloned DNA fragments. Science. 1985;229(4710):242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- 26.Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J. Mol. Biol. 1996;255(4):589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 27.Balashov S, Humayun MZ. Specificity of spontaneous mutations induced in mutA mutator cells. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2004;548(1–2):9–18. doi: 10.1016/j.mrfmmm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Patrick WM, Matsumura I. A study in molecular contingency: glutamine phosphoribosylpyrophosphate amidotransferase is a promiscuous and evolvable phosphoribosylanthranilate isomerase. J. Mol. Biol. 2008;377(2):323–336. doi: 10.1016/j.jmb.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roodveldt C, Tawfik DS. Shared promiscuous activities and evolutionary features in various members of the amidohydrolase superfamily. Biochemistry. 2005;44(38):12728–12736. doi: 10.1021/bi051021e. [DOI] [PubMed] [Google Scholar]
- 30.Rowe LA, Geddie ML, Alexander OB, Matsumura I. A comparison of directed evolution approaches using the β-glucuronidase model system. J. Mol. Biol. 2003;332(4):851–860. doi: 10.1016/s0022-2836(03)00972-0. [DOI] [PubMed] [Google Scholar]
- 31.DeSantis G, Wong K, Farwell B, et al. Creation of a productive, highly enantioselective nitrilase through gene site saturation mutagenesis (GSSM) J. Am. Chem. Soc. 2003;125(38):11476–11477. doi: 10.1021/ja035742h. [DOI] [PubMed] [Google Scholar]
- 32.Reidhaar-Olson JF, Sauer RT. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science. 1988;241(4861):53–57. doi: 10.1126/science.3388019. [DOI] [PubMed] [Google Scholar]
- 33.Reetz MT. Laboratory evolution of stereoselective enzymes: a prolific source of catalysts for asymmetric reactions. Angew. Chem., Int. Ed. 2011;50(1):138–174. doi: 10.1002/anie.201000826. [DOI] [PubMed] [Google Scholar]
- 34.Crameri A, Raillard SA, Bermudez E, Stemmer WP. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391(6664):288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 35.Stemmer WPC. DNA shuffling by random fragmentation and reassembly – in vitro recombination for molecular evolution. Proc. Natl Acad. Sci. USA. 1994;91(22):10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao HM, Giver L, Shao ZX, Affholter JA, Arnold FH. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 1998;16(3):258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- 37.Sieber V, Martinez CA, Arnold FH. Libraries of hybrid proteins from distantly related sequences. Nat. Biotechnol. 2001;19(5):456–460. doi: 10.1038/88129. [DOI] [PubMed] [Google Scholar]
- 38.Ostermeier M, Shim JH, Benkovic SJ. A combinatorial approach to hybrid enzymes independent of DNA homology. Nat. Biotechnol. 1999;17(12):1205–1209. doi: 10.1038/70754. [DOI] [PubMed] [Google Scholar]
- 39.Lutz S, Ostermeier M, Moore GL, Maranas CD, Benkovic SJ. Creating multiple-crossover DNA libraries independent of sequence identity. Proc. Natl Acad. Sci. USA. 2001;98(20):11248–11253. doi: 10.1073/pnas.201413698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox RJ, Davis SC, Mundorff EC, et al. Improving catalytic function by prosar-driven enzyme evolution. Nat. Biotechnol. 2007;25(3):338–344. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- 41.Taylor SV, Kast P, Hilvert D. Investigating and engineering enzymes by genetic selection. Ang. Chem. Int. Ed. 2001;40(18):3310–3335. doi: 10.1002/1521-3773(20010917)40:18<3310::aid-anie3310>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 42.Dietrich JA, McKee AE, Keasling JD. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu. Rev. Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- 43.Boersma YL, Droge MJ, Quax WJ. Selection strategies for improved biocatalysts. FEBS J. 2007;274(9):2181–2195. doi: 10.1111/j.1742-4658.2007.05782.x. [DOI] [PubMed] [Google Scholar]
