Abstract
Many plants respond to winter with epigenetic factors that gradually dampen repression of flowering so that they can flower in spring. The study of this process was important for the identification of the plant Polycomb group (PcG) of proteins and their role in the epigenetic control of plant gene expression. Fittingly, these studies continue to illuminate our understanding of PcG function. We discuss recent advances, particularly the role of noncoding RNA in the recruitment of PcG to target genes, and the role of the PcG in regulating the stem cell pool in flowers.
Introduction and context
Many plants that grow in climates with a cold winter require growth for several months at low temperatures—a process called vernalization—to promote flowering in spring, when days lengthen and temperatures increase. Without this period of cold, plants would grow leaves in the spring, but would fail to flower. It was noticed that this phenomenon, familiar to every horticulturist, had distinctive features; something occurred during those cold months that left a mark, which, in effect, released a switch that permitted flowering in spring. Experimental manipulation of temperature showed that this mark was very stable and could persist for at least a year after a vernalization treatment [1]. Although stable during a plant's life, the marks were erased between generations, as the progeny of vernalized plants themselves required vernalization in order to flower. In recent years, the field has looked beyond the genome and tested whether epigenetic changes (i.e., ones that aren't based on DNA sequence alteration) were involved. One epigenetic system involves DNA methylation, and in plants this is often used for genome defense; for example, to inactivate invading DNA such as transposons, viruses, and transgenes.
A second epigenetic system is implemented by two collections of genes known as the Polycomb group (PcG) and the trithorax group (trxG). Again, methylation is involved, but in this case it is not the DNA itself that is modified, it is the histone proteins that package DNA in the nucleus (Figure 1a). Unlike the changes wrought by DNA methylation, there is (so far) little clear-cut evidence that PcG- or trxG-mediated changes are passed between generations in plants. Rather, they are used to provide cells with memories of events that occur during the life of an organism. Typically this is a developmental memory, hence the PcG and trxG components were first identified on account of gross developmental abnormalities resulting from their mutation. However, they are also used to record transient environmental events that occur during the life of the plant. This can be used to predict and adapt to future environments and may be especially relevant in plants, which are sessile and so can't easily escape their circumstances. Here, we discuss important advances in our understanding of how PcG genes provide a memory of winter. Secondly, we describe the emerging role of PcG and trxG genes in controlling stem cell fate in flowers. This review is of necessity selective; for more comprehensive reviews see [2] or [3].
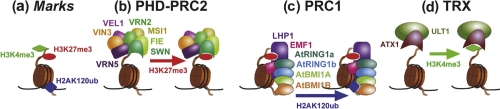
Figure 1. PcG and trxG proteins epigenetically control flowering and flower development.
(a) Histone modifications ‘mark’ nucleosomes at specific genes. The nucleosome is an octamer containing two molecules each of histone H2A, H2B, H3, and H4. For simplicity, only one of each of the two H2A and H3 tails are shown in the figure. H2AK120 resides on an exposed surface of the nucleosome core. (b,c,d) Likely components and functions of Arabidopsis PcG and trxG protein complexes equivalent to animal PRC2, PRC1, and TRX complexes are shown. (b) During vernalization in Arabidopsis, the PHD-PRC2 complex catalyzes H3K27me3 methylation through the SWN histone methyltransferase subunit. VIN3, VEL1, and VRN5 are plant-specific, whereas the other four members are homologs of the animal PRC2 core components. Because three of the four core components of animal PRC2 have been duplicated in Arabidopsis, it is likely that several related complexes exist that differ in components and target gene specificities [2]. For example, in some complexes, CLF may replace SWN as the histone methyltransferase unit. (c) LHP1, a component of Arabidopsis PRC1, binds H3K27me3. PRC1 may catalyze H2AK120Ub ubiquitination via its E3 ligase components AtRING1a, AtRING1b, AtBMI1A, and AtBMI1B. EMF1 is likely another (plant-specific) component whose precise function is unclear. (d) ULT1 and ATX1 may be components of an Arabidopsis trxG complex that catalyses H3K4me3. ATX1, ARABIDOPSIS HOMOLOG OF TRITHORAX 1; CLF, CURLY LEAF; EMF1, EMBRYONIC FLOWER 1; FIE, FERTILIZATION INDEPENDENT ENDOSPERM; LHP1, LIKE HETEROCHROMATIN PROTEIN 1; MSI1, MULTICOPY SUPPRESSOR OF IRA 1; PcG, Polycomb group; PHD-PRC2, PLANT HOMEO DOMAIN–POLYCOMB REPRESSIVE COMPLEX 2; PRC1, POLYCOMB REPRESSIVE COMPLEX 1; SWN, SWINGER; trxG, trithorax group; ULT1, ULTRAPETALA1; VEL1, VERNALIZATION5/VIN3-LIKE 1; VIN3, VERNALIZATION INSENSITIVE 3; VRN2, VERNALIZATION2; VRN5, VERNALIZATION5.
