Abstract
This article combines social and genetic epidemiology to examine the influence of self-reported ethnicity on body mass index (BMI) among a sample of adolescents and young adults. We use genetic information from more than 5,000 single nucleotide polymorphisms in combination with principal components analysis to characterize population ancestry of individuals in this study. We show that non-Hispanic white and Mexican-American respondents differ significantly with respect to BMI and differ on the first principal component from the genetic data. This first component is positively associated with BMI and accounts for roughly 3% of the genetic variance in our sample. However, after controlling for this genetic measure, the observed ethnic differences in BMI remain large and statistically significant. This study demonstrates a parsimonious method to adjust for genetic differences among individual respondents that may contribute to observed differences in outcomes. In this case, adjusting for genetic background has no bearing on the influence of self-identified ethnicity.
The goal of health-disparities research is to identify the mechanisms that are responsible for persistent differences in health status among members of different social groups (Banks, Marmot, Oldfield, and Smith 2006; Hayward, Crimmins, Miles, and Yang 2000). Within this body of work, there is a great deal of interest in racial/ethnic differences in obesity and overweight. According to estimates from the 2007 to 2008 NHANES, among U.S., adults of 32.8% of non-Hispanic whites, 44.1% of non-Hispanic blacks, and 39.3% of Mexican-Americans are either overweight or obese (Flegal, Carroll, Ogden, and Curtin 2010). Differences along racial and ethnic lines are also apparent among children and adolescents, with 15.3% of non-Hispanic whites, 20% of non-Hispanic blacks, and 20.8% of Mexican-Americans being in the ninety-fifth percentile for BMI (Ogden et al. 2010). Given the association between obesity and other illnesses such as hypertension, heart disease, and diabetes (Burke et al. 2008), racial and ethnic disparities in obesity bear important implications for the uneven distribution of disease among racial and ethnic groups.
Based on theoretical reasons and consistent empirical support (Williams 2003) the bulk of health disparities research has emphasized social and environmental mechanisms. Socioeconomic differences at the level of the family (Shrewsbury and Wardle 2008), school (Richmond and Subramanian 2008), and neighborhood (Gordon-Larsen, Adair, and Popkin 2003; Robert and Reither 2004, Boardman, Saint Onge, Rogers, and Denney 2005) consistently account for the bulk of the observed differences in BMI among white, black, and Latino adults and adolescents. Further, others have argued that small differences between racial and ethnic groups of shared understandings regarding health body sizes condition perceptions of desirable weight which may influence group differences in the prevalence of obesity (Allan, Mayo, and Michel 1993).
Though social factors clearly condition obesity, there is also evidence that obesity and related comorbidities are influenced by genetic and physiological characteristics (Nelson et al. 2006; Jacobsen and Rowe 1998; Saunders et al. 2007). Numerous biometric (Nelson, Gordon-Larsen, North, and Adair 2006; Jacobsen and Rowe 1998) and molecular studies (Saunders et al. 2007) consistently demonstrate that genetic factors have a strong influence on weight among adolescents and adults. Some of the strongest evidence for genetic influences on physical weight comes from studies comparing the correlation (.60 < r < . 74) of BMI among identical twins who are raised apart from one another (Price and Gottesman 1991; Stunkard et al. 1986) Jacobson and Rowe (1998), using data from Add Health, estimate the heritability of BMI among male adolescents to be roughly .80 (A = .65, D = .16) with no shared environmental influences; in this study, roughly 80% of the variation in BMI was shown to be genetically oriented. Other BMI heritability estimates have ranged between 0.30 to more than 0.90, varying as a function of study design and age (Schousboe et al, 2003; Ordonana et al, 2007; Cornes, Zhu, and Martin 2007; Silventoinen et al, 2007; Franz et al, 2007; Haberstick et al. 2010).
Linkage studies (Yang, Kelly, and He 2007) and genome-wide association studies have built on these twin and sibling studies by identifying specific locations on the human genome that are linked to BMI and waist circumference (Herbert et al. 2006; Fox et al. 2007; Frayling et al. 2007). To date, a single nucleotide polymorphism (SNP) in the FTO gene has demonstrated the most reliable associations with BMI; those homozygous for the A allele at rs9939609 on average weighed 3 pounds heavier than those with one or no copies of this allele (Frayling et al.). This positive result has been replicated in a number of other studies (Dina et al. 2007; Cecil et al. 2008; Andreasen et al. 2008; Meyre et al. 2009).
Both the socio-structural and physiological/genetic approaches to understanding obesity offer considerable insight into the etiology of the disease but, at the end of the day, researchers are often stuck with the scientific equivalent to the “nature-versus-nurture” debate (Hernandez, Blazer, and Institute of Medicine 2001). Though few contend that “nature” is unimportant, social scientists have been very reticent to incorporate genetic explanations for complex health-related phenotypes into their conceptual and analytic models. However, this is beginning to change. Recently there has been an interest among social scientists to include biological information in social surveys to improve our understanding of the biological pathways underpinning health disparities (Crimmins and Seeman 2004).
Though the use of biomarkers has yielded insight into the relationship between social characteristics and obesity (Goodman et al. 2005; Djuric et al. 2008), efforts to incorporate genetic information to shed light race/ethnic health differentials in obesity and overweight have been limited. Many efforts are under way to genotype participants and include genotypic data in many large social and demographic surveys. As psychosocial and genetic characteristics become available in the same surveys, researchers will have the opportunity to test the possibility that very small genetic differences between socially defined racial and ethnic groups may contribute to observed racial and ethnic differences in BMI, overweight, or obesity. Though the data are becoming available, the next logical question will be how these data can inform traditional statistical models.
In pursuit of this goal, we use a sample of non-Hispanic white and Mexican-American adolescents and young adults to apply a common data reduction technique, principal components analysis, to genome-wide data to characterize ancestry or ethnic background. We then examine the independent influences of self-reported ethnicity and genetically informed ancestry, the principal components, on BMI. Our study contributes to theoretical and empirical work in this area but, equally important, we also provide examples of efficient methods to incorporate large amounts of genetic data into social epidemiologic studies of health and health behaviors.
