Summary
The ER plays a fundamental role in storing cellular Ca2+, generating Ca2+ signals, and modulating Ca2+ in both the cytosol and mitochondria. Genetically encoded Ca2+ sensors can be explicitly targeted to the ER to directly define Ca2+ levels and monitor fluxes of Ca2+ within this organelle. In this study we use an ER-targeted Ca2+ sensor to define both the level and dynamics of ER Ca2+ in cells expressing mutant presenilin proteins. Growing evidence suggests the enigmatic presenilin-1 plays a role in regulating ER Ca2+. Presenilin-1 was initially identified in a screen for genetic causes of inherited familial Alzheimer's disease (fAD). The connection between presenilin-1, calcium regulation, and Alzheimer's disease may provide the key to understanding the long-observed, but poorly understood, link between Alzheimer's disease and Ca2+ dysregulation. In this study we examined seven fAD-causing mutations in presenilin-1 to define how they influence ER Ca2+ levels and dynamics. We observed that some, but not all, mutations in PS1 decrease the level of Ca2+ within the ER and this difference depends on the enzymatic activity of PS1. Two mutations tested altered the kinetics of Ca2+ release from the ER upon ATP stimulation, resulting in faster spiking. Combined, these results indicate that mutations in PS1 can alter the balance of Ca2+ in cells and have the potential to influence the nature of Ca2+ signals.
Introduction
Ca2+ is an essential and tightly regulated cellular second messenger, important for the maintenance of many cellular processes and functions. Ca2+ signals can be generated by influx across the plasma membrane or release of Ca2+ from the Endoplasmic Reticulum (ER), which serves as the storehouse and signaling hub of Ca2+. Recently, presenilin-1 (PS1) has emerged as a central player in the regulation of ER Ca2+1-3. An integral membrane protein predominantly localized to the ER, PS1 has been proposed to: form a Ca2+ leak channel in the ER 1; interact with and regulate the Sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) 2; modulate release of Ca2+ through the IP3 receptor (IP3R) 3; and interact with, change the expression levels of, and modify Ca2+ release through the Ryanodine Receptor (RyR) 4, 5.
PS1 was originally identified in a genetic screen for causative agents of familial Alzheimer's disease (fAD) 6. Since then, over 150 mutations in PS1 have been identified that give rise to fAD. The involvement of PS1 in Alzheimer's disease pathology has been primarily ascribed to altered processing of the amyloid precursor protein (APP) by gamma-secretase, a multi-protein complex containing PS1 7. Both sporadic and inherited cases of Alzheimer's disease are characterized by Ca2+ imbalance and growing evidence indicates that Ca2+ dysregulation is correlated with disease pathogenesis 8-10. To assess whether the Ca2+-regulatory function of PS1 is relevant to fAD, it is important to define how mutations in PS1 alter these functions. Though its exact role is unclear, mutations in PS1 have been suggested to cause both overloading and under-filling of ER Ca2+ stores, and to both increase and decrease Ca2+ release from the ER 1, 3, 11-15.
Due to the complexity of cellular Ca2+ regulation, live cell imaging of Ca2+ signaling events are greatly benefited by sensors targeted to specific subcellular compartments. However, many studies investigating the role of PS1 mutations on Ca2+ dyshomeostasis have inferred the impact on ER Ca2+ by measuring changes in cytosolic Ca2+. Such studies may be confounded by changes in cytosolic clearance mechanisms and discrepancies between ER Ca2+ load and Ca2+ release. Genetically encoded Ca2+ sensors, such as cameleons, can be specifically targeted to the ER, permitting investigators to monitor Ca2+ within the ER (i.e. ER Ca2+ load), directly observe the leak or release of Ca2+ from this organelle, and examine SERCA activity by measuring Ca2+ uptake. Despite these advantages, these sensors have remained underutilized tools in studies examining effects of disease on cellular Ca2+ homeostasis. In the present study, we apply one such sensor, D1ER 16, and examine a panel of fAD-associated mutations in PS1 to determine their effects on ER Ca2+ levels and dynamics. We found that mutations in PS1 indeed alter ER Ca2+, but not all mutations have the same effect. Alteration of ER Ca2+ levels by mutant PS1 could be reversed by addition of a gamma secretase inhibitor, suggesting that PS1 enzymatic activity impacts ER Ca2+ levels. In addition, because ER Ca2+ release can impact downstream signaling processes, we examined whether mutations in PS1 affected Ca2+ release from the ER upon stimulation with ATP. Two mutants tested led to an increase in the frequency of Ca2+ spikes, suggesting that mutations in PS1 affect the nature of ER Ca2+ release.
Results
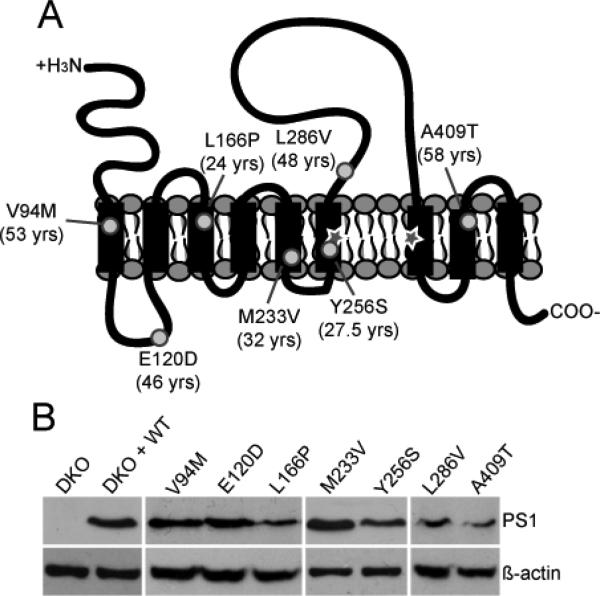
To examine whether mutations in PS1 affect Ca2+ in a universal manner we chose a range of mutations localized throughout PS1 that vary in their average age of Alzheimer's disease onset (Figure 1A). Mouse embryonic fibroblast (MEF) cells deficient in PS1 and PS2 (double knock out, DKO) were used as a model to eliminate potential effects of endogenous PS on the Ca2+ phenotype 17, 18. DKO MEF cells were reconstituted with WT or mutant PS1 by transient transfection and the presence of PS1 in cells was confirmed by GFP or mCherry fluorescence. Reconstitution of PS1 was corroborated by Western blot (Figure 1B).
Figure 1.
Verification of PS1 in cells. (A) Schematic of PS1 showing proposed membrane topology and relative locations of the mutations (circles) chosen for this study. Critical aspartates are denoted by a star. Average age at onset is indicated in parentheses. (B) Western blot showing relative expression levels of the PS1 variants upon transient transfection into DKO cells.
Mutations in PS1 have differential effects on ER Ca2+ store levels
The amount of Ca2+ within the ER is a balance of the ER buffering capacity, activity of the SERCA pump, release through channels and a passive “leak” of Ca2+ out of the ER. For this study, steady state levels of ER Ca2+ were determined by measuring the resting FRET ratio (R) using the D1ER cameleon sensor. Subsequent inhibition of SERCA with thapsigargin led to a decrease in R as Ca2+ slowly leaks out of the ER (Figure 2A), enabling us to determine relative ER Ca2+ levels. Under resting conditions, DKO cells exhibited a higher normalized ratio (ΔR), indicative of higher [Ca2+]ER when compared to WT and DKO cells reconstituted with WT PS1 (referred to as DKO + WT, Figure 2B). This is consistent with previous studies using these MEF cells in which the low affinity Ca2+ sensor MagFura-2 was used to measure [Ca2+]ER 1, 12.