Recent advances
Vernalization and flowering time
The mechanism of the vernalization response has been best studied in the model plant Arabidopsis thaliana, where it has been found to depend on the ability of the PcG system to silence the key gene mediating the vernalization response, FLOWERING LOCUS C (FLC). FLC is a potent repressor of flowering, most likely because its protein product binds and represses two genes, FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), which are central players in promoting flowering [4]. Thus in order for plants to become competent to respond to environmental factors promoting flowering (warm temperatures and long days), FLC must be switched off. During cold treatments, expression of FLC is progressively reduced and a vernalization-specific complex of PcG proteins (termed PHD-PRC2, which stands for ‘Plant Homeo Domain–Polycomb Repressive Complex 2’) (Figure 1b) is recruited to FLC. Biochemical purification of this complex [5] indicates that it contains the plant equivalents of the four core components of an animal PcG protein complex known as PRC2 (Polycomb repressive complex 2).
Recently, its function has become clearer. PRC2 acts as a histone methyltransferase, specifically catalyzing trimethylation of lysine 27 on the histone H3 tail (H3K27me3). In addition to the core PRC2 components, PHD-PRC2 contains several plant-specific components that are probably required to boost the histone methyltransferase activity of the complex. Consistent with this, H3K27me3 levels at FLC increase during cold treatments and persist afterwards when plants are moved to warm conditions. H3K27me3 is also necessary for maintenance of FLC silencing when plants are removed from the cold.
How then does H3K27me3 cause gene silencing? Although it is necessary for silencing of FLC, it is not sufficient: in vernalization1 (vrn1) mutants, FLC activity is restored after vernalization, despite the persistence of high H3K27 methylation levels [6]. H3K27me3 is not thought to directly cause chromatin compaction or inhibit transcription but is probably recognized by proteins that repress transcription of FLC. In animals, a second complex of PcG proteins, called PRC1, is also required for PcG-mediated silencing. Although the PRC1 complex is less well conserved than PRC2, an important recent finding is that several of its members are found in plants and play a similar role in PcG-mediated repression. One PRC1 member, POLYCOMB, binds H3K27me3, and in Arabidopsis the related LIKE HETEROCHROMATIN 1 (LHP1) protein has a similar function (Figure 1c) [7]. It is also likely that PRC2 itself binds H3K27me3, which may help in the stable propagation of this mark through cell division, by methylating newly synthesized (unmodified) histones as they are deposited into chromatin, for example [8,9]. Three other components of PRC1 in animals, RING1A, RING1B, and BMI1, form a ubiquitin ligase complex that monoubiquitinates histone H2A at lysine 119 (H2AK119Ub). The function of this modification is somewhat enigmatic. Although H2AK119Ub may help repress transcription directly by blocking RNA polymerase II transcriptional elongation from the promoters of PcG target genes [10,11], its importance is challenged by the observation that PRC1 can cause chromatin compaction and transcriptional repression independently of its H2A ubiquitination activity [12]. Be this as it may, two groups have now identified Arabidopsis homologs of RING1A, RING1B, and BMI1 (Figure 1c) and the similarity of their knockout phenotypes with those of mutants inactivating PRC2 is compelling evidence that they have a similar role to animal PRC1 [13,14]. In addition, the plant RING proteins can monoubiquitinate H2AK120 (the residue in plants that most closely corresponds to the animal H2AK119, although most of the 13 Arabidopsis H2A genes don't encode this residue) in in-vitro assays and are required for H2A ubiquitination in vivo in Arabidopsis [13]. Although so far only a handful of PcG targets have been shown to be misregulated in the Arabidopsis atring/atbmi1 mutants, it will obviously be interesting to see if the Arabidopsis ATRING/ATBMI genes have any role in the vernalization-induced repression of FLC.