Though the authors do not contend that these social categories are absolute biological realities, there is often some overlap between genetic ancestry and self-identified race or ethnicity, and research suggests that there is a small genetic basis to otherwise socially defined racial and ethnic groups. One line of research has been to use principal components analysis from genome-wide data spanning nearly hundreds of thousands of SNPs to characterize differences among individuals along continuous axes of variation (Price et al. 2006; Novembre et al. 2008; Reich, Price, and Patterson 2008). For example, Novembre et al. superimpose the first two principal components from 200,000 SNPs obtained from 1,387 European adults on a map of Europe, and they show surprising accuracy of the orthogonal X-Y genetic components with the X-Y geographic coordinates. With information from the first two components, the researchers could accurately predict the physical location (e.g., country of origin) of the respondent nearly 80% of the time. Similar results have also been shown with African populations (Reich et al. 2008).
As social scientists have long stated, racial and ethnic groups capture a very small proportion of the overall genetic variance in the population, and it is important to note that although these principal components can accurately predict an individual’s self-reported ethnicity or location, the first two components account for a very small proportion of the overall genetic variance. For example, in Novembre et al. (2008), these first components account for less than .5% of the total genetic variance in their sample. In multiracial studies, these first two components typically account for less than 5% of the genetic variance, which is in line with most common understandings of racial and ethnic differences. To date, the principal-components approach has been utilized primarily as a means to adjust for population stratification within the genetic research. The logic of genome-wide association studies (GWAS) is to measure the association between a particular SNP and an outcome, such as body weight. Population stratification poses a problem to GWAS in that spurious associations may be observed when minor allele frequencies differ among groups.
This same logic can (and should) be used by social demographers in their analysis of health disparities. Namely, it is a universally held assumption among social scientists that that genetic differences among racial and ethnic groups do not contribute to racial and ethnic differences in body weight but, to date, no existing study has examined this empirically. As genome wide data are becoming available, social scientists are able to answer this question fairly easily. Principal-components analysis (PCA) performed on genome-wide data provide a concise representation of population ancestry and, once derived, these components may be included in traditional quantitative models used to evaluate health disparities in general, and differences in BMI specifically. An additional benefit to the principal-components approach to measuring ancestry is that there is no assumption about the origins of the groups represented in the sample and, as such, components are sample-dependent and cannot be applied outside the study sample. We anticipate that a detailed accounting of ethnic background using the most current genetic epidemiologic methods will not alter the influence of self-identified ethnic status on BMI.
Methods
Data
Data come from an adolescent/young adult high-risk sibling sample (i.e., probands ages 14–19 selected for high-risk and their siblings ages 14–25) recruited into a study investigating genetic and environmental influences contributing to risk for substance use disorders and delinquency. High-risk probands were recruited from consecutive admissions (but see inclusion/exclusion criteria later) to three residential and outpatient treatment facilities in the Denver metropolitan area operated by the Division of Substance Dependence of the Department of Psychiatry, University of Colorado School of Medicine. Most of the probands were referred into treatment by juvenile justice or social service agencies; a small proportion (approximately 1%) were parental referrals. Additional probands were recruited into the study via clinical evaluations conducted by the Division of Substance Dependence for the Colorado Department of Corrections (i.e., probation evaluations).
A total of 695 respondents reported their height and weight. Among these, genotyping was performed using a number of chip platforms. To achieve consistency in genotyping, we used only those genotyped using the Affymetrix 10k chip (n = 349). Owing to the lack of racial and ethnic diversity within the sample, we decided to limit our sample to Mexican-Americans and non-Hispanic white respondents (n = 244). Further, owing to the household sampling procedures, we selected only one child per family so as not to bias our principle-components analysis. There were 120 families (i.e., 120 individuals after randomly selecting one sibling). We dropped those with missing information on family income (n = 9), leaving a completed sample of 111 unrelated individuals. These break down into 88 non-Hispanic whites and 23 Mexican Americans. We compared the BMI, age, gender, and poverty status of the reduced sample to the larger sample of 695 respondents. The full and reduced samples are virtually identical with respect to BMI and socio-demographics (age and gender), but we did observe a slight difference in the proportion of poor respondents in our reduced sample (p = .15) and the full sample (p = .22). This difference is statistically significant (p < .01) and is due to the increasing proportion of non-Hispanic white respondents in the reduced sample (~80%) compared to the full sample (~50%).
Genomic DNA was isolated from buccal cells using a modification of published procedures (Freeman et al., 1997). Briefly, our method involved collecting buccal cells by rubbing the cheeks with cotton swabs followed by a rinse with mouthwash. The DNA was isolated by solvent extraction, quantified with PicoGreen (Molecular Probes, Eugene, OR), and a working stock of 20 ng/ul was prepared in TE buffer. The average yield of DNA was 409/5 µg. Primer extension preamplification (PEP) was performed on 1 ml aliquots of the genomic DNA using a modification (Krauter et al. 2001) of the method of Zhang et al. (1992), which resulted in an approximately 100-fold amplification of the DNA. The PEP procedure was originally to be used only on those few samples that had poor yields. However, after confirming that allele calls for all markers were identical when comparing PEP DNA with DNA purified from cell lines in two CEPH individuals (Krauter et al.), PEP was routinely used for all of the DNA samples. Of the 11,000+ markers, we limited our analysis to SNPs with a minor allele frequency of at least 1% and SNPs in which missingness did not exceed 10% of the sample. In total, we use a sample of 5,490 SNPs. The RS numbers and MAF for these SNPs are available upon request.