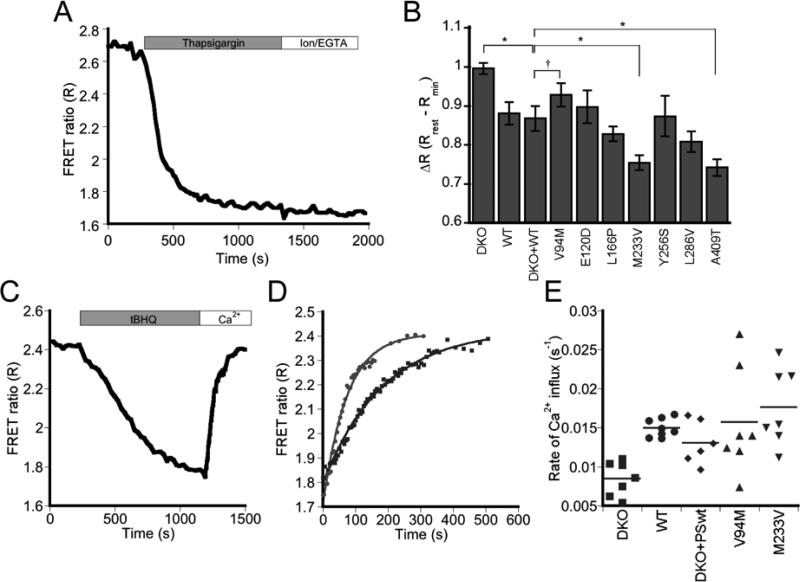
Figure 2.
Impact of PS1 on Ca2+ within the ER. (A) Representative experiment measuring ER Ca2+ load using the D1ER Ca2+ sensor. MEF cells transfected with the PS1 mutants were treated with thapsigargin in the absence of extracellular Ca2+ before calibrating the sensor with EGTA and ionomycin to obtain Rmin. (B) Bar graph showing ΔR at rest in the ER. Error bars indicate SEM. Asterisk: P < 0.05, ANOVA with Student-Newman-Keuls post-hoc test. Dagger: V94M appears to have more Ca2+ in the ER compared to control, though the difference is not statistically significant.WT: n = 17 (5 exps); DKO: n = 24 (9 exps); DKO + WT: n = 8 (6 exps), E120D: n = 8 (6 exps); V94M: n = 11 (7 exps); L166Pn = 13 (6 exps); M233V: n = 9 (4 exps); Y256S: n = 7 (3 exps); L286V: n = 13 (5 exps); A409T: n = 9 (5 exps). (C) Representative experiment examining the refilling of ER Ca2+ stores using D1ER. Cells were treated with tBHQ before adding excess Ca2+ to reload the ER. (D) Rates of ER refilling in WT (gray circles) and DKO (black squares). Data are fit to an exponential rise, A(t) = A0(1 – e -kt). (E) Plot of Ca2+ influx rate constants (k). Only DKO shows significant difference from WT (P < 0.015, ANOVA with Student-Newman-Keuls post-hoc test). Each point represents the k from an individual experiment and the horizontal line marks the mean k value for each data set. WT: n = 8 from 3 exps; DKO: n = 7 from 5 exps; DKO + WT: n = 6 from 4 exps; V94M: n = 7 from 4 exps; M233V: n = 7 from 3 exps.
Figure 2B also presents results for the PS1 variants examined in this study, illustrating that different mutants have differential effects on ER Ca2+ stores. WT cells consistently yielded a Ca2+ phenotype that was the same as DKO + WT. For statistical analysis, all mutant PS1 were compared to DKO + WT, allowing direct comparison of the impact of a specific mutation on Ca2+ within the same genetic background. Compared to DKO + WT, only M233V and A409T showed a statistically significant decrease in [Ca2+]ER (P < 0.05, ANOVA). V94M appeared to have a slightly increased Ca2+ level compared to reconstituted cells; however, this difference was not statistically significant. Our data support a role for PS1 in regulation of ER Ca2+ levels, though it is clear that not all mutations in PS1 affect the Ca2+ level within the ER.
Green, et al., recently proposed that PS1 regulates SERCA pump activity 2. Since SERCA is responsible for pumping Ca2+ into the ER, an over- or under-active SERCA could result in over- or under-filling of the ER store. To examine SERCA pump activity, cells were treated with the reversible SERCA inhibitor tBHQ leading to depletion of ER Ca2+. Once ER Ca2+ levels stabilized, tBHQ was washed out and Ca2+ added back, causing an influx of Ca2+ into internal stores through the SERCA pump. D1ER enabled us to directly monitor changes in [Ca2+]ER as shown in Figure 2C. The rate constant for re-filling was found by fitting the Ca2+ influx curve to a single exponential rise (Figure 2D), and was significantly higher in WT as compared to DKO cells (kWT = 15.0 × 10-3 ± 1.2 × 10-3 s-1, kDKO = 8.1 × 10-3 ± 2.2 × 10-3 s-1, P < 0.015, ANOVA), consistent with previous findings suggesting PS1 increases SERCA pump activity 2. To examine whether mutations in PS1 affect this regulatory function, we focused on PS1 variants that led to a detectable change in ER Ca2+ levels. However, we found that none of the mutants tested had an effect on Ca2+ influx through SERCA (Figure 2E), indicating that although PS1 impacts SERCA pump activity, the mutations in PS1 examined here do not appear to affect this function. Interestingly, the differences in the ER Ca2+ load can not be explained by SERCA activity alone, as cells containing WT PS1 had greater SERCA activity, but overall lower Ca2+ when compared to DKO.