Because the PcG components are mostly expressed fairly constitutively and seemingly lack specific DNA-binding activity, a major problem has been to explain how they are recruited to specific targets such as FLC. Numerous recent studies in animals have shown that noncoding RNA play a role in recruitment [3], and two noncoding RNAs have now been implicated in vernalization-induced recruitment of PHD-PRC2. One of these, termed COLDAIR, is a long (about 1 kb) noncoding sense RNA produced from sequences within the large first intron of FLC [15] (Figure 2). Expression of COLDAIR is cold-induced and several observations suggest that it is functionally important for FLC silencing. Firstly, two earlier studies using transgene reporters showed that sequences within the large first intron of FLC are needed for its stable silencing by cold [16,17]. In particular, transgenes with deletions that removed part or all of the COLDAIR gene showed fairly normal downregulation during cold treatments but regained activity when plants were returned to warm conditions. Secondly, several approaches showed that CURLY LEAF (CLF), the catalytic subunit of the PHD-PRC2 complex, binds COLDAIR RNA [15]. This may help recruit PHD-PRC2 to FLC; for example, if the COLDAIR transcript remains tethered to its site of transcription at FLC (Figure 2) or alternatively if the affinity of PHD-PRC2 for chromatin is altered by its binding COLDAIR. Consistent with this, downregulation of COLDAIR using RNA interference strongly reduces the recruitment of CLF to FLC and also impairs the maintenance of FLC silencing following cold treatments. A second noncoding RNA, termed COOLAIR, is also produced from FLC [18]. Like COLDAIR, COOLAIR expression is also strongly upregulated by cold temperatures. COOLAIR expression is driven by a promoter located downstream of the FLC coding region (Figure 2) and produces several antisense transcripts that differ according to their splicing or 3′ end. There is good evidence that COOLAIR also plays a role in regulating FLC expression. Two genes, FPA and FCA, promote flowering by downregulating FLC expression. Although FPA and FCA encode proteins that regulate mRNA processing, until recently it was problematic that they had no discernable effect on FLC mRNA processing. Two groups have now resolved this issue by demonstrating that they regulate the processing and relative amounts of the different COOLAIR transcripts [19,20]. However, FPA and FCA are not required for the vernalization response, and the function of COOLAIR in vernalization is less clear. Notably, FLC transgenes that lack sequences downstream of FLC that include the COOLAIR promoter retain a normal vernalization response [16]. Although this suggests that COOLAIR may not be necessary, interpretation is complicated because these plants retain an endogenous FLC gene that produces COOLAIR transcripts that could act in trans on the FLC transgene. It is also noteworthy that COLDAIR is not sufficient for a vernalization response; thus, the introduction of FLC intragenic sequences that contain COLDAIR into a heterologous reporter did not confer a vernalization response on the reporter [16]. One possibility, suggested by Heo and Sung [15], is that induction of COOLAIR by cold may reduce transcription from the normal promoter 5′ end of the FLC protein coding region and promote COLDAIR expression from its cryptic promoter within the first FLC intron.
Figure 2. The FLC locus produces at least two noncoding RNAs.
The open reading frame of FLC is shown in turquoise, with exons depicted as green boxes and introns as black lines. The antisense COOLAIR RNA is upregulated early after cold treatment. However, only the COLDAIR transcript is thought to be bound by PHD-PRC2 and so may be more important in recruiting PHD-PRC2 to the FLC locus. FLC, FLOWERING LOCUS C; PHD-PRC2, PLANT HOMEO DOMAIN–POLYCOMB REPRESSIVE COMPLEX 2; RNA Pol II, RNA polymerase II.