- 44.Arnold FH, Georgiou G. Directed Enzyme Evolution: Screening and Selection Methods. Totowa, NJ, USA: Humana Press; 2003. [Google Scholar]
- 45.Olsen M, Iverson B, Georgiou G. High-throughput screening of enzyme libraries. Curr. Opin. Biotechnol. 2000;11(4):331–337. doi: 10.1016/s0958-1669(00)00108-7. [DOI] [PubMed] [Google Scholar]
- 46.Wahler D, Reymond JL. Novel methods for biocatalyst screening. Curr. Opin. Chem. Biol. 2001;5(2):152–158. doi: 10.1016/s1367-5931(00)00184-8. [DOI] [PubMed] [Google Scholar]
- 47.Akanuma S, Yamagishi A, Tanaka N, Oshima T. Serial increase in the thermal stability of 3-isopropylmalate dehydrogenase from Bacillus subtilis by experimental evolution. Protein Sci. 1998;7(3):698–705. doi: 10.1002/pro.5560070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rothman SC, Kirsch JF. How does an enzyme evolved in vitro compare to naturally occurring homologs possessing the targeted function? Tyrosine aminotransferase from aspartate aminotransferase. J. Mol. Biol. 2003;327(3):593–608. doi: 10.1016/s0022-2836(03)00095-0. [DOI] [PubMed] [Google Scholar]
- 49.Yano T, Oue S, Kagamiyama H. Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc. Natl Acad. Sci. USA. 1998;95(10):5511–5515. doi: 10.1073/pnas.95.10.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castle LA, Siehl DL, Gorton R, et al. Discovery and directed evolution of a glyphosate tolerance gene. Science. 2004;304(5674):1151–1154. doi: 10.1126/science.1096770. [DOI] [PubMed] [Google Scholar]
- 51.Cho CMH, Mulchandani A, Chen W. Altering the substrate specificity of organophosphorus hydrolase for enhanced hydrolysis of chlorpyrifos. Appl. Environ. Microbiol. 2004;70(8):4681–4685. doi: 10.1128/AEM.70.8.4681-4685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Claren J, Malisi C, Hocker B, Sterner R. Establishing wild-type levels of catalytic activity on natural and artificial (βα) (8)-barrel protein scaffolds. Proc. Natl Acad. Sci. USA. 2009;106(10):3704–3709. doi: 10.1073/pnas.0810342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gulick AM, Fahl WE. Forced evolution of glutathione-s-transferase to create a more efficient drug detoxication enzyme. Proc. Natl Acad. Sci. USA. 1995;92(18):8140–8144. doi: 10.1073/pnas.92.18.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoseki J, Yano T, Koyama Y, Kuramitsu S, Kagamiyama H. Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for thermus thermophilus. J. Biochem. 1999;126(5):951–956. doi: 10.1093/oxfordjournals.jbchem.a022539. [DOI] [PubMed] [Google Scholar]
- 55.Landis DM, Loeb LA. Random sequence mutagenesis and resistance to 5-fluorouridine in human thymidylate synthases. J. Biol. Chem. 1998;273(40):25809–25817. doi: 10.1074/jbc.273.40.25809. [DOI] [PubMed] [Google Scholar]
- 56.Walter KU, Vamvaca K, Hilvert D. An active enzyme constructed from a 9-amino acid alphabet. J. Biol. Chem. 2005;280(45):37742–37746. doi: 10.1074/jbc.M507210200. [DOI] [PubMed] [Google Scholar]
- 57.Zhang KC, Li H, Cho KM, Liao JC. Expanding metabolism for total biosynthesis of the nonnatural amino acid l-homoalanine. Proc. Natl Acad. Sci. USA. 2010;107(14):6234–6239. doi: 10.1073/pnas.0912903107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Black ME, Newcomb TG, Wilson HM, Loeb LA. Creation of drug-specific herpes simplex virus type 1 thymidine kinase mutants for gene therapy. Proc. Natl Acad. Sci. USA. 1996;93(8):3525–3529. doi: 10.1073/pnas.93.8.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christians FC, Scapozza L, Crameri A, Folkers G, Stemmer WP. Directed evolution of thymidine kinase for AZT phosphorylation using DNA family shuffling. Nat. Biotechnol. 1999;17(3):259–264. doi: 10.1038/7003. [DOI] [PubMed] [Google Scholar]