Analysis
We use two measures obtained from genome-wide data to characterize each individual in our study. We first perform PCA on the 5,490 SNPs described earlier. We use an additive coding method of genotype in which individuals receive a score of 0 if they are homozygous for the minor allele, 1 if they are heterozygous, and 2 if they are homozygous for the major allele. Because of the unusually large amount of information from genetic data sources, PCA provides an effective and efficient method to distill multidimensional and complicated data into continuous axes of variation, thereby reducing the dimensionality to the number of observations in the data. As shown elsewhere (Price et al. 2006; Novembre et al. 2008; Reich et al. 2008), these components provide important information regarding population ancestry that can be used in statistical analysis to purge the parameter estimates of genetic influences due to group and subgroup membership. We use the same approach as Price et al (2006) to compute the PCAs in our study. Let mij be a matrix of genotypes for SNP i and individual j. Each individual’s genotype at that location is subtracted by the row mean to create a matrix with row sums equal to zero. As with Price et al. (2006), we then normalize each row by dividing each genotype by
where pi is the allele frequency at location i. This new matrix (X) is used to estimate a new 111×111 covariance matrix ψ where ψjj' is the covariance of column j' and column of X. Axis of variation is distilled into eigenvectors with the largest eigenvalues. As such, ajk captures the genetic background of individual j for the kth axis of variation.
We will examine the top PCs and use any PC that significantly differs between white and Mexican-American respondents. All analyses were performed using the prcomp routine in R (R Development Core Team, 2009). As one of the stated goals of this project was to increase the use of genetic data by social demographers, the R syntax for these runs is available to interested researchers upon request.
Components are dependent on the heterogeneity of the sample population and, as such, have little utility outside the relevant study. Although these components do not represent a universal measure of ancestry, they are robust to very selective samples, such as the one currently used. Because these components are reflective of ancestry within the sample, we believe the use of principal components to control for latent genetic influences on BMI to be unbiased by the use of a highly selective sample. Although the choice of sample may jeopardize the generalizability of the findings to all adolescents, the use of this sample should not bias the components or the demonstration of their utility within regression models.
We then estimate regression models with and without controls for population ancestry. And second, we then calculate a summary measure of total genetic distance using a Mahalanobis distance (Mahalnobis 1936) calculation. These measures are then used as controls in the linear model predicting BMI. This measure reports departure from the sample mean of all of the SNPs: a person-specific difference in genetic space from a fictitious person who is average on all of the SNPs.
Results
Table 1 presents the descriptive statistics for all variables used in the analysis. Non-Hispanic whites in this study are significantly taller than the Mexican-American respondents (roughly 2.6 inches, p < .001), and they are slightly lighter (~ 6 pounds, n.s.) These differences are also evidenced in the significant difference in BMI among white () and Mexican-American () respondents. Mexican-American respondents are also nearly three times more likely than whites to report relatively low family income.
Table 1.
Descriptive statistics for all variables used in the analysis
| Variable | Total | White | Mexican American | Pr (w-m) = 0 |
|---|---|---|---|---|
| Height (in) | 67.50 | 68.05 | 65.39 | 0.001 |
| Weight (lb) | 150.09 | 148.81 | 155.00 | 0.414 |
| BMI | 23.10 | 22.51 | 25.35 | 0.004 |
| Age | 16.62 | 16.53 | 16.95 | 0.432 |
| Female | 27.93 | 30.68 | 17.39 | 0.209 |
| Poor (yearly income < $15k) | 15.31 | 11.36 | 30.43 | 0.023 |
| Genetic measures | ||||
| PC1 | 0.00 | 1.804 | −0.472 | 0.000 |
| Mahalanobis Distance | 44.59 | 37.68 | 71.01 | 0.000 |
| Sample size (n) | 111 | 88 | 23 | — |
BMI = body mass index; PC1 = First principal component.
Note: See Methods for a detailed description of the sample and the measures described here. p-values describe the probability of a type-I error regarding differences between white and Mexican-American respondents across the measured items.
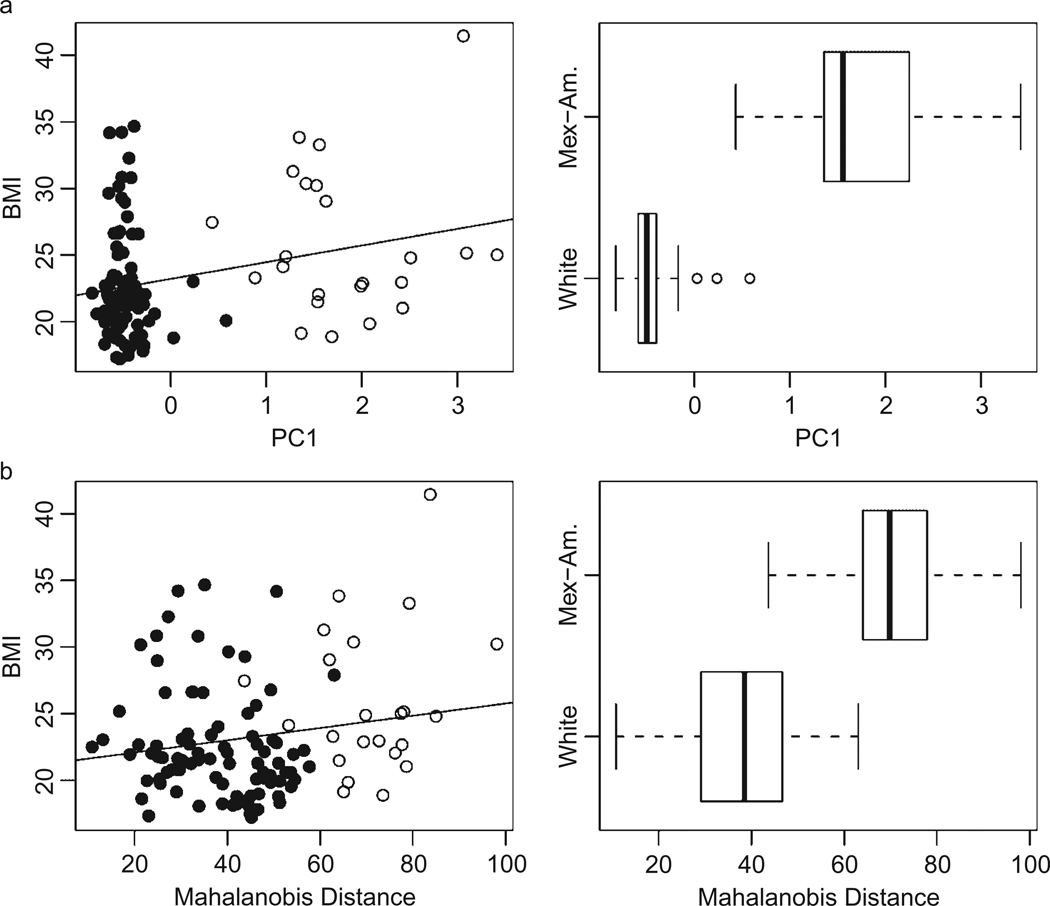
Importantly, we find large and significant ethnic differences in the first principal component (PC1) and the Mahalanobis distance (MD) based on the first 45 principal components. The full description of the race and BMI associations with the first 10 principal components is provided in Table 2. This first component (PC1) accounts for 3% of the overall genetic variance in the sample. Given that this is the only significant PC in these models, it is fair to characterize the between-group differences in genetic variance as accounting for no more than 3% of the total variance, and 97% occurs within each self-identified group. The next two columns of data present the association between each PC, BMI, and ethnicity. We show a positive and significant association between PC1 and BMI and, as shown in Table 1, a strong association between PC1 and self-reported ethnicity. These associations are presented graphically in Figure 1.