Although many plants show a vernalization response with epigenetic features, the target genes and even the mechanisms may be different than those in Arabidopsis. In sugar beet, vernalization results in stable repression of a floral repressor, which, although it is unrelated to FLC, acts in a similar fashion to repress an FT homolog [21]. It is not clear yet if this is also PcG mediated. In cereals, vernalization causes the stable upregulation of VRN1, which promotes flowering. Here, the epigenetic memory is of an active state, and is correlated with H3K4me3 methylation at VRN1 [22]. It is likely, although not yet proven, that this will involve the trxG genes as these are known to catalyze H3K4me3 methylation and mediate stable gene activation.
Shaping floral architecture: stemming the stem cell pool
Once flower induction occurs, the indeterminate vegetative shoot apical meristem at the shoot apex changes its identity to become an inflorescence meristem, which makes floral meristems on its flanks. The floral meristem produces whorls of floral organ primordia, and so gives rise to the flowers. Like all meristems, the continued production of organ primordia requires the activity of a pool of stem cells in the centre of the meristem. Unlike the other meristems, the floral meristem is determinate so that organ production ceases once the carpels are initiated in the innermost whorl of the flower. Mutations in PcG and trxG genes are well known to cause changes in floral organ identity, reflecting their role in controlling floral homeotic gene activity. However, a second less well emphasized aspect of their mutant phenotype is an increase in floral organ number. This reflects an emerging role in controlling when the flower stem cell pool is terminated.
In Arabidopsis shoots, the stem cells at the top of the meristem secrete the CLAVATA3 (CLV3) signal peptide, which restricts the expression of the transcription factor WUSCHEL (WUS) in the organizing centre beneath the stem cells. In turn, WUS promotes stem cell fate and as a result also promotes CLV3 expression non-cell-autonomously in cells above the organizing centre. This CLV3/WUS feedback loop mediates homeostasis of the stem cells via a rapid response to fluctuations in either CLV3 or WUS expression [23-25].
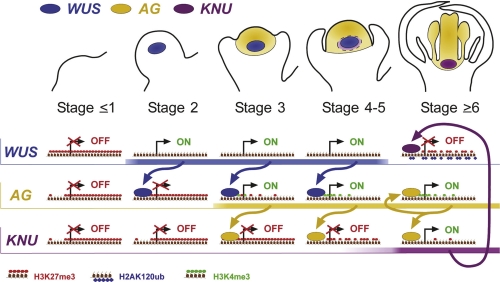
However, flowers are determinate structures and as a consequence, a second feedback loop terminates the stem cell pool of floral meristems. In contrast to the fast and continuously operating CLV3/WUS system, this feedback loop, built by WUS and the MADS-box transcription factor AGAMOUS (AG), occurs just once. Early in flower development (stage 3), WUS and another transcription factor, LEAFY, together activate expression of AG in the centre of the floral meristem. In turn, AG represses WUS expression but there is a delay of about two days before WUS expression terminates at stage 6 [26,27]. In addition, the first step (the activation of AG expression by WUS) also exhibits a delay: WUS expression occurs during floral stages 2-5. By contrast, AG is not detectable in floral meristem until stage 3. The reason for these delays has been unclear but two recent publications [28,29] reveal the relevance of epigenetic regulation by the PcG and trxG (Figure 3).
Figure 3. Mechanistic model for transcriptional and epigenetic control of the floral meristem stem cell pool.
Serial floral stages with the expression of WUS, AG, and KNU are shown. Underneath, the genomic regions of these genes are shown schematically with likely binding of transcription factors, expression status, and histone modifications. At the initial stage of flower development (stage ≤1), the KNU locus is covered by the repressive mark H3K27me3. WUS is activated at stage 2, but the transcriptional activation of AG by WUS is inhibited by H3K27me3. At stage 3, the AG locus undergoes activation by progressive ULT1/ATX1-dependent H3K4me3 and likely demethylation of H3K27. WUS binds in the large second intron of AG. AG expression is maintained by autoactivation as well as by H3K4me3. WUS expression terminates at stage 6. The activation of KNU by AG is initially inhibited by H3K27me3 at the KNU locus between stages 3 to 5. The level of H3K27me3 at the KNU locus is gradually depleted, perhaps by dilution via successive rounds of cell division, and at stage 6 KNU is expressed, causing WUS repression. The punctual termination of WUS expression needs additional PRC1 activity, which suggests that the WUS locus accumulates H3K27me3 and H2AK120Ub. AG, AGAMOUS; ATX1, ARABIDOPSIS HOMOLOG OF TRITHORAX 1; KNU, KNUCKELS; ULT1, ULTRAPETALA1; WUS, WUSCHEL.
