Abstract
Intramembrane proteases are responsible for a number of regulated proteolysis events occurring within or near the plasma and intracellular membranes. Members of one large and diverse family of putative intramembrane metalloproteases are widely distributed in all domains of life, including the type II CAAX prenyl proteases and their prokaryotic homologs with putative bacteriocin-related functions. We used sensitive sequence similarity searches to expand this large CPBP (CAAX Proteases and Bacteriocin-Processing enzymes) family to include more than 5,800 members, and infer its homologous relationships to several other protein families, including the PrsW proteases, the DUF2324 family and the γ-secretase subunit APH-1 proteins. They share four predicted core transmembrane segments and possess similar, yet distinct sets of sequence motifs. Remote similarity between APH-1 and membrane proteases sheds light on APH-1’s evolutionary origin and raises the possibility that APH-1 may possess proteolytic activity in the current or ancestral form of γ-secretase.
Keywords: type II CAAX protease, APH-1, γ-secretase, PrsW, DUF2324, intramembrane protease
A number of key cellular processes involve the intramembrane proteolytic cleavage of proteins 1-2. Several distinct families of membrane proteases that carry out peptide bond cleavage inside lipid bilayers have been characterized. Recent structural studies for two of these families (site-2 protease and rhomboid) have provided insights into their catalytic mechanisms 3-4. Eukaryotic site-2 protease (S2P) is an intramembrane zinc metalloprotease that releases a transcription factor domain from the membrane-bound SREBP protein to regulate cholesterol and fatty acid biosynthesis 5-6. Similar reactions important for transcriptional regulation are carried out by bacterial homologs of S2P, such as Eep from Enterococcus faecalis 7, SpoIVFB from Bacillus subtilis 8 and RseP from Escherichia coli 9. Sequence analysis revealed that S2P homologs are widely distributed in archaeal, bacterial and eukaryotic species 10. S2P contains a conserved ‘HExxH’ motif resembling the ones found in many soluble metalloproteases, albeit this motif in S2P is located in a transmembrane region. Recently, the crystal structure of an archaeal S2P homolog showed that this motif together with a couple of other conserved residues form the membrane-embedded metal-binding active site that overlays well with those from soluble metalloproteases 11, suggesting a similar catalytic mechanism. Another intramembrane protease, rhomboid, has been characterized as a serine protease functioning in signaling pathways of Drosophila development 12. Structural studies of select bacterial rhomboid homologs uncovered a catalytic Ser-His diad embedded in transmembrane regions 3,13-15.
Another important event of intramembrane proteolysis is performed by γ-secretase 16-17, which is a large membrane-bound protein complex consisting of four core subunits: presenilin, APH-1, nicastrin and PEN-2. Its substrates are type I membrane-spanning proteins such as the cell surface receptor NOTCH and the Alzheimer disease-related amyloid precursor protein. Presenilin, an aspartyl protease, is the catalytic subunit of γ-secretase. Homologs of presenilin also include signal peptide peptidases 18 and bacterial type 4 prepilin proteases 19. Nicastrin is a type I membrane protein and may function as the substrate receptor 20. PEN-2 is a small protein with two transmembrane segments. APH-1 is a seven-transmembrane spanning protein suggested to have a stabilizing role for γ-secretase. The precise function and the evolutionary origin of APH-1 remain unclear.
Two types of membrane-bound proteases are involved in the prenylation process that covalently attaches lipid molecules to proteins in order to facilitate their membrane localization 21. These prenyl proteases, located in the ER 22, perform cleavage within the CAAX motif of the substrate proteins to release the ‘AAX’ tripeptide (A: a typically aliphatic residue, X: one of several allowed residue types that can help determine the specificity of prenyltransferases 23). The new C-terminal residue of the processed protein is the prenylated cysteine. Type I CAAX prenyl proteases, namely ‘α-factor converting enzyme’ (AFC1, also called Ste24p in yeast), are metalloproteases with a conserved ‘HExxH’ motif 21,24-25. Type II CAAX proteases, ‘Ras and a-factor converting enzyme’ (RCE1), lack this sequence motif and their catalytic type has been debated in the literature. Early work suggested them to be cysteine proteases based on the activity of cysteine, histidine, and glutamate mutations and inhibitor analysis 26. Mutation of these residues all lead to loss of catalytic activity. However, the suspected catalytic cysteine residue was not conserved in the bacterial homologs. It was suggested that type II CAAX proteases were metalloproteases based on a set of conserved glutamates and histidines 27. These conserved residues were later shown to be important for catalysis in a site-directed mutagenic study of yeast Rce1p 28. The most probable explanation for the early suggestion that CAAX proteases were cysteine proteases is that there is a cysteine near the active site. Upon mutation or modification by inhibitors, the enzyme is deactivated although the residue does not directly participate in catalysis 29. Beyond these yeast type II CAAX protease studies, some bacterial homologs are involved in the proteolytic maturation of bacteriocins 30. Recently, the PrsW protease from Bacillus subtilis was found to be a distant homolog of type II CAAX proteases 31. PrsW was shown to mediate site-1 cleavage of anti-σ factor RsiW 31.