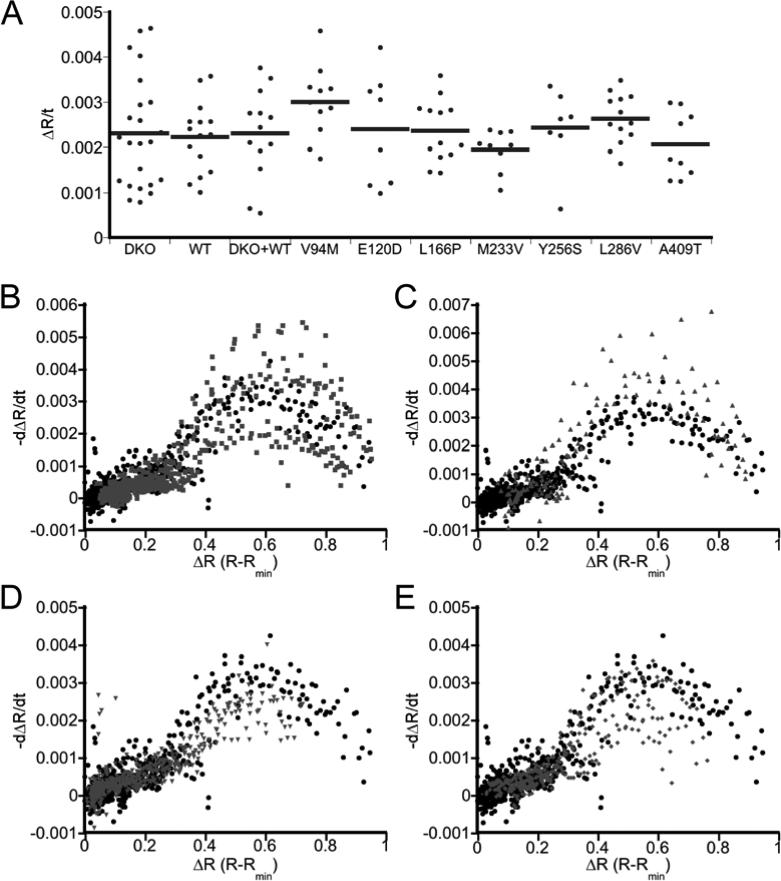
In addition to regulating ER Ca2+ stores, previous studies have suggested PS1 forms a leak channel in the ER 1 or contributes to the rate at which Ca2+ leaks out of the ER 3. To examine the leak rate, the change in R within the ER was monitored upon inhibition of SERCA with thapsigargin. Figure 3 presents the initial rate of decay (linear fit to the first 150 s of the R curve upon thapsigargin treatment) and the rate of change as a function of the Ca2+ level (dΔR/dt). Because PS mutations differentially affect the level of Ca2+ in the ER (Figure 2B), we felt it was important to compare the leak rate as a function of Ca2+ to ensure that any potential differences in leak rate were not simply reflections of the different amount of Ca2+ at a given point in time. No difference in Ca2+ leak rate was observed between DKO and WT cells (Figure 3A and 3B); however, three of the mutants tested exhibited an altered leak rate compared to WT cells. V94M exhibited a greater leak for both the initial rate and the leak rate as a function of ER Ca2+ level, while M233V and A409T both exhibited a lower leak (Figure 3). These results indicate that mutations in PS1 can affect the rate at which Ca2+ leaks out of the ER, with some mutations causing an increase in the leak and some causing a decrease. However, it is clear that the ER Ca2+ load is not defined by the leak rate, but rather the altered leak rate may be a consequence of altered Ca2+ load as mutations that cause a decrease in the ER Ca2+ load yield a lower leak rate, and vice versa.
Figure 3.
Effect of PS1 on ER Ca2+ leak rates. (A) Initial Ca2+ leak rate, defined as the change in ratio per unit time (ΔR/t). Rates were determined from the first 150 s after treatment of cells with 4 μM thapsigargin using a linear fit. (B – E) Comparison of Ca2+ leak rate as a function of Ca2+ concentration, reported as the rate of change of the ratio (dΔR/dt) as a function of the ΔR for WT (black) versus DKO (B), V94M (C), M233V (D) and A409T (E) in gray.
ER levels are regulated by PS1 activity
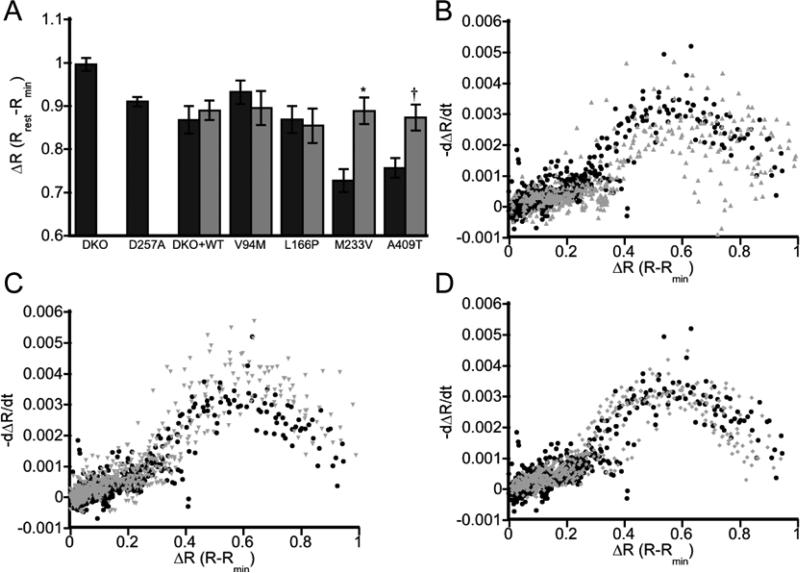
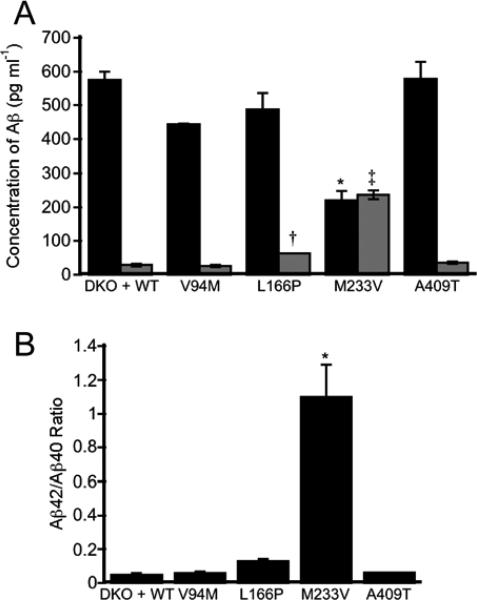
In addition to its Ca2+ regulatory functions, PS1 serves as the catalytic subunit of the multi-protein gamma-secretase complex, which cleaves a number of substrates in the cell, including APP. APP is a single pass transmembrane protein that is cleaved by gamma-secretase into two fragments: an intracellular domain and an extracellular peptide called amyloid beta (Aβ). The activity of our PS1 mutants within the gamma-secretase complex was verified using an APP-C99-mCherry probe to show cleavage of APP in cells (Supplementary Figure S1). As altered ER Ca2+ levels could not be explained by SERCA activity or the leak rate of Ca2+ out of the ER, we next examined whether the activity of PS1 as part of the gamma-secretase complex affected ER Ca2+ homeostasis by using the small molecule gamma-secretase inhibitor N-[N-(3,5-Difluorophenacetyl-L-alanyl)]-S-phenylglycine t-Butyl Ester (DAPT). Treatment with DAPT had no effect on [Ca2+]ER in DKO + WT, V94M, and L166P cells. However, inhibition of gamma-secretase activity resulted in an enhancement of ER Ca2+ levels for both M233V and A409T, raising them to DKO + WT levels (Figure 4A). Moreover the leak rates for all mutants examined were comparable to WT, consistent with leak rate being a consequence of the ER Ca2+ load (Figure 4B – D). To assess whether changes in Ca2+ could be attributed to the gamma-secretase mediated cleavage products of APP, the relative amounts of two cleavage products (Aβ40 and Aβ42) were determined by ELISA (Figure 5). While the observed changes in Aβ cleavage products are intriguing and may have implications for Alzheimer's disease progression (L166P and M233V have the highest Aβ42/40 ratios and the earliest ages of onset in 24 and 32 yrs, respectively), these changes do not correlate with changes in ER Ca2+. For example, M233V and A409T are the two mutants with reduced ER Ca2+ and inhibition of gamma secretase increases Ca2+ to WT levels. However these two mutants have very different Aβ profiles. Therefore while gamma secretase activity plays a role in influencing ER Ca2+ levels, this can not be easily ascribed to Aβ cleavage products. Overall our data reveal that there may be multiple mechanisms by which PS1 affects ER Ca2+ levels, one dependent on and another independent of gamma secretase activity. Comparison of DKO and DKO + WT reveals that PS1 lowers ER Ca2+ levels regardless of its enzymatic activity. This is confirmed by measurement of the ER Ca2+ load in DKO cells expressing a catalytically inactive PS1 mutant (D257A) which phenocopies the DAPT-inhibited DKO + WT cells. However AD-causing mutations in PS1 can lead to additional alterations in ER Ca2+ (i.e. a further decrease for M233V and A409T compared to WT PS1), and this additional perturbation is clearly dependent on gamma secretase activity as the difference could be reversed by DAPT inhibition.
