Abstract
Oestrogen receptor α (ERα) is key player in the progression of breast cancer. Recently, the cistrome and interactome of ERα were mapped in breast cancer cells, revealing the importance of spatial organization in oestrogen-mediated transcription. However, the underlying mechanism of this process is unclear. Here, we show that ERα binding sites (ERBS) identified from the Chromatin Interaction Analysis-Paired End DiTag of ERα are enriched for AP-2 motifs. We demonstrate the transcription factor, AP-2γ, which has been implicated in breast cancer oncogenesis, binds to ERBS in a ligand-independent manner. Furthermore, perturbation of AP-2γ expression impaired ERα DNA binding, long-range chromatin interactions, and gene transcription. In genome-wide analyses, we show that a large number of AP-2γ and ERα binding events converge together across the genome. The majority of these shared regions are also occupied by the pioneer factor, FoxA1. Molecular studies indicate there is functional interplay between AP-2γ and FoxA1. Finally, we show that most ERBS associated with long-range chromatin interactions are colocalized with AP-2γ and FoxA1. Together, our results suggest AP-2γ is a novel collaborative factor in ERα-mediated transcription.
Keywords: AP-2γ, chromatin immunoprecipitation and sequencing, FoxA1, long-range chromatin interaction, oestrogen receptor
Introduction
Oestrogens have important roles in both female development and reproductive function, as well as in regulating the growth and progression of breast and endometrial cancers (Ali and Coombes, 2000). The effects of oestrogens are mediated by two receptors, oestrogen receptor α (ERα) and ERβ, which are members of the nuclear hormone receptor superfamily (Nilsson et al, 2001). Upon oestrogen stimulation, ER is recruited to the cis-regulatory element of target genes either directly via oestrogen response elements (EREs) or indirectly by interacting with DNA bound transcription factors such as AP-1 and Sp1 (Paech et al, 1997; Porter et al, 1997; Qin et al, 1999; Webb et al, 1999). ER then recruits in a temporal- and spatial-specific manner a combination of general transcription factors, coregulator proteins (co-activators and corepressors), and collaborative factors that ultimately leads to either the activation or repression of target genes (Horwitz et al, 1996; Glass et al, 1997; Metivier et al, 2003). Therefore, identifying these factors and understanding how they function together with ER will constitute a key source of potential novel therapeutic targets in ER-related diseases such as breast cancer.
Genomic analyses of ERα binding sites (ERBS) by chromatin immunoprecipitation (ChIP)-based technologies such as ChIP-chip and ChIP-seq have recently led to the discovery and a better understanding of the function of ERα collaborative factors (Carroll et al, 2005; Lin et al, 2007; Welboren et al, 2009; Cheung and Kraus, 2010). For example, the pioneer factor FoxA1, which is overexpressed in breast cancer, was found occupied at ERBS before oestrogen stimulation (Carroll et al, 2005). More importantly, the pre-bound FoxA1 appears to be required for facilitating ERα recruitment and modifying chromatin structure at the regulatory regions of oestrogen target genes (Carroll et al, 2005; Lupien et al, 2008; Eeckhoute et al, 2009). PAX2, a factor previously implicated in endometrial cancer, was also shown localized at ERBS but mainly in response to tamoxifen treatment (Hurtado et al, 2008). PAX2 blocked the recruitment of co-activators such as SRC3, resulting in the repression of transcription and ultimately tamoxifen response in breast cancer cells (Hurtado et al, 2008). More recently, two studies showed ERα and the retinoic acid receptor-α (RARα), another member of the nuclear hormone receptor superfamily and a direct target of ERα, shared a large number of genomic binding sites (Hua et al, 2009; Ross-Innes et al, 2010). Interestingly, depending on the mechanism involved RARα can have both a positive and negative effect on the transcriptional activity of ERα (Hua et al, 2009; Ross-Innes et al, 2010). Other collaborative factors of ERα that have been identified include NKX3.1, LEF1, and GATA3 (Eeckhoute et al, 2007; Holmes et al, 2008). Considering the complexity of transcriptional regulation, it is likely there are many more transcription factors that are required for coordinating ERα function, which currently remains unknown.
To obtain a better understanding of ERα-regulated transcription, we recently developed a novel technique, Chromatin Interaction Analysis-Paired End DiTag (ChIA-PET), for the de novo and high-throughput identification of long-range chromatin interactions mediated by protein factors (Fullwood et al, 2009). This method couples ChIP, chromosome conformation capture (3C), and paired-end cloning, as well as massive-parallel sequencing to capture interactions between distant DNA fragments brought in close proximity. A single ChIA-PET analysis of a transcription factor provides two sets of genome-wide information: transcription factor binding site and long-range chromatin interaction. Using this new approach, we mapped the binding sites and long-range chromatin interactions mediated by ERα under oestrogen stimulation in the breast cancer cell line, MCF-7. In all, 14 468 ERBS and 1451 long-range chromatin interactions were identified. The majority of high confidence ERBS (genomic regions that have at least 50 or more self-ligation PET counts per cluster) are involved in long-range chromatin interactions and these interactions are strongly correlated with oestrogen-regulated genes. With extensive maps of both the ERα cistrome and interactome in breast cancer cells, we next turned our efforts to determine the factors that are involved in regulating ERα binding and long-range chromatin interactions as well as the underlying mechanism(s) mediating these processes. Herein, we used a combination of molecular, genomic, and computational approaches to identify and functionally characterize in detail a novel collaborative factor of ERα. Our results suggest AP-2γ, a transcription factor involved in breast cancer oncogenesis, facilitates ERα binding, long-range chromatin interactions, and transcription by working together with FoxA1.
Results
AP-2γ is recruited to ERBS
Recent studies showed that transcription factors such as FoxA1, PAX2, and RARα are important in the activation and repression of ERα-dependent transcription (Carroll et al, 2005; Hurtado et al, 2008; Hua et al, 2009; Ross-Innes et al, 2010). To discover novel factors that function with ERα, we examined the ERBS (both high confidence sites and all ERBS) identified from our recent ChIA-PET of ERα by scanning for over-represented DNA binding motifs of TFs from the TRANSFAC database. As expected, our screen identified binding sites for previously reported collaborative factors of ERα such as AP1, forkhead factors, and GATA (Supplementary Figure S1A). Moreover, we found sequences that were enriched for the AP-2 family of transcription factors. In fact, our result showed that AP-2 motifs were one of the highest enriched sequences in ERBS, ranking higher than motifs for forkhead factors. We characterized the AP-2 motifs further by determining their location with respect to ERBS. Consistent with previous findings on cofactors of ERα (Carroll et al, 2006), AP-2 motifs were preferentially distributed near the centre of ERBS (Supplementary Figure S1B). Finally, we reasoned that if members of the AP-2 family are potentially involved in ERα-mediated transcription then a large fraction of the ERα ChIA-PET binding sites should contain AP-2 motifs. Indeed, ∼40% of all ERBS identified from ChIA-PET were predicted to harbour AP-2 motifs (Supplementary Figure S2).