- 60.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99(17):11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292(5516):498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 62.Schwimmer LJ, Rohatgi P, Azizi B, Seley KL, Doyle DF. Creation and discovery of ligand-receptor pairs for transcriptional control with small molecules. Proc. Natl Acad. Sci. USA. 2004;101(41):14707–14712. doi: 10.1073/pnas.0400884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc. Natl Acad. Sci. USA. 2005;102(32):11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hida K, Hanes J, Ostermeier M. Directed evolution for drug and nucleic acid delivery. Adv. Drug Delivery Rev. 2007;59(15):1562–1578. doi: 10.1016/j.addr.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 65.Fernandez-Gacio A, Uguen M, Fastrez J. Phage display as a tool for the directed evolution of enzymes. Trends Biotechnol. 2003;21(9):408–414. doi: 10.1016/S0167-7799(03)00194-X. [DOI] [PubMed] [Google Scholar]
- 66.Jose J. Autodisplay: efficient bacterial surface display of recombinant proteins. Appl. Microbiol. Biotechnol. 2006;69(6):607–614. doi: 10.1007/s00253-005-0227-z. [DOI] [PubMed] [Google Scholar]
- 67.Bosma T, Damborsky J, Stucki G, Janssen DB. Biodegradation of 1,2,3-trichloropropane through directed evolution and heterologous expression of a haloalkane dehalogenase gene. Appl. Environ. Microbiol. 2002;68(7):3582–3587. doi: 10.1128/AEM.68.7.3582-3587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carter BT, Lin H, Goldberg SD, Althoff EA, Raushel J, Cornish VW. Investigation of the mechanism of resistance to third-generation cephalosporins by class c β-lactamases by using chemical complementation. ChemBioChem. 2005;6(11):2055–2067. doi: 10.1002/cbic.200500058. [DOI] [PubMed] [Google Scholar]
- 69.Cheon YH, Park HS, Kim JH, Kim Y, Kim HS. Manipulation of the active site loops of d-hydantoinase, a (β/α)(8)-barrel protein, for modulation of the substrate specificity. Biochemistry. 2004;43(23):7413–7420. doi: 10.1021/bi036330o. [DOI] [PubMed] [Google Scholar]
- 70.Delagrave S, Murphy DJ, Pruss JLR, et al. Application of a very high-throughput digital imaging screen to evolve the enzyme galactose oxidase. Protein Eng. 2001;14(4):261–267. doi: 10.1093/protein/14.4.261. [DOI] [PubMed] [Google Scholar]
- 71.Iffland A, Gendreizig S, Tafelmeyer P, Johnsson K. Changing the substrate specificity of cytochrome c peroxidase using directed evolution. Biochem. Biophys. Res. Commun. 2001;286(1):126–132. doi: 10.1006/bbrc.2001.5366. [DOI] [PubMed] [Google Scholar]
- 72.Lin L, Meng X, Liu PF, et al. Improved catalytic efficiency of endo-β-1,4-glucanase from Bacillus subtilis BME-15 by directed evolution. Appl. Microbiol. Biotechnol. 2009;82(4):671–679. doi: 10.1007/s00253-008-1789-3. [DOI] [PubMed] [Google Scholar]
- 73.Nakagawa Y, Hasegawa A, Hiratake J, Sakata K. Engineering of pseudomonas aeruginosa lipase by directed evolution for enhanced amidase activity: mechanistic implication for amide hydrolysis by serine hydrolases. Protein Eng. Des. Sel. 2007;20(7):339–346. doi: 10.1093/protein/gzm025. [DOI] [PubMed] [Google Scholar]
- 74.Nakazawa H, Okada K, Onodera T, Ogasawara W, Okada H, Morikawa Y. Directed evolution of endoglucanase III (Cel12A) from Trichoderma reesei. Appl. Microbiol. Biotechnol. 2009;83(4):649–657. doi: 10.1007/s00253-009-1901-3. [DOI] [PubMed] [Google Scholar]
- 75.Park SH, Park HY, Sohng JK, et al. Expanding substrate specificity of GT-B fold glycosyltransferase via domain swapping and high-throughput screening. Biotechnol. Bioeng. 2009;102(4):988–994. doi: 10.1002/bit.22150. [DOI] [PubMed] [Google Scholar]