Figure 4.
Gamma-secretase activity affects ER Ca2+ levels. (A) Bar graph depicting ΔR at rest in the ER for cells with (light gray) and without (dark gray) 24 h treatment with the gamma-secretase inhibitor DAPT. Error bars indicate SEM. Asterisk: P < 0.0005, unpaired t-test; Dagger: P < 0.005, unpaired t-test. DKO + WT: n = 6 (3 exps); V94M: n = 14 (2 exps); L166P: n = 13 (4 exps); M233V: n = 23 (4 exps); A409T: n = 16 (4 exps). (B – D) Comparison of the ER Ca2+ leak rate as a function of relative Ca2+ concentration. Leak rates are reported as a rate of change in the normalized ratio (ΔR) versus ΔR for V94M (B), M233V (C) and A409T (D). Graphs depict mutant leak rates (gray) compared to leaks rates in WT not treated with DAPT (black)
Figure 5.
Examination of gamma-secretase mediated cleavage of APP. (A) Relative amounts of Aβ40 (dark gray) and Aβ42 (light gray) in cells expressing a subset of the PS1 mutants. Both L166P and M233V led to an increase in Aβ42 levels. M233V also led to a significant decrease in Aβ40 levels. Asterisk: P < 0.005; Dagger: P < 0.05; Double dagger: P < 0.0001. (B) Relative ratios of Aβ42/40 for a subset of the PS1 mutants. Asterisk: P < 0.002. All statistics were calculated using an ANOVA with Student-Newman-Keuls post-hoc test. Error bars indicate SEM.
Mutations in PS1 affect the kinetics of ER Ca2+ release
As the central storehouse of Ca2+ the ER plays a central role in generation of Ca2+ signals. For example activation of P2Y G-protein coupled receptors by ATP leads to the production of IP3 and release of Ca2+ from the ER. Repetitive release leads to the generation of Ca2+ oscillations in the cytosol, the duration, amplitude and frequency of which affect downstream processes such as cell cycle regulation, gene transcription, and differentiation 19. A recent report indicates that PS1 interacts with the IP3R to modulate the open probability and hence release of Ca2+ from the ER 3. Moreover, an fAD-associated mutation (M146L) was shown to alter release of Ca2+ through the IP3R independent of Ca2+ load. For this study, we chose to examine the L166P, M233V, and A409T mutants as they showed a change in Aβ production and/or lowered [Ca2+]ER levels. Cells stimulated with 5 μM ATP exhibited two categories of response: immediate response with rapid Ca2+ spiking followed by slower irregular transients or a delayed response with no repetitive Ca2+ fluctuations. Investigation into the nature of ATP-induced Ca2+ release resulted in no observable difference in ATP sensitivity for L166P and A409T, but a decrease in sensitivity (as defined by decreased responsiveness to ATP) for M233V (Supplementary Figure S2A and B). A general decrease in the amplitude of Ca2+ release was observed for L166P and M233V, but not A409T, even though it contains less Ca2+ in the ER than WT (Supplementary Figure S2C – H).
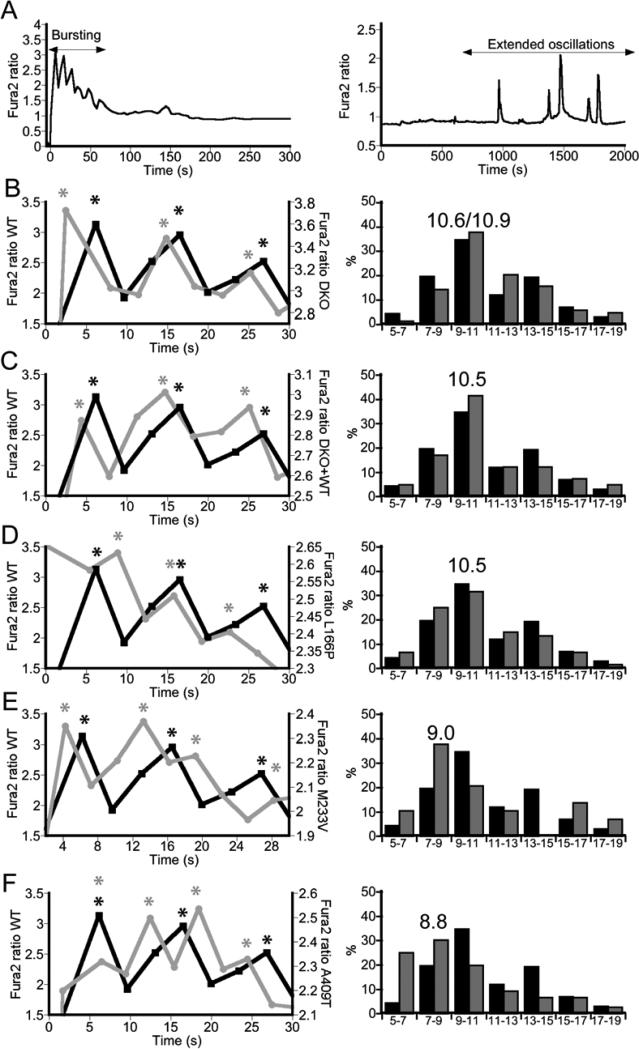
Given the versatility of Ca2+ signaling, cells can interpret changes in frequency of Ca2+ oscillations in order to differentially control multiple cellular processes 19. Because of this, changes in the kinetics of Ca2+ release through the IP3R could alter the fate of a cell, contributing to a diseased state. We therefore examined how the mutations studied here affected the frequency of the ATP-induced Ca2+ signals. Treatment of cells with 5 μM ATP lead to a rapid burst of Ca2+ spikes superimposed on a broad Ca2+ decay curve (Figure 6A left side). Longer imaging revealed irregular slower Ca2+ transients in a small subset of cells (Figure 6A right side). Attempts to elicit more regular extended oscillations in a larger fraction of cells using different stimuli (ATP, UTP, histamine, and carbachol) and lower doses of stimuli (i.e. 1 μM ATP vs. 5 μM ATP) were unsuccessful. Therefore we focused our analysis on the rapid initial bursting of Ca2+, as this bursting is still dependent on release from the IP3R and allows comparison of the kinetics of release between the different mutants. Figure 6 shows representative traces which reveal that cells expressing M233V and A409T, but not L166P, exhibited faster spiking than either WT or DKO cells. In an attempt to quantify the differences between cells expressing mutant PS1, we calculated the spike period (i.e. time between spikes from peak to peak) for every cell that exhibited repetitive spiking (see experimental section). These data are presented in the histograms (Figure 6, right side) which show that cells expressing M233V or A409T exhibit a greater percentage of faster spike periods (histogram shifted to lower time intervals). Given that many downstream effectors of Ca2+ signaling “read-out” both the amplitude and frequency of the Ca2+ signal, the fact that mutations in PS1 can alter the kinetics of ER Ca2+ release may play a significant role in eliciting Ca2+ dyshomeostasis.
Figure 6.