In this study, we conducted extensive sequence similarity searches of type II CAAX prenyl proteases in current sequence database using transitive PSI-BLAST 32 iterations. The CPBP (type II CAAX Proteases and Bacteriocin-Processing enzymes) family of putative metalloproteases was expanded to encompass more than 5,800 members. Remote similarities between type II CAAX proteases and other protein families were also investigated by the sensitive profile-profile search method HHpred 33. Weak yet significant similarities, highlighted by conserved sequence motifs, were identified between the CPBP family, the PrsW proteases, Pfam 34 family DUF2324 (DUF: Domain of Unknown Function), and surprisingly, the γ-secretase subunit APH-1. The protease origin of APH-1 raises interesting possibility that it may possess proteolytic activity in the current or ancestral form of γ-secretase.
Sequence similarity searches for homologs of type II CAAX prenyl peptidases
PSI-BLAST 32 was used to search for homologs of the type II CAAX protease family starting with the yeast Rce1p (NCBI gene identification (gi) number: 6323930) against the non-redundant (nr) database (12,322,590 sequences and 4,211,205,364 letters) with an e-value inclusion cutoff of 1e-4. To make transitive searches, found PSI-BLAST hits were grouped by BLASTCLUST (with option -S 1) and a representative sequence from each group was selected to initiate new PSI-BLAST searches. More than 5,800 members of the CPBP (type II CAAX Proteases and Bacteriocin Processing enzymes) family were found. Weaker similarities between members of the CPBP family, the PrsW protease family and the DUF2324 family were occasionally found during transitive PSI-BLAST searches (e-value inclusion cutoff: 1e-4, more stringent than the recommended cutoff value of web PSI-BLAST (0.005) to prevent profile corruption). For example, a PSI-BLAST search using a bacterial CPBP member from Atopobium vaginae (gi|303233059) found a significant hit to a PrsW protease family member from Sorangium cellulosum (gi| 162453351) with an e-value of 8e-5 in the fifth iteration. A PSI-BLAST search using a DUF2324 protein from Ruminococcus gnavus (gi| 154504350) found a significant hit to a PrsW protease family member from Flavobacteriales bacterium (gi| 163788689) with an e-value of 8e-5 in the ninth iteration.
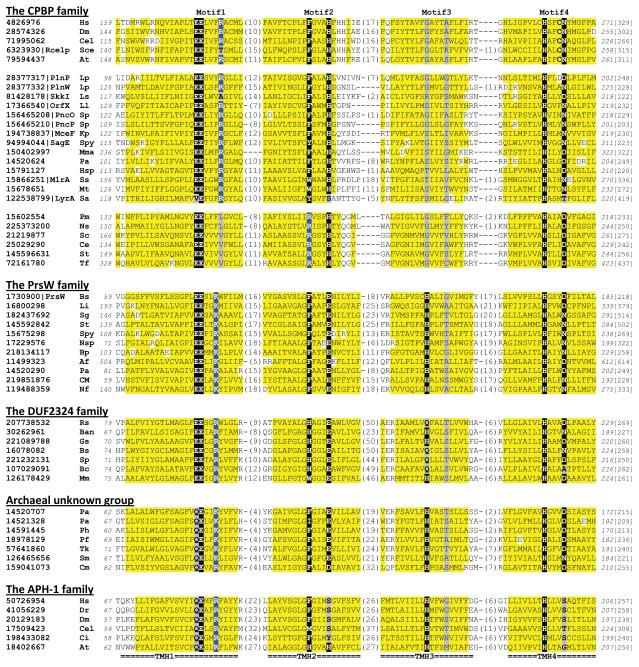
Transitive PSI-BLAST searches (e-value inclusion cutoff 1e-4) using members from the CPBP family, PrsW protease family and the DUF2324 family did not identify APH-1 proteins with statistical significance. Similarly, PSI-BLAST searches initiated with the APH-1 family representatives converged to about 150 eukaryotic members and did not find CPBP members, PrsW proteases or DUF2324 members as significant hits. However, the similarity between APH-1 and the other families can be found and statistically supported by the sensitive profile-profile method, HHpred 33. For example, an HHpred search using the human APH-1A (gi| 117606362) detected Pfam family DUF2324 (PF10086) with a probability score of 98.1 and an e-value of 4e-6. HHpred searches using PrsW and DUF2324 members also frequently identified Pfam domain Aph-1 (PF06015) among the top hits with probability scores above 90. Furthermore, their homologous relationships are supported by the presence of a set of similar sequence motifs harboring semi-invariant residues in four predicted core transmembrane segments (Figure 1, discussed below).
