Abstract
Successful reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) ushered in a new era of regenerative medicine. Human iPSCs provide powerful new approaches for disease modeling, drug testing, developmental studies, and therapeutic applications. Investigating iPSC behavior in vivo and the ultimate feasibility of cell transplantation therapy necessitates the development of novel imaging techniques to longitudinally monitor iPSC localization, proliferation, integration, and differentiation in living subjects. At this five year mark of initial iPSC discovery, we review the current status of imaging iPSCs which ranges from in vitro studies, where imaging was used to study the processes/mechanisms of cellular reprogramming, to in vivo imaging of the survival of transplanted cells. To date, most imaging studies of iPSCs have been based on optical techniques, which include fluorescence and bioluminescence imaging. Since each imaging technique has its advantages and limitations, a combination of multiple imaging modalities may provide complementary information. The ideal imaging approach for tracking iPSCs or their derivatives in patients requires the imaging tag to be non-toxic, biocompatible, and highly specific to reduce perturbation of these cells. In few other scenarios can “personalized medicine” be better illustrated than the use of individual patient-specific iPSCs. Much future effort will be required before this can become a reality and clinical routine, where imaging will play an indispensible role in many facets of iPSC-based research and therapies.
Keywords: Induced pluripotent stem cells (iPSCs), molecular imaging, regenerative medicine, cell tracking, bioluminescence imaging (BLI), fluorescence imaging, positron emission tomography (PET), teratoma
Introduction
In 2006, the pioneering report on the reprogramming of mouse somatic cells into induced pluripotent stem cells (iPSCs) opened new horizons for regenerative medicine [1]. It was demonstrated that iPSCs can be reprogrammed from mouse fibroblasts by introducing two transcription factors (i.e. Oct4 and Sox2) and two proto-oncogenes (i.e. c-Myc and Klf4). One year later, human iPSCs were also generated independently by two groups, one from Japan [2] and the other from the United States [3]. Subsequently, a wide variety of new methods have been used to generate iPSCs [4].
Human iPSCs create fascinating options with regard to disease modeling, drug testing, developmental studies, and therapeutic applications. However, many obstacles have to be overcome and more efficient protocols need to be established before iPSCs can enter clinical investigation. For example, c-Myc is an oncogene and its reactivation may give rise to tumor formation [5]. Therefore, human and murine iPSCs have been established from fibroblasts with only Oct4, Sox2, and Klf4 [6]. However, the efficiency is significantly lower than when c-Myc was used. Another major concern is that most early reports of direct reprogramming were achieved by forced expression of defined factors using multiple viral vectors [1-3], which could cause unpredictable genetic dysfunction by viral vector integrations as well as persistent transgene expression. To generate iPSCs free of integrated transgenes, new approaches have been investigated to introduce reprogramming factors using non-viral transfection of a single multiprotein expression vector [7], a piggyBac transposon/transposase system [8], viral vectors that do not integrate into the host genome [9], DNA vectors [10-11], small molecules [12], direct protein delivery [13], among others.
The biggest concern in the clinical implementation of iPSC transplantation therapy, similar to that based on human embryonic stem cells (ESCs), is the possibility of teratoma formation [14-15]. To better understand the in vivo behavior and true therapeutic power of iPSCs and/or iPSC-derived cells, an accurate and sensitive tool for monitoring these cells is needed. Investigating iPSC behavior in vivo and the ultimate feasibility of cell transplantation therapy necessitates the development of novel imaging techniques to longitudinally monitor iPSC localization, proliferation, integration, and differentiation in living subjects [16]. In addition, imaging techniques can be powerful tools for understanding the fundamental biology of iPSCs [17].
