Abstract
The NIMA-family kinases Nek9/Nercc1, Nek6 and Nek7 form a signalling module required for mitotic spindle assembly. Nek9, the upstream kinase, is activated during prophase at centrosomes although the details of this have remained elusive. We now identify Plk1 as Nek9 direct activator and propose a two-step activation mechanism that involves Nek9 sequential phosphorylation by CDK1 and Plk1. Furthermore, we show that Plk1 controls prophase centrosome separation through the activation of Nek9 and ultimately the phosphorylation of the mitotic kinesin Eg5 at Ser1033, a Nek6/7 site that together with the CDK1 site Thr926 we establish contributes to the accumulation of Eg5 at centrosomes and is necessary for subsequent centrosome separation and timely mitosis. Our results provide a basis to understand signalling downstream of Plk1 and shed light on the role of Eg5, Plk1 and the NIMA-family kinases in the control of centrosome separation and normal mitotic progression.
Keywords: centrosome, Eg5, kinase, Nek, Plk1
Introduction
The cyclin-dependent kinase CDK1 orchestrates the onset of mitosis through the regulation of multiple proteins either directly or in collaboration with a number of helper kinases (Nigg, 2001), among them the Polo-like kinase Plk1 (Petronczki et al, 2008; Archambault and Glover, 2009) and different members of the NIMA family (O’Connell et al, 2003). Plk1 is involved in the complex mechanism that culminates in CDK1 activation during mitotic entry and is crucial for different mitotic events including the formation of the spindle. The molecular basis for some of Plk1 functions is only beginning to be understood and relies on the recognition of previously phosphorylated proteins by the Polo-box domain (PBD) of Plk1, which also targets the kinase to different sites of action such as the centrosomes and centromeres (Elia et al, 2003). The functions of the various members of the NIMA family of protein kinases (Nek1-11 in mammalian cells) are not so well known as these of Plk1. Aspergillus nidulans NIMA, the founding member of the family, is necessary for entry into mitosis and has several roles during mitotic progression, including the regulation of chromosome condensation and spindle formation, although it is not clear whether all these functions are shared with its mammalian counterparts. Different Neks have been implicated in the control of the centrosome and microtubule cytoskeleton (Quarmby and Mahjoub, 2005). Among them Nek2, active in S through G2, regulates premitotic centrosome disjunction, while Nek9 (also known as Nercc1) and the ∼80% identical Nek6 and Nek7, active during mitosis, are involved in the control of spindle structure and function (O’Regan et al, 2007).
Nek9 is activated at centrosomes during early mitosis, interacts with both Nek6 and Nek7 and directly phosphorylates and activates them (Roig et al, 2002, 2005; Belham et al, 2003). Microinjection of anti-Nek9 antibodies into prophase cells induces prometaphase arrest and in some cases aberrant chromosome segregation, resulting in mitotic catastrophes or aneuploidy (Roig et al, 2002), while Nek9 depletion from Xenopus meiotic egg extracts results in delayed spindle assembly, reduced number of bipolar spindles and appearance of aberrant microtubule structures (Roig et al, 2005). Downregulation of either Nek6 or Nek7 by RNAi delays cells at metaphase with fragile mitotic spindles (O’Regan and Fry, 2009) and for Nek7 has been shown to result in an increased incidence of multipolar spindle phenotypes (Yissachar et al, 2006). Mice lacking Nek7 die during late embryogenesis or at early postnatal stages, and Nek7(−/−) cells show increased tendency for chromosomal lagging as well as abnormalities in primary cilia number (Salem et al, 2010). Nek6-deficient mice die early during embryogenesis (our unpublished results). Thus, Nek9 together with Nek6/7 form a mitotically activated module with key roles during mitotic progression and more specifically spindle organization (Nek6 and Nek7 seem to be functionally equivalent in most instances, thus when adequate the two kinases will be collectively referred to as Nek6/7). Nevertheless, to this date a clear picture of the module activation mechanism, integration with other mitotic signalling systems and precise functions during mitosis has been missing.
We have previously suggested that Nek9 could be controlling spindle organization in part through the action of Nek6/7 and their ability to phosphorylate the kinesin Eg5 at a site necessary for normal mitotic progression (Rapley et al, 2008). Here, we present data showing that Plk1, in conjunction with CDK1, activates Nek9 early in mitosis, and that downstream of Plk1, Nek9 and Nek6/7 are responsible for centrosome separation during prophase through the control of Eg5 recruitment to centrosomes. Our results emphasize Nek9, Nek6 and Nek7 importance in mitotic signalling and describe the molecular mechanism controlling the separation of the centrosomes during prophase.