Effect of mutations in PS1 on kinetics of ER Ca2+ release. (A) Cells exhibited either a burst in Ca2+ from the ER (left) or, less frequently, irregular extended Ca2+ oscillations (right) in response to ATP. Bursting activity was examined to compare mutations in these studies. (B – F) Representative traces of Ca2+ bursts (left) compare differences between WT (black) and transfected DKO (gray) in initial release of Ca2+ from the ER. Peaks of each spike are indicated by an asterisk. Histograms (right) show the distribution of Ca2+ spike periods compared to WT (black; n = 274, 34 cells, 3 exps) for (B) DKO (n = 211, 25 cells, 3 exps), (C) DKO + WT (n = 41, 5 cells, 3 exps), (D) DKO + L166P (n = 60, 5 cells, 4 exps), (E) DKO + M233V (n = 29, 4cells, 2 exps) and (F) DKO + A409T (n = 76, 6 cells, 2 exps) after treatment with 5 μM ATP. The median period for spiking is given above the histogram.
Discussion
Calcium homeostasis is tightly regulated by dynamic interplay between channels, pumps, and transporters that control Ca2+ levels in the ER, mitochondria, and cytosol. Genetically targeted sensors provide a powerful way of monitoring Ca2+ directly within each of these locales. This feature may be especially important because cells appear to be adept at compensating for perturbations to Ca2+ homeostatic mechanisms. If a perturbation, such as expression of mutant presenilin-1, alters Ca2+ homeostasis the cell may compensate to minimize the perturbation, such as upregulating or downregulating channels or pumps. Indeed, in this study we found that the ER Ca2+ leak is altered by changes in ER Ca2+ levels such that lower ER Ca2+ levels lead to a decreased leak rate. This compensation is dynamic and could be reversed by elevating ER Ca2+ levels using DAPT. These results reinforce the importance of directly measuring [Ca2+]ER to investigate ER Ca2+ homeostasis, as measurement of cytosolic Ca2+ to interpret ER load may be confounded by compensatory changes in channels and pumps that exist to maintain cytosolic Ca2+.
Deletion of PS1 caused a significant increase in ER Ca2+, consistent with some, but not all studies which have measured Ca2+ directly within the ER. There are three methods for measuring Ca2+ within the ER: the D1ER sensor employed in this study, the low affinity Ca2+ indicator MagFura-2, and an ER-targeted aequorin-based sensor. It should be noted that a fourth method has been used to indirectly infer the ER Ca2+ load by measuring how much Ca2+ is released into the cytosol upon treatment with the ionophore ionomycin 1, 12. This approach suffers from two drawbacks: it measures release from all internal compartments, not just the ER, and it cannot account for potential differences in cytosolic clearance mechanisms that may arise if Ca2+ homeostasis is perturbed. Given that PS1 alters many aspects of Ca2+ homeostasis, we suspect direct measurement of ER Ca2+ is likely to be a more accurate measure of the ER Ca2+ load and for this, D1ER possesses some advantages. First, both MagFura-2 and aequorin require additional cellular manipulation for loading the sensors into internal compartments. MagFura-2 relies on permeabilization, ER store depletion, and addition of MgATP to activate store refilling, while aequorin requires ER store depletion, reconstitution of cells with coelenterazine, and store re-filling upon addition of extracellular Ca2+. Second, both of these approaches infer resting Ca2+ levels by monitoring store re-filling after depletion, which may be complicated by differences in SERCA pump activity. For example, in a study using MagFura-2, Cheung et al. found that untransfected DT40 cells filled to a higher level than cells transfected with PS1, consistent with our findings for DKO vs. WT cells 3. However, two studies using ER-targeted aequorin found that PS1 deficient cells filled to a lower level than cells containing PS1 13, 20. These experiments inadvertently rely on SERCA activity for measuring ER load, and one interpretation of the reported discrepancies is that different conditions for re-loading intracellular stores may promote differential re-filling of the ER.
By using the genetically encoded Ca2+ sensor D1ER, we were able to directly monitor ER Ca2+ levels and dynamics, including leak rates of Ca2+ out of the ER and uptake into the ER. We discovered that fAD-associated mutations in PS1 differentially affect levels of Ca2+ in the ER and leak from the ER. This helps put into context the numerous, but often conflicting studies that have examined individual mutations in PS1. Here we found one mutation that appeared to increase both ER Ca2+ and the leak rate (V94) and two that decreased ER Ca2+ and the leak rate (M233V and A409T). However, a number of mutations had no effect on ER Ca2+ levels, further suggesting that there is not a universal phenotype for perturbation of ER Ca2+ by mutant PS1. Unfortunately, the mechanism(s) by which PS1 mutations alter ER Ca2+ levels remain enigmatic. In particular, changes in the leak rate appeared to be a consequence rather than a cause of altered ER Ca2+. Likewise, PS1 increases SERCA activity, but leads to a decrease in ER Ca2+ load. Intriguingly, inhibition of the proteolytic activity of PS1 (as part of the gamma secretase complex) affected cells expressing mutant PS1 but not WT PS1 strongly suggesting there are multiple mechanisms by which PS1 can influence ER Ca2+. Combined, these factors (compensatory changes and the possibility of multiple mechanisms) make it difficult for us to define the molecular mechanism by which f-AD causing mutations in PS1 alter ER Ca2+, and thus these mechanism(s) remain elusive. On the other hand, our results do help provide a framework for understanding the sometimes conflicting studies on PS1 and fAD-causing mutations. In particular, they highlight that different mutations in PS1 give rise to different Ca2+ phenotypes and that many aspects of Ca2+ regulation may be altered in mutant PS1 cells, some of which are compensatory changes that result from PS1 expression.
The two mutations that lead to a significant decrease in ER Ca2+ (M233V and A409T) also gave rise to altered kinetics of Ca2+ release from the ER upon ATP stimulation, exhibiting an increase in the frequency of Ca2+ bursting. To our knowledge, M146L is the only other PS1 mutation examined for its ability to influence release kinetics. Cheung et al3 found that M146L lead to enhanced Ca2+ oscillation frequency and this was due to altered gating of the IP3R. Interestingly, this mutant also gave rise to a lower ER Ca2+ load. Our results are consistent with this finding, further supporting the notion that PS1 interacts with and modulates the IP3R. Given that release of Ca2+ from the ER results in signals that are decoded by the cell to control cellular function, an important next step will be to identify the consequences of these altered signals and what role, if any, these changes play in Alzheimer's disease. A growing body of evidence including the results presented here, indicate that in addition to modulating ER Ca2+ levels, fAD-causing mutations in PS1 contribute to an overall dysregulation of Ca2+, including alteration of fundamental Ca2+ signals.
In summary, our study demonstrates the power of an ER-targeted Ca2+ sensor by illustrating that the sensor can be used to directly monitor Ca2+ levels, Ca2+ leak from the ER, and uptake into the ER. Overall, we show that mutations in PS1 give rise to numerous and varied changes in Ca2+ which we hope will provide a framework for interpreting reports on ER Ca2+ in AD as well as other diseases.
Experimental
Cell culture and transfections
Wild type (WT) and presenilin-1/2- double knock out (DKO) mouse embryonic fibroblasts (MEFs) were obtained from Dr. Bart de Strooper, Katholieke Universiteit Leuven, Leuven, Netherlands 17, 18 and were cultured in DMEM supplemented with 10% (v/v) FBS and 1% (v/v) penicillin and streptomycin. Due to the propensity for MEFs to accumulate mutations over time, cells were only used through passage number 20. DKO cells were reconstituted with either mutant or WT PS1 by transient transfection using TransIT (Mirus), and the resulting Ca2+-phenotype was compared to WT MEF cells. Cells were imaged 24-48 h post-transfection.
