Abstract
Cyclophilin (CyP) is a peptidyl prolyl cis/trans isomerase, catalyzing the cis-trans isomerization of proline residues in proteins. CyP plays key roles in several different aspects of cellular physiology including the immune response, transcription, mitochondrial function, cell death, and chemotaxis. In addition to these cellular events, a number of reports demonstrated that CyP plays a critical role in the life cycle of viruses, especially human immunodeficiency virus (HIV) and hepatitis C virus (HCV). These two viruses are significant causes of morbidity and mortality worldwide, but current therapies are often insufficient. CyP may provide a novel therapeutic target for the management and/or cure of these diseases, in particular HCV.
Keywords: cyclosporin, HIV, HCV, virus, replication, MPTP
Immunophilins and Immunosuppressants
Cyclophilin (CyP) and FK506 binding protein (FKBP) are peptidyl-prolyl cis-trans isomerases (PPIases), enzymes that catalyze the cis-trans interconversion of peptide bonds amino terminal to proline residues (Fischer et al. 1989; Harding et al. 1989; Takahashi, 1999; Takahashi et al. 1989). CyP and FKBP are originally identified as cellular factors that bind CsA and FK506, respectively, both of which are immunosuppressants used clinically for the prevention of graft rejection following organ transplantation (Handschumacher et al. 1984; Harding et al. 1989). Therefore, these PPIases are also called immunophilin. The action of PPIases leads to changes in protein conformation (Takahashi, 1999), but the binding of CsA and FK506 to CyP and FKBP, respectively, inhibits the activity of these enzymes (Fischer et al. 1989; Rosen et al. 1990; Takahashi et al. 1989). However, the inhibition of PPIase activity by CsA and FK506 is an insufficient requirement for their immunosuppressive function (Bierer et al. 1990; Schreiber, 1991). The CsA/CyP or FK506/FKBP complex, subsequently interacts with and inhibits calcineurin (CN), a phosphatase involved in the activation of the transcription factor NF-AT. Proper NF-AT function is essential for the generation of a productive T cell response (Clipstone and Crabtree, 1992; Fruman et al. 1992; Liu et al. 1991). In the absence of immunosuppressants, CN dephosphorylates cytoplasmic NF-AT, leading to NF-AT nuclear translocation and transactivation of downstream genes participating in the immune response (Liu et al. 1992; McCaffrey et al. 1993). CsA and FK506 prevent the dephosphorylation and subsequent nuclear translocation of NF-AT leading to immunosuppression.
Role of CyP Family Members in Cellular Events
More than 10 CyP subtypes are found in mammals (Table 1). The subcellular localization of CyPs varies. CyPA is primarily found in the cytoplasm, while CyPB, CyPD, CyPE, and RanBP2 are distributed in the endoplasmic reticulum (ER), mitochondria, nucleus, and nuclear pore, respectively. Members of the CyP family play roles in a variety of cellular processes including the immune response, transcription, mitochondrial function, cell death, and chemotaxis, as described below. While a number of CyP family members have been ideitified, intensive functional analysis has been performed on only a few including CyPA, CyPB, CyPD, and CyP40.
Table 1.
Human cyclophilin subtypes.
| Protein name | length | GenBank accession no. | Reference |
|---|---|---|---|
| CyPA | 165 aa | NM_021130 | Liu et al. 1990 |
| CyPB | 216 aa | NM_000942 | Price et al. 1991 |
| CyPC | 212 aa | NM_000943 | Friedman et al. 1991 |
| CyP40 | 370 aa | NM_005038 | Kieffer et al. 1992 |
| CyPE, CyP33 | 301 aa | NM_006112 | Mi et al. 1996 |
| CyPD, CyPF, CyP3 | 207 aa | NM_005729 | Bergsma et al. 1991 |
| CyPG, CARS-CyP, SRcyp | 754 aa | NM_004792 | Nestel et al. 1996 |
| CyPH, USA-CyP, SnuCyP-20 | 177 aa | NM_006347 | Horowitz et al. 1997 |
| PPI-L1 | 166 aa | NM_016059 | Ozaki et al. 1996 |
| PPI-L2, CyP60 | 520 aa | NM_014337 | Wang et al. 1996 |
| PPI-L3 | 165 aa | NM_032472 | Zhou et al. 2001 |
| PPI-L4 | 492 aa | NM_139126 | Zeng et al. 2001 |
| PPI-L5, LRR-1 | 414 aa | NM_152329 | Jang et al. 2001 |
| RanBP2 | 3224 aa | NM_006267 | Yokoyama et al. 1995 |
CyPA is the most abundant CyP subtype found in the cells (Waldmeier et al. 2003), and it is the primary factor mediating the immunosuppressive effects of CsA (Colgan et al. 2005). However, even in the absence of CsA, CyPA plays an important role in regulating the immune responses as seen in CyPA-deficient mice. CyPA-knockout mice have an “allergic” phenotype with increased serum IgG1 and IgE levels and tissue infiltration by mononuclear cells, eosinophils, and mast cells (Colgan et al. 2004), related to increased and dysregulated activity of Th2 CD4+ T cells. In CyPA-knockout cells, interleukin-2 tyrosine kinase (Itk), a signaling molecule crucial for the development of a Th2 response, is constitutively activated. Itk is a member of the Tec family of SH2/SH3-containing tyrosine kinases, and it participates in the signal transduction cascade leading to T cell activation. CyPA can bind Itk, and this negatively regulates Itk activity (Brazin et al. 2002). Thus, CyPA plays a suppressive role in the development of CD4+ T cell responses through its interaction with Itk.