Figure 1. Multiple sequence alignments of representative members from protein families homologous to the type II CAAX prenyl proteases.
Four predicted core transmembrane helical segments (labeled TMH1-4 below the sequences) with conserved motifs (labeled Motif1-4 above the sequences) are shown for representative sequences of the CPBP family, the PrsW proteases, the DUF2324 family, a group of archaeal proteins with unknown function, and APH-1 proteins. The three separated groups in the CPBP family are eukaryotic type II CAAX proteases, prokaryotic homologs including putative bacteriocin-processing enzymes, and a group of bacterial proteins with the conserved arginine residues missing from the first motif but present in the second motif. Putative active site residues are shown on black background, except for the conserved positively charged residues, which are shown on blue background. Substitutions in these positions are on grey background. Non-charged residues in mainly hydrophobic positions are on yellow background. Small residues (residue types: G, A, S, C, V, T, N, D and P) in positions with mainly small residues are shown in red letters. NCBI gene identification numbers, along with common names for some proteins, are shown before the species name abbreviations. Numbers of residues in between the segments are shown in parentheses. Starting/ending residue numbers and sequence lengths are shown in italic font and in brackets, respectively. Species name abbreviations are: At: Arabidopsis thaliana, Af: Archaeoglobus fulgidus, Ba: Bacillus anthracis, Bs: Bacillus subtilis, Bp: Bacteroides pectinophilus, Bc: Burkholderia cenocepacia, Cel: Caenorhabditis elegans, Cm: Caldivirga maquilingensis, Ci: Ciona intestinalis, Ce: Corynebacterium efficiens, Dr: Danio rerio, Dm: Drosophila melanogaster, Gs: Geobacillus sp., Hsp: Halobacterium sp., Hs: Homo sapiens, Kp: Klebsiella pneumoniae, Lp: Lactobacillus plantarum, Ll: Lactococcus lactis, Li: Listeria innocua, Ls: Lactobacillus sakei, Mma: Methanococcus maripaludis, Mm: Methanoculleus marisnigri, Mp: Methanosphaerula palustris, Mt: Methanothermobacter thermautotrophicus, Ns: Neisseria subflava, Nf: Neosartorya fischeri, Nsp: Nostoc sp., Pm: Pasteurella multocida, Pa: Pyrococcus abyssi, Pf: Pyrococcus furiosus, Ph: Pyrococcus horikoshii, Rs: Ralstonia solanacearum, Sce: Saccharomyces cerevisiae, St: Salinispora tropica, Ss: Sphingomonas sp., Sa: Staphylococcus aureus, Sm: Staphylothermus marinus, Sp: Streptococcus pneumoniae, Spy: Streptococcus pyogenes, Sc: Streptomyces coelicolor, Sg: Streptomyces griseus, Tf: Thermobifida fusca, and Tk: Thermococcus kodakarensis. Domain color-coding: bacteria - black, archaea - orange, and eukaryotes - blue. This alignment was made by PROMALS 63 followed by manual adjustment.
Sequence families of proteins homologous to type II CAAX prenyl proteases
Four major families of proteins homologous to type II CAAX prenyl proteases exist: the CPBP family (type II CAAX Proteases and Bacteriocin Processing enzymes), PrsW proteases, DUF2324, and APH-1 (Figure 1). They share several similar sequence motifs with distinct variations.
The CPBP family
This family includes eukaryotic type II CAAX prenyl proteases and their related bacterial and archaeal homologs that can be repeatedly found in transitive PSI-BLAST searches and often share similar sequence motifs. This family is widely represented in the current sequence database with more than 5,800 members. CPBPs are present in all domains of life with most being bacterial. This family corresponds to Pfam entry Abi (PF02517, CAAX amino terminal protease family). A member of this family was originally identified as the product of an open reading frame (orfX) upstream of two genes (abiGi and abiGii) involved in an abortive infection system 35. However, inactivation of this CPBP member (OrfX, gi|17366540) suggests that it is not involved in the abortive infection mechanism. Thus it is inappropriate to name this family ‘Abi’ and annotate CPBP members as ‘abortive infection protein’ (about one fifth of the CPBP members (>1,000) are currently annotated in this way).
To obtain a clear picture of distribution of CPBPs in prokaryotes, we searched for CPBPs in 1190 fully sequenced prokaryotic genomes (downloaded from NCBI genome ftp site). 61 out of 90 archaeal genomes and 793 out of 1100 bacterial genomes have at least one CPBP (supplementary Table S2). The number of CPBPs among different prokaryotic phyla shows high variation (supplementary Table S3). About 50% of proteobacteria do not contain any identifiable CPBP. Conversely, the representation of CPBP family members in many other phyla is remarkably higher, with all sequenced cyanobacteria, bacteroidetes, chylamydiae, chlorobi, thermotogae, and deinococcus having at least one CPBP member. Within the firmicutes, actinobacteria, and euryarchaeaota phyla, it is not uncommon to find 5-20 CPBP family members within a single genome (supplementary Table S2). In one notable case, 21 copies are found within the genome of Streptococcus sanguinis SK36, which is currently the highest known. CPBP members in eukaryotes also have a wide phylogenetic distribution, with members from metazoans, fungi, plants and various protists.