Strategies for imaging/tracking of cells
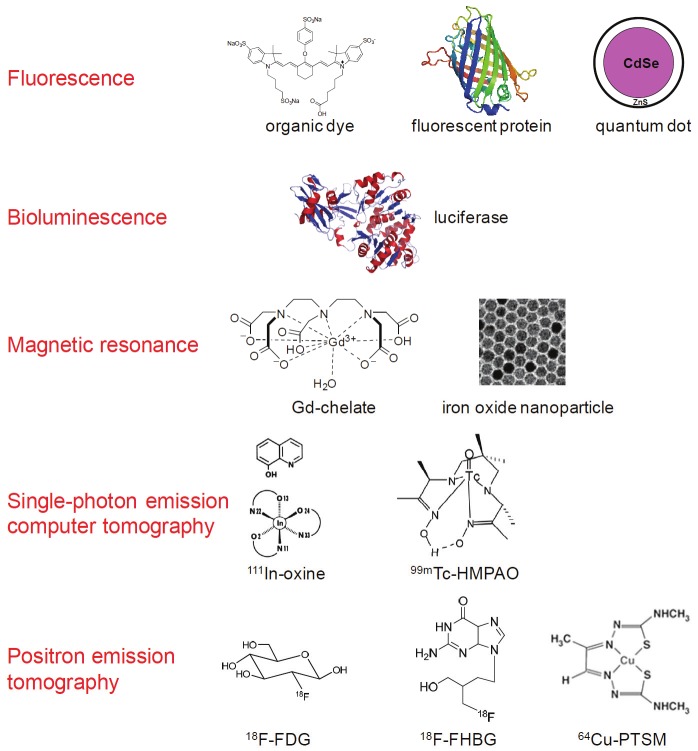
In general, there are two approaches for cell tracking: direct and indirect cell labeling [18]. Although the cell types can vary which include iPSCs, ESCs, immune cells, and cancer cells, the labeling strategies are essentially the same (Figure 1) [14,19]. Direct labeling of cells with image tags (e.g. magnetic nanoparticles, fluorescent dyes, 111In-oxine, etc.) is relatively easy, inexpensive, and well-established. In most cases, the safety profiles of direct cell labeling are quite acceptable. However, the disadvantage of these techniques is that the label itself is detected rather than the live cells of interest. The labels may leak out of the cells when the cells are alive or be taken up by other cells when the labeled cells die. Therefore, care must be taken when interpreting the experimental results and rigorous validation is certainly needed to obtain more robust and reliable data.
Figure 1.
Many imaging labels and techniques can be used to label stem cells and track them in vivo. Some involve direct labeling of the cells while others require genetic modification of the cells.
Indirect labeling typically uses a reporter gene approach which can allow for non-invasive visualization of live cells (which have been transfected with the reporter gene construct before transplantation), after administration of a reporter probe that can be detected by certain imaging techniques [20-21]. With reporter gene techniques, only live cells are detected thus they can provide more insights about the cell migration, differentiation, and proliferation in vivo. Since it has only been less than 5 years from the initial report of iPSCs [1], the current focus of iPSC research is mostly on the fundamental biology (e.g. how to generate iPSCs with high efficiency and good safety profile) instead of long term tracking of the survival, engraftment, proliferation, differentiation, and function of iPSCs after transplantation into animal models. In this review, we will summarize the current status on the imaging of iPSCs, which will provide guidance for future research endeavor in this extremely dynamic area.
To date, most imaging studies of iPSCs have been based on optical techniques, which include fluorescence and bioluminescence imaging (BLI). Optical imaging is less expensive and more convenient than other imaging modalities such as magnetic resonance imaging (MRI) and positron emission tomography (PET) [22-23]. Therefore, it can serve as an attractive alternative in preclinical imaging studies since light penetration in small animals is less of a concern than in humans. In small animal models, optical imaging (including BLI and fluorescence) is able to longitudinally detect transplanted cells sensitively and reliably which has significantly facilitated the understanding of the spatial-temporal kinetics of stem cell engraftment, proliferation, and teratoma formation in living subjects [24-25]. Generally speaking, fluorescence imaging is more useful in tagging individual cells or evaluation of the transfection efficiency in the cell population. On the other hand, BLI is very sensitive since it has virtually non-existent background signal, which is a major limitation of fluorescence imaging even in the near-infrared (700-900 nm) range [26-27].
Imaging iPSCs with fluorescence
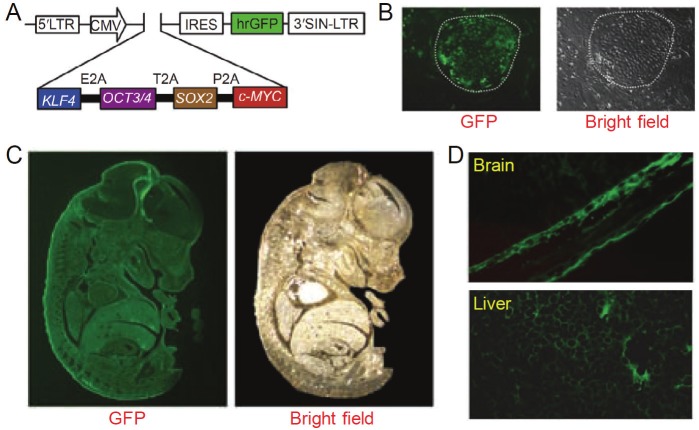
IPSCs were generated using a lentivirus reprogramming system, where the four defined factors (i.e. Klf4, Oct4, Sox2, and c-Myc, termed “KOSM”) were fused in-frame into a single open reading frame via self-cleaving 2A sequences [28]. A green fluorescent protein (GFP) marker was placed downstream of KOSM to track its expression in the resulting iPSCs. Generation of iPSCs was achieved quite efficiently, which exhibited normal karyotypes similar to mouse ESCs in terms of both morphology and gene expression. When these GFP-labeled iPSCs were injected into blastocysts, it was found that donor iPSCs contributed significantly to the development of chimeric embryos (Figure 2), which is the gold standard for pluripotency [5]. Detailed examination of the chimeric embryos revealed that GFP expression was evident in the brain and liver, as well as many other tissues, suggesting that the iPSCs generated by expression of the KOSM fusion gene possess full capacity to differentiate into the three embryonic germ layers in vivo.