The AP-2 family of transcription factors consists of five members, AP-2α–ε (Eckert et al, 2005). To date, little is known on the interaction between AP-2 transcription factors and ERα; however, previous studies have shown that AP-2γ is a key player in mammary oncogenesis, a predictor of poor survival outcome in breast cancer patients, and its protein level is elevated in human mammary carcinomas (Jager et al, 2003, 2005; Woodfield et al, 2007; Gee et al, 2009; Williams et al, 2009). In MCF-7 cells, AP-2γ is the predominantly expressed member of the AP-2 family at both the mRNA and protein levels (Woodfield et al, 2007). Based on these findings, we examined whether AP-2γ is recruited to ERBS in MCF-7 cells. As shown in Supplementary Figure S1C, using a specific antibody raised against AP-2γ, our ChIP assay revealed AP-2γ was enriched at 10 selected ERBS harbouring predicted AP-2 motifs. Taken together, our bioinformatics findings combined with our ChIP results suggest that AP-2γ is colocalized at ERBS.
AP-2γ is essential for efficient transcription of oestrogen-regulated genes
Our previous results showed that ERα-mediated long-range chromatin interactions are preferentially associated with oestrogen upregulated genes (Fullwood et al, 2009). To understand the role of AP-2γ in ERα-mediated long-range chromatin interaction and transcription, we scanned for the presence of AP-2 motifs in ERBS that are involved in long-range chromatin interactions and associated with 17β-estradiol (E2) responsive genes. In general, we found predicted AP-2 motifs frequently occurring in many ERBS at model E2 responsive genes such as GREB1 (Supplementary Figure S3A). In addition, we identified AP-2 motifs in a cluster of ERBS within the regulatory region of the RET (REarranged after Transfection) proto-oncogene (Figure 1A). The RET gene was recently shown to be regulated by oestrogen stimulation (Boulay et al, 2008). We confirmed this in time course analyses of RET expression at both transcript and protein levels in MCF-7 cells. Our results showed that RET (both the RET9 and RET51 isoforms) is upregulated by E2 (Figure 1B; Supplementary Figure S4A). Moreover, cycloheximide treatment and knockdown of ERα in MCF-7 cells indicate RET is likely a direct target of ERα (Figure 1C; Supplementary Figures S4B, C and S5).
Figure 1.
AP-2γ is required for transcription of oestrogen-regulated genes. (A) Screenshot of ERα ChIA-PET analysis showing ERα binding and long-range chromatin interactions at the RET gene locus. ERBS are represented as density histogram (red) and long-range chromatin interactions are represented as intra-chromosomal interaction PETs (magenta). RET-associated ERBS are denoted by numbers (blue). (B) MCF-7 cells were stimulated with or without E2 for 0, 3, 6, 12, and 24 h and then analysed by reverse transcription and real-time RT–PCR for the level of RET 9 and 51 mRNA expressions. (C) MCF-7 cells were transfected with control, ERα or AP-2γ siRNA, stimulated with or without E2 for 12 h and then analysed for RET 9 and 51 mRNA levels. (D) Gene expression profiling was performed on MCF-7 cells that were transfected with control or AP-2γ siRNA and stimulated with or without E2 for 12 h. The heatmap represents all E2-regulated genes and fold change in expression is indicated below. E2-upregulated genes and E2-downregulated genes that are no longer activated and repressed due to AP-2γ knockdown are marked by the red and blue asterisks, respectively. All results represent the average of three independent experiments ±s.e.m.
The colocalized binding of AP-2γ with ERα at ERBS suggests AP-2γ may be important in regulating ERα-dependent gene transcription. To test this, we examined the effect of siRNA-mediated knockdown of AP-2γ on RET expression. As shown in Figure 1C and Supplementary Figure S4B, knockdown of AP-2γ in MCF-7 cells completely reduced the transcript and protein levels of both RET isoforms, but had no effect on ERα protein level (Supplementary Figure S6). Similar effects were also observed for the transcription of the GREB1 gene after AP-2γ knockdown (data not shown). We expanded our knockdown studies by performing microarray analysis on MCF-7 cells transfected with AP-2γ siRNAs, in the presence and absence of E2 (Figure 1D). Our analysis revealed 676 E2 responsive genes (1.5-fold change cutoff), of which 349 were upregulated while 327 were downregulated by E2. Interestingly, the depletion of AP-2γ affected the expression of a significant number of E2-induced (59.8%/209) and E2-repressed genes (29.1%/95), suggesting that AP-2γ can function as either a transcriptional activator or repressor to regulate ERα-mediated transcription. Taken together, our results demonstrate that AP-2γ is required for efficient transcription of ERα target genes.
AP-2γ binds to RET-associated ERBS in a ligand-independent manner
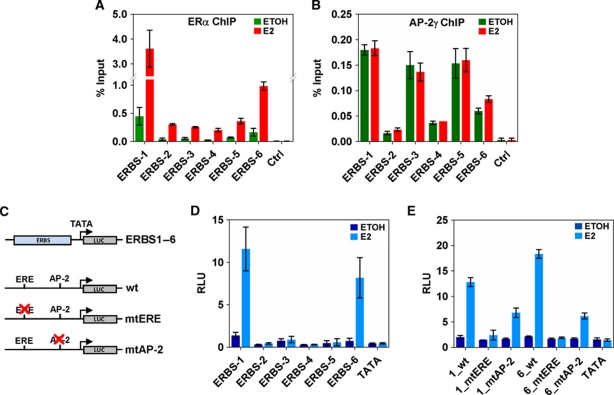
To examine the detailed mechanism of AP-2γ function in ERα-dependent transcription, we focused our study on the regulation of the RET gene. RET encodes for a tyrosine kinase receptor that has been shown to be involved in thyroid carcinoma development and breast cancer proliferation (Santoro et al, 2002; Arighi et al, 2005; Boulay et al, 2008). However, how it is regulated by E2 signalling is unclear. As shown in Figure 1A, the RET gene is associated with multiple ERBS (ERBS-1 to -6) which are part of ‘a complex’ chromatin interaction that spans from −50 to +35 kb of transcription start site. In ChIP assays, we showed AP-2γ was enriched at all six binding sites with the strongest recruitment at ERBS-1, -3, -5, and -6 (Figure 2A and B). Interestingly, AP-2γ binding was present at these ERBS before any stimulation and remained constant after the addition of E2. The ligand-independent binding of AP-2γ was not only observed at the ERBS of RET, but also detected at randomly selected ERBS and at ERBS associated with GREB1 (Supplementary Figures S1C and S3B).
Figure 2.