Members of this family share four predicted core transmembrane segments, each of which contains a signature sequence motif (Figure 1). Most of the members contain several additional predicted transmembrane segments at the N-terminus and some members possess an additional predicted transmembrane segment at the C-terminus. For the majority of the CPBP family, the sequence motifs are ‘EExxxR’, ‘FxxxH’, ‘sxxxs’, and ‘HxxxB’ (‘x’: any amino acid residue, usually a hydrophobic residue; ‘s’: a small residue; ‘B’: asparagine or aspartate) from N- to C-termimus. While the presence of two conserved glutamates and two histidines matches the active site composition of several well-studied metal-dependent hydrolases (e.g. thermolysin 36, bovine carboxypeptidase A 37, D-Ala-D-Ala carboxypeptidase 38 and S2P-like proteases 11), the linear motifs used in metal-binding and catalysis are unlike the CPBP family. Accordingly, these enzymes have different structural folds and belong to different clans in the MEROPS peptidase database 39. Three residues are used for metal binding and one residue (usually glutamate) activates a metal-bound water molecule as the nucleophile to attack the substrate peptide bond. Presence of such a set of conserved residues in the CPBP family supports the prediction that most members in this family are metal-dependent proteases. The positively charged sidechain of the conserved arginine residue in the ‘EExxxR’ motif may contribute to the oxyanion hole that accommodates the carbonyl group of the substrate’s scissile bond. The third motif (‘sxxxs’), frequently found in transmembrane proteins 40, may be involved in helical packing, similar to the one observed in the second predicted core transmembrane segment in S2P 10.
Limited experiments have been carried out to investigate the function of the conserved glutamates and histidines in CPBP members. However, their functional importance is suggested by the results of several site-directed mutagenesis experiments performed on yeast type II CAAX protease Rce1p 26,28 and a bacterial CPBP member SkkI from Lactobacillus sakei that confers immunity to bacteriocins 41. In one site-directed mutagenesis study of yeast Rce1p 28, mutation of any one of the conserved glutamates and histidines (E156A or E157A in the first motif, H194A in the second motif, or H248A in the fourth motif) inactivated Rce1p’s enzymatic activity according to an in vitro assay. The same results for E156A, H194A and H248A were also obtained in a previous mutagenesis study 26. Mutation of the conserved aromatic residue in the second motif (F190A) or the asparagine in the fourth motif (N252A) resulted in reduced enzymatic activity 28. An in vivo assessment of Rce1p’s activity based on a mating assay exhibited similar results to the in vitro assessment 28. The importance of the negative charge in the first conserved glutamate in the first motif was suggested as the E156Q mutant had no in vivo activity under normal or over-expressed conditions (E156D had marginal activity though) 28. Mutations of several residues that are not conserved across the CPBP family did not affect the activity of Rce1p 28. For the L. sakei immunity protein SkkI, double mutants of the two conserved glutamates (E133A/E134A or E133Q/E134Q) in the first motif or a single mutant of the conserved histidine in the fourth motif (H214D) lost their ability to confer immunity 41. Further experimental studies are required to elucidate the catalytic mechanism of CPBP family members and the exact functional roles of the conserved residues.
A sequence clustering by the BLASTCLUST program was made on a non-redundant set of domains in this family (sequence redundancy removed at 90% level by CD-HIT 42). 34 out of 76 clusters with five or more members (including two largest clusters and capturing most of the CPBP members in this non-redundant set) contain the ‘EExxxR’, ‘FxxxH’, ‘sxxxs’ and ‘HxxxB’ motifs in the first, second, third and fourth predicted core transmembrane segments respectively (supplementary Table S1). The other clusters with five or more members show variations in at least one of these sequence motifs (supplementary Table S1). For example, several BLASTCLUST clusters have a ‘WxxxH’ motif instead of the ‘FxxxH’ motif in the second predicted core transmembrane helix. In the CLANS 43 visualization results (supplementary Figure S1), most of the ‘WxxxH’-containing members are close to each other and form a distinct sequence group. In two BLASTCLUST clusters (clusters 3 and 22 in supplementary Table S1), the second motif, instead of the first motif, harbors a conserved arginine (the first and second motifs become ‘EExxxx’ and ‘RxxxH’ respectively in these two clusters, Figure 1). Some clusters of proteins in this family (supplementary Table S1) have one or several amino acid replacements at the conserved glutamate and histidine positions, most probably rendering such family members proteolytically inactive (e.g., the LyrA protein from Staphylococcus aureus 44).