Cloning and constructs
For PS1 constructs, the I-467 isomer of human PS1 (NM_000021) was purchased from OriGene Technologies, Inc. AD-causing mutations were generated using QuikChange® Site-Directed Mutagenesis (Stratagene). PS1 variants were cloned into pRex-IRES-GFP and pRex-IRES-mCherry vectors (Dr. Xuedong Liu, University of Colorado-Boulder) between BamHI and NotI sites. These vectors encode an internal ribosomal entry site between the PS1 and fluorescent protein gene, enabling the selection of PS1-expressing cells on the basis of fluorescence.
Western blotting
Cells transfected with mutant PS1 in the pRex-IRES-GFP vector were washed in ice-cold Phosphate Buffered Saline (PBS) before being scraped into 1 mL PBS and spun down at 3000 g for 5 minutes. To lyse cells, pellets were re-suspended in STEN buffer (50mM Tris-HCl, pH 7.6, 150mM NaCl, 2mM EDTA, 1% TritonX-100 and 0.2% NP-40, plus protease inhibitor cocktail) and incubated on ice for 30 minutes. Lysis suspensions were spun down at 21000 g for 20 minutes to remove cell debris. Total protein concentration was determined using the BCA Protein Assay kit (Pierce) prior to separation on a 4 - 20% Tris-HEPES-SDS polyacrylimide gel and transfer to a PVDF membrane. Membranes were blocked in Tris-buffered saline supplemented with 0.1% (v/v) Tween-20 (TBS-T) and 5% (w/v) milk for 1 h at room temperature. To probe membranes, we used the anti-PS1 antibody APS18 (3μg/mL; Novus Biologicals) and anti-β-actin antibody (1:5000; Sigma). Proteins were detected by incubating in a Rabbit anti-mouse HRP conjugated secondary antibody (Zymed). Although we attempted numerous times, we could not get the APS18 antibody to detect the endogenous mouse PS1 in MEF cells. Therefore we cannot directly compare the level of human PS1 in reconstituted cells, to endogenous levels of mouse PS1.
Instrumentation for fluorescence microscopy
For imaging experiments, cells were washed and placed in Hank's Balanced Salt Solution with HEPES (20mM HEPES, 1X HBSS (Gibco), and 2g/L D-glucose, pH 7.2) or Ca2+-free HHBSS (20mM HEPES, 1X HBSS without Ca2+, Mg2+, or sodium bicarbonate, 2g/L D-glucose, 490μM MgCl2, 450μM MgSO4, pH 7.2). Imaging experiments were conducted at room temperature (25 °C). Fluorescence imaging was performed on an Axiovert 200M wide-field microscope (Zeiss) equipped with a Lambda 10-3 filter changer (Sutter Instruments) and Cascade 512B camera (Photometrics) for rapid acquisition of ratio images. Images were acquired using METAFLUOR software (Universal Imaging). All experiments were performed using a 1.3 NA 40X objective (Zeiss). The filter combinations used were as follows: Fura-2: 340/26 (excitation), 380/10 (excitation), 535/40 (emission), 455 (dichroic); YFP FRET: 430/24 (excitation), 535/25 (emission), 455 (dichroic); CFP: 430/24 (excitation), 470/24 (emission), 455 (dichroic); YFP: 495/10 (excitation), 535/25 (emission), 515 (dichroic); GFP: 480/20 (excitation), 510/20 (emission), 495 (dichroic); mCherry: 577/20 (excitation), 630/60 (emission), 595 (dichroic).
Calcium imaging
MEF DKO cells reconstituted with PS1 (WT or mutant) were identified by mCherry or GFP fluorescence. In general we selected cells expressing a similar amount of mCherry (or GFP) fluorescence as this would indicate a similar amount of PS1 expression. Over the range of intensities, we did not see a correlation between expression level (as assessed by mCherry or GFP) and calcium phenotypes.
For Fura-2 studies, cells were incubated at room temperature with 5μM Fura-2-AM and 3μM Pluronic® F-127 (Invitrogen) for 45 minutes followed by a 15 minute incubation in 1 mL HHBSS to allow for cleavage of the AM-ester. Cells were placed in fresh HHBSS (1 mL) before imaging. Calcium release from the ER was induced by 5 μM ATP (Sigma). At the end of each experiment, Fura-2 was calibrated by adding 5 mM EGTA and 5 μM ionomycin to obtain an Rmin before adding 5 μM ionomycin and 10 mM Ca2+ to obtain an Rmax. Concentrations of Ca2+ were determined using the formula [Ca2+] = Kd[(Rmax-R)/(R-Rmin)]*Sf/Sb, where Kd is 220nM, R is the ratio of emission intensity at 535 nm upon excitation at 340 nm divided by the emission intensity at 535 nm upon excitation at 380 nm at each time point, Sf is the emission intensity upon excitation at 380nm in the Ca2+-depleted state, and Sb is the emission intensity upon excitation at 380nm in the maximum Ca2+ state 21. Rmax, Rmin, Sf, and Sb were determined individually for each cell. All images were background corrected prior to determining the ratio (R).
For studies using D1ER, cells were transfected 48 h prior to imaging. Cells were either treated with 4 μM thapsigargin or 3 μM 2,5-Di-(t-butyl)-1,4-hydroquinone (tBHQ) (Calbiochem) in Ca2+-free HHBSS. These concentrations were chosen to obtain robust release of Ca2+ from the ER. For SERCA activity experiments, cells were washed in Ca2+-free HHBSS to remove tBHQ before adding Ca2+. The D1ER sensor was calibrated in cells at the end of each experiment using 5 μM ionomycin and 5 mM EGTA to obtain an Rmin. The FRET ratio (R) is proportional to the amount of Ca2+ and is defined as the emission intensity in the FRET channel (CFP excitation, YFP emission) divided by the emission intensity in the CFP channel (CFP excitation, CFP emission). We attempted to find the maximum ratio of the sensor (Rmax) when saturated with Ca2+ using established methods 22; however, the cells did not survive the calibration procedure. Because Rmax is necessary to convert the FRET ratio into a [Ca2+], we instead report the ΔR (R- Rmin), which is proportional to [Ca2+] and allows comparison of the relative amount of Ca2+ in cells expressing the various PS1 mutants. We find this is more accurate than simply comparing R because it incorporates the calibration for Rmin in each individual cell. All intensities were background corrected before ratioing.
Analysis of calcium spiking
To quantify the period between individual Ca2+ spikes, the broad Ca2+ decay curves were fit to a single exponential, which was then subtracted from the response curve. The obtained residuals correspond to the oscillatory component of the signal (Supplementary Figure S3). The spiking period was then calculated using the peak finding routine in IGOR Pro (Wavemetrics Inc.). The number of spikes in each time period was normalized to the total number of spikes to obtain the percent of spikes that fell in each time period. The total number of spikes for each cell condition was as follows: DKO 211 spikes (25 cells from 3 exps); WT 274 spikes (34 cells from 3 exps); DKO + WT 41 spikes (5 cells from 3 exps); DKO + L166P 60 spikes (5 cells from 4 exps); DKO + M233V 29 spikes (4 cells from 2 exps); DKO + A409T 76 spikes (6 cells from 2 exps). Because images were acquired every 3 sec oscillations with a period faster than 6 sec could not be measured due to the Nyquist criterion.
Statistical Analysis
Statistical analysis on data was performed using Kaleidagraph 4.0 (Synergy Software). Data are represented as the mean ± SEM. Differences between the means were compared using an ANOVA with Student-Newman-Keuls post-hoc test to determine statistical significance (P < 0.05).