Other studies have reported several non-immune system roles for CyPA. CyPA interacts with apoptosis-inducing factor (AIF) and promotes AIF-mediated chromatinolysis during apoptosis (Cande et al. 2004). Additionally, CyPA interacts with membrane-bound guanylate cyclase-A (GC-A), a receptor for atrial natriuretic factor (ANF) (Chen et al. 2004). GC-A and ANF are involved in cardiovascular homeostasis, and CyPA appears to function as an endogenous inhibitor of GC-A activation by competing for ANF binding. Further interactions of CyPA with prolactin receptor (Syed et al. 2003) and transcription factor YY1 (Yang et al. 1995) have been observed, but the exact role of CyPA in these processes remains unclear. CypA was also observed to bind DNA in a zinc-dependent manner in a mouse macrophage cell line (Krummrei et al. 1995). However, the best-characterized role identified for CyPA is not in normal cellular physiology, but rather as co-factor during the human immunodeficiency virus-1 (HIV-1) viral life cycle (See below).
CyPB was originally identified as a CyP family member bearing a signal sequence leading to the ER lumen or the secretory pathway (Price et al. 1991), but the specific function of CyPB is poorly understood. A yeast two-hybrid screening using CyPB as a bait identified an interaction with calcium-signal modulating cyclophilin ligand (CAML) (Bram and Crabtree, 1994). CAML is located on the cytoplasmic face of the ER membrane (Holloway and Bram, 1998). CAML participates in calcium signal transduction pathway and it is essential for peripheral T cell development (Tran et al. 2005). However, the importance of CyPB binding to CAML function remains unknown. CyPB also enhances prolactin-driven cell proliferation (Rycyzyn et al. 2000) and promotes the nuclear retrotranslocation of prolactin through a direct interaction. Additionally, CyPB potentiates prolactin-induced STAT5 transactivation by promoting the dissociation of PIAS3, a STAT5 repressor (Rycyzyn and Clevenger, 2002). CyPB can also associate with interferon regulatory factor (IRF)-3 (Obata et al. 2005). Extracellular CyPB can bind platelets (Allain et al. 1999) and this initiates a transmembranous influx of calcium ion, kinase activation, and platelet adhesion to collagen. Accumulating evidence suggests that CyPs, in particular CyPA and CyPB, can mediate intercellular communication similar to cytokines. CyPs are secreted from cells in response to inflammatory stimuli or oxidative stress (Jin et al. 2000; Seko et al. 2004; Sherry et al. 1992; Xu et al. 1992) and they can act as potent chemoattractants for neutrophils (Sherry et al. 1992), eosinophils (Xu et al. 1992), and T cells (Allain et al. 2002). CyPA and CyPB are recognized by the cell surface receptor CD147, and CyP binding leads to ERK activation and chemotaxis (Pushkarsky et al. 2001; Yurchenko et al. 2001; Yurchenko et al. 2002).
CyPD plays a critical role in mitochondrial function and cell death (Tanveer et al. 1996). During ischemia-induced necrosis, e.g. following a heart attack or stroke, the accumulation of calcium and increase of reactive oxygen species (ROS) trigger the opening of a pore in the inner mitochondrial membrane, known as the membrane permeability transition pore (MPTP) (Halestrap, 1999). Calcium overload and ROS induce a conformational change in adenine nucleotide translocase (ANT), a key component regulating the opening of MPTP at the inner mitochondrial membrane. The opening of MPTP leads to mitochondrial swelling, rupture of the outer membrane, and the release of small molecules (Waldmeier et al. 2003). CyPD is located within the matrix of the mitochondria and it binds ANT facilitating its conformational change (Crompton et al. 1998; Woodfield et al. 1998). In CyPD-knockout cells, necrosis induced by calcium and ROS was decreased, but apoptotic cell death induced by cytokines or anti-cancer agents was unaffected (Baines et al. 2005; Nakagawa et al. 2005). CyPD-knockout mice also experienced reduced cardiac injury following reperfusion after ischemia. Thus, CyPD is a key molecule involved in the cell death process.
CyP40 regulates the activity of steroid receptors (SRs) (Duina et al. 1996; Owens-Grillo et al. 1995; Ratajczak et al. 1993). SRs including the glucocorticoid receptor, estrogen receptor, androgen receptor and progesterone receptor are nuclear hormone receptors that exert transcriptional activity following steroid ligand binding and nuclear translocation. In the absence of steroid ligands, SRs form complexes with heat shock protein 90 (HSP90) together with the immunophilins CyP40, FKBP51, or FKBP52 in the cytoplasm. These immunophilins control SR activity by increasing receptor avidity for hormone ligands through PPIase-dependent conformational changes. Upon hormone binding, this SR/HSP90/ immunophilin complex dissociates, leaving homodimeric SR, which then translocates into the nucleus to transactivate downstream genes.
Although there are some reports on other CyP subtypes (Table 1), the precise functions and significances of them are largely unknown.
Viruses Requiring CyPs
As described above, CyPs play essential roles in diverse cellular processes. Interestingly, several viruses have evolved to use CyPs during their life cycles. In particular, CyPs are demonstrated to be involved in the proliferation of HIV-1 and hepatitis C virus (HCV). Other viruses using CyPs during their life cycle include vaccinia virus (VV), vesicular stomatitis virus (VSV), and SARS-coronavirus.
Vaccinia virus
A significant role of CyP in VV replication was first identified through the analysis of several CsA analogs. The ability of cyclosporins to suppress VV replication correlated with the inhibition of CyP function (Damaso and Moussatche, 1998). VV infection stabilizes CyPA, leading to the accumulation of CyPA (Castro et al. 2003). In VV infected cells, CyPA relocalizes to the peripheral region of the nucleus, colocalizing with sites of virus production. CyPA is incorporated into viral particles and is located in the viral core.
Vesicular stomatitis virus
CyPA interacted with the nucleocapsid protein of VSV (Bose et al. 2003), and, like VV, CyPA is incorporated into VSV viral particles. Although the binding and incorporation of CyPA occurred beyond the virus serotypes, the functional role of CyPA in the viral life cycle appears to be strain-dependent. Inhibition of CyP activity by CsA reduced primary transcription of VSV-New Jersey (VSV-NJ) but not VSV-Indiana (VSV-IND) serotype, and CyPA activity was required for the replication of VSV-NJ to a greater extent than VSV-IND. The authors suggest that differential requirements of CyPA are likely the results of evolutionary pressure during lineage development.