Figure 2.
Imaging iPSCs with GFP. A. A lentiviral vector which includes the four defined transcription factors and GFP, separated by an internal ribosome entry site (IRES) sequence. B. Tracking transgene expression with GFP during reprogramming. C. Significant contribution of iPSCs in a chimeric embryo, where GFP expression from donor iPSCs is evident. D. Sections of a chimeric embryo showed contribution of iPSCs to the brain and liver. Adapted from [28].
Through serial live imaging of human fibroblasts undergoing reprogramming, distinct colonies that resemble ESCs morphologically yet differ in molecular phenotype and differentiation potential were identified [29]. The four standard transcription factors were introduced into fibroblasts with retroviruses carrying the GFP gene, which was used to monitor proviral silencing. Since FACS analysis disrupts individual colonies which precludes precise lineage tracing, serial live cell imaging of emerging colonies was carried out after staining in situ with antibodies against a number of pluripotency markers such as SSEA-4, TRA-1-60, CD13, NANOG, etc. The same area of cells underwent reprogramming was scanned by automated fluorescence microscopy and images were reviewed retrospectively for regions where colonies of bona fide human iPSCs had formed. It was determined that only one colony type represents iPSCs, whereas the others represent reprogramming intermediates.
Since the majority of reprogrammed cells do not become pluripotent [30-31], studying induced pluripotency based on average measurements across a large population of cells is inadequate. Therefore, high-resolution time-lapse imaging was employed to trace the reprogramming process from single mouse embryonic fibroblasts (MEFs) to pluripotency factor-positive colonies [32]. Continuous single-cell imaging of GFP- or yellow fluorescent protein (YFP)-labeled MEFs was achieved. Retrospective analysis revealed that successfully reprogrammed cells underwent a rapid shift in their proliferative rate that coincided with a reduction in cellular area, which occurred as early as the first cell division and exhibited similar kinetics in all cells that formed iPSC colonies, suggesting that the reprogramming process likely follows defined rather than stochastic steps. In a recent report, fluorescence microscopy with long-term single cell tracking was also employed to analyze the emergence and composition of early iPSC clusters [33]. Using engineered lentiviral vectors, where codon-optimized reprogramming factors are co-expressed by a strong retroviral promoter that is rapidly silenced in iPSCs, it was demonstrated that vector silencing typically occurs prior to or simultaneously with the induction of Oct4. Tracking of single cell-derived iPSC colonies supported the concept that stochastic epigenetic changes are necessary for reprogramming.
Recently, a microRNA-regulated lentiviral system was used to visualize and segregate differentiating neuronal cells in pluripotent cultures [34]. Efficient suppression of transgene expression in undifferentiated pluripotent cells was monitored with a GFP-expressing lentiviral vector regulated by microRNA-292, which is highly expressed in murine ESCs but not in murine neural stem cells (NSCs) [35]. With this strategy, progenies from murine/human ESCs and iPSCs were successfully tracked as they differentiated toward the neural lineage [34]. Furthermore, the presence of GFP also enabled purification of neuronal progenitors with FACS for molecular analysis and transplantation, which reduced tumor formation and increased survival of ESC-derived neuronal progenitors after transplantation. This study suggested that the microRNA-292 system may be broadly applicable to different pluripotent cell lines of both human and murine origin.
Although human iPSCs can potentially be an abundant source of blood cells, how to select the best iPSC clones from the large cohort that can be simultaneously established from an identical source is not clear [36]. In one study, it was shown that c-Myc reactivation after reprogramming could affect platelet generation from human iPSCs [37]. During differentiation, reduction of c-Myc expression after its initial reactivation in selected human iPSC clones was associated with more efficient generation of CD41a+CD42b+ platelets. In addition, intravital imaging was performed to visually investigate the function of iPSC-derived platelets during normal circulation and thrombus formation in the mesentery of living mice. FITC-dextran was intravenously injected to delineate blood vessels, which enabled the visualization of host blood cell dynamics, while human iPSC-derived platelets were labeled with a rhodamine-based dye before injection. It was found that these platelets were present in thrombi after laser-induced vessel wall injury, indicating that human iPSC-derived platelets are capable of mediating hemostasis and thrombosis in vivo (Figure 3).