Eukaryotic type II CAAX proteases proteolytically remove the C-terminal tripeptide ‘AAX’. This tripeptide lies directly C-terminal to a cysteine residue modified with a farnesyl (C15) or geranylgeranyl (C20) prenyl chain, which facilitates membrane localization 26. These proteases form a distinct sequence group in the CLANS diagram (supplementary Figure S1). Despite the fact that most CPBP members are from bacteria, only a few bacterial members have been studied experimentally and details of their function remain elusive. Based on available data, the function of many of the uncharacterized bacterial CPBP members is likely related to bacteriocin maturation. Bacteriocins are polypeptide toxins secreted by virtually all bacteria to inhibit the growth of competing bacterial strains/species 45-46. The proposed protease activity of bacterial CPBP members could be utilized in the maturation and secretion process of bacteriocins, and/or help conferring immunity against self-produced bacteriocins. For example, a number of CPBP members (PlnI, PlnL, PlnT, PlnP, PlnV, PlnU and PlnW) were found in operons that produce bacteriocins in various strains of Lactobacillus plantarum 30,47. Recently, it was shown that CPBP members SkkI from L. actobacillus and PlnI and PlnL from L. plantarum confer immunity to their cognate bacteriocins 41. In Streptococcus pneumoniae, CPBP members PncO and PncP were found in the pnc locus encoding bacteriocins, and mutation of PncO abolished bacteriocin production 48. A CPBP member, SagE from Streptococcus pyogenes, is encoded by a biosynthetic operon that produces streptolysin S, a toxin generated by post-translational modification of a bacteriocin-like precursor 49-51. Toxins similar to streptolysin S have been found in several other Gram-positive pathogens, such as Staphylococcus aureus, Listeria monocytogenes, and Clostridium botulinum. In all cases, a type II CAAX protease (SagE homolog) is found in the biosynthetic gene cluster 52. In the Gram-negative Klebsiella pneumoniae, a CPBP member MceF has been shown to be important in the export of microcin E492 53. Another CPBP member MlrA from Sphingomunas sp., which has a second motif ‘WxxxH’ instead of ‘FxxxH’, was characterized as an endopeptidase (microcystinase) responsible for the degradation of the cyanobacterial toxin microcystin LR 54. Inhibition of MlrA by EDTA is consistent with the prediction that CPBP members are metalloproteases. The four conserved glutamates and histidines within the sequence motifs are present in most of the above-mentioned examples of bacterial CPBP members. Some of these examples, such as PlnP, SkkI, PncO, SagE and MlrA, are included in the alignment shown in Figure 1 with conserved residues highlighted.
Except for the protease domain, most of the CPBP members appear to lack other known domains as revealed by profile HMM-based 55 searches against the Pfam database 34 (about 96% of CPBP members have no more than one Pfam domain hits at the default e-value cutoff of 0.01). Domain composition analysis offers insights into the function of some multi-domain CPBPs. For example, some CPBP members from Bacillus species (e.g., gi| 206968619) are annotated as AbrB family transcriptional regulators as they also contain a DNA-binding AbrB domain at the C-terminus, suggesting that the protease activity may be involved in transcriptional regulation. Functional association with AbrB domains is also supported by a recent study of an AbrB family protein CalA in Nostoc sp., which was shown to be co-transcribed with a CPBP member 56. In some CPBP members, the protease domains are fused with components of ABC transporters into a single protein (e.g., gi| 254975247). In other cases, some genes encoding CPBP members are neighbors of genes encoding components of ABC transporters, as revealed in the bacteriocin operon clusters 30 as well as by the STRING functional association server (e.g., gi| 118476928) 57. Co-expression of CPBP members with ABC transporters were found in a recent study of the two-component system BfrAB involved in biofilm formation in Streptococcus species 58. These lines of evidence suggest that some CPBP members function to couple leader peptide cleavage with cellular export in a similar fashion to certain bacteriocin ABC transporters with protease domains 59.
The PrsW protease family
This group includes close homologs of the experimentally characterized PrsW protease from Bacillus subtilis 31,60. PrsW mediates site-1 cleavage of anti-sigma factor RsiW, one the two steps of proteolysis events that lead to activation of transcription factor σW in response to antimicrobial peptides such as bacteriocins and cell envelop stress. Several noticeable differences in sequence motifs exist between PrsW proteases and the CPBP family (Figure 1). First, the most N-terminal motif of interest from PrsW proteases bears the consensus signature of ‘EExxK’ instead of ‘EExxxR’ as in most others CPBPs. Second, the PrsW proteases have the second motif ‘FxxxE’ in place of ‘FxxxH’ as in the CPBP family. Third, PrsW proteases possess a conserved histidine in the third motif, which is absent in the CPBP family. The fourth motif, ‘HxxxB’, is shared by the PrsW proteases and the CPBP family. Limited experiments have been performed on PrsW members and the only site-directed mutagenesis study was available for the PrsW member from B. subtilis 31. Either double point mutation of the two conserved glutamates in the first motif (E75A/E76A), or a single mutation of the conserved histidine in the fourth motif (H175A), was not able to complement the phenotype caused by the prsW-null mutation 31, suggesting the functional importance of these conserved residues for B. subtilis PrsW. Further experimental studies are needed to determine the functional roles of the conserved residues in PrsW members.