SARS coronavirus
The nucleocapsid protein (NP) of severe acute respiratory syndrome coronavirus (SARS-CoV) binds CyPA (Luo et al. 2004), and another group reported that CyPA is incorporated into SARS-CoV particles (Chen et al. 2005). Extracellular CyPA binds CD147 on the cell surface, and treatment with a peptide that blocks CD147 binding inhibits viral infection. Thus, CyPA may be involved in SARS-CoV invasion into host cells through interaction with NP and CD147, respectively.
CyPA and HIV-1
CyPA plays an important role in the viral life cycle of HIV-1. In 1993, CyPA was found to interact with HIV-1 Gag (Luban et al. 1993), and in 1994, CyPA was reportedly incorporated into viral particles (Franke et al. 1994; Thali et al. 1994). A gene targeting study demonstrated that only CyPA among Cyp subtypes was essential for HIV-1 proliferation (Braaten and Luban, 2001). Within the HIV-1 life cycle, CyPA plays multiple roles through different interaction partners, including an early step prior to reverse transcription (Braaten et al. 1996; Mlynar et al. 1997; Steinkasserer et al. 1995). Although CyPA is incorporated into virions through binding to the CA domain of the Gag polyprotein (Franke et al. 1994; Ott et al. 1995; Thali et al. 1994), this incorporation is not required for viral infection. Instead, target cell expressed CyPA is important for productive infection and viral replication (Hatziioannou et al. 2005; Sokolskaja et al. 2004). It has been known for several decades that host cells express different restriction factors to prevent infection by certain retroviruses (Cullen, 2003), and several recent studies have suggested that CyPA modulates sensitivity to such a restriction factor early in the HIV-1 life cycle prior to reverse transcription. TRIM5α is a host restriction factor originally identified using expression cloning that recognizes CA limiting retrovirus proliferation (Stremlau et al. 2004). Towers et al. showed that CyPA regulates the activity of a host restriction factor (Towers et al. 2003). Disruption of CyPA-CA binding by introducing of point mutation into CA or treating human cells with CsA decreases HIV-1 infectivity. Conversely, the loss of CyPA-CA binding greatly enhanced HIV-1 infectivity in simian cells (Berthoux et al. 2005; Kootstra et al. 2003; Sayah et al. 2004). From the results, the hypothesis was proposed by Luban et al. that CA binding by CyPA prevented normal antiviral effects mediated by TRIM5α during HIV-1 infection of human cells, but this same interaction mediated HIV-1 restriction in nonhuman primate cells (Sokolskaja et al. 2006; Luban, in press). Both the mechanism of TRIM5α restriction of HIV-1 and the modulation of CA recognition by CyPA remain unclear, and further studies are clearly needed to resolve these important issues in the HIV-1 life cycle and the host response to HIV-1 infection.
CyPA may be important for other aspects of HIV-1 infection. CyPA interacts with CD147 (Pushkarsky et al. 2001), heparans (Saphire et al. 1999), Vpr (Zander et al. 2003), and envelope glycoprotein gp120 (Endrich and Gehring, 1998), although their relevances of the interactions should be further verified.
CyPB and HCV
Current therapy against HCV
HCV is a major causative agent of chronic liver diseases such as chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (Liang et al. 1993). HCV infection is a serious health problem affecting approximately 170 million individuals worldwide (Poynard et al. 2003). The current standard therapy for HCV is restricted to interferon (IFN) or pegylated-IFN either alone or in combination with ribavirin. Because treatment with these agents, however, fails to produce sustained virus elimination in about one half of the patients (Di Bisceglie et al. 2002), alternative and effective strategies to combat against HCV are greatly needed.
HCV encodes a single polypeptide that is cleaved by host and HCV-encoded protease including NS3 to generate a set of functional proteins. Its genome is replicated by the HCV-encoded RNA-dependent RNA polymerase (RdRp) NS5B. Both of these proteins, NS3 and NS5B, are essential for HCV genome replication and are possible targets for the development of anti-HCV therapeutics (Di Bisceglie et al. 2002). Small molecule compounds targeting NS3 and NS5B have been developed, and their efficacy has been examined in clinical trials (Di Bisceglie et al. 2002). In addition to these viral enzymes, host cell factors are required for viral replication, and these may provide other options for the development of novel anti-viral agents. Disrupting the function of host cell derived factors is particularly appealing as the mutation rate of host proteins is much less than that of viral encoded proteins and should less give rise to drug-resistant viruses. However, while some host factors required for viral genome replication have been identified, other host proteins need to be identified to develop optimal anti-viral therapies with few side effects.
At this time, only a limited number of host proteins have been found to be involved in HCV genome replication with biological relevance. hVAP-33 is one of SNARE family proteins that regulate vesicle biogenesis, protein sorting, and membrane fusion. hVAP-33 binds HCV NS5A and NS5B, and this interaction regulates the presence of HCV proteins in the subcellular compartment performing viral genome replication (Evans et al. 2004; Gao et al. 2004; Tu et al. 1999). FBL2 is a member of the F-box protein family, involved in the ubiquitination pathway. FBL2 is geranylgeranylated in cells (Wang et al. 2005), and associates with NS5A in a geranylgeranylation-dependent manner to regulate HCV genome replication. However, the mechanism of action of FBL2 in HCV genome replication is not known. Additionally, we recently found that CyPB is a cofactor for HCV replication in host cells, and this may represent a new target for anti-HCV therapeutics (Watashi et al. 2005). We will first discuss the role of CyPB in HCV genome replication followed by the therapeutic implications of this discovery in HCV treatment.