Supplementary Material
Supporting Information for:
Using a genetically targeted sensor to investigate the role of presenilin-1 in ER Ca2+ levels and dynamics
Janet E. McCombs 1, Emily A. Gibson 2, Amy E. Palmer 1*
Supplementary Methods
Supplementary Figure S1: Demonstration of gamma-secretase activity in MEF cells.
Supplementary Figure S2: The effect of PS1 on ATP-induced ER Ca2+ release.
Supplementary Figure S3: Analysis of calcium spiking data using IGOR Pro.
Supplementary Methods
Cloning and constructs
To confirm gamma-secretase activity in individual cells, a fluorescently-tagged APP substrate (termed C99-mCherry) was designed. C99-mCherry was generated by PCR amplification of the APP signal sequence (residues 1 – 21) and its post β-secretase cleaved C-terminus (residues 653 – 751) from Gene Pool™ cDNA Human Normal Adult Brain library (Invitrogen). The signal sequence was ligated between HindIII and KpnI into the multiple cloning site of pcDNA3 (Invitrogen). The C99-APP fragment was subsequently ligated into the same vector between KpnI and NotI. The mCherry fluorescent protein was ligated into the APP-pcDNA3 vector between NotI and XbaI in order to tag the C-terminus of C99-APP.
AβELISA
MEF cells were doubly transfected with full-length APP and PS1 mutants 72 hours prior to carrying out the assay and media was changed 18 hours post-transfection. Analysis was performed using the BetaMark x-40 and BetaMark x-42 ELISA Kits (Covance). Secreted Aβ protein in the media was concentrated using Amicon Ultra 3K centrifugal filter devices (Millipore). Samples diluted 1:2 in working incubation buffer were run in duplicate according to manufacturer recommended protocol.
Figure S1. Demonstration of gamma-secretase activity in MEF cells. (A) Schematic representation of the C99-mCherry probe consisting of post-β-secretase cleaved APP with the mCherry fluorophore on its C-terminus. Upon recognition by the gamma-secretase complex, PS cleaves the probe into its intracellular domain (AICD) and Aβ fragments. (B) The C99-mCherry probe in WT MEF cells displaying diffuse cytosolic fluroescence. (C) WT MEF cells treated with the gamma-secretase inhibitor DAPT and expressing the C99-mCherry probe displaying punctate fluorescence. (D) DKO MEF cells expressing the C99-mCherry probe. Fluorescence is comparable to WT + DAPT cells, as there is no PS and thus no cleavage of the probe in cells. Scale bar is 10μm.
Figure S2. The effect of PS1 on ATP-induced ER Ca2+ release. (A) Upon treatment with ATP, Ca2+ release from cells either occurred immediately (blue) or displayed a noticeable delay (red), with a comparatively smaller Ca2+ response. (B) Percent of responding cells that showed immediate (dark blue) or delayed (light blue) Ca2+ release. Cells showed similar responsiveness (~ 80%) except for DKO + M233V, which had decreased ATP sensitivity. WT: n = 42; DKO: n = 56; DKO + WT: 14; L166P: n = 8; M233V: n = 18; A409T: n = 12. (C – F) Representative oscillation curves for the amount of Ca2+ released in immediately responding cells for DKO (red), WT (blue) and DKO + WT (green); (D – F) Comparison of the amount of Ca2+ released upon stimulation with ATP for DKO + WT (green) versus L166P (purple; D), M233V (pink; E), and A409T (orange; F). (G) Maximum [Ca2+] peak heights upon addition of 5μM ATP. Asterisk: P < 0.05, ANOVA with Student-Newman-Keuls post-hoc test. WT: n = 27; DKO: n = 25; DKO + WT: n = 6; L166P: n = 3; M233V: n = 2; A409T: n = 4. (H) Amount of Ca2+ released from the ER upon ATP treatment. Data represent area under the oscillation curve. DKO cells released more Ca2+ upon stimulation with ATP than DKO + WT, consistent with levels of ER Ca2+. However, though not significant, L166P appeared to release less Ca2+ upon treatment with ATP though the ER concentration of Ca2+ was similar to WT. M233V also appeared to release less Ca2+ than WT, consistent with the lower level of ER Ca2+, though differences were not significant. Interestingly, the Ca2+ response for the A409T mutant was similar to WT despite this mutant having a lower ER Ca2+ load. Asterisk: P < 0.05, ANOVA with Student-Newman-Keuls post-hoc test. Error bars indicate SEM.
Figure S3. Analysis of calcium spiking data using IGOR Pro. (A) Representative curve (blue) fit to an exponential decay (red), A(t) = A0e –(t – t0)k. (B) The exponential decay fit from A is subtracted from the data curve to enhance the peaks. Peak values (depicted by red dots) are determined using the Peak AutoFind macro in IGOR Pro.
Acknowledgements
We would like to thank the following sources for financial support: University of Colorado Signaling and Cell Cycle Regulation Training Grant (NIH T32 GM08759), NIH GM084027 to A. E. P., and the University of Colorado.
Abbreviations
- APP
amyloid precursor protein
- fAD
familial Alzheimer's disease
- FRET
Fluorescence Resonance Energy Transfer
- R
FRET ratio
- IP3
Inositol-1,4,5-trisphosphate
- IP3R
Inositol-1,4,5-trisphosphate receptor
- PS1
Presenilin-1
- PS2
presenilin-2
- MEF
mouse embryonic fibroblast
- DKO
presenilin-1 and presenilin-2 double knock out
- PM
Plasma Membrane
- SERCA
Sarco-Endoplasmic Reticulum ATPase
Footnotes
Online supplemental materials
The following supplemental materials are available online: Supplementary Methods, Supplementary Figure S1: Demonstration of γ-secretase activity in MEF cells, Supplementary Figure S2: The effect of PS1 on ATP-induced ER Ca2+ release, and Supplementary Figure S3: Analysis of calcium spiking data using IGOR Pro.