Figure 3.
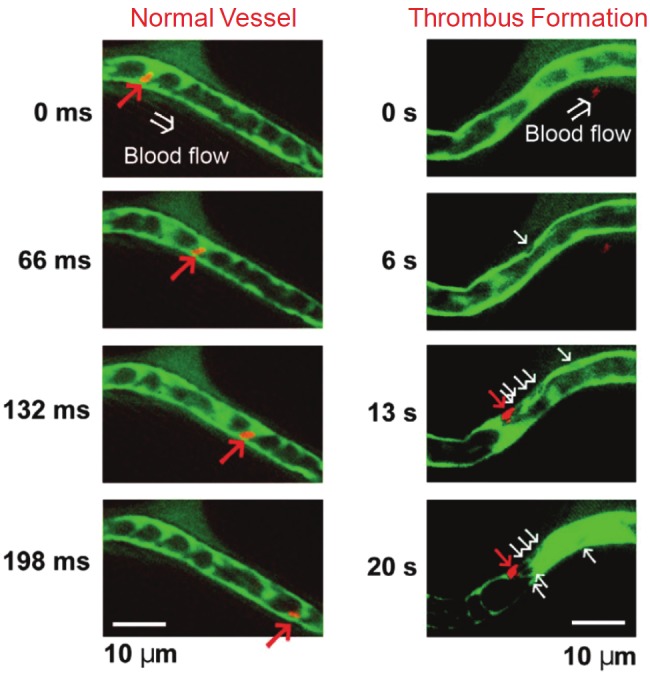
Human iPSC-derived platelets adhere to blood vessels during thrombus formation in vivo after intravenous injection. Left: normal vessels. Right: Representative sequential images of thrombus formation by human iPSC-derived platelets in a blood vessel. Red arrows indicate human iPSC-derived platelets and white arrows indicate host mouse platelets. Green: FITC-dextran; Red: fluorescent dye-labeled human iPSC-derived platelets. Adapted from [37].
Imaging iPSCs with bioluminescence
During ontogenesis, the molecular oscillations underlying the generation of circadian rhythmicity gradually evolve in mammals [38]. However, the developmental process of mammalian cellular circadian oscillator formation remains unknown. In one study, BLI was employed to monitor the clock gene expression [39].The circadian bioluminescence rhythm was detected in differentiated cells but not in murine ESCs or iPSCs, suggesting that an intrinsic program controls the formation of the circadian oscillator during the differentiation process. This study may shed new light on the future design of new strategies to understand the key mechanisms responsible for the organization of the molecular oscillator in mammals.
Similar to ESCs, iPSCs are attractive sources for tissue regeneration and transplantation therapies because they can differentiate into virtually any cell types in the adult body [40-41]. However, a major obstacle facing the engraftment of ESCs or iPSCs is transplant rejection by the immune system. Recently, it was demonstrated that immunosuppressive therapy can promote engraftment of a variety of pluripotent cell types, including iPSCs [42]. For long term in vivo tacking, both murine and human iPSCs were transduced with a Fluc-GFP (where Fluc denotes firefly luciferase) double-fusion construct by lentiviral techniques. After injection of iPSCs into the gastrocnemius muscle of recipient mice, survival of the transplanted cells was longitudinally monitored via BLI, upon intraperitoneal injection of D-luciferin.