Results
Nek9 mitotic phosphorylation sites
Nek9 is phosphorylated at unknown sites during mitosis resulting in a change in electrophoretical mobility (Roig et al, 2002). This does not directly result in Nek9 activation, a process that occurs at centrosomes during prophase, involves only a small (<5%) fraction of Nek9 and requires further phosphorylation of Nek9 activation loop (Roig et al, 2005). To better understand this two-step activation mechanism and identify the protein kinases responsible for the described modifications, we immunoprecipitated endogenous Nek9 from exponentially growing and mitotic HeLa cells and identified the sites of phosphorylation present by mass spectrometry (MS) analysis (Supplementary Figure S1). Approximately 80% of sequence coverage was obtained in each case, leading to the identification of four Nek9 phosphosites from exponentially growing cells and six from mitotic cells. None of these sites corresponded to known Nek9 activation loop or autophosphorylation sites (Roig et al, 2005), thus indicating that the analysed sample contained mostly inactive Nek9. All sites modified in exponential cells (Ser29, Thr333, Ser750 and Ser869) were also present in mitotic cells, although a higher phosphorylated/unphosphorylated peptide ratio indicated that the corresponding phosphosites were more abundant in mitotic cells (see Supplementary Figure S1). Additionally, phosphorylated Ser827 and Thr885 were only detected in Nek9 from mitotic cells. All identified sites but Thr333 conform to a [ST]P sequence, and thus are putative phosphorylation sites for CDK1, a protein kinase that we have shown is able to readily phosphorylate Nek9 in vitro (Roig et al, 2002 and also see below).
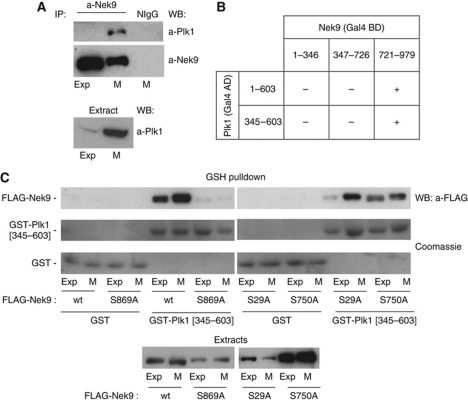
Plk1 interacts with Nek9 through the PBD
Three of the Nek9 phosphorylation sites identified, Ser29, Ser750 and Ser869, conform to a S[S/T]P sequence, a motif that when phosphorylated at the serine/threonine immediately preceding the proline (usually by proline-directed protein kinases such as CDK1) can be recognized by Plk1 PBD (Elia et al, 2003). Thus, we tested whether Plk1 could interact with Nek9 in exponentially growing and mitotic cells. Figure 1A shows that Plk1 specifically coimmunoprecipitates with Nek9 in mitosis in HeLa cells (similar results were obtained with mouse embryo fibroblasts, see Supplementary Figure S2A). This interaction could also be detected using the yeast two-hybrid system (Figure 1B; Supplementary Figure S2B), allowing us to map the Nek9–Plk1 interaction to the C-terminal tail of Nek9 (where two of the three Nek9 putative PBD-binding sites reside), and the PBD domain of Plk1. To confirm Nek9 binding to the PBD and to identify the Nek9 S[S/T]P phosphorylation sites responsible for interaction, we tested whether bacterially expressed Plk1 PBD fused to GST (GST-Plk1[345–603]) could bind different recombinant forms of FLAG-tagged Nek9 from cell extracts. Figure 1C shows that FLAG-Nek9 wild type was able to interact with GST-Plk1 PBD but not GST beads. The Nek9–PBD interaction was increased in mitotic extracts and was totally abrogated by mutation of Nek9 Ser869 to the non-phosphorylable residue alanine. Mutation of Nek9 Ser29 to alanine did not have any effect (consistently with our two-hybrid results), while mutation of Ser750 had only a minor effect on the binding. We concluded that Nek9 specifically binds Plk1 during mitosis through an interaction between phosphorylated Nek9[Ser869] and Plk1 PBD. This is further supported by additional experiments showing that in contrast to wild-type Nek9, Nek9[S869] does not interact with endogenous Plk1 in mitosis (see Supplementary Figure S2C).
Figure 1.