Transitive PSI-BLAST searches found about 500 PrsW members, most of which are from the Gram-positive bacterial phyla of Firmicutes and Actinobacteria. Some PrsW members (e.g., gi|298242689) contain an FHA (Forkhead-associated) domain 61 at the N-terminus. Sequence clustering by the CLANS program (supplementary Figure S2) revealed two distinct groups of archaeal PrsW members. One archaeal group is closely related to and clustered with members from bacterial species. The second archaeal group forms a separate cluster and has relatively high sequence divergence among its members. Proteins in the second archaeal group also contain an ABC-transporter permease domain, suggesting their roles in membrane transportation. The distribution of PrsW proteases in eukaryotes is dramatically restricted to only a few unicellular organisms, suggesting massive gene loss in most eukaryotic lineages. PrsW proteases were found in the diatom species Thalassiosira pseudonana and Phaeodactylum tricornutum, Apicomplexa from genera Plasmodium and Cryptosporidium, the percolozoan Naegleria gruberi, and two fungal species Neosartorya fischeri and Aspergillus fumigatus.
The DUF2324 family
This family includes proteins from Pfam family DUF2324 (DUF: Domain of Unknown Function), with about 200 proteins in the current sequence database. They are mainly from Firmicutes and Proteobacteria. Motif variation from the CPBP family and the PrsW family is observed in DUF2324 (Figure 1), with its first, second and third motifs being ‘EExxR’, ‘HxxxE’ and ‘[HQ]xxxs’, respectively. The fourth motif ‘HxxxB’ remains the same. The function of these proteins has not been revealed by experimental studies, and many of the proteins in this group are annotated as hypothetical proteins. PSI-BLAST searches using DUF2324 members as queries also identified a distantly related group of archaeal proteins with another version of sequence motifs (‘QExxK’, ‘FxxxE’, ‘Hxxxs’ and ‘HxxxB’) similar to that of the PrsW proteases (Figure 1).
The APH-1 family
This family includes the APH-1 subunits of γ-secretases. APH-1 proteins are restricted to eukaryotes. Widely distributed in metazoans and plants, APH-1s were also found in amoebozoans such as Dictyostelium discoideum, two parasitic euglenozoan genera Trypanosoma and Leishmania, and several stramenopiles such as Phaeodactylum tricornutum (a diatom), Ectocarpus siliculosus (a brown algae), Phytophthora infestans (an oomycete) and Blastocystis hominis. The gene encoding APH-1 appears to have been lost in fungi and may also be absent in many other unicellular eukaryotic species (fast evolutionary rates in certain lineages could prevent detection of some APH-1s). APH-1 proteins have a similar but different set of motifs as compared to other families. Their four motifs are ‘QExxR’, ‘Fxxxx’, ‘Hxxxs’ and ‘Hxxxs’ respectively (Figure 1).
The remote similarity between APH-1 and the other protease families is an interesting finding. The presenilin subunit has been the center in the study of γ-secretase as it is regarded as the catalytic subunit 62. On the other hand, much less is known about the exact function and evolutionary origin of the subunit APH-1. Given the inferred homology between APH-1 and other membrane proteases such as type II CAAX proteases and PrsW proteases, we speculate that APH-1 may possess protease activity, thus expanding the range of substrates for γ-secretase. It should be noted that another subunit of γ-secretase, nicastrin, also possesses a protease domain, which is located in the extracellular region and has uncharacterized function. As no protease activity of APH-1 has been shown experimentally, it is likely that APH-1 lost the protease activity and has other functional roles in the γ-secretase complex. In this scenario, it is plausible that APH-1 in the ancestral form of γ-secretase was catalytically active and performed proteolytic reactions along with the presenilin subunit.
Supplementary Material
The CPBP intramembrane proteases are widely distributed in all domains of life.
CPBP, PrsW, DUF2324 and γ-secretase subunit APH-1 are distantly related.
APH-1 may possess proteolytic activity in the extant or ancestral form of γ-secretase.