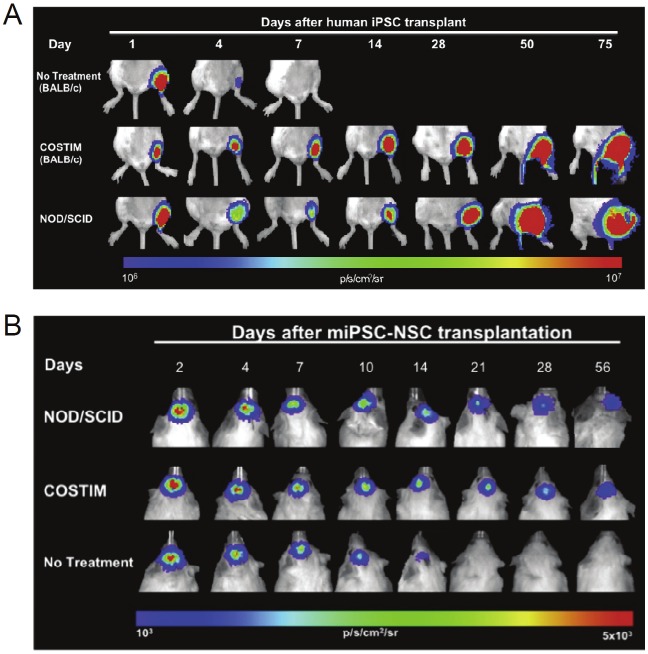
Survival of human iPSCs in immunocompetent mice was significantly lower than mice treated with three costimulatory receptor-blocking agents: cytotoxic T-lymphocyte-associated antigen 4-Ig, anti-CD40 ligand, and anti-lymphocyte function-associated antigen 1 [42]. The BLI signal dropped to background levels in untreated animals within 7 days after transplantation, whereas engraftment with steadily increasing BLI signal and teratoma formation were observed in costimulatory blockade-treated animals (Figure 4). Since iPSC-based therapy will likely utilize a differentiated rather than undifferentiated cell population, mouse iPSC-derived NSCs were generated and their survival was also investigated in untreated and costimulatory blockade-treated allogeneic recipients. Similar to iPSCs, survival of iPSC-derived NSCs was significantly limited in untreated compared to costimulatory blockade-treated mice. Taken together, this study demonstrated that a short course of costimulatory blockade treatment is sufficient to induce engraftment of iPSCs as well as their differentiated derivates. It was suggested that costimulatory blockade permits transplanted cell engraftment by decreasing the expression of pro-inflammatory cytokines and polarization of naive T cells toward a type I phenotype, increasing the establishment of a proapoptotic phenotype, and inducing clonal anergy.
Figure 4.
Short-term immunosuppression promotes engraftment of iPSCs and iPSC-derived cells. A. Serial bioluminescence images representing the survival of human iPSCs transplanted into the gastrocnemius muscle of immunodeficient (i.e. NOD/SCID) and immunocompetent (i.e. BALB/c) mice receiving costimulatory blockade (COSTIM) or no treatment. B. Serial bioluminescence images of murine iPSC derived neural stem cells (miPSC-NSCs) transplanted into the subcortical area of the brain in immunodeficient and immunocompetent mice. Adapted from [42].
Dual-modality imaging of iPSCs
Each imaging technique has its advantages and limitations. Combination of multiple imaging modalities may provide complementary information than a single modality alone [43-44]. Recently, various transcription factor genes and packaging plasmids were co-transfected into different cells using dendrimer-modified magnetic nanoparticles (dMNPs) as the delivery system [45]. It was found that dMNPs could efficiently deliver all vectors into certain cells. Specific surface markers of ESCs such as SSEA-3/4 and Tra-1-60/81 were positive in the resulting iPSCs, which formed terotomas when implanted into immunodeficient mice. The prepared iPSCs were subsequently labeled with fluorescent magnetic nanoparticles (FMNPs), where the fluorescence signal of labeled iPSCs was examined under microscopy while the magnetic property enabled detection by MRI. Such dual-modality labeling of iPSCs laid the foundation for isolating, in vivo imaging, and tracking of iPSCs in the future.
Conclusion and future perspectives
Each imaging modality has its advantages and disadvantages in terms of sensitivity, tissue penetration, spatial resolution, and clinical potential [22]. Optical imaging is mostly applicable to preclinical studies where light penetration is less of an issue than in patients. BLI cannot be used in human studies while tracking of labeled cells with MRI and/or PET may potentially be performed in patients. Combination of various imaging modalities can give complementary information. As a matter of fact, many of the reporter gene-based studies of pluripotent stem cells (e.g. ESCs) incorporated multiple reporter genes [24]. For example, fluorescent genes (e.g. GFP) can facilitate cell sorting, BLI (with luciferases) can enable in vivo long term monitoring of these cells in a quantitative manner in small animal models, and PET can allow for more clinically relevant, highly sensitive detection of the injected cells and/or the daughter cells. Future development and validation of various iPSC labeling/tracking techniques will further strengthen the arsenal for iPSC-based therapy of various diseases.
Safety of cell labeling is always a major concern in clinical studies since introduction of foreign substances (e.g. image labels or genes) may cause unpredictable alterations in cells. Based on the available literature data, labeling of stem cells with magnetic nanoparticles appears to be safe and is in active clinical development [46-47]. For indirect cell labeling, one of the most intensively studied reporter genes, HSV1-tk, is also a suicide gene which adds an extra layer of control to ensure safety [48]. The ideal imaging approach for tracking iPSCs or their derivatives in patients requires the imaging tag(s) to be non-toxic, biocompatible, and highly specific to reduce perturbation to these cells. Much future effort will be required before this can become a reality and clinical routine.