Ligand-independent recruitment of AP-2γ at RET ERBS. ChIP assays were performed in MCF-7 cells treated with or without E2 for 45 min using antibodies against (A) ERα and (B) AP-2γ. Binding was assessed by real-time RT–PCR at RET-associated ERBS as described in Figure 1A. (C) Schematic diagram showing the reporter constructs that were generated and used in transient transfection analysis. (D) MCF-7 cells were transfected with reporter constructs and treated with or without E2 for 24 h. Luciferase assays were performed using a dual-luciferase system with Renilla as an internal control. The six RET-associated ERBS, ERBS-1–6, were each cloned into pGL4-TATA and assessed in transient transfection analysis. (E) ERE and AP-2 motifs predicted in ERBS-1 and -6 were mutated and compared with their equivalent wild-type versions in transient transfection analysis. All results represent the average of three independent experiments ±s.e.m.
To explore the potential interplay between ERα and AP-2γ, we cloned the six ERBS associated with RET into luciferase reporter constructs containing a minimal TATA box (Figure 2C) and transiently transfected them into MCF-7 cells with or without E2 stimulation. Luciferase constructs containing ERBS-1 and -6, but not ERBS-2 to -5, were efficiently activated and regulated in an E2-dependent manner (Figure 2D). Interestingly, the luciferase results are consistent with the binding of ERα but not the binding of AP-2γ. This is likely due to the limitations of the assay which can be indicative of transcriptional mechanisms (e.g., for sites such as ERBS-1 and -6), but does not always reflect real mechanisms for endogenous proteins and genes (e.g., for sites such as ERBS-2 to -5). Nevertheless, since only ERBS-1 and -6 exhibited enhancer activity and E2 response, we used constructs containing these two binding sites for further mutagenesis analysis. The sequences of both binding sites contain an imperfect ERE and an AP-2 motif. As expected, mutating the ERE motif in the ERBS abolished the E2 response and transcriptional activity of both constructs (Figure 2E). Similarly, mutating the AP-2 motif also significantly reduced the reporter activity of the constructs, but this effect was not as drastic as mutating the EREs (Figure 2E). Together, our results suggest that AP-2γ binds to chromatin in a ligand-independent manner and functions together with ERα to induce maximal transcriptional activation.
AP-2γ is required for ERα-mediated long-range chromatin interactions
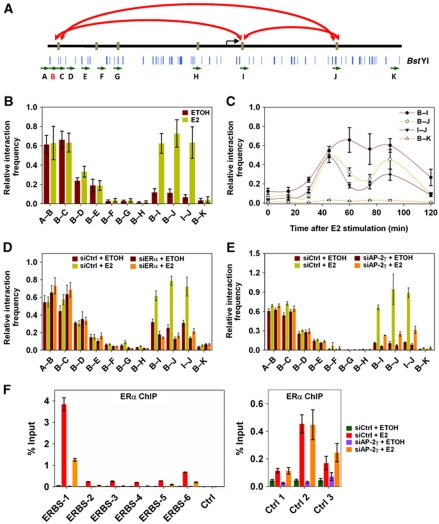
The ChIA-PET map of ERα revealed the ERBS spanning the regulatory region of the RET gene is in close proximity with one another forming ‘a complex’ chromatin interaction (Figure 1A). Since our results showed colocalization of AP-2γ at these ERBS, we asked if AP-2γ is required in the formation of the complex interaction. To determine this, we used the 3C assay (Dekker et al, 2002). With this approach, we were able to detect three of the duplex chromatin interactions that were identified from the ERα ChIA-PET analysis (Figure 3A and B). These duplex chromatin interactions involved regions from ERBS-1, -5, and -6. Moreover, the three duplex chromatin interactions were enhanced upon E2 stimulation, occurred in a cyclical manner, and dependent on ERα (Figure 3B–D). Although we were able to detect some but not all of the interactions at the RET region by 3C, likely due to differences in sensitivity of the two assays, we nevertheless proceeded to examine whether AP-2γ is involved in the formation of this complex interaction. As shown in Figure 3E, knockdown of AP-2γ disrupted the formation of the three long-range duplex chromatin interactions between the ERBS at the RET gene but not at the control regions. Thus, our results suggest that AP-2γ is required for the formation of ERα-mediated long-range chromatin interactions.
Figure 3.
AP-2γ is required for efficient ERα binding and long-range chromatin interactions. (A) Schematic diagram showing the location of primers (green arrows) and BstYI restriction enzyme cutting sites (blue lines) at the regulatory region of the RET gene. Primer B (red) was used as the main ‘anchor’ region for the 3C assay. Long-range chromatin interactions detected by the 3C assay are indicated by red arrows. (B) 3C assay was performed on MCF-7 cells treated with or without E2 for 45 min. Interactions were detected by real-time PCR using primers indicated in (A). (C) MCF-7 cells were exposed to E2 for 0, 15, 30, 45, 60, 75, 90, and 120 min and then examined by 3C analysis. MCF-7 cells were transfected with (D) ERα, (E) AP-2γ, and control siRNA, treated with or without E2 for 45 min, and then subjected to 3C analysis. (F) ChIP assay using ERα antibody was performed on MCF-7 cells transfected with control or AP-2γ siRNA and treated with or without E2 for 45 min. ERα binding was assessed at the RET-associated ERBS and at control ERBS that do not coincide with AP-2γ binding (right panel). All results represent the average of three independent experiments ±s.e.m.
AP-2γ is required for the efficient recruitment of ERα to ERBS
The observation that AP-2 motifs are close to the centre of ERBS and AP-2γ binding at ERBS occurs before E2 stimulation (Figure 2B; Supplementary Figure S1B) led us to speculate that AP-2γ might be involved in stabilizing ERα binding to chromatin, which would explain why the depletion of AP-2γ had a drastic effect on the long-range chromatin interactions at RET. To explore this possibility, we tested the effect of AP-2γ knockdown on ERα binding. As shown in Figure 3F (left panel), reduction of AP-2γ protein levels caused a significant drop in ERα recruitment at all six ERBS associated with the RET gene. We also observed similar trends at the ERBS of GREB1 (Supplementary Figure S3C). In contrast, knockdown of AP-2γ had no effect on the binding of ERα at ERBS that do not harbour predicted AP-2 motifs or coincide with AP-2γ binding (Figure 3F, right panel). Taken together, our results suggest that AP-2γ is required for facilitating the recruitment of ERα to ERBS.
Majority of AP-2γ binding in the breast cancer genome is independent of oestrogen signalling
Thus far the collaborative effects of AP-2γ on ERα binding and its importance in long-range chromatin interactions were examined on a limited number of ERBS and at the ‘complex’ chromatin interaction of the RET gene. In MCF-7 cells, we had identified over 14 000 ERBS and >1400 ERα-mediated long-range chromatin interactions. To what extent is AP-2γ involved in ERα binding and long-range chromatin interactions is unclear. Therefore, to begin addressing whether the two factors collaborate on a genome-wide level, we performed ChIP-seq analysis of AP-2γ in MCF-7 cells before and after E2 stimulation and integrated these results together with our ERα ChIA-PET map. A summary of the ChIP-seq analysis is shown in Supplementary Table S1.