Table 2.
Principal components from genome-wide data: associations with BMI and self-reported ethnicity
| Variance explained |
PC-BMI association |
PC-ethnicity association |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Unique | Cumulative | r (PC,BMI) | Pr (r = 0) | Pr (w-b = 0) | |||||
| PC1 | 0.030 | 0.030 | 0.216 | 0.030 | 1.804 | −0.472 | 0.000 | ||
| PC2 | 0.014 | 0.044 | −0.183 | 0.067 | −0.043 | 0.011 | 0.768 | ||
| PC3 | 0.013 | 0.057 | 0.037 | 0.710 | 0.130 | −0.034 | 0.364 | ||
| PC4 | 0.013 | 0.070 | 0.010 | 0.923 | 0.027 | −0.007 | 0.843 | ||
| PC5 | 0.012 | 0.083 | −0.021 | 0.838 | −0.044 | 0.011 | 0.892 | ||
| PC6 | 0.012 | 0.095 | 0.116 | 0.249 | 0.108 | −0.028 | 0.720 | ||
| PC7 | 0.012 | 0.106 | −0.063 | 0.531 | −0.145 | 0.038 | 0.607 | ||
| PC8 | 0.012 | 0.118 | 0.119 | 0.237 | 0.096 | −0.025 | 0.718 | ||
| PC9 | 0.011 | 0.129 | 0.145 | 0.148 | −0.201 | 0.053 | 0.482 | ||
| PC10 | 0.011 | 0.141 | −0.015 | 0.881 | −0.032 | 0.008 | 0.906 | ||
Note: Principal components were estimated from the 111 people across 5,490 single nucleotide polymorphisms. See the Methods section for a more detailed description of these techniques.
Figure 1.
Genome-wide measures of population ancestry and risk of obesity compared to self-reported ethnicity.
Note: Figures describe the association between the three genomic control measures and BMI by self-identified ethnicity (dark circles are non-Hispanic whites). The distribution for each measure is shown for each group in the corresponding figures on the right. As shown in Table 3, the bivariate linear associations are estimated to be bPC1 = .92 (p < .05), bm.dist = .03 (n.s.).
As is evident in Figure 1 and Table 2, PC1 (r = .22, p < .03) and BMI are positively associated with each other. We do not show any association between the MD measure and BMI. It is also clear in Figure 1 and Table 1 that there are clear ethnic differences in both PC1 and MD. As such, it is possible that PC1 may account for some of the observed ethnic differences in BMI. To examine this possibility, we present a series of multivariate models in Table 3. The first models present the unadjusted effects of the two genetic measures. PC1 (b = .92, p < .05) is positively and significantly associated with BMI and accounts for roughly 4% of the variance in BMI. Model 3 presents the first social demographic model in which ethnicity (b = 2.72, p < .01) and age (b = .44, p < .05) are both associated with BMI. In total, these factors explain 11% of the variance in BMI. Model 4 enters a control for socioeconomic status (income < $15k/year), and relatively low levels of income are positively associated with BMI (b = 2.91, p < .01). These results also show that the influence of income and ethnicity operate independent of each other; they both remain statistically significant. Models 5 and 6 add genetic measures independently to the full model (Model 4). If genetic differences in ethnic groups accounted for the observed ethnic differences in BMI, these controls should eliminate the effect of ethnicity. In both cases, it is clear that these controls do not account for the observed ethnic differences in BMI.
Table 3.
OLS parameter estimates: genetic and social influences on BMI
| Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | Model 6 | |
|---|---|---|---|---|---|---|
| (Intercept) | 23.10*** | 21.66*** | 15.05*** | 14.67*** | 14.05*** | 16.47*** |
| (.40) | (1.10) | (2.83) | (2.75) | (2.81) | (2.96) | |
| PC1 | 0.92* | −1.07 | ||||
| (.40) | (.99) | |||||
| Mahalanobis Distance | 0.03 | −0.05 | ||||
| (.02) | (.03) | |||||
| Mexican-American | 2.72** | 2.15* | 4.56+ | 3.87** | ||
| (.95) | (.95) | (2.44) | (1.43) | |||
| Age | 0.44* | 0.45** | 0.46** | 0.47** | ||
| (.17) | (.17) | (.17) | (.17) | |||
| Female | 0.52 | 0.42 | 0.31 | 0.07 | ||
| (.88) | (.85) | (.86) | (.87) | |||
| Low Income | 2.91** | 2.90** | 3.10** | |||
| (1.05) | (1.05) | (1.05) | ||||
| R-squared | 0.04 | 0.01 | 0.11 | 0.17 | 0.17 | 0.18 |
Note: Cell entries represent unstandardized OLS regression estimates with standard errors in parentheses. The dependent variable in all models is BMI obtained from self-reported height and weight.
p < .05;
p < .01;
p < .001;
p < .10.