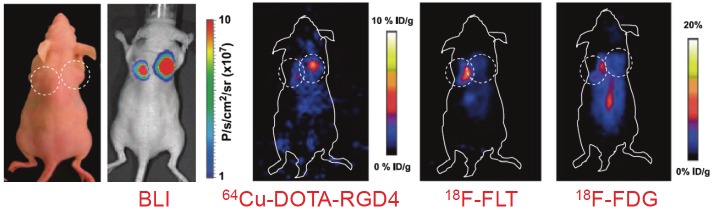
IPSC-based therapy has tremendous therapeutic potential. However, numerous questions still remain unanswered. Non-invasive imaging techniques have proven to be of great value in preclinical and clinical studies for tracking transplanted stem cells, and will continue to guide the development of future cell-based therapies. With non-invasive imaging techniques, we will eventually be able to determine which cell type is preferable for a given disease (e.g. which cell types to use for iPSC generation and which cell types to transplant) as well as choose the right delivery methods of the cells (e.g. intravenous, intracoronary, or local injection). Detection and correction of iPSC misbehavior (e.g. teratoma formation) is also an important task for imaging. For example, studies have shown that two of the most widely used PET tracers in the clinic, 2-deoxy-2-18F-fluoro-D-glucose (18F-FDG, which images glucose metabolism [49]) and 3'-deoxy-3'-18F-fluorothymidine (18F-FLT, which detects cell proliferation [50]), failed to detect human ESC-derived teratomas since the growth rate of teratomas is quite slow [51]. However, another tracer, 64Cu-DOTA-RGD4 which binds to integrin αvβ3 [52-53], enabled non-invasive visualization of the teratomas with PET (Figure 5). This and other studies suggested that imaging integrin αvβ3 expression, instead of imaging the metabolic activity and/or cellular proliferation, may have potential clinical applicability in monitoring the tumorigenicity after stem cell transplantation [51,54].
Figure 5.
Bioluminescence imaging (BLI) and positron emission tomography of human ESC-derived teratoma. The photograph and BLI shows teratoma formation by human ESCs at the right flank and tumor formation by a control cell line at the left flank of mice. Specific and prominent uptake of 64Cu-DOTA-RGD4 was observed in the teratoma (high integrin αvβ3 expression) but not in the control tumor (low integrin αvβ3 expression). In contrast, the teratoma had significantly lower uptake of 18F-FDG and 18F-FLT than the control tumor. Adapted from [51].
The requirement for iPSC tracking techniques depends on the clinical scenario. In some cases, only short term tracking is needed while in other cases, long term survival and proliferation would also need to be monitored. For example, transplantation of iPSCs or their derivatives in heart diseases needs to be targeted to the ischemic but not viable myocardium, if the therapy is aimed at improving vascularization. Effective treatment of many diseases may benefit from repeat dosing, where non-invasive imaging can inform clinical decisions regarding the need for repeat dosing and direct where repeat doses are needed. MRI, radionuclide-based imaging techniques, and reporter gene-based approaches will each have their own niches in imaging of iPSCs. Rather than identifying and optimizing one technique applicable for all clinical scenarios, it is probably more appropriate to optimize how a certain imaging technique/modality can be best used to serve the purpose in a specific situation. The continued evolvement of non-invasive imaging techniques will undoubtedly contribute to significant advances in understanding iPSC biology and mechanisms of action. The various imaging modalities, complementary rather than competitive, have the same ultimate goal: personalized medicine for patients. In few other scenarios can such “personalized medicine” be better illustrated than the use of patient-specific iPSCs.