Plk1 interacts with Nek9 through the PBD. (A) a-Nek9 or normal IgG (NIgG) immunoprecipitates from exponentially growing (Exp) or nocodazole-arrested mitotic (M) HeLa cell extracts were analysed by western blot (WB) using the indicated antibodies. Plk1 in the corresponding extracts is shown in the lower panel. (B) The ability of the full-length Plk1 (1–603) or Plk1 PBD (345–603) to interact with the different domains of Nek9 (kinase domain: 1–346; RCC1 domain: 347–726; C-terminal tail:721–979) was assessed using the two-hybrid assay (see Supplementary Figure S2B). Gal4 AD/BD, Gal4 activation/binding domains. (C) In vitro binding of different Nek9 forms to GST-Plk1 PBD. Extracts of exponentially growing (Exp) or nocodazole-arrested mitotic (M) HeLa cells expressing the indicated FLAG-tagged forms of Nek9 were incubated with GST or GST-PBD (GST-Plk1[345–603]) bound to GSH beads. After repeated washes, bound Nek9 was detected by WB with a-FLAG antibody, and GST-fusion proteins by Coomassie staining. FLAG-Nek9 in the corresponding extracts is shown in the lower panel.
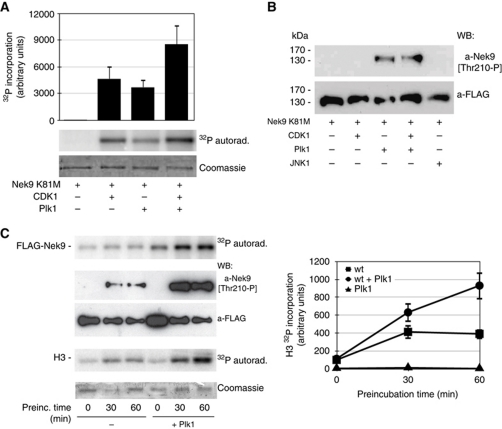
Plk1 phosphorylates and activates Nek9 in vitro
We next tested whether Plk1, alone or in combination with CDK1, could phosphorylate Nek9. For this we used purified kinase-deficient FLAG-Nek9[K81M] (Roig et al, 2002). As expected from our previous results (Roig et al, 2002), FLAG-Nek9[K81M] was phosphorylated by purified CDK1/cyclin B complexes (Figure 2A). Purified Plk1 readily phosphorylated FLAG-Nek9[K81M] to a similar extent (up to ∼6 mol of phosphate/mol of protein), and in the in vitro conditions used showed only slight or no synergy with CDK1. Phosphorylation of Nek9 at Thr210 in the kinase activation loop is required for Nek9 activation (Roig et al, 2005); using a phosphospecific antibody that recognizes Nek9[Thr210-P] (Roig et al, 2005), we determined that Plk1 was able to modify this site in vitro (Figure 2B; Supplementary Figure S3). Under identical conditions, CDK1/cyclin B and the non-relevant kinase JNK1 were not able to phosphorylate Nek9[Thr210]. To determine whether, as expected from previous data, Nek9[Thr210] phosphorylation by Plk1 resulted in Nek9 activation, we incubated purified FLAG-Nek9 without or with Plk1 in the presence of [γ-32P]ATP/Mg2+. After different times, the model substrate histone H3 was added, and 32P incorporation into H3 quantified (Figure 2C). The ATP/Mg2+ concentration used (100 μM) supports Plk1 phosphorylation of Nek9, but also Nek9 autoactivation through autophosphorylation (Roig et al, 2005); thus, in the absence of Plk1, Nek9 phosphorylation and activity towards histone H3 increased with time. Nevertheless, Plk1 induced a further increase in Nek9 phosphorylation when present, including Nek9 phosphorylation in the activation loop at Thr210, and a concomitant increase of activity towards histone H3 (that it is not a substrate of Plk1). We therefore concluded that in vitro Plk1 is able to directly activate Nek9.
Figure 2.