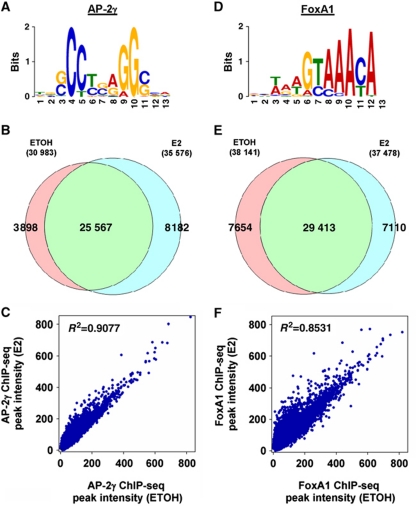
We identified 30 983 and 35 576 AP-2γ binding sites (AP2GBS) in MCF-7 cells before and after E2 stimulation, respectively (Supplementary Table S1). De novo motif analysis of AP2GBS revealed an enriched sequence that is similar to an AP-2 motif found in the TRANSFAC database (Figure 4A). To compare the binding of AP-2γ in E2 stimulated and unstimulated MCF-7 cells, we overlapped the peaks from the two data sets. While we observed a large number of AP2GBS appearing upon E2 stimulation (8182), the majority of sites (25 567) appears to be present in both conditions (Figure 4B; Supplementary Figure S7). Moreover, we also observed a strong correlation (R2=0.9077) between the peak intensities of AP2GBS in the E2 stimulated and unstimulated states, indicating that the majority of AP2GBS are already pre-bound before any stimulation and the occupancy of these sites remain relatively unchanged after E2 induction (Figure 4C). Our data therefore suggest that AP-2γ is a pioneering factor of ERα with similar characteristics as FoxA1 (Carroll et al, 2005). Since currently there is no publically available data of FoxA1 ChIP-seq before and after E2 stimulation, we decided to generate ChIP-seq of FoxA1 under the same conditions as AP-2γ for global comparisons of the two factors. The overall binding profiles of AP-2γ and FoxA1 are remarkably similar even at the global level (Figure 4D–F; Supplementary Figure S8). Taken together, our genomic analyses reveal the majority of AP-2γ binding across the genome is independent of ligand and may function in a similar manner to FoxA1.
Figure 4.
Global analysis of AP-2γ and FoxA1 binding events in MCF-7 cells. (A, D) De novo identification of the AP-2 and FoxA1 binding motif with the top 500 AP2GBS and FoxA1BS (±50 bp of sequence from the ChIP-seq peak) using MEME. (B, E) Comparison of AP2GBS and FoxA1BS overlap (with a window size of ±250 bp) under vehicle or E2 conditions. (C, F) Scatter plots representing the correlation of peak intensities of AP-2γ and FoxA1 before and after E2 stimulation.
AP-2γ is colocalized with FoxA1 at ERBS across the genome in breast cancer cells
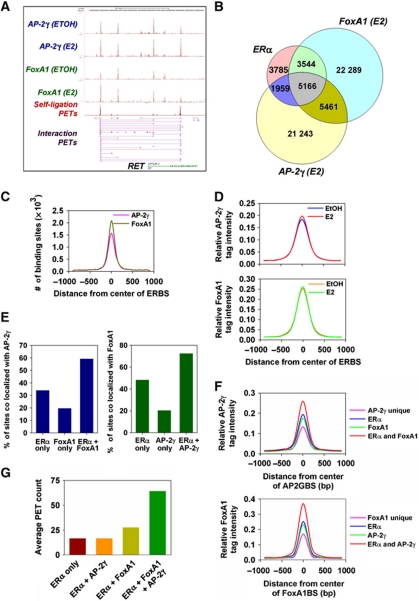
Next, we examined the binding of AP-2γ and FoxA1 with respect to ERα-mediated long-range chromatin interactions. At the RET gene locus, we observed that AP-2γ and FoxA1 binding coincide with ERBS that are involved in long-range chromatin interactions (Figure 5A). To determine whether this also occurs at the global scale for the ERα interactome, we overlapped the AP-2γ and FoxA1 ChIP-seq peaks after E2 stimulation with the ERBS identified from the ChIA-PET library of ERα. As shown in Figure 5B, almost half of the ERBS are colocalized with AP2GBS (7125 out of 14 468). Furthermore, most regions containing ERα and AP-2γ overlapping binding sites (5166 out of 7125) also contain FoxA1BS in close proximity (Figure 5B). This large overlap of the three factors was observed under various ChIP-seq peak thresholds (Supplementary Figure S9). We also compared the binding of AP-2γ and FoxA1 with ERBS identified from our recent ChIP-seq of ERα (Joseph et al, 2010). Similar to our analysis with ERBS from the ERα ChIA-PET, we found AP-2γ and FoxA1 colocalizing with ERα at a large number of binding sites (4406) (Supplementary Figure S10B). More importantly, 87% (3844 out of 4406) of these triple overlapping binding sites can also be found in the triple overlapping binding sites with the ERα ChIA-PET (Supplementary Figure S10A and C). Taken together, our results show that a large fraction of ERBS in the MCF-7 genome are co-occupied with AP-2γ and FoxA1.
Figure 5.
AP-2γ, FoxA1, and ERα are colocalized at a large fraction of ERBS. (A) Screenshot showing tracks from the ChIA-PET of ERα (magenta) and ChIP-seq profiles of AP-2γ (blue) and FoxA1 (green) at the RET gene locus. (B) Venn diagram showing overlap of ChIA-PET ERBS, AP2GBS, and FoxA1BS within ±250 bp of each respective peak binding location. (C) Frequency of AP-2γ and FoxA1 peak distribution with respect to the centre of ERBS (50 bp bin size). (D) Distribution of the average AP-2γ and FoxA1 ChIP-seq tag intensity before and after E2 stimulation was examined with respect to the centre of ERBS (±1 kb with 100 bp bin size). ChIA-PET ERBS, AP2GBS, and FoxA1BS were clustered and different overlapping regions were analysed for (E) the frequency of AP2GBS and FoxA1BS occurrence, (F) the average tag intensity distribution of AP-2γ and FoxA1 (±1 kb with 100 bp bin size), and (G) the average PET count from the ERα ChIA-PET data set.
An analysis of the AP-2γ and FoxA1 ChIP-seq peak distribution and average ChIP-seq tag intensity at colocalized ERα binding regions showed that the majority of both factors are found near the centre of ERBS (Figure 5C and D), which is consistent with our earlier co-motif prediction analysis (Supplementary Figure S1A). Furthermore, the average binding intensities of both AP-2γ and FoxA1 at the overlapped regions are nearly identical before and after E2 induction (Figure 5D). Thus, our results indicate that a large proportion of ERBS already have AP-2γ and FoxA1 pre-bound together before E2 stimulation.