Anti-HCV activity of cyclosporin
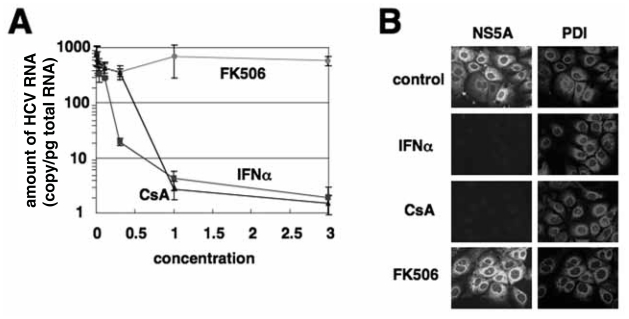
We identified CsA as an anti-HCV agent using a HCV replicon system, a cell culture system supporting HCV genome replication (Lohmann et al. 1999), and CsA inhibits HCV genome replication as potently as IFNα (Watashi et al. 2003). Since that time, several groups have made similar observations (Firpi et al. 2006; Nakagawa et al. 2004; Paeshuyse et al. 2006). As shown in Fig. 1A, cellular treatment with 1 μg/ml CsA decreases HCV RNA levels by approximately 1/500 (Watashi et al. 2003). CsA also reduces the expression of HCV-encoded proteins to undetectable levels (Fig. 1B). In contrast, FK506 has no effect on the production of HCV RNA or proteins (Fig. 1A and B). The differences between CsA and FK506 suggest that CsA prevents viral genome replication independently of CN, an effector common to both CsA and FK506 mediated immunosuppression. And the anti-HCV effects of CsA are mediated by pathway(s) distinct from those of IFNα (Watashi et al. 2003).
Figure 1.
CsA suppresses HCV genome replication. (A) HCV RNA was quantified in total RNA isolated from HCV replicon-bearing cells treated with various concentrations of CsA, FK506, or IFNα for 7 days. The amount of HCV RNA per 1 pg total RNA was plotted against the concentration of CsA (μg/ml), FK506 (μg/ml), or IFNα (× 100 IU/ml). (B) The expression of HCV NS5A and protein disulfide isomerase (PDI) as a cellular protein was examined in the HCV replicon-bearing cells treated without (control) or with 100 IU/ml IFNα, 1_ μg/ml CsA, or 1_ μg/ml FK506 for 7 days.
CyPB as a cellular cofactor of HCV genome replication
The ability of CsA to inhibit HCV genome replication correlates with the inhibition of CyP activity (Watashi et al. 2005). Moreover, an alternative CyP inhibitor, sanglifehrin, also decreases the levels of HCV RNA. Thus, the inhibition of CyP activity is essential for the anti-HCV effect of CsA, and this strongly suggests that CyP plays a direct, important role in HCV genome replication. Moreover, the specific knockdown of CyPB by RNAi reduced HCV RNA titer, but knockdown of CyPA, CyPC, CyPE, or CyPH had no effect on HCV replication activity. These data indicate that CyPB plays a critical role in HCV genome replication.
Regulation of NS5B by CyPB
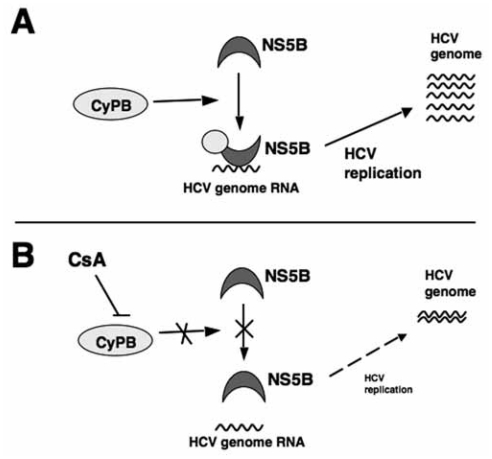
The effects of CyPB on HCV genome replication are mediated through a direct interaction with NS5B as demonstrated in both in vitro and in cells (Watashi et al. 2005) (Fig. 2A). CyPB do not bind any other HCV proteins involved in viral replication. NS5B binds HCV genome RNA in order to function as a RdRp. CyPB but not CyPA promotes the RNA binding activity of NS5B and stimulates HCV genome replication in cells (Fig. 2A). This functional support by CyPB to NS5B is essential for the efficient replication of the HCV genome, and CsA blocks the interaction of CyPB with NS5B, leading to reduced RNA binding (Fig. 2B). Thus, CyPB serves as a cellular cofactor for HCV genome replication.
Figure 2.
CyPB regulates the activity of NS5B. (A) In the absence of CsA (normal conditions), NS5B associates with cellular CyPB to efficiently bind to the HCV genome RNA and drive genome replication. (B) In the presence of CsA, CyPB does not interact with NS5B. Free NS5B less functions, and viral genome replication is impaired in the absence of functional CyPB.
Therapeutic Implications of CyP Inhibition for the Treatment for HCV
The anti-HCV activity of CsA analogs correlates with their ability to inhibit CyP function (Watashi et al. 2005). The dissociation of CyPB and NS5B greatly reduces the extent of HCV genome replication. These observations suggest that the inhibition of CyPB may represent a novel therapeutic strategy against HCV. This possibility has been examined by two reports using stronger CyP inhibitors than CsA. Paeshuyse et al. used the CsA analog DEBIO-025 to inhibit CyP activity (Paeshuyse et al. 2006), and this compound inhibited HCV replication 10-fold more potently than CsA. The authors speculated that DEBIO-025 might be an attractive drug candidate for the treatment of individuals with HCV/HIV coinfection because CsA derivatives should also inhibit HIV-1 replication. We used the non-immunosuppressive CsA derivative NIM811 to target CyP (Goto et al. 2006; Ishii et al. 2006). NIM811 inhibits CyP enzymatic activity two-fold more than CsA (Rosenwirth et al. 1994), and this increased inhibition correlates with greater suppression of HCV genome replication than CsA, especially at lower doses. Cotreatment of cells with NIM811 and IFNα led to a synergistic anti-HCV effect at higher doses of NIM811. Treatment of NIM811 for three weeks eliminated HCV RNA from host cells to under detectable level. Because the immunosuppression in patients during a viral infection is undesirable, these non-immunosuppressive variants of CsA that inhibit CyP activity are likely to offer great promise for the treatment of patients with chronic HCV infection.