References
- 1.Tu H, Nelson O, Bezprozvanny A, Wang Z, Lee SF, Hao YH, Serneels L, De Strooper B, Yu G, Bezprozvanny I. Cell. 2006;126:981–993. doi: 10.1016/j.cell.2006.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green KN, Demuro A, Akbari Y, Hitt BD, Smith IF, Parker I, LaFerla FM. J Cell Biol. 2008;181:1107–1116. doi: 10.1083/jcb.200706171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung KH, Shineman D, Muller M, Cardenas C, Mei L, Yang J, Tomita T, Iwatsubo T, Lee VM, Foskett JK. Neuron. 2008;58:871–883. doi: 10.1016/j.neuron.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith IF, Hitt B, Green KN, Oddo S, LaFerla FM. J Neurochem. 2005;94:1711–1718. doi: 10.1111/j.1471-4159.2005.03332.x. [DOI] [PubMed] [Google Scholar]
- 5.Chan SL, Mayne M, Holden CP, Geiger JD, Mattson MP. J Biol Chem. 2000;275:18195–18200. doi: 10.1074/jbc.M000040200. [DOI] [PubMed] [Google Scholar]
- 6.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature. 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 7.Duff K, Eckman C, Zehr C, Yu X, Prada CM, Perez-tur J, Hutton M, Buee L, Harigaya Y, Yager D, Morgan D, Gordon MN, Holcomb L, Refolo L, Zenk B, Hardy J, Younkin S. Nature. 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 8.Marx J. Science. 2007;318:384–385. doi: 10.1126/science.318.5849.384. [DOI] [PubMed] [Google Scholar]
- 9.LaFerla FM. Nat Rev Neurosci. 2002;3:862–872. doi: 10.1038/nrn960. [DOI] [PubMed] [Google Scholar]
- 10.Bezprozvanny I, Mattson MP. Trends Neurosci. 2008;31:454–463. doi: 10.1016/j.tins.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stutzmann GE, Caccamo A, LaFerla FM, Parker I. J Neurosci. 2004;24:508–513. doi: 10.1523/JNEUROSCI.4386-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson O, Tu H, Lei T, Bentahir M, de Strooper B, Bezprozvanny I. J Clin Invest. 2007;117:1230–1239. doi: 10.1172/JCI30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zatti G, Burgo A, Giacomello M, Barbiero L, Ghidoni R, Sinigaglia G, Florean C, Bagnoli S, Binetti G, Sorbi S, Pizzo P, Fasolato C. Cell Calcium. 2006;39:539–550. doi: 10.1016/j.ceca.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Leissring MA, Akbari Y, Fanger CM, Cahalan MD, Mattson MP, LaFerla FM. J Cell Biol. 2000;149:793–798. doi: 10.1083/jcb.149.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leissring MA, Paul BA, Parker I, Cotman CW, LaFerla FM. J Neurochem. 1999;72:1061–1068. doi: 10.1046/j.1471-4159.1999.0721061.x. [DOI] [PubMed] [Google Scholar]
- 16.Palmer AE, Jin C, Reed JC, Tsien RY. Proc Natl Acad Sci U S A. 2004;101:17404–17409. doi: 10.1073/pnas.0408030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herreman A, Hartmann D, Annaert W, Saftig P, Craessaerts K, Serneels L, Umans L, Schrijvers V, Checler F, Vanderstichele H, Baekelandt V, Dressel R, Cupers P, Huylebroeck D, Zwijsen A, Van Leuven F, De Strooper B. Proc Natl Acad Sci U S A. 1999;96:11872–11877. doi: 10.1073/pnas.96.21.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herreman A, Van Gassen G, Bentahir M, Nyabi O, Craessaerts K, Mueller U, Annaert W, De Strooper B. J Cell Sci. 2003;116:1127–1136. doi: 10.1242/jcs.00292. [DOI] [PubMed] [Google Scholar]
- 19.Berridge MJ, Bootman MD, Roderick HL. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 20.Kasri NN, Kocks SL, Verbert L, Hebert SS, Callewaert G, Parys JB, Missiaen L, De Smedt H. Cell Calcium. 2006;40:41–51. doi: 10.1016/j.ceca.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Grynkiewicz G, Poenie M, Tsien RY. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 22.Palmer AE, Tsien RY. Nat Protoc. 2006;1:1057–1065. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information for:
Using a genetically targeted sensor to investigate the role of presenilin-1 in ER Ca2+ levels and dynamics
Janet E. McCombs 1, Emily A. Gibson 2, Amy E. Palmer 1*
Supplementary Methods
Supplementary Figure S1: Demonstration of gamma-secretase activity in MEF cells.
Supplementary Figure S2: The effect of PS1 on ATP-induced ER Ca2+ release.
Supplementary Figure S3: Analysis of calcium spiking data using IGOR Pro.
Supplementary Methods
Cloning and constructs
To confirm gamma-secretase activity in individual cells, a fluorescently-tagged APP substrate (termed C99-mCherry) was designed. C99-mCherry was generated by PCR amplification of the APP signal sequence (residues 1 – 21) and its post β-secretase cleaved C-terminus (residues 653 – 751) from Gene Pool™ cDNA Human Normal Adult Brain library (Invitrogen). The signal sequence was ligated between HindIII and KpnI into the multiple cloning site of pcDNA3 (Invitrogen). The C99-APP fragment was subsequently ligated into the same vector between KpnI and NotI. The mCherry fluorescent protein was ligated into the APP-pcDNA3 vector between NotI and XbaI in order to tag the C-terminus of C99-APP.
AβELISA
MEF cells were doubly transfected with full-length APP and PS1 mutants 72 hours prior to carrying out the assay and media was changed 18 hours post-transfection. Analysis was performed using the BetaMark x-40 and BetaMark x-42 ELISA Kits (Covance). Secreted Aβ protein in the media was concentrated using Amicon Ultra 3K centrifugal filter devices (Millipore). Samples diluted 1:2 in working incubation buffer were run in duplicate according to manufacturer recommended protocol.
Figure S1. Demonstration of gamma-secretase activity in MEF cells. (A) Schematic representation of the C99-mCherry probe consisting of post-β-secretase cleaved APP with the mCherry fluorophore on its C-terminus. Upon recognition by the gamma-secretase complex, PS cleaves the probe into its intracellular domain (AICD) and Aβ fragments. (B) The C99-mCherry probe in WT MEF cells displaying diffuse cytosolic fluroescence. (C) WT MEF cells treated with the gamma-secretase inhibitor DAPT and expressing the C99-mCherry probe displaying punctate fluorescence. (D) DKO MEF cells expressing the C99-mCherry probe. Fluorescence is comparable to WT + DAPT cells, as there is no PS and thus no cleavage of the probe in cells. Scale bar is 10μm.
Figure S2. The effect of PS1 on ATP-induced ER Ca2+ release. (A) Upon treatment with ATP, Ca2+ release from cells either occurred immediately (blue) or displayed a noticeable delay (red), with a comparatively smaller Ca2+ response. (B) Percent of responding cells that showed immediate (dark blue) or delayed (light blue) Ca2+ release. Cells showed similar responsiveness (~ 80%) except for DKO + M233V, which had decreased ATP sensitivity. WT: n = 42; DKO: n = 56; DKO + WT: 14; L166P: n = 8; M233V: n = 18; A409T: n = 12. (C – F) Representative oscillation curves for the amount of Ca2+ released in immediately responding cells for DKO (red), WT (blue) and DKO + WT (green); (D – F) Comparison of the amount of Ca2+ released upon stimulation with ATP for DKO + WT (green) versus L166P (purple; D), M233V (pink; E), and A409T (orange; F). (G) Maximum [Ca2+] peak heights upon addition of 5μM ATP. Asterisk: P < 0.05, ANOVA with Student-Newman-Keuls post-hoc test. WT: n = 27; DKO: n = 25; DKO + WT: n = 6; L166P: n = 3; M233V: n = 2; A409T: n = 4. (H) Amount of Ca2+ released from the ER upon ATP treatment. Data represent area under the oscillation curve. DKO cells released more Ca2+ upon stimulation with ATP than DKO + WT, consistent with levels of ER Ca2+. However, though not significant, L166P appeared to release less Ca2+ upon treatment with ATP though the ER concentration of Ca2+ was similar to WT. M233V also appeared to release less Ca2+ than WT, consistent with the lower level of ER Ca2+, though differences were not significant. Interestingly, the Ca2+ response for the A409T mutant was similar to WT despite this mutant having a lower ER Ca2+ load. Asterisk: P < 0.05, ANOVA with Student-Newman-Keuls post-hoc test. Error bars indicate SEM.
Figure S3. Analysis of calcium spiking data using IGOR Pro. (A) Representative curve (blue) fit to an exponential decay (red), A(t) = A0e –(t – t0)k. (B) The exponential decay fit from A is subtracted from the data curve to enhance the peaks. Peak values (depicted by red dots) are determined using the Peak AutoFind macro in IGOR Pro.