Carles and Fletcher [28] investigated whether the activation of AG is dependent on the Arabidopsis SAND (Sp100, AIRE-1, NucP41/75, DEAF-1) domain protein ULTRAPETALA1 (ULT1) and furthermore, whether ULT1 is involved in trxG function. The authors were inspired by the gain-of-function phenotypes of 35S::ULT1 transgenic plants, which resemble clf mutants in showing increased AG expression in leaves and inflorescences [28,30]. Furthermore, ult1 mutants suppress the clf mutant phenotype in ult1 clf double mutants and show reduced AG expression [28]. This fits with idea that ULT1 is a trxG factor and works antagonistically to the PcG to promote AG activity, and may also explain why WUS is misexpressed in ult1 mutants [30]. Consistent with this antagonism, 35S::ULT1 seedlings, similar to clf mutants, have reduced levels of the repressive mark H3K27me3 at AG, whereas ult1 mutants have increased H3K27me3 levels. The structure of ULT1 makes it unlikely that it is itself an H3K4me3 methyltransferase, rather it may interact with such an enzyme as part of a trxG complex or promote trxG recruitment to targets. Consistent with this, ULT1 interacts physically with the trxG protein ARABIDOPSIS HOMOLOG OF TRITHORAX 1 (ATX1), which is responsible for local H3K4me3 deposition (Figure 1d) [28,32]. ATX1 cannot be the only factor for ULT1-dependent H3K4me3 at the AG locus because atx1 mutants, unlike ult1 mutants, do not show indeterminacy of floral meristems [28] and also retain 85% of global H3K4me3 levels [33]. Furthermore, although the ULT1-dependent activation of AG by a slow increase of H3K4me3 may provide a time buffer that the transcription factor WUS on its own may not provide, it remains unclear whether ULT1 and WUS work in parallel or in the same pathway.
Intriguingly, the delay in the second step of the AG/WUS feedback loop—the final suppression of WUS by AG—may also involve epigenetic control. Sun et al. [29] identified KNUCKELS (KNU), which encodes a C2H2-type zinc finger transcription factor, as a direct target of AG and a key intermediate in the suppression of WUS expression. Genetic analysis and overexpression experiments imply that KNU is activated by AG and represses WUS expression [29]. Curiously, although AG binds the KNU promoter directly, there is a two day lag between the activation of AG and that of KNU. This time, buffering may be caused by PcG regulation of KNU. Several observations indicate that KNU is a PcG target: first, early in floral meristem development the KNU locus is strongly decorated with H3K27me3; second, in various PcG mutants KNU is misexpressed. Using an elegant system for synchronizing floral development, Sun et al. show that between floral development stage 3, when AG is activated, and stage 6, when KNU is activated and WUS expression terminated, H3K27me3 is gradually removed from the KNU locus. But how can the MADS domain transcription factor AG cause histone demethylation? One possibility is that AG recruits H3K27me3 demethylases, which have been identified in animals but not yet in plants. A simple alternative is that AG somehow inhibits the binding of PRC1 and PRC2 complexes to the KNU locus and the H3K27me3 mark is gradually diluted as unmodified histones are incorporated onto replicated DNA during cell division [34]. Either way, the H3K27me3 at KNU is not sufficient to prevent KNU activation by AG but does delay it. Similarly, two other recent studies that looked at the activation of another PcG target, in this case by cold, also conclude that H3K27me3 is important for slowing the kinetics of induction [35,36]. Although the tardy induction of KNU in flowers suggests a mechanism for the delayed suppression of WUS by AG, it cannot be the only factor that terminates WUS expression. Thus, knu mutants have much weaker effects on floral determinacy than ag mutants, leaving space for other AG-dependent mechanisms. As WUS is itself a target of H3K27me3 [37], it is also possible that its repression by KNU is just the initial step that facilitates a long-term epigenetic repression by PRC1. Supporting this thesis, WUS is misexpressed in mutants of components of PRC1, which bear extra floral organs [13,38].