Acknowledgements
We would like to thank David Baker, Vladimir Yarov-Yarovoy and Gang Yu for helpful discussions and Lisa Kinch for critical reading of the manuscript. This work was supported in part by National Institute of Health (1R01GM094575-01 to NVG and 2R01GM090328-06A1 to JED) and the Welch Foundation (I-1505 to NVG). DAM would like to thank the Department of Chemistry and Institute for Genomic Biology at University of Illinois at Urbana-Champaign for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100(4):391–8. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe MS, Kopan R. Intramembrane proteolysis: theme and variations. Science. 2004;305(5687):1119–23. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci U S A. 2007;104(2):462–6. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urban S, Shi Y. Core principles of intramembrane proteolysis: comparison of rhomboid and site-2 family proteases. Curr Opin Struct Biol. 2008;18(4):432–41. doi: 10.1016/j.sbi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77(1):53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 6.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci U S A. 1999;96(20):11041–8. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An FY, Sulavik MC, Clewell DB. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J Bacteriol. 1999;181(19):5915–21. doi: 10.1128/jb.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci U S A. 1999;96(26):14765–70. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 2002;16(16):2156–68. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinch LN, Ginalski K, Grishin NV. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci. 2006;15(1):84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318(5856):1608–12. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 12.Urban S. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 2006;20(22):3054–68. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444(7116):179–80. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 14.Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13(12):1084–91. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 15.Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc Natl Acad Sci U S A. 2007;104(3):750–4. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dries DR, Yu G. Assembly, maturation, and trafficking of the gamma-secretase complex in Alzheimer’s disease. Curr Alzheimer Res. 2008;5(2):132–46. doi: 10.2174/156720508783954695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiner H, Fluhrer R, Haass C. Intramembrane proteolysis by gamma-secretase. J Biol Chem. 2008;283(44):29627–31. doi: 10.1074/jbc.R800010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296(5576):2215–8. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 19.LaPointe CF, Taylor RK. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J Biol Chem. 2000;275(2):1502–10. doi: 10.1074/jbc.275.2.1502. [DOI] [PubMed] [Google Scholar]
- 20.Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE, 3rd, Sudhof T, Yu G. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122(3):435–47. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275(5307):1796–800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt WK, Tam A, Fujimura-Kamada K, Michaelis S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc Natl Acad Sci U S A. 1998;95(19):11175–80. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurer-Stroh S, Washietl S, Eisenhaber F. Protein prenyltransferases. Genome Biol. 2003;4(4):212. doi: 10.1186/gb-2003-4-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt WK, Tam A, Michaelis S. Reconstitution of the Ste24p-dependent N-terminal proteolytic step in yeast a-factor biogenesis. J Biol Chem. 2000;275(9):6227–33. doi: 10.1074/jbc.275.9.6227. [DOI] [PubMed] [Google Scholar]
- 25.Tam A, Schmidt WK, Michaelis S. The multispanning membrane protein Ste24p catalyzes CAAX proteolysis and NH2-terminal processing of the yeast a-factor precursor. J Biol Chem. 2001;276(50):46798–806. doi: 10.1074/jbc.M106150200. [DOI] [PubMed] [Google Scholar]
- 26.Dolence JM, Steward LE, Dolence EK, Wong DH, Poulter CD. Studies with recombinant Saccharomyces cerevisiae CaaX prenyl protease Rce1p. Biochemistry. 2000;39(14):4096–104. doi: 10.1021/bi9923611. [DOI] [PubMed] [Google Scholar]
- 27.Pei J, Grishin NV. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci. 2001;26(5):275–7. doi: 10.1016/s0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- 28.Plummer LJ, Hildebrandt ER, Porter SB, Rogers VA, McCracken J, Schmidt WK. Mutational analysis of the ras converting enzyme reveals a requirement for glutamate and histidine residues. J Biol Chem. 2006;281(8):4596–605. doi: 10.1074/jbc.M506284200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dechert AM, MacNamara JP, Breevoort SR, Hildebrandt ER, Hembree NW, Rea AC, McLain DE, Porter SB, Schmidt WK, Dore TM. Modulation of the inhibitor properties of dipeptidyl (acyloxy)methyl ketones toward the CaaX proteases. Bioorg Med Chem. 2010;18(17):6230–7. doi: 10.1016/j.bmc.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diep DB, Havarstein LS, Nes IF. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J Bacteriol. 1996;178(15):4472–83. doi: 10.1128/jb.178.15.4472-4483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellermeier CD, Losick R. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 2006;20(14):1911–22. doi: 10.1101/gad.1440606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–60. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 34.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–22. doi: 10.1093/nar/gkp985. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connor L, Coffey A, Daly C, Fitzgerald GF. AbiG, a genotypically novel abortive infection mechanism encoded by plasmid pCI750 of Lactococcus lactis subsp. cremoris UC653. Appl Environ Microbiol. 1996;62(9):3075–82. doi: 10.1128/aem.62.9.3075-3082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews BW, Colman PM, Jansonius JN, Titani K, Walsh KA, Neurath H. Structure of thermolysin. Nat New Biol. 1972;238(80):41–3. doi: 10.1038/newbio238041a0. [DOI] [PubMed] [Google Scholar]