Conclusion
CyPs are cellular PPIases that catalyze conformational changes in proteins, but the role(s) and substrates for this protein family in cells are not well-characterized. However, discoveries of the significance of CyP in life cycles for several viruses make this protein virologically notable and present novel therapeutic anti-viral targets. Further analyses of the role of CyPs in viral life cycles should reveal novel functions for these proteins as well as provide mechanistic insight into possible therapeutic targets.
References
- Allain F, Durieux S, Denys A, Carpentier M, Spik G. Cyclophilin B binding to platelets supports calcium-dependent adhesion to collagen. Blood. 1999;94:976–983. [PubMed] [Google Scholar]
- Allain F, Vanpouille C, Carpentier M, Slomianny MC, Durieux S, Spik G. Interaction with glycosaminoglycans is required for cyclophilin B to trigger integrin-mediated adhesion of peripheral blood T lymphocytes to extracellular matrix. Proc Natl Acad Sci USA. 2002;99:2714–2719. doi: 10.1073/pnas.052284899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- Bergsma DJ, Eder C, Gross M, Kersten H, Sylvester D, Appelbaum E, Cusimano D, Livi GP, McLaughlin MM, Kasyan K, et al. The cyclophilin multigene family of peptidyl-prolyl isomerases. Characterization of three separate human isoforms. J Biol Chem. 1991;266:23204–23214. [PubMed] [Google Scholar]
- Berthoux L, Sebastian S, Sokolskaja E, Luban J. Cyclophilin A is required for TRIM5{alpha}-mediated resistance to HIV-1 in Old World monkey cells. Proc Natl Acad Sci USA. 2005;102:14849–14853. doi: 10.1073/pnas.0505659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierer BE, Somers PK, Wandless TJ, Burakoff SJ, Schreiber SL. Probing immunosuppressant action with a nonnatural immunophilin ligand. Science. 1990;250:556–559. doi: 10.1126/science.1700475. [DOI] [PubMed] [Google Scholar]
- Bose S, Mathur M, Bates P, Joshi N, Banerjee AK. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J Gen Virol. 2003;84:1687–1699. doi: 10.1099/vir.0.19074-0. [DOI] [PubMed] [Google Scholar]
- Braaten D, Franke EK, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaten D, Luban J. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. Embo J. 2001;20:1300–1309. doi: 10.1093/emboj/20.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram RJ, Crabtree GR. Calcium signalling in T cells stimulated by a cyclophilin B-binding protein. Nature. 1994;371:355–358. doi: 10.1038/371355a0. [DOI] [PubMed] [Google Scholar]
- Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci USA. 2002;99:1899–1904. doi: 10.1073/pnas.042529199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cande C, Vahsen N, Kouranti I, Schmitt E, Daugas E, Spahr C, Luban J, Kroemer RT, Giordanetto F, Garrido C, et al. AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene. 2004;23:1514–1521. doi: 10.1038/sj.onc.1207279. [DOI] [PubMed] [Google Scholar]
- Castro AP, Carvalho TM, Moussatche N, Damaso CR. Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles. J Virol. 2003;77:9052–9068. doi: 10.1128/JVI.77.16.9052-9068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Mi L, Xu J, Yu J, Wang X, Jiang J, Xing J, Shang P, Qian A, Li Y, et al. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J Infect Dis. 2005;191:755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Vetter M, Chang GD, Liu S, Che D, Ding Y, Kim SS, Chang CH. Cyclophilin A functions as an endogenous inhibitor for membrane-bound guanylate cyclase-A. Hypertension. 2004;44:963–968. doi: 10.1161/01.HYP.0000145859.94894.23. [DOI] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Colgan J, Asmal M, Neagu M, Yu B, Schneidkraut J, Lee Y, Sokolskaja E, Andreotti A, Luban J. Cyclophilin A regulates TCR signal strength in CD4+ T cells via a proline-directed conformational switch in Itk. Immunity. 2004;21:189–201. doi: 10.1016/j.immuni.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Colgan J, Asmal M, Yu B, Luban J. Cyclophilin A-deficient mice are resistant to immunosuppression by cyclosporine. J Immunol. 2005;174:6030–6038. doi: 10.4049/jimmunol.174.10.6030. [DOI] [PubMed] [Google Scholar]
- Crompton M, Virji S, Ward JM. Cyclophilin-D binds strongly to complexes of the voltage-dependent anion channel and the adenine nucleotide translocase to form the permeability transition pore. Eur J Biochem. 1998;258:729–735. doi: 10.1046/j.1432-1327.1998.2580729.x. [DOI] [PubMed] [Google Scholar]
- Cullen BR. HIV-1 infection: fooling the gatekeeper. Nat Med. 2003;9:1112–1113. doi: 10.1038/nm0903-1112. [DOI] [PubMed] [Google Scholar]
- Damaso CR, Moussatche N. Inhibition of vaccinia virus replication by cyclosporin A analogues correlates with their affinity for cellular cyclophilins. J. Gen. Virol. 1998;79(Pt 2):339–346. doi: 10.1099/0022-1317-79-2-339. [DOI] [PubMed] [Google Scholar]
- Di Bisceglie AM, McHutchison J, Rice CM. New therapeutic strategies for hepatitis C. Hepatology. 2002;35:224–231. doi: 10.1053/jhep.2002.30531. [DOI] [PubMed] [Google Scholar]