Future directions
It is likely that whole genome profiling studies will soon clarify how generally noncoding RNAs are involved in PcG recruitment; for example, by sequencing the RNAs that are associated with PRC2 components, as has recently been done in animals [39]. It will be important to test whether these RNAs can act in trans, that is, to recruit PRC2 to loci that are not adjacent to the site of their production. If so, the mechanism of recruitment is an issue, as it is unlikely to be simply by sequence homology between the RNA and the target gene. Whether or not PRC2 binds specific RNA sequences or secondary structures is also an interesting question as, at least in in-vitro assays, components such as CLF bind RNA and single-strand DNA quite generally [40]. It will be important to know whether the AtRING1/AtBMI1 proteins and their putative mark (H2AK120Ub) colocalize with PRC2 targets, or rather are restricted to a subset of targets, and if the latter, whether these targets show more stable silencing. Lastly, it will be interesting to learn whether the PcG and trxG play a more general role in recording transient environmental events than simply the vernalization response.
Acknowledgments
We thank the anonymous reviewers for helpful comments on the manuscript. The work of RM and JG is supported by the Biotechnology and Biological Sciences Research Council (BBRSC) and the European Research Area Network (ERA-NET).
Abbreviations
- AG
AGAMOUS
- ATX1
ARABIDOPSIS HOMOLOG OF TRITHORAX 1
- CLF
CURLY LEAF
- CLV3
CLAVATA3
- FLC
FLOWERING LOCUS C
- FT
FLOWERING LOCUS T
- KNU
KNUCKELS
- PcG
Polycomb group
- PHD-PRC2
PLANT HOMEO DOMAIN–POLYCOMB REPRESSIVE COMPLEX 2
- PRC2
Polycomb repressive complex 2
- SOC1
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1
- trxG
trithorax-group
- ULT1
ULTRAPETALA1
- VRN1
VERNALIZATION1
- WUS
WUSCHEL
Competing Interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/3/13
References
- 1.Lang A. Physiology of flower initiation. In: Ruhland W, editor. Encyclopedia of Plant Physiology. Berlin: Springer-Verlag; 1965. pp. 1371–536. [Google Scholar]
- 2.Alvarez-Venegas R. Regulation by Polycomb and Trithorax Group Proteins in Arabidopsis. The Arabidopsis Book. 2010;8:1–20. doi: 10.1199/tab.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Vivian Irish 11 May 2006
- 5.De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci U S A. 2008;105:16831–6. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Justin Goodrich 08 Feb 2011
- 6.Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–7. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Detlef Weigel 05 Feb 2004 Thomas Jenuwein 05 Jan 2005
- 7.Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Phil Wigge 14 Jun 2007
- 8.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Leonie Ringrose 06 Nov 2008 Andrew Feinberg 25 Nov 2008
- 9.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, III, Voigt P, Martin SR, Taylor WR, De MV, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–7. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–35. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Brad Bernstein 11 Dec 2007
- 11.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Cheng-Ming Chiang 05 Feb 2008
- 12.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–64. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Rob White 16 Jun 2010
- 13.Bratzel F, Lopez-Torrejon G, Koch M, Del Pozo JC, Calonje M. Keeping cell identity in arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol. 2010;20:1853–9. doi: 10.1016/j.cub.2010.09.046. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Jennifer Fletcher 25 Oct 2010
- 14.Chen D, Molitor A, Liu C, Shen WH. The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 2010;20:1332–44. doi: 10.1038/cr.2010.151. [DOI] [PubMed] [Google Scholar]
- 15.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–9. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]; F1000 Factor 14Evaluated by Fyodor Urnov 16 Dec 2010, Elizabeth Dennis 23 Dec 2010, Ilha Lee 04 Jan 2011, Kan Wang 31 Jan 2011, Jae-Hyuk Yu 11 Feb 2011
- 16.Sheldon CC, Conn AB, Dennis ES, Peacock WJ. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell. 2002;14:2527–37. doi: 10.1105/tpc.004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet. 2006;38:706–10. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- 18.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Vitaly Citovsky 18 Jan 2010