Conclusion
Because childhood and adolescent obesity is strongly linked to adult obesity (Whitaker, Wright, Pepe, Seidel, and Dietz 1997) and because obesity is linked to an increased risk of chronic health problems including type-II diabetes and hypertension among relatively young adults, understanding individual differences in current weight and weight gain is a critical public health issue. This is of particular concern to health-disparities researchers because of persistent and large differences in BMI across racial and ethnic groups. The purpose of this study is to introduce social demographers to a relatively simple method to incorporate genome-wide data into their studies with the explicit aim to eliminate potentially confounding effects due to very small genetic differences across socially defined racial and ethnic groups. We use the phenotype of BMI because of distributional characteristics and the clear disparities that exist in this phenotype across racial and ethnic groups.
To illustrate this technique, we used a very small existing sample in which we have data on height/weight, self-reported ethnicity, and genome-wide data. However, there are two important limitations that should be considered when interpreting the results of this study. First, this is not a representative sample of adolescents and, as such, these findings should not necessarily be generalized to the larger population. This convenience sample was not designed as a cohort or a case control-study, and far more elaborate research designs would be needed to characterize population ancestry at a larger level. Selection in this study may have to do with drug use and body mass, which may further complicate the interpretation of the parameter estimates of this study. This is particularly problematic because the PCs are identified from the sample and not the population. Ideally, these methods would be used on a random sample of the population. However, our methods adequately differentiate subtle genetic differences among individuals in our sample of high-risk adolescents. Second, the relatively small sample size reduces our power to detect differences, and it increases our risk of a type II error. For example, this is evident in the sensitivity of the parameter estimate for PC1 in Model 5 of Table 3. The sign for this estimate switches signs after controlling for background characteristics. This is owing in large part to the strong correlation between PC1 and Mexican-American identification; however, it is also likely that this instability is due to the small sample sizes in this study. The same can be said for the variability seen in the coefficient for ethnicity. These limitations should be considered when interpreting the results of this study.
With these limitations in mind, our article nevertheless makes an important contribution to this literature because it demonstrates that these ethnic differences in our sample are independent of any small genetic differences that may exist between the groups. We also demonstrate relatively parsimonious statistical methods to incorporate large amounts of genetic information into social epidemiologic studies of BMI and obesity without compromising the overall aims of this body of work. These genomic controls help to rule out alternate explanations regarding the influence of racial and ethnic ancestry on this important health outcome. These findings make an important comment on the social and genetic epidemiology of obesity.
Thus far, genome-wide information on population ancestry has been used exclusively to adjust parameter estimates in genome-wide association studies for population stratification, and some efforts have been made to infer immigration patterns from molecular data (Novembre et al. 2008). To date, however, the information from genome-wide principal components has not been fully incorporated into social scientific studies. We argue that this rich information about individual differences has the prospect of sharpening social epidemiologic analysis and bolstering the precision and consistency of parameter estimates for social structural variables.
In many ways, the inclusion of genome-wide data into existing social demographic studies turns our typical study design problem on its head. That is, demographers usually have very large samples with a fairly limited number of items to characterize the sample (our rows far outweigh our columns). However, with the addition of hundreds of thousands of columns, with the columns surpassing the rows by a factor 105, typical approaches to multivariable analyses are not possible. We have far more parameters than observations, and efforts need to be made to reduce this information to valid measures that capture meaningful genetic variation. To date, although there is much interest in this line of work (Finch, Vaupel, and Kinsella 2001), social epidemiologic researchers still have very few methodological options to incorporate large amounts of genetic information from their respondents into standard quantitative models. One of the purposes of this study was to demonstrate the usefulness of genome-wide data for health-disparities researches and to provide several useful strategies for researchers involved in studies with genetic information from their respondents. For purposes of illustrating the usefulness of genome-wide data for sociological purposes, we focused on race/ethnic differences in obesity. We argue that a sociologically informed study of race and ethnicity provides an important bridge that allows for genetic factors to be included within the health-disparities literature related to obesity. We use a relatively small data set and a limited number of SNPs (~6,000) to demonstrate the usefulness of this approach; however, this approach can be extended to a significantly larger number of SNPs (>1m) for the type of data that are currently available. We focus on racial/ethnic differences in BMI, but this understanding can be applied to a number of different outcomes or social groupings.
With limited dissent (Shiao et al. 2008), the collective demographic and sociological voice has been unequivocally opposed to scientific efforts to examine the possibility that genetic factors may play some part in persistent health disparities, and there are many good reasons for this reticence (Duster 1990). Much of this skepticism is well founded, but some criticisms have not been supported as we learn more about the human genome. The basic logic of this line of inquiry relies on three hypotheses regarding genes, race, and body weight: (1) that genetic factors have considerable influence on an individual’s body weight, (2) that socially defined racial and ethnic groups differ from one another genetically, and (3) that genetic differences among racial and ethnic groups contribute to racial and ethnic differences in body weight. Each of these ideas has received a great deal of criticism from the social scientific community (Duster 2005, 2006). Though there is significant support of the genetic epidemiology of body weight, there is still a great deal of skepticism when it comes to discussing the genetic origins of race and ethnicity. Our study demonstrates that the genetic differences across racial and ethnic groups are evident but quite small in magnitude. Given this small contribution to overall genetic variation, it is no surprise that genes do not account for observed ethnic differences in BMI; however, this important empirical question remained untested prior to this current study.
As stated throughout this article, these results bolster the position of social epidemiologists who look to social context (e.g., friends, schools, workplace, residential areas) to account for important differences in the prevalence of obesity among racial and ethnic groups. Social epidemiologists have focused on normative and institutional factors that may increase the risk of obesity for racial and ethnic minorities through lifestyles and access to a healthy environment (Faith and Kral 2006). This manifests through an emphasis on health behaviors and conditions stemming from the probability that various minority groups are likely to be of lower socioeconomic status and cultural differences between racial and ethnic groups in regard to food preferences and beliefs about a healthy body weight.