We characterized the binding of AP-2γ in further detail by comparing the fraction of ERα, FoxA1, and shared ERα/FoxA1 binding sites that colocalize with AP-2γ. We found that binding regions containing both ERα and FoxA1 showed the highest frequency for AP-2γ binding compared with binding regions containing only ERα or FoxA1 unique sites (Figure 5E, left panel). Taking the average ChIP-seq tag intensities of AP-2γ at these regions as an indicator of overall binding strength, we found that not only are shared ERα/FoxA1 binding sites more likely to contain AP-2γ binding events but they also exhibit the strongest AP-2γ binding (Figure 5F, top panel). Similar findings were also obtained from our analysis of the ChIP-seq of FoxA1 (Figure 5E and F). In addition to AP-2γ and FoxA1, we found ERα binding was also the strongest at overlapped ERBS (Figure 5G).
Next, we determined whether AP-2γ and FoxA1 are indeed colocalized at ERBS at the same time by performing sequential ChIP–qPCR and examining for DNA enrichment at the ERBS of RET. Consistent with our ChIP–qPCR and ChIP-seq data at the RET ERBS (Figures 2A, B, 3F, and 5A), we found significant enrichment of ERα, AP-2γ, and FoxA1 at ERBS-1, -3, and -5 after E2 stimulation (Supplementary Figure S11), suggesting these three factors can be found together on chromatin as a complex. Furthermore, since the formation of the complex is enhanced by E2, our results would indicate that the presence of AP-2γ and FoxA1 at ERBS may facilitate the binding of ERα to chromatin.
AP-2γ and FoxA1 stabilize the binding of each other at ERBS
FoxA1 is a pioneering factor that is necessary for the recruitment of ERα to chromatin (Carroll et al, 2005); however, the underlying mechanism is currently unclear. The molecular and genomic results from above suggest that AP-2γ may have an important role in this process. This is based on the observations that (1) AP-2γ is also required for ERα binding (Figure 3F) and (2) AP-2γ and FoxA1 recruitment to chromatin are mostly independent of ligand stimulation (Figure 4) and the two factors are colocalized together at a large fraction of ERBS (Figure 5B). From these findings, we speculated that there could be potential functional interplay between AP-2γ and FoxA1.
Our binding analysis of FoxA1 and AP-2γ showed that both factors are bound at most of the ERBS associated with the RET gene (Figures 2B, 5A, and 6A). We observed that FoxA1 was recruited to mainly ERBS-1, -3, and -5 of RET, sites where AP-2γ can also be found (Figure 2B). To examine the interplay between AP-2γ and FoxA1, we first asked whether FoxA1 binding requires AP-2γ at the ERBS of RET. Our results showed that depletion of AP-2γ by siRNA resulted in a significant decrease of FoxA1 recruitment at ERBS-1, -3, and -5 (Figure 6B, left panel; Supplementary Figure S6). We also observed similar effects at other ERBS where both factors are colocalized (Supplementary Figure S13A), but not at control ERBS where AP-2γ is not present (Figure 6B, right panel). We explored this interplay further and next examined whether the binding of AP-2γ to chromatin requires FoxA1. We found that when we knocked down FoxA1 there was also a decrease in AP-2γ binding at the ERBS of RET and other ERBS, but there was no effect on control ERBS where FoxA1 is not present (Figure 6C; Supplementary Figures S12 and S13B). Taken together, our results indicate that AP-2γ and FoxA1 are ‘co-pioneering’ factors that are required for the stable binding of each other to chromatin in order to recruit ERα to ERBS.
Figure 6.
Mutual requirement of AP-2γ and FoxA1 recruitment at ERBS. (A) ChIP for FoxA1 was performed on MCF-7 cells treated with or without E2 for 45 min. (B) ChIP for FoxA1 was performed on MCF-7 cells transfected with control or AP-2γ siRNA. (C) ChIP for AP-2γ was performed on MCF-7 cells transfected with control or FoxA1 siRNA. FoxA1 and AP-2γ binding were examined at RET-associated ERBS and at control ERBS that FoxA1 and AP-2γ do not colocalize. All results represent the average of three independent experiments ±s.e.m.
AP-2γ and FoxA1 are associated with ERBS involved in ERα-mediated interactome
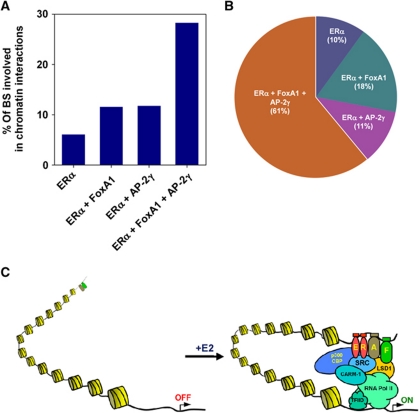
Finally, we addressed the global regulation of ERα-mediated long-range chromatin interactions by AP-2γ and FoxA1. For this, we asked what fraction of the different overlapped ERBS as depicted in Figure 5B is involved in long-range chromatin interactions. As shown in Figure 7A, only ∼10% of ERBS that are colocalized with either AP-2γ or FoxA1 are associated with long-range chromatin interactions. However, the frequency was increased by almost three-fold when ERBS contain both AP-2γ and FoxA1, indicating that ERBS containing all three factors are more likely to be involved in long-range chromatin interactions. Finally, we found that over 72% of ERBS that are involved in long-range chromatin interactions are colocalized with AP-2γ, with 61% of ERBS containing both AP-2γ and FoxA1 (Figure 7B). Overall, our results further show that both AP-2γ and FoxA1 likely function as partners in regulating the global binding of ERα and long-range chromatin interactions in breast cancer cells.
Figure 7.
ERα collaborates with AP-2γ and FoxA1 to promote long-range chromatin interactions across the genome. (A) Percentages of different overlapping ERBS regions that are involved in chromatin interactions. (B) Pie chart showing the proportion of interacting ERBS that are unique or colocalized with AP-2γ and/or FoxA1 binding. (C) Schematic diagram illustrating how AP-2γ and FoxA1 facilitates and coordinate ERα transcriptional activity. AP-2γ and FoxA1 are pre-recruited to the ERBS where they both work cooperatively to promote ERα binding and subsequent chromatin looping, finally stimulating transcription. AP-2γ and FoxA1 are denoted by ‘A’ and ‘F’, respectively.
Discussion
We recently reported the cistrome and interactome of ERα in the breast cancer cell line, MCF-7 (Fullwood et al, 2009). Our results indicate that ERα directly regulates global gene expression in large part by communicating with the transcription machinery over long genomic distances. However, the molecular determinants and the mechanism(s) involved in this regulatory process are unclear. Herein, using a combination of molecular, genomic, and bioinformatic approaches, we have identified AP-2γ as a novel player in the oestrogen signalling pathway that is critical for the efficient formation of long-range intra-chromosomal interactions, as well as regulating ERα-mediated gene transcription. Globally, we showed that AP-2γ is pre-bound across the genome before oestrogen stimulation, with striking similar binding characteristics as the pioneer factor, FoxA1. Furthermore, ERBS that are involved in mediating long-range chromatin interactions are more likely to contain both AP-2γ and FoxA1, suggesting that collaborative action of these two factors can promote recruitment of ERα which is necessary for the subsequent formation of long-range chromatin interactions. Overall, our results implicate AP-2γ as a novel collaborative factor of ERα.