Acknowledgements
The authors acknowledge financial support from the University of Wisconsin Carbone Cancer Center, NCRR 1UL1RR025011, NIH R01 HL08416 (TJK), NIH U01 HL099773 (TJK), and a DOD PCRP IDEA Award.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Chun YS, Chaudhari P, Jang YY. Applications of patient-specific induced pluripotent stem cells; focused on disease modeling, drug screening and therapeutic potentials for liver disease. Int J Biol Sci. 2010;6:796–805. doi: 10.7150/ijbs.6.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 7.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Scholer HR, Duan L, Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong H, Yang Y, Zhang Y, Cai W. Noninvasive imaging of human embryonic stem cells. Curr Pharm Biotechnol. 2010;11:685–692. doi: 10.2174/138920110792246500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fong CY, Gauthaman K, Bongso A. Teratomas from pluripotent stem cells: A clinical hurdle. J Cell Biochem. 2010;111:769–781. doi: 10.1002/jcb.22775. [DOI] [PubMed] [Google Scholar]
- 16.Sun N, Longaker MT, Wu JC. Human iPS cell-based therapy: considerations before clinical applications. Cell Cycle. 2010;9:880–885. doi: 10.4161/cc.9.5.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kooreman NG, Wu JC. Tumorigenicity of pluripotent stem cells: biological insights from molecular imaging. J R Soc Interface. 2010;7(Suppl 6):S753–763. doi: 10.1098/rsif.2010.0353.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ponomarev V. Nuclear imaging of cancer cell therapies. J Nucl Med. 2009;50:1013–1016. doi: 10.2967/jnumed.109.064055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong H, Yang Y, Zhang Y, Cai W. Noninvasive cell tracking in cancer and cancer therapy. Curr Top Med Chem. 2010;10:1237–1248. doi: 10.2174/156802610791384234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang JH, Chung JK. Molecular-genetic imaging based on reporter gene expression. J Nucl Med. 2008;49(Suppl 2):164S–179S. doi: 10.2967/jnumed.107.045955. [DOI] [PubMed] [Google Scholar]
- 21.Serganova I, Mayer-Kukuck P, Huang R, Blasberg R. Molecular imaging: reporter gene imaging. Handb Exp Pharmacol. 2008:167–223. doi: 10.1007/978-3-540-77496-9_8. [DOI] [PubMed] [Google Scholar]
- 22.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 23.Cai W, Niu G, Chen X. Multimodality imaging of the HER-kinase axis in cancer. Eur J Nucl Med Mol Imaging. 2008;35:186–208. doi: 10.1007/s00259-007-0560-9. [DOI] [PubMed] [Google Scholar]
- 24.Cao F, Lin S, Xie X, Ray P, Patel M, Zhang X, Drukker M, Dylla SJ, Connolly AJ, Chen X, Weissman IL, Gambhir SS, Wu JC. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–1014. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson K, Yu J, Lee A, Wu JC. In vitro and in vivo bioluminescence reporter gene imaging of human embryonic stem cells. J Vis Exp. 2008 doi: 10.3791/740. ID=740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dothager RS, Flentie K, Moss B, Pan MH, Kesarwala A, Piwnica-Worms D. Advances in bioluminescence imaging of live animal models. Curr Opin Biotechnol. 2009;20:45–53. doi: 10.1016/j.copbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai W, Hsu AR, Li ZB, Chen X. Are quantum dots ready for in vivo imaging in human subjects? Nanoscale Res Lett. 2007;2:265–281. doi: 10.1007/s11671-007-9061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao L, Feng W, Sun Y, Bai H, Liu J, Currie C, Kim J, Gama R, Wang Z, Qian Z, Liaw L, Wu WS. Generation of iPS cells using defined factors linked via the self-cleaving 2A sequences in a single open reading frame. Cell Res. 2009;19:296–306. doi: 10.1038/cr.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, Daley GQ, Schlaeger TM. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 30.Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301–312. doi: 10.1016/j.stem.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, Liu Y, Yong J, Zhang P, Cai J, Liu M, Li H, Li Y, Qu X, Cui K, Zhang W, Xiang T, Wu Y, Liu C, Yu C, Yuan K, Lou J, Ding M, Deng H. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warlich E, Kuehle J, Cantz T, Brugman MH, Maetzig T, Galla M, Filipczyk AA, Halle S, Klump H, Scholer HR, Baum C, Schroeder T, Schambach A. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19:782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sachdeva R, Jonsson ME, Nelander J, Kirkeby A, Guibentif C, Gentner B, Naldini L, Bjorklund A, Parmar M, Jakobsson J. Tracking differentiating neural progenitors in pluripotent cultures using microRNA-regulated lentiviral vectors. Proc Natl Acad Sci U S A. 2010;107:11602–11607. doi: 10.1073/pnas.1006568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miura K, Okada Y, Aoi T, Okada A, Takahashi K, Okita K, Nakagawa M, Koyanagi M, Tanabe K, Ohnuki M, Ogawa D, Ikeda E, Okano H, Yamanaka S. Variation in the safety of induced pluripotent stem cell lines. Nat Biotechnol. 2009;27:743–745. doi: 10.1038/nbt.1554. [DOI] [PubMed] [Google Scholar]