- Duina AA, Chang HC, Marsh JA, Lindquist S, Gaber RF. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- Endrich MM, Gehring H. The V3 loop of human immunodeficiency virus type-1 envelope protein is a high-affinity ligand for immunophilins present in human blood. Eur J Biochem. 1998;252:441–446. doi: 10.1046/j.1432-1327.1998.2520441.x. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Rice CM, Goff SP. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci USA. 2004;101:13038–13043. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firpi RJ, Zhu H, Morelli G, Abdelmalek MF, Soldevila-Pico C, Machicao VI, Cabrera R, Reed AI, Liu C, Nelson DR. Cyclosporine suppresses hepatitis C virus in vitro and increases the chance of a sustained virological response after liver transplantation. Liver Transpl. 2006;12:51–57. doi: 10.1002/lt.20532. [DOI] [PubMed] [Google Scholar]
- Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid FX. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature. 1989;337:476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Friedman J, Weissman I. Two cytoplasmic candidates for immunophilin action are revealed by affinity for a new cyclophilin: one in the presence and one in the absence of CsA. Cell. 1991;66:799–806. doi: 10.1016/0092-8674(91)90123-g. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Klee CB, Bierer BE, Burakoff SJ. Calcineurin phosphatase activity in T lymphocytes is inhibited by FK 506 and cyclosporin A. Proc Natl Acad Sci USA. 1992;89:3686–3690. doi: 10.1073/pnas.89.9.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Watashi K, Murata T, Hishiki T, Hijikata M, Shimotohno K. Evaluation of the anti-hepatitis C virus effects of cyclophilin inhibitors, cyclosporin A and NIM811. Biochem Biophys Res Commun. 2006;343:879–884. doi: 10.1016/j.bbrc.2006.03.059. [DOI] [PubMed] [Google Scholar]
- Halestrap AP. The mitochondrial permeability transition: its molecular mechanism and role in reperfusion injury. Biochem Soc Symp. 1999;66:181–203. doi: 10.1042/bss0660181. [DOI] [PubMed] [Google Scholar]
- Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- Harding MW, Galat A, Uehling DE, Schreiber SL. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J Virol. 2005;79:176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway MP, Bram RJ. Co-localization of calcium-modulating cyclophilin ligand with intracellular calcium pools. J Biol Chem. 1998;273:16346–16350. doi: 10.1074/jbc.273.26.16346. [DOI] [PubMed] [Google Scholar]
- Horowitz DS, Kobayashi R, Krainer AR. A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. Rna. 1997;3:1374–1387. [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Watashi K, Hishiki T, Goto K, Inoue D, Hijikata M, Wakita T, Kato N, Shimotohno K. Diverse effects of cyclosporine on hepatitis C virus strain replication. J Virol. 2006;80:4510–4520. doi: 10.1128/JVI.80.9.4510-4520.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang LK, Lee ZH, Kim HH, Hill JM, Kim JD, Kwon BS. A novel leucine-rich repeat protein (LRR-1): potential involvement in 4-1BB-mediated signal transduction. Mol Cells. 2001;12:304–312. [PubMed] [Google Scholar]
- Jin ZG, Melaragno MG, Liao DF, Yan C, Haendeler J, Suh YA, Lambeth JD, Berk BC. Cyclophilin A is a secreted growth factor induced by oxidative stress. Circ Res. 2000;87:789–796. doi: 10.1161/01.res.87.9.789. [DOI] [PubMed] [Google Scholar]
- Kieffer LJ, Thalhammer T, Handschumacher RE. Isolation and characterization of a 40-kDa cyclophilin-related protein. J Biol Chem. 1992;267:5503–5507. [PubMed] [Google Scholar]
- Kootstra NA, Munk C, Tonnu N, Landau NR, Verma IM. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc Natl Acad Sci USA. 2003;100:1298–1303. doi: 10.1073/pnas.0337541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummrei U, Bang R, Schmidtchen R, Brune K, Bang H. Cyclophilin-A is a zinc-dependent DNA binding protein in macrophages. FEBS Lett. 1995;371:47–51. doi: 10.1016/0014-5793(95)00815-q. [DOI] [PubMed] [Google Scholar]
- Liang TJ, Jeffers LJ, Reddy KR, De Medina M, Parker IT, Cheinquer H, Idrovo V, Rabassa A, Schiff ER. Viral pathogenesis of hepatocellular carcinoma in the United States. Hepatology. 1993;18:1326–1333. [PubMed] [Google Scholar]
- Liu J, Albers MW, Chen CM, Schreiber SL, Walsh CT. Cloning, expression, and purification of human cyclophilin in Escherichia coli and assessment of the catalytic role of cysteines by site-directed mutagenesis. Proc Natl Acad Sci USA. 1990;87:2304–2308. doi: 10.1073/pnas.87.6.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Albers MW, Wandless TJ, Luan S, Alberg DG, Belshaw PJ, Cohen P, MacKintosh C, Klee CB, Schreiber SL. Inhibition of T cell signaling by immunophilin-ligand complexes correlates with loss of calcineurin phosphatase activity. Biochemistry. 1992;31:3896–3901. doi: 10.1021/bi00131a002. [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- Luban J. Cyclophilin A, TRIM5 and resistance to HIV-1 infection. J Virol (in press) [Google Scholar]
- Luo C, Luo H, Zheng S, Gui C, Yue L, Yu C, Sun T, He P, Chen J, Shen J, et al. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem Biophys Res Commun. 2004;321:557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey PG, Perrino BA, Soderling TR, Rao A. NF-ATp, a T lymphocyte DNA-binding protein that is a target for calcineurin and immunosuppressive drugs. J Biol Chem. 1993;268:3747–3752. [PubMed] [Google Scholar]
- Mi H, Kops O, Zimmermann E, Jaschke A, Tropschug M. A nuclear RNA-binding cyclophilin in human T cells. FEBS Lett. 1996;398:201–205. doi: 10.1016/s0014-5793(96)01248-3. [DOI] [PubMed] [Google Scholar]