AP-2γ is a member of the developmentally regulated AP-2 transcription factor family that was originally identified from a MCF-7 cDNA library (McPherson et al, 1997). It is an important regulator of embryogenesis and tumourigenesis (Pellikainen and Kosma, 2007). Recently, AP-2γ has been implicated in mammary carcinogenesis, where it is required for oestrogen-mediated proliferation of breast cancer cells and tumour growth (Woodfield et al, 2007). Interestingly, the genomic region 20q13.2, where AP-2γ is located, is often amplified in breast cancers (Nikolsky et al, 2008). This is in concordance with high expression level of the gene in breast tumours which is correlated with resistance to anti-oestrogen, oestrogen deprivation, and poor patient survival outcome (Gee et al, 2009). AP-2γ has been characterized as a transcriptional activator in breast cancer cells, directly upregulating genes such as HER2 (Perissi et al, 2000). However, a recent report showed that AP-2γ can also function as a transcriptional repressor, directly inhibiting the expression of the growth inhibitory CDKN1A gene in breast cancer cells (Williams et al, 2009). Our current data unveil a new mechanism by which AP-2γ regulates transcription in breast cancer cells, namely by collaborating with ERα to directly control the expression of oestrogen target genes. In this study, we have only examined the transcriptional activating properties of AP-2γ with ERα; however, ERBS containing AP-2γ are also found to be associated with oestrogen-repressed genes (Figure 1D). This observation raises the possibility that AP-2γ may also function as a repressor in the oestrogen signalling pathway by inhibiting the activity of ERα.
FoxA1 is a pioneer factor that facilitates the recruitment of ERα to activate transcription (Carroll et al, 2005). In this report, our results indicate that AP-2γ may also possess pioneering properties similar to FoxA1. Our work shows that AP-2γ, like FoxA1, is pre-bound at ERBS before E2 stimulation (Figures 2B and 4B, C; Supplementary Figure S1C). We show this at selected sites including the ERBS associated with the RET gene, and also globally by ChIP-seq (Figure 4B and C). Surprisingly, we found that AP-2γ is required for the binding of FoxA1 at ERBS that contain both factors (Figure 6B). Initially, we thought this suggested the binding of AP-2γ to ERBS was upstream of FoxA1. However, as our data show, the binding of AP-2γ is also dependent on FoxA1, thus it could be that both AP-2γ and FoxA1 are recruited to ERBS at the same time, possibly as a ‘pioneering complex’ (Figure 6C). Clearly, further investigation is required to fully understand the mechanism of action by these two factors at ERBS.
Although this work only examined ERBS containing both AP-2γ and FoxA1, a large subset of ERBS is colocalized with only AP-2γ but not FoxA1 (1959) (Figure 5B). This finding suggests that AP-2γ may be able to facilitate the recruitment of ERα to ERBS on its own, acting as the sole pioneering factor independent of FoxA1. How AP-2γ functions by itself at these ERBS is unclear. It is possible there could be another pioneer factor at these ERBS that has not been identified. Thus, additional analyses will be necessary to determine whether AP-2γ can function alone at these FoxA1-independent ERBS. Furthermore, to understand the transcriptional network of AP-2γ and FoxA1 with ERα, it would be interesting to determine what group of genes are regulated by ERBS that harbour different combinations of these factors.
Using ChIA-PET, we were able for the first time capture of long-range chromatin interactions mediated by ERα on a global scale. The results from our current study suggest that AP-2γ could be an essential factor in establishing ERα-mediated chromatin loops. To extend our knowledge in the mechanism of oestrogen-mediated long-range chromatin interactions, it would be important in future studies to generate an AP-2γ ChIA-PET and compare the long-range chromatin interaction maps of AP-2γ and ERα. However, in preliminary studies we find the AP-2γ antibody that we used for ChIP-seq may not be ideal for our current ChIA-PET protocol. This is likely because ChIA-PET requires a large amount of starting material and is highly dependent on the strength as well as the specificity of antibody. To optimize the antibody for an AP-2γ ChIA-PET, we are currently modifying the ChIA-PET protocol such as adding protein–protein crosslinkers to enhance the capture of long-range chromatin interactions.
Long-range transcriptional regulation has been described in numerous pathways and biological systems including immune response (Spilianakis and Flavell, 2004; Tsytsykova et al, 2007), development (Drissen et al, 2004; Vakoc et al, 2005; Jing et al, 2008), pluripotency in stem cells (Levasseur et al, 2008), and nuclear hormone signalling (Carroll et al, 2005; Pan et al, 2008; Fullwood et al, 2009; Cheung and Kraus, 2010). The factors that are important in modulating long-range chromatin interactions are beginning to emerge. For example, GATA1 and EKLF are required for DNA looping and the specific transcription of genes at the β-globin locus (Drissen et al, 2004; Vakoc et al, 2005). Furthermore, differential regulation of the Kit gene by GATA1 and GATA2 requires the presence of the cofactor, FOG-1, for proper long-range chromatin conformation (Jing et al, 2008). With regard to oestrogen-regulated transcription, ERα and FoxA1, and more recently Rad21 have been shown to be important for the formation of long-range chromatin interactions (Schmidt et al, 2010). From this study, we now include AP-2γ as another critical determinant of oestrogen-mediated long-range chromatin interaction. We propose a model (Figure 7C) where ERBS associated with long-range chromatin interactions are pre-bound with AP-2γ (A) and FoxA1 (F). These factors possibly then recruit chromatin modifiers and generate a local environment that is more accessible and conducive for ERα binding (Lupien et al, 2009). Once extracellular stimuli triggers the recruitment of ERα to ERBS, interaction surfaces generated through protein complex formation at the enhancer will interact with other protein complex at the promoter regions, forming a stable chromatin loop for transcription activation.
Materials and methods
Reagents and antibodies
17β-estradiol (E2) was purchased from Sigma. The following antibodies were used for ChIP and western blot analyses: ERα (HC-20, sc-543), AP-2γ (H-77, sc-8977), and RET (C-20, sc-1290) from Santa Cruz Biotechnologies; FOXA1 (ab5089) from Abcam; and GAPDH (Mab374) from Chemicon.
Cell culture and transient transfection reporter assays
MCF-7 cells (ATCC, Manassas, VA) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum (FBS), penicillin, streptomycin, and gentamycin in a 37°C incubator with 5% CO2. Before oestrogen stimulation, cells were transferred into phenol red-free DMEM/F-12 containing 5% charcoal stripped FBS with penicillin, streptomycin, and gentamycin for 3 days. Cells were transfected with reporter constructs using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer. Vehicle (EtOH) or 17β-estradiol (E2) was added the next day to a final concentration of 10 nM. Twenty-four hours after drug treatment, the cells were washed with PBS twice, treated with lysis buffer, and the extract was analysed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) as recommended by the manufacturer. All the primers used for cloning and mutagenesis of reporter constructs can be found in Supplementary Table S2.