Plk1 phosphorylates and activates Nek9. (A) Kinase-defective FLAG-Nek9[K81M] was expressed and purified from 293T cells and incubated with the indicated kinases for 30 min at 30°C in the presence of [γ-32P]ATP/Mg2+. After SDS–PAGE, Nek9 was visualized by Coomassie staining, and 32P incorporation was visualized by autoradiograph (lower and middle panels) and quantified by PhosphorImager (upper graph, mean±s.e.m. of three independent experiments). CDK1, CDK1/cyclin B. (B) FLAG-Nek9[K81M] obtained as in (A) was incubated with the indicated kinases for 60 min at 30°C in the presence of ATP/Mg2+ and analysed by western blot (WB) using the indicated antibodies. CDK1, CDK1/cyclin B. (C) FLAG-Nek9 was expressed and purified from 293T cells and incubated with or without purified Plk1 in the presence of ATP/Mg2+ for the indicated times at 25°C. After incubation, [γ-32P]ATP/Mg2+ and histone H3 were added to the reactions and further incubated for 10 min. 32P incorporation into Nek9 and H3 was visualized by autoradiograph. In parallel, total Nek9 and Nek9[Thr210-P] were visualized by WB using the indicated antibodies (left, lower panels), and H3 was visualized by Coomassie staining (left). 32P incorporation into H3 was quantified by PhosphorImager (right graph, mean±s.e.m. of three independent experiments).
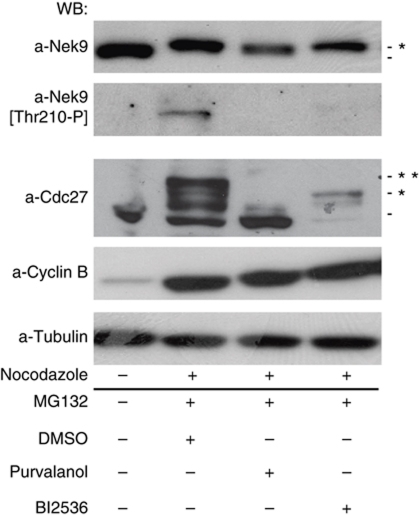
CDK1 and Plk1 are necessary for Nek9 activation in vivo during mitosis
We next determined whether CDK1 and Plk1 activities were necessary for Nek9 activation in vivo during mitosis. We arrested cells in metaphase with the proteasome inhibitor MG132. As expected from previous data (Roig et al, 2005), this induced a shift in Nek9 electrophoretical mobility (a result of Nek9 mitotic phosphorylation) as well as Nek9[Thr210] phosphorylation (Figure 3). The APC/C subunit Cdc27 was used as readout for CDK1 and Plk1 activities, as it changes its apparent MW in response to changes in phosphorylation by both kinases (van Vugt et al, 2004). When under these conditions CDK1 was inhibited with Purvalanol A (Gray et al, 1998), neither Nek9 reduced electrophoretical mobility nor phosphorylation at Thr210 could be observed. Inhibition of Plk1 with BI2536 (Lénárt et al, 2007) although not affecting Nek9 reduced electrophoretical mobility completely abrogated the Nek9[Thr210-P] signal. Thus, while CDK1 activity is necessary for Nek9 phosphorylation in mitosis and the resulting change in electrophoretical mobility, Nek9 Thr210 phosphorylation and mitotic activation requires both CDK1 and Plk1.
Figure 3.
CDK1 and Plk1 are necessary for Nek9 activation during mitosis. HeLa cells were arrested in mitosis with nocodazole. Mitotic cells were collected, washed and released in media containing MG132 (20 μM) plus DMSO, Purvalanol A (20 μM) or Bi2536 (100 nM) for 2 h, and cell extracts were analysed by western blot (WB) using the indicated antibodies. Untreated cells are shown in the first lane as a control. Asterisks mark protein bands with altered mobility due to phosphorylation.
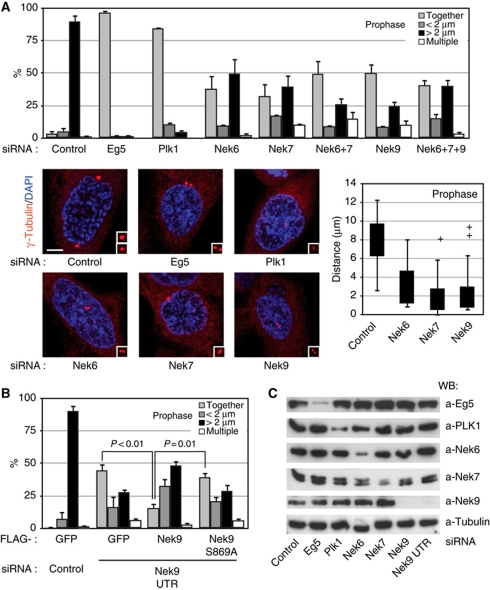
Plk1, Nek9, Nek6, Nek7 and the mitotic kinesin Eg5 are necessary for normal centrosome separation during prophase
One of the most conspicuous functions of Plk1 is the control of centrosome separation during early mitosis (Lane and Nigg, 1996), although how the kinase performs this function remains unknown. In mammalian cells (Whitehead and Rattner, 1998; Tanenbaum et al, 2008; Woodcock et al, 2010), prophase centrosome separation depends on the activity of Eg5 (kinesin-5), a BimC-family kinesin that is like Plk1 involved in the assembly and maintenance of a bipolar spindle during mitosis by sliding anti-parallel microtubules apart (Sawin et al, 1992; Blangy et al, 1995; Kapitein et al, 2005). Eg5 is a substrate for Nek6 (Rapley et al, 2008) and Nek7 (our unpublished data), and therefore we sought to determine whether the Nek9/Nek6/7 module could provide a link connecting Plk1 and Eg5 in the context of centrosome separation. For this we analysed the effects of Plk1, Eg5, Nek9, Nek6 or Nek7 downregulation by RNAi on the extent of separation of duplicated centrosomes in prophase cells (Figure 4).