- Mlynar E, Bevec D, Billich A, Rosenwirth B, Steinkasserer A. The non-immunosuppressive cyclosporin A analogue SDZ NIM 811 inhibits cyclophilin A incorporation into virions and virus replication in human immunodeficiency virus type 1-infected primary and growth-arrested T cells. J. Gen. Virol. 1997;78(Pt 4):825–835. doi: 10.1099/0022-1317-78-4-825. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Sakamoto N, Enomoto N, Tanabe Y, Kanazawa N, Koyama T, Kurosaki M, Maekawa S, Yamashiro T, Chen CH, et al. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem Biophys Res Commun. 2004;313:42–47. doi: 10.1016/j.bbrc.2003.11.080. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- Nestel FP, Colwill K, Harper S, Pawson T, Anderson SK. RS cyclophilins: identification of an NK-TR1-related cyclophilin. Gene. 1996;180:151–155. doi: 10.1016/s0378-1119(96)00436-2. [DOI] [PubMed] [Google Scholar]
- Obata Y, Yamamoto K, Miyazaki M, Shimotohno K, Kohno S, Matsuyama T. Role of cyclophilin B in activation of interferon regulatory factor-3. J Biol Chem. 2005;280:18355–18360. doi: 10.1074/jbc.M501684200. [DOI] [PubMed] [Google Scholar]
- Ott DE, Coren LV, Johnson DG, Sowder RC, 2nd, Arthur LO, Henderson LE. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res Hum Retroviruses. 1995;11:1003–1006. doi: 10.1089/aid.1995.11.1003. [DOI] [PubMed] [Google Scholar]
- Owens-Grillo JK, Hoffmann K, Hutchison KA, Yem AW, Deibel MR, Jr, Handschumacher RE, Pratt WB. The cyclosporin A-binding immunophilin CyP-40 and the FK506-binding immunophilin hsp56 bind to a common site on hsp90 and exist in independent cytosolic heterocomplexes with the untransformed glucocorticoid receptor. J Biol Chem. 1995;270:20479–20484. doi: 10.1074/jbc.270.35.20479. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Fujiwara T, Kawai A, Shimizu F, Takami S, Okuno S, Takeda S, Shimada Y, Nagata M, Watanabe T, et al. Cloning, expression and chromosomal mapping of a novel cyclophilin-related gene (PPIL1) from human fetal brain. Cytogenet Cell Genet. 1996;72:242–245. doi: 10.1159/000134199. [DOI] [PubMed] [Google Scholar]
- Paeshuyse J, Kaul A, De Clercq E, Rosenwirth B, Dumont JM, Scalfaro P, Bartenschlager R, Neyts J. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43:761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- Poynard T, Yuen MF, Ratziu V, Lai CL. Viral hepatitis C. Lancet. 2003;362:2095–2100. doi: 10.1016/s0140-6736(03)15109-4. [DOI] [PubMed] [Google Scholar]
- Price ER, Zydowsky LD, Jin MJ, Baker CH, McKeon FD, Walsh CT. Human cyclophilin B: a second cyclophilin gene encodes a peptidyl-prolyl isomerase with a signal sequence. Proc Natl Acad Sci USA. 1991;88:1903–1907. doi: 10.1073/pnas.88.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarsky T, Zybarth G, Dubrovsky L, Yurchenko V, Tang H, Guo H, Toole B, Sherry B, Bukrinsky M. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc Natl Acad Sci USA. 2001;98:6360–6365. doi: 10.1073/pnas.111583198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak T, Carrello A, Mark PJ, Warner BJ, Simpson RJ, Moritz RL, House AK. The cyclophilin component of the unactivated estrogen receptor contains a tetratricopeptide repeat domain and shares identity with p59 (FKBP59) J Biol Chem. 1993;268:13187–13192. [PubMed] [Google Scholar]
- Rosen MK, Standaert RF, Galat A, Nakatsuka M, Schreiber SL. Inhibition of FKBP rotamase activity by immunosuppressant FK506: twisted amide surrogate. Science. 1990;248:863–866. doi: 10.1126/science.1693013. [DOI] [PubMed] [Google Scholar]
- Rosenwirth B, Billich A, Datema R, Donatsch P, Hammerschmid F, Harrison R, Hiestand P, Jaksche H, Mayer P, Peichl P, et al. Inhibition of human immunodeficiency virus type 1 replication by SDZ NIM 811, a nonimmunosuppressive cyclosporine analog. Antimicrob Agents Chemother. 1994;38:1763–1772. doi: 10.1128/aac.38.8.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycyzyn MA, Clevenger CV. The intranuclear prolactin/cyclophilin B complex as a transcriptional inducer. Proc Natl Acad Sci USA. 2002;99:6790–6795. doi: 10.1073/pnas.092160699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycyzyn MA, Reilly SC, O’Malley K, Clevenger CV. Role of cyclophilin B in prolactin signal transduction and nuclear retrotranslocation. Mol Endocrinol. 2000;14:1175–1186. doi: 10.1210/mend.14.8.0508. [DOI] [PubMed] [Google Scholar]
- Saphire AC, Bobardt MD, Gallay PA. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. Embo J. 1999;18:6771–6785. doi: 10.1093/emboj/18.23.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- Schreiber SL. Chemistry and biology of the immunophilins and their immunosuppressive ligands. Science. 1991;251:283–287. doi: 10.1126/science.1702904. [DOI] [PubMed] [Google Scholar]
- Seko Y, Fujimura T, Taka H, Mineki R, Murayama K, Nagai R. Hypoxia followed by reoxygenation induces secretion of cyclophilin A from cultured rat cardiac myocytes. Biochem Biophys Res Commun. 2004;317:162–168. doi: 10.1016/j.bbrc.2004.03.021. [DOI] [PubMed] [Google Scholar]