Short interfering RNA studies
siRNA against ERα transcript was purchased from 1st Base. 1st Base synthesized siRNA (siAP-2γ(1)) and ON-TARGETplus SMARTpool L-005238-00 (siAP-2γ(2)) were used to target AP-2γ. siAP-2γ(1) was used in all knockdown experiments, while siAP-2γ(2) was used only in the knockdown microarray experiment. Dharmacon ON-TARGETplus SMARTpool L-010319-00 was used to target FoxA1. siRNA was transfected into hormone-depleted MCF-7 using Lipofectamine RNAi Max (Invitrogen) and treated with vehicle or E2 (10 nM) for 12 h. For gene expression analysis, total RNA was collected using TRI-reagent (Sigma) followed by purification with PureLink™ RNA Mini Kit (Invitrogen). Reverse transcription of RNA was performed with M-MLV reverse transcriptase (Promega), Oligo p(dT)15 (Roche), and dNTP (Fermentas). Real-time PCR was used to quantify specific RNA level and expression level was normalized against GAPDH. The siRNAs sequences used were as follows: siERα sense 5′-UCAUCGCAUUCCUUGCAAAdTdT-3′ and antisense 5′-UUUGCAAGGAAUGCGAUGAdTdT-3′; siAP-2γ-1 sense 5′-GGUACUGAAGCUUUAUUUGdTdT-3′ and antisense 5′-CAAAUAAAGCUUCAGUACCdTdT-3′.
ChIP and sequential ChIP-qPCR
ChIP was performed as described previously (Pan et al, 2008). Briefly, hormone-depleted MCF-7 cells were treated with either vehicle or E2 (100 nM) for 45 min and subjected to crosslinking with a final concentration of 1% formaldehyde (Sigma) for 10 min at room temperature and stopped with 0.25 M glycine for 5 min. Nuclei were isolated and resuspended in SDS lysis buffer and sonicated for 8 min in a Biorupter (Diagenode) to generate DNA fragments of 500–1000 bp. Chromatin extracts were diluted in dilution buffer, pre-cleared and immunoprecipitated with specific antibodies. After immunoprecipitation, beads were washed and eluted with 1% SDS elution buffer. Eluate was subjected to reverse crosslinking at 65°C overnight followed by DNA purification.
Chromatin extracts used for sequential ChIP–qPCR were prepared as described above, except the first immunoprecipitation was performed with antibody crosslinked to protein A or protein A/G Sepharose beads using Dimethyl pimelimidate•2 HCl (DMP) (Pierce). Before the second round of immunoprecipitation, washed beads were eluted with 10 mM DTT at 37°C for 30 min. Second round of immunoprecipitation was carried out as described above.
Chromosome conformation capture
3C was performed as described previously (Fullwood et al, 2009). To compare signal intensities obtained with different primer sets in a quantitative manner, a control template containing all possible ligation products in equimolar amounts was used to correct for the PCR efficiency of each primer set. For this purpose, we used a BAC spanning each locus of interest and digested it with the corresponding restriction enzyme before ligation and column purification. The ligated control fragments were diluted to appropriate concentration and mixed with genomic DNA to obtain a final working concentration of total DNA (∼12.5–50 ng/μl) that was similar to that of the 3C templates. Quantitative real-time PCR was carried out to determine all dissociation curves for the primer pairs that produced only a single peak in their melting curves with the control template. The dissociation curves for all 3C templates were checked against that of the confirmed control templates ran concurrently each time to ensure they give the same single peak at the correct melting temperature. The identities of the positive PCR products were also confirmed through direct sequencing. To obtain data points for normalized relative interaction in the final results, Ct values of 3C template were first normalized with values from an internal primer to account for quantity. Next, the values were normalized with values for each corresponding primer pair to account for relative primer efficiency. Each qPCR was carried out in duplicates and 3C validations were repeated between three and six times independently for each interaction.
Expression array experiment
Total RNA was isolated and purified from MCF-7 cells as described above. Total RNA from three biological replicates was processed to cRNA using the Illumina® TotalPrep™-96 RNA Amplification Kit (Ambion) according to the manufacturer's protocol. cRNA was hybridized directly onto the microarray using the Sentrix® HumanRef-8 v3 Expression BeadChip Kit (Illumina) and BeadChips were scanned using the BeadArray Reader. Image data were processed using GenomeStudio and gene expression data were analysed using GeneSpring GX 11.0 software.
Solexa sequencing and binding site determination
ChIP DNA was quantified using Quant-iT™ PicoGreen® dsDNA assay kit (Invitrogen, Molecular Probes) and libraries were prepared from 5 to 10 ng of ChIP DNA using Illumina's ChIP-seq DNA sample preparation kit with some modifications. Adaptor-ligated DNA was amplified with Pfx DNA polymerase (Invitrogen) and Illumina primers for 15 cycles. Amplified products of 200–300 bp were excised and purified from agarose gel stained with SYBR® Green I nucleic acid gel stain (Invitrogen, Molecular Probes). ChIP-seq reads were aligned to the reference human genome (UCSC, hg18). Binding peaks were determined using Control based ChIPSeq Analysis Tools with reference to a set of input reads as negative control (Xu et al, 2010). Peaks with a stringent cutoff of FDR 0.005 were considered.
Motif enrichment screen of ChIA-PET ERBS
The entire genome was first scanned with the PWM of motifs from Transfac and the locations of the top-scoring 300 000 motif hits or predictions were recorded (such that the expected occurrence of hits is 1 in 10 000 bp). Next, we intersected the 2513 ERBS with the motif hits to obtain the histogram of the hits occurrence around the ERBS within ±5 kb window. In order to ensure the validity of motif enrichment, we attempt to account for the false positive motifs by subtracting the number of motif hits around the ERBS (approximately ±250 bp) by the expected false positive motifs estimated in the flanking background. This in turn gives us the number of true predictions and is represented as the ‘score’ in Supplementary Figure S1A. Under the assumption that each binding site is associated with at most one true prediction, the ‘score’ may be interpreted as the number of binding sites containing one ‘true prediction’.
Real-time PCR
All real-time PCR primer sequences can be found in Supplementary Table S3. Quantitative real-time PCR was carried out with KAPA SYBR green master mix on the ABI PRISM 7900.
Data deposition
Gene expression and ChIP-seq data are deposited at NCBI GEO repository under accession number GSE26741.
Supplementary Material
Acknowledgments
We thank Mei Hui Liu for technical assistance with the 3C assay. We also thank the Genome Technology and Sequencing group and the IT group at GIS for sequencing support. This work was supported by the Biomedical Research Council/Science and Engineering Research Council of A*STAR (Agency for Science, Technology and Research), Singapore. ZHL is supported by a NUS graduate scholarship. SKT is supported by an NUS Graduate School (NGS) for Integrative Sciences and Engineering scholarship. KRC and CWC, are supported by A*STAR graduate scholarships.
Author contributions: SKT and ZHL designed and performed all the experiments. SKT, CWC, VV, KRC, and WKS performed the bioinformatic analyses. YFP performed the ERα ChIP-seq experiment. ELY provided suggestions and comments. EC conceived and supervised the study. SKT and EC analysed the data and wrote the manuscript.