Having a lower socioeconomic status is predictive of health behaviors and lifestyle choices, such as fast-food consumption, skipping breakfast, and physical inactivity (Faith and Kral 2006; Gordon-Larsen, McMurray, and Popkin 2000; Certain and Kahn 2002), and these behaviors have been linked to increased BMI (Niemeier et al. 2006). As Popkin, Duffy, and Gordon-Larsen (2005) show, these differences in behavior and BMI are strongly influenced by differential access to healthy foods and physical activity outlets. Similar work is shown by others (Estabrooks, Lee, and Gyurcsik 2003; Gordon-Larsen et al. 2000). For example, studies show that neighborhoods composed of racial and ethnic minorities have more than twice the number of fast-food restaurants (Block, Scribner, and DeSalvo 2004; Ball. Timperio, and Crawford 2009). The proximity to fast-food restaurants is not balanced by easy access to chain grocery stores (Chung and Myers 1999). Further, many have cited government policies and market conditions that make high-calorie, low-nutrient foods, such as sodas, candy, and fast food, cheaper than low-calorie, high-nutrient foods, such as fruits and vegetables (Andrieu, Darmon, and Drewnowski 2006). Thus, many minorities living in a low socioeconomic environment adopt poor eating habits to cope with limited resources.
In addition to these institutional factors affecting health behavior, normative influences are also believed to influence BMI because of the meaning(s) that people ascribe to body size and group differences regarding the perception of overweight (Flynn and Fitzgibbon 1998). Because socioeconomic and racial characteristics are linked with differences in BMI, de facto racial and class segregation (Massey and Denton 1993; Jargowsky 1997) structures shared understandings of health body sizes and otherwise small differences in norms about body among friends are further exacerbated (Boardman et al. 2005). The notion that cultural beliefs about food and ideal bodies are embedded within groups provides one explanation for why one’s own weight is predicted by his or her associates. For example, Chistiakis and Fowler (2007) model the probability of weight gain as a function of friendship networks. They show that individuals’ risk of becoming obese increased 57% if they had a friend who became obese during the observed interval. Importantly, the social influences of one’s peers were larger in magnitude than the corresponding risk associated with a person’s sibling (40% increase).
In sum, there is consistent evidence that social and demographic factors are strongly associated with physical weight because of access to health-related environments and energy-balance behaviors and lifestyles. Excess caloric intake and increased sedentary lifestyles as a result of environmental and cultural contexts place persons of disadvantaged backgrounds at a greater risk of obesity and overweight. Based on the results of this study, a critical examination of the built environment and cultural milieu of racial and ethnic groups will yield far more insights into body size differences across socially defined groups of people than a corresponding exploration of small (and apparently meaningless) genetic differences for this important phenotype.
Acknowledgment
This research was funded in part by grants from the Eunice Kennedy Shriver NICHD (K01HD50336) and NIDA (DA 11015 & DA012845).
Footnotes
Publisher's Disclaimer: Full terms and conditions of use: http://www.tandfonline.com/page/terms-and-conditions
This article may be used for research, teaching and private study purposes. Any substantial or systematic reproduction, re-distribution, re-selling, loan, sub-licensing, systematic supply or distribution in any form to anyone is expressly forbidden.
The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. The accuracy of any instructions, formulae and drug doses should be independently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings, demand or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with or arising out of the use of this material.
References
- Allan JD, Mayo K, Michel Y. Body size values of white and black women. Res Nurs Health. 1993;16:323–333. doi: 10.1002/nur.4770160503. [DOI] [PubMed] [Google Scholar]
- Andreasen Camilla H, Stender-Petersen Kirstine L, Mogensen Mette S, Torekov Signe S, Wegner Lise, Andersen Gitte, Nielsen Arne L, Albrechtsen Anders, Borch-Johnsen Knut, Rasmussen Signe S, et al. Low physical activity accentuates the effect of the FTO Rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57:95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- Andrieu E, Darmon N, Drewnowski A. Low-cost diets: more energy, fewer nutrients. Eur J Clin Nutr. 2006;60:434–436. doi: 10.1038/sj.ejcn.1602331. [DOI] [PubMed] [Google Scholar]
- Ball K, Timperio A, Crawford D. Neighbourhood socioeconomic inequalities in food access and affordability. Health Place. 2009;15:578–585. doi: 10.1016/j.healthplace.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Banks James, Marmot Michael, Oldfield Zoe, Smith James P. Disease and disadvantage in the United States and in England. JAMA. 2006;295:2037–2045. doi: 10.1001/jama.295.17.2037. [DOI] [PubMed] [Google Scholar]
- Block JP, Scribner RA, DeSalvo KB. Fast food, race/ethnicity, and income. A geographic analysis Am J Prev Med. 2004;27:211–217. doi: 10.1016/j.amepre.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Boardman Jason D, Saint Onge Jarron M, Rogers Richard G, Denney Justin T. Race differentials in obesity: The impact of place. J Health Soc Behav. 2005;46:229–243. doi: 10.1177/002214650504600302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke GL, Bertoni AG, Shea S, Tracy R, Watson KE, Blumenthal RS, Chung H, Carnethon MR. The impact of obesity on cardiovascular disease risk factors and subclinical vascular disease: The Multi-Ethnic Study of Atherosclerosis. Arch Intern Med. 2008;168:928–935. doi: 10.1001/archinte.168.9.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil Joanne E, Tavendale Roger, Watt Peter, Hetherington Marion M, Palmer Colin NA. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359:2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- Certain Laura K, Kahn Robert S. Prevalence, correlates, and trajectory of television viewing among infants and toddlers. Pediatrics. 2002;109(4):634–624. doi: 10.1542/peds.109.4.634. [DOI] [PubMed] [Google Scholar]