- 76.Zhang ZG, Liu Y, Guengerich FP, Matse JH, Chen J, Wu ZL. Identification of amino acid residues involved in 4-chloroindole 3-hydroxylation by cytochrome P450 2A6 using screening of random libraries. J. Biotechnol. 2009;139(1):12–18. doi: 10.1016/j.jbiotec.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antipov E, Cho AE, Wittrup KD, Klibanov AM. Highly l and d enantioselective variants of horseradish peroxidase discovered by an ultrahigh-throughput selection method. Proc. Natl Acad. Sci. USA. 2008;105(46):17694–17699. doi: 10.1073/pnas.0809851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aharoni A, Amitai G, Bernath K, Magdassi S, Tawfik DS. High-throughput screening of enzyme libraries: thiolactonases evolved by fluorescence-activated sorting of single cells in emulsion compartments. Chem. Biol. 2005;12(12):1281–1289. doi: 10.1016/j.chembiol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 79.Griswold KE, Aiyappan NS, Iverson BL, Georgiou G. The evolution of catalytic efficiency and substrate promiscuity in human theta class 1–1 glutathione transferase. J. Mol. Biol. 2006;364(3):400–410. doi: 10.1016/j.jmb.2006.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Griswold KE, Kawarasaki Y, Ghoneim N, Benkovic SJ, Iverson BL, Georgiou G. Evolution of highly active enzymes by homology-independent recombination. Proc. Natl Acad. Sci. USA. 2005;102(29):10082–10087. doi: 10.1073/pnas.0504556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu LF, Li YF, Liotta D, Lutz S. Directed evolution of an orthogonal nucleoside analog kinase via fluorescence-activated cell sorting. Nucleic Acids Res. 2009;37(13):4472–4481. doi: 10.1093/nar/gkp400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mastrobattista E, Taly V, Chanudet E, Treacy P, Kelly BT, Griffiths AD. High-throughput screening of enzyme libraries: in vitro evolution of a β-galactosidase by fluorescence-activated sorting of double emulsions. Chem. Biol. 2005;12(12):1291–1300. doi: 10.1016/j.chembiol.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 83.Droge MJ, Boersma YL, van Pouderoyen G, et al. Directed evolution of Bacillus subtilis lipase a by use of enantiomeric phosphonate inhibitors: crystal structures and phage display selection. ChemBioChem. 2006;7(1):149–157. doi: 10.1002/cbic.200500308. [DOI] [PubMed] [Google Scholar]
- 84.Sunbul M, Marshall NJ, Zou YK, Zhang KY, Yin J. Catalytic turnover-based phage selection for engineering the substrate specificity of Sfp phosphopantetheinyl transferase. J. Mol. Biol. 2009;387(4):883–898. doi: 10.1016/j.jmb.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 85.Matsumura I, Wallingford JB, Surana NK, Vize PD, Ellington AD. Directed evolution of the surface chemistry of the reporter enzyme β-glucuronidase. Nat. Biotechnol. 1999;17(7):696–701. doi: 10.1038/10910. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt-Dannert C. Engineering novel carotenoids in microorganisms. Curr. Opin. Biotechnol. 2000;11(3):255–261. doi: 10.1016/s0958-1669(00)00093-8. [DOI] [PubMed] [Google Scholar]
- 87.Schmidt-Dannert C, Umeno D, Arnold FH. Molecular breeding of carotenoid biosynthetic pathways. Nat. Biotechnol. 2000;18(7):750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- 88.Hibbert EG, Senussi T, Costelloe SJ, et al. Directed evolution of transketolase activity on non-phosphorylated substrates. J. Biotechnol. 2007;131(4):425–432. doi: 10.1016/j.jbiotec.2007.07.949. [DOI] [PubMed] [Google Scholar]