The authors declare that they have no conflict of interest.
07/06/2011
This article has been corrected and a corrigendum is also printed in this issue.
References
- Ali S, Coombes RC (2000) Estrogen receptor alpha in human breast cancer: occurrence and significance. J Mammary Gland Biol Neoplasia 5: 271–281 [DOI] [PubMed] [Google Scholar]
- Arighi E, Borrello MG, Sariola H (2005) RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev 16: 441–467 [DOI] [PubMed] [Google Scholar]
- Boulay A, Breuleux M, Stephan C, Fux C, Brisken C, Fiche M, Wartmann M, Stumm M, Lane HA, Hynes NE (2008) The Ret receptor tyrosine kinase pathway functionally interacts with the ERalpha pathway in breast cancer. Cancer Res 68: 3743–3751 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M (2006) Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Cheung E, Kraus WL (2010) Genomic analyses of hormone signaling and gene regulation. Annu Rev Physiol 72: 191–218 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W (2004) The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D, Buhl S, Weber S, Jager R, Schorle H (2005) The AP-2 family of transcription factors. Genome Biol 6: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M (2007) Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 67: 6477–6483 [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Lupien M, Meyer CA, Verzi MP, Shivdasani RA, Liu XS, Brown M (2009) Cell-type selective chromatin remodeling defines the active subset of FOXA1-bound enhancers. Genome Res 19: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY et al. (2009) An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee JM, Eloranta JJ, Ibbitt JC, Robertson JF, Ellis IO, Williams T, Nicholson RI, Hurst HC (2009) Overexpression of TFAP2C in invasive breast cancer correlates with a poorer response to anti-hormone therapy and reduced patient survival. J Pathol 217: 32–41 [DOI] [PubMed] [Google Scholar]
- Glass CK, Rose DW, Rosenfeld MG (1997) Nuclear receptor coactivators. Curr Opin Cell Biol 9: 222–232 [DOI] [PubMed] [Google Scholar]
- Holmes KA, Song JS, Liu XS, Brown M, Carroll JS (2008) Nkx3-1 and LEF-1 function as transcriptional inhibitors of estrogen receptor activity. Cancer Res 68: 7380–7385 [DOI] [PubMed] [Google Scholar]
- Horwitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L (1996) Nuclear receptor coactivators and corepressors. Mol Endocrinol 10: 1167–1177 [DOI] [PubMed] [Google Scholar]
- Hua S, Kittler R, White KP (2009) Genomic antagonism between retinoic acid and estrogen signaling in breast cancer. Cell 137: 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, Jiang J, Howat WJ, Ali S, Carroll JS (2008) Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature 456: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager R, Friedrichs N, Heim I, Buttner R, Schorle H (2005) Dual role of AP-2gamma in ErbB-2-induced mammary tumorigenesis. Breast Cancer Res Treat 90: 273–280 [DOI] [PubMed] [Google Scholar]
- Jager R, Werling U, Rimpf S, Jacob A, Schorle H (2003) Transcription factor AP-2gamma stimulates proliferation and apoptosis and impairs differentiation in a transgenic model. Mol Cancer Res 1: 921–929 [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA (2008) Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, Pan YF, Li G, Lim M, Thomsen JS, Ruan Y, Clarke ND, Prabhakar S, Cheung E, Liu ET (2010) Integrative model of genomic factors for determining binding site selection by estrogen receptor-alpha. Mol Syst Biol 6: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH (2008) Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes Dev 22: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y et al. (2007) Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet 3: e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Krum SA, Rhodes DR, Liu XS, Brown M (2009) Coactivator function defines the active estrogen receptor alpha cistrome. Mol Cell Biol 29: 3413–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson LA, Baichwal VR, Weigel RJ (1997) Identification of ERF-1 as a member of the AP2 transcription factor family. Proc Natl Acad Sci USA 94: 4342–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F (2003) Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115: 751–763 [DOI] [PubMed] [Google Scholar]
- Nikolsky Y, Sviridov E, Yao J, Dosymbekov D, Ustyansky V, Kaznacheev V, Dezso Z, Mulvey L, Macconaill LE, Winckler W, Serebryiskaya T, Nikolskaya T, Polyak K (2008) Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res 68: 9532–9540 [DOI] [PubMed] [Google Scholar]
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA (2001) Mechanisms of estrogen action. Physiol Rev 81: 1535–1565 [DOI] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS (1997) Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science 277: 1508–1510 [DOI] [PubMed] [Google Scholar]
- Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, Lim KS, Bourque G, Liu ET, Cheung E (2008) Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem 283: 32977–32988 [DOI] [PubMed] [Google Scholar]
- Pellikainen JM, Kosma VM (2007) Activator protein-2 in carcinogenesis with a special reference to breast cancer—a mini review. Int J Cancer 120: 2061–2067 [DOI] [PubMed] [Google Scholar]
- Perissi V, Menini N, Cottone E, Capello D, Sacco M, Montaldo F, De Bortoli M (2000) AP-2 transcription factors in the regulation of ERBB2 gene transcription by oestrogen. Oncogene 19: 280–288 [DOI] [PubMed] [Google Scholar]
- Porter W, Saville B, Hoivik D, Safe S (1997) Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol Endocrinol 11: 1569–1580 [DOI] [PubMed] [Google Scholar]
- Qin C, Singh P, Safe S (1999) Transcriptional activation of insulin-like growth factor-binding protein-4 by 17beta-estradiol in MCF-7 cells: role of estrogen receptor-Sp1 complexes. Endocrinology 140: 2501–2508 [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS (2010) Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev 24: 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M, Melillo RM, Carlomagno F, Fusco A, Vecchio G (2002) Molecular mechanisms of RET activation in human cancer. Ann NY Acad Sci 963: 116–121 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT (2010) A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA (2004) Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol 5: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Tsytsykova AV, Rajsbaum R, Falvo JV, Ligeiro F, Neely SR, Goldfeld AE (2007) Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers. Proc Natl Acad Sci USA 104: 16850–16855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA (2005) Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Valentine C, Lopez GN, Kwok GR, McInerney E, Katzenellenbogen BS, Enmark E, Gustafsson JA, Nilsson S, Kushner PJ (1999) The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol 13: 1672–1685 [DOI] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG (2009) ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28: 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CM, Scibetta AG, Friedrich JK, Canosa M, Berlato C, Moss CH, Hurst HC (2009) AP-2gamma promotes proliferation in breast tumour cells by direct repression of the CDKN1A gene. EMBO J 28: 3591–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfield GW, Horan AD, Chen Y, Weigel RJ (2007) TFAP2C controls hormone response in breast cancer cells through multiple pathways of estrogen signaling. Cancer Res 67: 8439–8443 [DOI] [PubMed] [Google Scholar]
- Xu H, Handoko L, Wei X, Ye C, Sheng J, Wei CL, Lin F, Sung WK (2010) A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics 26: 1199–1204 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.