- 37.Christianson DW, David PR, Lipscomb WN. Mechanism of carboxypeptidase A: hydration of a ketonic substrate analogue. Proc Natl Acad Sci U S A. 1987;84(6):1512–5. doi: 10.1073/pnas.84.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dideberg O, Charlier P, Dive G, Joris B, Frere JM, Ghuysen JM. Structure of a Zn2+-containing D-alanyl-D-alanine-cleaving carboxypeptidase at 2.5 A resolution. Nature. 1982;299(5882):469–70. doi: 10.1038/299469a0. [DOI] [PubMed] [Google Scholar]
- 39.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38(Database issue):D227–33. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296(3):911–9. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 41.Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. The abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol. 2010;192(8):2068–76. doi: 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 43.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20(18):3702–4. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 44.Grundling A, Missiakas DM, Schneewind O. Staphylococcus aureus mutants with increased lysostaphin resistance. J Bacteriol. 2006;188(17):6286–97. doi: 10.1128/JB.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3(10):777–88. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 46.Drider D, Fimland G, Hechard Y, McMullen LM, Prevost H. The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev. 2006;70(2):564–82. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rojo-Bezares B, Saenz Y, Navarro L, Jimenez-Diaz R, Zarazaga M, Ruiz-Larrea F, Torres C. Characterization of a new organization of the plantaricin locus in the inducible bacteriocin-producing Lactobacillus plantarum J23 of grape must origin. Arch Microbiol. 2008;189(5):491–9. doi: 10.1007/s00203-007-0342-6. [DOI] [PubMed] [Google Scholar]
- 48.Lux T, Nuhn M, Hakenbeck R, Reichmann P. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J Bacteriol. 2007;189(21):7741–51. doi: 10.1128/JB.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datta V, Myskowski SM, Kwinn LA, Chiem DN, Varki N, Kansal RG, Kotb M, Nizet V. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol Microbiol. 2005;56(3):681–95. doi: 10.1111/j.1365-2958.2005.04583.x. [DOI] [PubMed] [Google Scholar]
- 50.Lee SW, Mitchell DA, Markley AL, Hensler ME, Gonzalez D, Wohlrab A, Dorrestein PC, Nizet V, Dixon JE. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci U S A. 2008;105(15):5879–84. doi: 10.1073/pnas.0801338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitchell DA, Lee SW, Pence MA, Markley AL, Limm JD, Nizet V, Dixon JE. Structural and functional dissection of the heterocyclic Peptide cytotoxin streptolysin s. J Biol Chem. 2009;284(19):13004–12. doi: 10.1074/jbc.M900802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cotter PD, Draper LA, Lawton EM, Daly KM, Groeger DS, Casey PG, Ross RP, Hill C. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008;4(9):e1000144. doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagos R, Baeza M, Corsini G, Hetz C, Strahsburger E, Castillo JA, Vergara C, Monasterio O. Structure, organization and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol Microbiol. 2001;42(1):229–43. doi: 10.1046/j.1365-2958.2001.02630.x. [DOI] [PubMed] [Google Scholar]
- 54.Saito T, Okano K, Park HD, Itayama T, Inamori Y, Neilan BA, Burns BP, Sugiura N. Detection and sequencing of the microcystin LR-degrading gene, mlrA, from new bacteria isolated from Japanese lakes. FEMS Microbiol Lett. 2003;229(2):271–6. doi: 10.1016/S0378-1097(03)00847-4. [DOI] [PubMed] [Google Scholar]
- 55.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23(1):205–11. [PubMed] [Google Scholar]
- 56.Agervald A, Zhang X, Stensjo K, Devine E, Lindblad P. CalA, a cyanobacterial AbrB protein, interacts with the upstream region of hypC and acts as a repressor of its transcription in the cyanobacterium Nostoc sp. strain PCC 7120. Appl Environ Microbiol. 2010;76(3):880–90. doi: 10.1128/AEM.02521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–6. doi: 10.1093/nar/gkn760. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Whiteley M, Kreth J, Lei Y, Khammanivong A, Evavold JN, Fan J, Herzberg MC. The two-component system BfrAB regulates expression of ABC transporters in Streptococcus gordonii and Streptococcus sanguinis. Microbiology. 2009;155(Pt 1):165–73. doi: 10.1099/mic.0.023168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Havarstein LS, Diep DB, Nes IF. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16(2):229–40. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 60.Heinrich J, Wiegert T. YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol Microbiol. 2006;62(2):566–79. doi: 10.1111/j.1365-2958.2006.05391.x. [DOI] [PubMed] [Google Scholar]
- 61.Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4(3):387–94. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 62.Ahn K, Shelton CC, Tian Y, Zhang X, Gilchrist ML, Sisodia SS, Li YM. Activation and intrinsic gamma-secretase activity of presenilin 1. Proc Natl Acad Sci U S A. 2010;107(50):21435–40. doi: 10.1073/pnas.1013246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23(7):802–8. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.