- 89.Ohki T, Shibata N, Higuchi Y, et al. Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a β-lactamase fold: x-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase. Protein Sci. 2009;18(8):1662–1673. doi: 10.1002/pro.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei CL, Yang YB, Deng CH, et al. Directed evolution of Streptomyces clavuligerus deacetoxycephalosporin C synthase for enhancement of penicillin G expansion. Appl. Environ. Microbiol. 2005;71(12):8873–8880. doi: 10.1128/AEM.71.12.8873-8880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bhuiya M-W, Liu C-J. Engineering monolignol 4-O-methyltransferases to modulate lignin biosynthesis. J. Biol. Chem. 2010;285(1):277–285. doi: 10.1074/jbc.M109.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hsu CC, Hong ZY, Wada M, Franke D, Wong CH. Directed evolution of d-sialic acid aldolase to l-3-deoxy-manno-2-octulosonic acid (l-KDO) aldolase. Proc. Natl Acad. Sci. USA. 2005;102(26):9122–9126. doi: 10.1073/pnas.0504033102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Williams GJ, Domann S, Nelson A, Berry A. Modifying the stereochemistry of an enzyme-catalyzed reaction by directed evolution. Proc. Natl Acad. Sci. USA. 2003;100(6):3143–3148. doi: 10.1073/pnas.0635924100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Woodhall T, Williams G, Berry A, Nelson A. Creation of a tailored aldolase for the parallel synthesis of sialic acid mimetics. Angew. Chem. Int. Ed. 2005;44(14):2109–2112. doi: 10.1002/anie.200462733. [DOI] [PubMed] [Google Scholar]
- 95.Fujii R, Nakagawa Y, Hiratake J, Sogabe A, Sakata K. Directed evolution of Pseudomonas aeruginosa lipase for improved amide-hydrolyzing activity. Protein Eng. Des. Sel. 2005;18(2):93–101. doi: 10.1093/protein/gzi001. [DOI] [PubMed] [Google Scholar]
- 96.Hawwa R, Larsen SD, Ratia K, Mesecar AD. Structure-based and random mutagenesis approaches increase the organophosphate-degrading activity of a phosphotriesterase homologue from deinococcus radiodurans. J. Mol. Biol. 2009;393(1):36–57. doi: 10.1016/j.jmb.2009.06.083. [DOI] [PubMed] [Google Scholar]
- 97.Morley KL, Kazlauskas RJ. Improving enzyme properties: when are closer mutations better? Trends Biotechnol. 2005;23(5):231–237. doi: 10.1016/j.tibtech.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 98.Paramesvaran J, Hibbert EG, Russell AJ, Dalby PA. Distributions of enzyme residues yielding mutants with improved substrate specificities from two different directed evolution strategies. Protein Eng. Des. Sel. 2009;22(7):401–411. doi: 10.1093/protein/gzp020. [DOI] [PubMed] [Google Scholar]
- 99.Parikh MR, Matsumura I. Site-saturation mutagenesis is more efficient than DNA shuffling for the directed evolution of β-fucosidase from β-galactosidase. J. Mol. Biol. 2005;352(3):621–628. doi: 10.1016/j.jmb.2005.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fasan R, Meharenna YT, Snow CD, Poulos TL, Arnold FH. Evolutionary history of a specialized P450 propane monooxygenase. J. Mol. Biol. 2008;383(5):1069–1080. doi: 10.1016/j.jmb.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Larock RC. Comprehensive Organic Transformations: A Guide to Functional Group Preparations (Second Edition) NY, USA: Wiley-VCH; 1999. [Google Scholar]
- 102.Nannemann DP, Kaufmann KW, Meiler J, Bachmann BO. Design and directed evolution of a dideoxy purine nucleoside phosphorylase. Protein Eng. Des. Sel. 2010;23(8):607–616. doi: 10.1093/protein/gzq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zanghellini A, Jiang L, Wollacott AM, et al. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15(12):2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang L, Althoff EA, Clemente FR, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319(5868):1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Siegel JB, Zanghellini A, Lovick HM, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels–Alder reaction. Science. 2010;329(5989):309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Koder RL, Anderson JL, Solomon LA, Reddy KS, Moser CC, Dutton PL. Design and engineering of an O2 transport protein. Nature. 2009;458(7236):305–309. doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.