- Christakis Nicholas A, Fowler James H. The spread of obesity in a large social network over 32 years. N Engl J Med. 2007;357:370–379. doi: 10.1056/NEJMsa066082. [DOI] [PubMed] [Google Scholar]
- Chung Chanjin, Myers Samuel L., Jr Do the poor pay more for food? An analysis of grocery store availability and food price disparities. J Consumer Affairs. 1999;33(2):276–296. [Google Scholar]
- Cornes BK, Zhu G, Martin NG. Sex differences in genetic variation in weight: A longitudinal study of body mass index in adolescent twins. Behav Genet. 2007;37:648–660. doi: 10.1007/s10519-007-9165-0. [DOI] [PubMed] [Google Scholar]
- Crimmins Eileen M, Seeman Teresa E. Integrating biology into the study of health disparities. Popul Dev Rev. 2004;30:89–107. [Google Scholar]
- Dina C, Meyre D, Samson C, Tichet J, Marre M, Jouret B, Charles MA, Balkav B, Froguel P. Comment on “A common genetic variant is associated with adult and childhood obesity.”. Science. 2007;315:187. doi: 10.1126/science.1129402. [DOI] [PubMed] [Google Scholar]
- Djuric Zora, Bird Chloe E, Furumoto-Dawson Alice, Rauscher Garth H, Ruffin Mack T, IV, Stowe Raymond P, Tucker Katherine L, Masi Christopher M. Open Biomark J. 2008;1:7–19. doi: 10.2174/1875318300801010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duster Troy. Backdoor to eugenics. New York: Routledge; 1990. [Google Scholar]
- Duster Troy. 2005 Presidential address—comparative perspectives and competing explanations: taking on the newly configured reductionist challenge to sociology. Am Sociol Rev. 2006;71(1):1. [Google Scholar]
- Duster Troy. Medicine: Enhanced: Race and reification in science. Science. 2005;307(5712):1050–1051. doi: 10.1126/science.1110303. [DOI] [PubMed] [Google Scholar]
- Estabrooks Paul A, Lee Rebecca E, Gyurcsik Nancy C. Resources for physical activity participation: Does availability and accessibility differ by neighborhood socioeconomic status? Ann Behav Med. 2003;25(2):1532–4796. doi: 10.1207/S15324796ABM2502_05. [DOI] [PubMed] [Google Scholar]
- Faith Myles S, Kral Tanja VE. Institute of Medicine. Social environmental and genetic influences on obesity and obesity-promoting behaviors: Fostering research integration. In: Hernandez Lyla M, Blazer Dan G, editors. Appendix C In Genes, behavior, and the social environment: Moving beyond the nature/nurture debate. Washington, DC: National Academies Press; 2006. pp. 236–280. [Google Scholar]
- Finch CE, Vaupel JW, Kinsella KG, editors. Cells and surveys: Should biological measures be included in social science research? Washington, DC: National Academy Press; 2001. [PubMed] [Google Scholar]
- Flegal Katherine M, Carroll Margaret D, Ogden Cynthia L, Curtin Lester R. Prevalence and trends in obesity among U.S. adults, 1999–2008. JAMA. 2010;303(3):235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- Flynn Kristin J, Fitzgibbon Marian. Body images and obesity risk among black females: A review of the literature. Ann Behav Med. 1998;20:13–24. doi: 10.1007/BF02893804. [DOI] [PubMed] [Google Scholar]
- Fox CS, Heard-Costa N, Cupples LA, Dupuis J, Vasan RS, Atwood LD. Genome-wide association to body mass index and waist circumference: The Framingham Heart Study 100K project. BMC Med Genet. 2007;8 Suppl:1. doi: 10.1186/1471-2350-8-S1-S18. S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz CE, Grand MD, Jacobson KC, Kremen WS, Eisen SA, Xian H, Romeis J, Thompson-Brenner H, Lyons MJ. Genetics of body mass index stability and risk for chronic disease: A 28-year longitudinal study. Twin Res Hum Genet. 2007;10:537–545. doi: 10.1375/twin.10.4.537. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman Bernard, Powell John, Ball David, Hill Linzey, Craig Ian W, Plomin Rorbert J. DNA by mail: An inexpensive and noninvasive method for collecting DNA samples from widely dispersed populations. Behav Genet. 1997;27:251–257. doi: 10.1023/a:1025614231190. [DOI] [PubMed] [Google Scholar]
- Goodman Elizabeth, McEwen Bruce S, Huang Bin, Dolan Lawrence M, Adler Nancy E. Social inequalities in biomarkers of cardiovascular risk in adolescence. Psychosom Med. 2005;67:9–15. doi: 10.1097/01.psy.0000149254.36133.1a. [DOI] [PubMed] [Google Scholar]
- Gordon-Larsen Penny, Adair Linda S, Popkin Barry M. The relationship of ethnicity, socioeconomic factors, and overweight in US adolescents. Obesity Res. 2003;11:121–129. doi: 10.1038/oby.2003.20. [DOI] [PubMed] [Google Scholar]
- Gordon-Larsen P, McMurray RG, Popkin BM. Determinants of adolescent physical activity and inactivity patterns. Pediatrics. 2000;105:E83. doi: 10.1542/peds.105.6.e83. [DOI] [PubMed] [Google Scholar]
- Haberstick BC, Lessem JM, McQueen M, Boardman JD, Hopfer CJ, Smolen A, Hewitt JK. Stable genes and changing environments: Body mass index across adolescence and young adulthood. Behav Genet. 2010;40:495–504. doi: 10.1007/s10519-009-9327-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward Mark D, Crimmins Eileen M, Miles Toni P, Yang Yu. The significance of socioeconomic status in explaining the racial gap in chronic health conditions. Am Sociol Rev. 2000;65:910–929. [Google Scholar]
- Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- Hernandez Lyla M, Blazer Dan G Institute of Medicine. Genes, behavior, and the social environment: Moving beyond the nature/nurture debate. National Academies Press; Washington, DC: 2006. [PubMed] [Google Scholar]
- Jacobson Kristen C, Rowe David C. Genetic and shared environmental influences on adolescent BMI: Interactions with race and sex. Behav Genet. 1998;28(4):265–278. doi: 10.1023/a:1021619329904. [DOI] [PubMed] [Google Scholar]