- Sherry B, Yarlett N, Strupp A, Cerami A. Identification of cyclophilin as a proinflammatory secretory product of lipopolysaccharide-activated macrophages. Proc Natl Acad Sci USA. 1992;89:3511–3515. doi: 10.1073/pnas.89.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolskaja E, Sayah DM, Luban J. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J Virol. 2004;78:12800–12808. doi: 10.1128/JVI.78.23.12800-12808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolskaja E, Luban J. Cyclophilin A, TRIM5, and innate immunity to HIV-1. Curr Opin Microbiol. 2006;9:404–408. doi: 10.1016/j.mib.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Steinkasserer A, Harrison R, Billich A, Hammerschmid F, Werner G, Wolff B, Peichl P, Palfi G, Schnitzel W, Mlynar E, et al. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus type 1 (HIV-1): interference with early and late events in HIV-1 replication. J Virol. 1995;69:814–824. doi: 10.1128/jvi.69.2.814-824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Syed F, Rycyzyn MA, Westgate L, Clevenger CV. A novel and functional interaction between cyclophilin A and prolactin receptor. Endocrine. 2003;20:83–90. doi: 10.1385/ENDO:20:1-2:83. [DOI] [PubMed] [Google Scholar]
- Takahashi N. Pharmacodynamics of ciclosporin A (cyclosporine) Clin Exp Nephrol. 1999;3:S16–26. [Google Scholar]
- Takahashi N, Hayano T, Suzuki M. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature. 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- Tanveer A, Virji S, Andreeva L, Totty NF, Hsuan JJ, Ward JM, Crompton M. Involvement of cyclophilin D in the activation of a mitochondrial pore by Ca2+ and oxidant stress. Eur J Biochem. 1996;238:166–172. doi: 10.1111/j.1432-1033.1996.0166q.x. [DOI] [PubMed] [Google Scholar]
- Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- Tran DD, Edgar CE, Heckman KL, Sutor SL, Huntoon CJ, van Deursen J, McKean DL, Bram RJ. CAML is a p56Lck-interacting protein that is required for thymocyte development. Immunity. 2005;23:139–152. doi: 10.1016/j.immuni.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Tu H, Gao L, Shi ST, Taylor DR, Yang T, Mircheff AK, Wen Y, Gorbalenya AE, Hwang SB, Lai MM. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology. 1999;263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- Waldmeier PC, Zimmermann K, Qian T, Tintelnot-Blomley M, Lemasters JJ. Cyclophilin D as a drug target. Curr Med Chem. 2003;10:1485–1506. doi: 10.2174/0929867033457160. [DOI] [PubMed] [Google Scholar]
- Wang BB, Hayenga KJ, Payan DG, Fisher JM. Identification of a nuclear-specific cyclophilin which interacts with the proteinase inhibitor eglin c. Biochem. J. 1996;314(Pt 1):313–319. doi: 10.1042/bj3140313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Gale M, Jr, Keller BC, Huang H, Brown MS, Goldstein JL, Ye J. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell. 2005;18:425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Watashi K, Hijikata M, Hosaka M, Yamaji M, Shimotohno K. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology. 2003;38:1282–1288. doi: 10.1053/jhep.2003.50449. [DOI] [PubMed] [Google Scholar]
- Watashi K, Ishii N, Hijikata M, Inoue D, Murata T, Miyanari Y, Shimotohno K. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol Cell. 2005;19:111–122. doi: 10.1016/j.molcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Woodfield K, Ruck A, Brdiczka D, Halestrap AP. Direct demonstration of a specific interaction between cyclophilin-D and the adenine nucleotide translocase confirms their role in the mitochondrial permeability transition. Biochem. J. 1998;336(Pt 2):287–290. doi: 10.1042/bj3360287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Leiva MC, Fischkoff SA, Handschumacher RE, Lyttle CR. Leukocyte chemotactic activity of cyclophilin. J Biol Chem. 1992;267:11968–11971. [PubMed] [Google Scholar]
- Yang WM, Inouye CJ, Seto E. Cyclophilin A and FKBP12 interact with YY1 and alter its transcriptional activity. J Biol Chem. 1995;270:15187–15193. doi: 10.1074/jbc.270.25.15187. [DOI] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- Yurchenko V, O’Connor M, Dai WW, Guo H, Toole B, Sherry B, Bukrinsky M. CD147 is a signaling receptor for cyclophilin B. Biochem Biophys Res Commun. 2001;288:786–788. doi: 10.1006/bbrc.2001.5847. [DOI] [PubMed] [Google Scholar]
- Yurchenko V, Zybarth G, O’Connor M, Dai WW, Franchin G, Hao T, Guo H, Hung HC, Toole B, Gallay P, et al. Active site residues of cyclophilin A are crucial for its signaling activity via CD147. J Biol Chem. 2002;277:22959–22965. doi: 10.1074/jbc.M201593200. [DOI] [PubMed] [Google Scholar]
- Zander K, Sherman MP, Tessmer U, Bruns K, Wray V, Prechtel AT, Schubert E, Henklein P, Luban J, Neidleman J, et al. Cyclophilin A interacts with HIV-1 Vpr and is required for its functional expression. J Biol Chem. 2003;278:43202–43213. doi: 10.1074/jbc.M305414200. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhou Z, Xu J, Zhao W, Wang W, Huang Y, Cheng C, Xu M, Xie Y, Mao Y. Molecular cloning, structure and expression of a novel nuclear RNA-binding cyclophilin-like gene (PPIL4) from human fetal brain. Cytogenet Cell Genet. 2001;95:43–47. doi: 10.1159/000057015. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Ying K, Dai J, Tang R, Wang W, Huang Y, Zhao W, Xie Y, Mao Y. Molecular cloning and characterization of a novel peptidylprolyl isomerase (cyclophilin)-like gene (PPIL3) from human fetal brain. Cytogenet Cell Genet. 2001;92:231–236. doi: 10.1159/000056909. [DOI] [PubMed] [Google Scholar]