- 19.Hornyik C, Terzi LC, Simpson GG. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell. 2010;18:203–13. doi: 10.1016/j.devcel.2009.12.009. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Ilha Lee 14 May 2010, Motoaki Seki 13 Jul 2010
- 20.Liu F, Marquardt S, Lister C, Swiezewski S Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–7. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 21.Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJ, Nilsson O. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science. 2010;330:1397–400. doi: 10.1126/science.1197004. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9Evaluated by Ilha Lee 04 Feb 2011, Justin Goodrich 08 Feb 2011
- 22.Oliver SN, Finnegan EJ, Dennis ES, Peacock WJ, Trevaskis B. Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc Natl Acad Sci U S A. 2009;106:8386–91. doi: 10.1073/pnas.0903566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–9. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 24.Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–44. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 25.Müller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell. 2006;18:1188–98. doi: 10.1105/tpc.105.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Christiaan van der Schoot 12 Jan 2007
- 26.Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–14. doi: 10.1016/S0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Philip Benfey 05 Nov 2001
- 27.Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/S0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]; F1000 Factor 15Evaluated by Richard M Amasino 18 Oct 2001, Philip Benfey 05 Nov 2001, David Baum 21 May 2002
- 28.Carles CC, Fletcher JC. The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 2009;23:2723–8. doi: 10.1101/gad.1812609. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 12Evaluated by Vivian Irish 09 Dec 2009, Elena Alvarez-Buylla 17 Dec 2009, Kay Schneitz 05 Jan 2010, Justin Goodrich 08 Feb 2011
- 29.Sun B, Xu Y, Ng KH, Ito T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009;23:1791–804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 13Evaluated by Kay Schneitz 13 Aug 2009, David Smyth 15 Sep 2009, Jennifer Fletcher 19 Oct 2009, Justin Goodrich 08 Feb 2011
- 30.Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- 31.Carles CC, Lertpiriyapong K, Reville K, Fletcher JC. The ULTRAPETALA1 gene functions early in Arabidopsis development to restrict shoot apical meristem activity and acts through WUSCHEL to regulate floral meristem determinacy. Genetics. 2004;167:1893–903. doi: 10.1534/genetics.104.028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol. 2003;13:627–37. doi: 10.1016/S0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Marjori Matzke 05 Jun 2003
- 33.Alvarez-Venegas R, Avramova Z. Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 2005;33:5199–207. doi: 10.1093/nar/gki830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito T, Sun B. Epigenetic regulation of developmental timing in floral stem cells. Epigenetics. 2009;4:564–7. doi: 10.4161/epi.4.8.10351. [DOI] [PubMed] [Google Scholar]
- 35.Kim DH, Zografos BR, Sung S. Vernalization-mediated VIN3 Induction Overcomes the LIKE-HETEROCHROMATIN PROTEIN1/POLYCOMB REPRESSION COMPLEX2-mediated epigenetic repression. Plant Physiol. 2010;154:949–57. doi: 10.1104/pp.110.161083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finnegan J, Bond D, Buzas D, Goodrich J, Helliwell C, Tamada Y, Yun J, Amasino R, Dennis E. Polycomb proteins regulate the quantitative induction of VERNALIZATION INSENSITIVE 3 in response to low temperatures. Plant J. 2011;65:382–91. doi: 10.1111/j.1365-313X.2010.04428.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 10Evaluated by Gregory Copenhaver 25 Apr 2007, Elizabeth Dennis 27 Apr 2007
- 38.Xu L, Shen WH. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol. 2008;18:1966–71. doi: 10.1016/j.cub.2008.11.019. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6Evaluated by Elena Alvarez-Buylla 22 Jan 2009
- 39.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8Evaluated by Michael Cole 07 Mar 2011
- 40.Krajewski WA, Nakamura T, Mazo A, Canaani E. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol. 2005;25:1891–9. doi: 10.1128/MCB.25.5.1891-1899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]