- Jargowsky PA. Poverty and place: Ghettos, barrios, and the American city. New York: Russell Sage Foundation; 1997. [Google Scholar]
- Krauter KS, Hutchinson J, Hewitt JK, Stallings MC, Smolen A, Crowley TJ. Large-scale marker screening using minute quantities of DNA. Drug Alcohol Depend. 2001;63 Suppl 1:S84. [Google Scholar]
- Mahalanobis PC. On the generalised distance in statistics. Proc Natl Inst Sci India. 1936;2(1):49–55. [Google Scholar]
- Massey Douglas S, Denton Nancy A. American apartheid: Segregation and the making of the underclass. Cambridge MA: Harvard University Press; 1993. [Google Scholar]
- Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proenca C, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- Nelson Melissa C, Gordon-Larsen Penny, North Kari E, Adair Linda S. Body mass index gain, fast food, and physical activity: Effects of shared environments over time. Obesity Res. 2006;14:701–709. doi: 10.1038/oby.2006.80. [DOI] [PubMed] [Google Scholar]
- Niemeier Heather M, Raynor Hollie A, Lloyd-Richardson Elizabeth E, Rogers Michelle L, Wing Rena R. Fast food consumption and breakfast skipping: Predictors of weight gain from adolescence to adulthood in a nationally representative sample. J Adolesc Health. 2006;39:842–849. doi: 10.1016/j.jadohealth.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Novembre John, Johnson Toby, Bryc Katarzyna, Kutalik Zoltan, Boyko Adam R, Auton Adam, Indap Amit, King Karen S, Bergmann Sven, Nelson Matthew R, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden Cynthia L, Carroll Margaret D, Curtin Lester R, Lamb Molly M, Flegal Katherine M. Prevalence of high body mass index in US children and adolescents, 2007–2008. JAMA. 2010;303(3):242–249. doi: 10.1001/jama.2009.2012. [DOI] [PubMed] [Google Scholar]
- Ogden Cynthia L, Carroll Margaret D, Curtin Lester R, McDowell Margaret A, Tabak Carolyn J, Flegal Katherine M. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006;295:1549–1555. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- Ordonana JR, Rebollo-Mesa I, Gonzalez-Javier R, Perez-Riquelme F, Martinez-Selva JM, Willemsen G, Boomsma DI. Heritability of body mass index: A comparison between the Netherlands and Spain. Twin Res Hum Genet. 2007;10:749–756. doi: 10.1375/twin.10.5.749. [DOI] [PubMed] [Google Scholar]
- Popkin Barry M, Duffey Kiyah, Gordon-Larsen Penny. Environmental influences on food choice, physical activity and energy balance. Physiol Behav. 2005;86:603–613. doi: 10.1016/j.physbeh.2005.08.051. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price RA, Gottesman II. Body fat in identical twins reared apart: Roles for genes and environment. Behav Genet. 1991;21:1–7. doi: 10.1007/BF01067662. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2009. [accessed October 10, 2009]. URL http://www.R-project.org. [Google Scholar]
- Reich David, Price Alkes L, Patterson Nick. Principal component analysis of genetic data. Nat Genet. 2008;40:491–492. doi: 10.1038/ng0508-491. [DOI] [PubMed] [Google Scholar]
- Richmond Tracy K, Subramanian SV. School level contextual factors are associated with the weight status of adolescent males and females. Obesity. 2008;16:1324–1330. doi: 10.1038/oby.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert SA, Reither EN. A multilevel analysis of race, community disadvantage, and body mass index among adults in the US. Soc Sci Med. 2004;59:2421–2434. doi: 10.1016/j.socscimed.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Rosenbaum P, Rubin D. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70(1):41–55. [Google Scholar]
- Saunders CL, Chiodini BD, Sham P, Lewis CM, Abkevich V, Adeyemo AA, de Andrade M, Arya R, Berenson GS, Blangero J, et al. Meta-analysis of genome-wide linkage studies in BMI and obesity. Obesity. 2007;15(9):2263–2275. doi: 10.1038/oby.2007.269. [DOI] [PubMed] [Google Scholar]
- Schousboe K, Willemsen G, Kyvik KO, Mortensen J, Boomsma DI, Cornes BK, Davis CJ, Fagnani C, Hjelmborg J, Kaprio J, et al. Sex differences in heritability of BMI: A comparative study of results from twin studies in eight countries. Twin Res Hum Genet. 2003;6:409–421. doi: 10.1375/136905203770326411. [DOI] [PubMed] [Google Scholar]
- Shrewsbury Vanessa, Wardle Jane. Socioeconomic status and adiposity in childhood: A systematic review of cross-sectional studies 1990–2005. Obesity. 2008;16:275–284. doi: 10.1038/oby.2007.35. [DOI] [PubMed] [Google Scholar]
- Shiao Jiannbin Lee, Bode Thomas, Beyer Amber, Fox with Alison, Selvig Daniel, Tandingan Justin. The genomic challenge to the social construction of race. Paper presented at the annual meetings of the American Sociological Association.2008. [Google Scholar]
- Silventoinen K, Pietlainen KH, Tynelius P, Sorensen TIA, Kaprio J, Rasmussen F. Genetic and environmental factors in relative weight from birth to age 18: The Swedish Young Male Twins Study. Int J Obesity. 2007;31:615–621. doi: 10.1038/sj.ijo.0803577. [DOI] [PubMed] [Google Scholar]
- Stunkard AJ, Sorensen TI, Hanis C, Teasdale TW, Chakraborty R, Schull WJ, Schulsinger F. An adoption study of human obesity. N Engl J Med. 1986;314:193–198. doi: 10.1056/NEJM198601233140401. [DOI] [PubMed] [Google Scholar]
- United States Department of Health and Human Services. Healthy people 2010. Washington, DC: Author; 2000. [Google Scholar]
- Whitaker RC, Wright JA, Pepe MS, Seidel KD, Dietz WH. Predicting obesity in young adulthood from childhood and parental obesity. N Engl J Med. 1997;337:869–873. doi: 10.1056/NEJM199709253371301. [DOI] [PubMed] [Google Scholar]
- Williams David R. The health of men: Structured inequalities and opportunities. Am J Public Health. 2003;93:724–731. doi: 10.2105/ajph.93.5.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Kelly T, He J. Genetic epidemiology of obesity. Epidemiol Rev. 2007;29:49–61. doi: 10.1093/epirev/mxm004. [DOI] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: Implications for genetic analysis. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]