- 37.Takayama N, Nishimura S, Nakamura S, Shimizu T, Ohnishi R, Endo H, Yamaguchi T, Otsu M, Nishimura K, Nakanishi M, Sawaguchi A, Nagai R, Takahashi K, Yamanaka S, Nakauchi H, Eto K. Transient activation of c-MYC expression is critical for efficient platelet generation from human induced pluripotent stem cells. J Exp Med. 2010;207:2817–2830. doi: 10.1084/jem.20100844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yagita K, Horie K, Koinuma S, Nakamura W, Yamanaka I, Urasaki A, Shigeyoshi Y, Kawakami K, Shimada S, Takeda J, Uchiyama Y. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci USA. 2010;107:3846–3851. doi: 10.1073/pnas.0913256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishikawa S, Goldstein RA, Nierras CR. The promise of human induced pluripotent stem cells for research and therapy. Nat Rev Mol Cell Biol. 2008;9:725–729. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- 41.Seifinejad A, Tabebordbar M, Baharvand H, Boyer LA, Salekdeh GH. Progress and promise towards safe induced pluripotent stem cells for therapy. Stem Cell Rev. 2010;6:297–306. doi: 10.1007/s12015-010-9121-x. [DOI] [PubMed] [Google Scholar]
- 42.Pearl JI, Lee AS, Leveson-Gower DB, Sun N, Ghosh Z, Lan F, Ransohoff J, Negrin RS, Davis MM, Wu JC. Short-term immunosuppression promotes engraftment of embryonic and induced pluripotent stem cells. Cell Stem Cell. 2011;8:309–317. doi: 10.1016/j.stem.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai W, Chen X. Nanoplatforms for targeted molecular imaging in living subjects. Small. 2007;3:1840–1854. doi: 10.1002/smll.200700351. [DOI] [PubMed] [Google Scholar]
- 44.Cai W, Chen X. Multimodality molecular imaging of tumor angiogenesis. J Nucl Med. 2008;49(Suppl 2):113S–128S. doi: 10.2967/jnumed.107.045922. [DOI] [PubMed] [Google Scholar]
- 45.Ruan J, Shen J, Wang Z, Ji J, Song H, Wang K, Liu B, Li J, Cui D. Efficient preparation and labeling of human induced pluripotent stem cells by nanotechnology. Int J Nanomedicine. 2011;6:425–435. doi: 10.2147/IJN.S16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Budde MD, Frank JA. Magnetic tagging of therapeutic cells for MRI. J Nucl Med. 2009;50:171–174. doi: 10.2967/jnumed.108.053546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau JF, Anderson SA, Adler E, Frank JA. Imaging approaches for the study of cell-based cardiac therapies. Nat Rev Cardiol. 2010;7:97–105. doi: 10.1038/nrcardio.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao F, Drukker M, Lin S, Sheikh AY, Xie X, Li Z, Connolly AJ, Weissman IL, Wu JC. Molecular imaging of embryonic stem cell misbehavior and suicide gene ablation. Cloning Stem Cells. 2007;9:107–117. doi: 10.1089/clo.2006.0E16. [DOI] [PubMed] [Google Scholar]
- 49.Gambhir SS, Czernin J, Schwimmer J, Silverman DH, Coleman RE, Phelps ME. A tabulated summary of the FDG PET literature. J Nucl Med. 2001;42:1S–93S. [PubMed] [Google Scholar]
- 50.Shields AF, Grierson JR, Dohmen BM, Machulla HJ, Stayanoff JC, Lawhorn-Crews JM, Obradovich JE, Muzik O, Mangner TJ. Imaging proliferation in vivo with [F-18] FLT and positron emission tomography. Nat Med. 1998;4:1334–1336. doi: 10.1038/3337. [DOI] [PubMed] [Google Scholar]
- 51.Cao F, Li Z, Lee A, Liu Z, Chen K, Wang H, Cai W, Chen X, Wu JC. Noninvasive de novo imaging of human embryonic stem cell-derived teratoma formation. Cancer Res. 2009;69:2709–2713. doi: 10.1158/0008-5472.CAN-08-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li ZB, Cai W, Cao Q, Chen K, Wu Z, He L, Chen X. 64Cu-labeled tetrameric and octameric RGD peptides for small-animal PET of tumor αvβ3 integrin expression. J Nucl Med. 2007;48:1162–1171. doi: 10.2967/jnumed.107.039859. [DOI] [PubMed] [Google Scholar]
- 53.Cai W, Niu G, Chen X. Imaging of integrins as biomarkers for tumor angiogenesis. Curr Pharm Des. 2008;14:2943–2973. doi: 10.2174/138161208786404308. [DOI] [PubMed] [Google Scholar]
- 54.Aide N, Briand M, Bohn P, Dutoit S, Lasnon C, Chasle J, Rouvet J, Modzelewski R, Vela A, Deslandes E, Vera P, Poulain L, Carreiras F. alphavbeta3 imaging can accurately distinguish between mature teratoma and necrosis in 18F-FDG-negative residual masses after treatment of non-seminomatous testicular cancer: a preclinical study. Eur J Nucl Med Mol Imaging. 2011;38:323–333. doi: 10.1007/s00259-010-1624-9. [DOI] [PubMed] [Google Scholar]