Figure 4.
Plk1, Nek9, Nek6, Nek7 and Eg5 are necessary for normal centrosome separation in prophase. (A) HeLa cells were transfected with the indicated siRNAs, and after 24 (Eg5, Plk1) or 48 (control, Nek6, Nek7, Nek9) hours, fixed and stained with antibodies against γ-tubulin (red) and DAPI (blue). Cells showing condensed chromosomes and intact nuclei (assessed by the shape of the DNA and a γ-tubulin exclusion from the nucleus) were scored as in prophase (these cells were 100% positive for histone H3[Ser10] phosphorylation, thus confirming the cell-cycle phase assignation, data not shown). The percentage of prophase cells showing two unseparated centrosomes (together), two centrosomes separated <2 μm (<2 μm), fully separated centrosomes (>2 μm) or more than two centrosomes (multiple centrosomes) is shown in the upper graphic (mean±s.e.m. of three independent experiments; ∼50 cells counted in each experiment). Additionally, the distribution of distances from the centre of the duplicated centrosomes in each case is shown as a box plot (boxes show the first and third quartiles, whiskers mark minimum and maximum values unless these exceed 1.5 × interquartile range and crosses correspond to outliers; 20 cells counted for each experimental condition). Representative examples of the observed phenotypes are shown (bar, 5 μm). In each case, insets show magnified centrosomes. (B) HeLa cells were cotransfected with either control or Nek9 3′ UTR siRNAs plus expression plasmids for the indicated FLAG-tagged proteins, and 48 h latter processed and FLAG-positive cells scored as in (A) (mean±s.e.m. of three independent experiments; ∼40 cells counted in each experiment; statistical significance was determined using the standard Student's t-test). Levels of endogenous and recombinant Nek9 as determined by western blot are shown in Supplementary Figure S4C. (C) Efficiency of the different RNAi treatments used in (A) or (B) as determined by western blot of total cell extracts.
Our results confirmed the requirement for Eg5 and Plk1 for mitotic centrosome separation prior to nuclear envelope breakdown in HeLa cells. While prophase cells transfected with control siRNA mainly contained centrosomes that were separated >2 μm (and frequently located at opposite sides of the nucleus), in cells with reduced amounts of Eg5 and Plk1 centrosomes were for the most part overlapping or separated <2 μm. Similarly, Nek6, Nek7 or Nek9 depletion, as well as combined Nek6 and Nek7 or Nek6, Nek7 and Nek9 depletion, resulted in a diminished number of prophase cells with centrosomes separated >2 μm and in the appearance of a significant number of prophase cells with either unseparated centrosomes or centrosomes separated but closer than 2 μm (Figure 4A and C). Furthermore, even when centrosomes were separated >2 μm, intercentrosomal distances were greatly diminished when compared to these of control cells, and in almost no cases reached 9 μm, the control median centrosomal separation in prophase (Figure 4A, box plot). Downregulation of Nek6, Nek7 or Nek9 resulted in some cases in the appearance of cells that contained more than two centrosomes. Costaining with anti-centrin antibody (Supplementary Figure S4A) confirmed that this was not the result of PCM fragmentation and suggested that supernumerary centrosomes could be the result of abortive mitosis, a hypothesis that is supported by the frequent observation of multiple nuclei associated to the existence of more than two centrosomes.
Our observations could be repeated with alternative siRNAs against Nek6 and Nek7 (Supplementary Figure S4B) as well as against Nek9 (Figure 4B), thus confirming the specificity of our results. Furthermore, the use of siRNAs directed against Nek9 mRNA 3′ UTR allowed us to downregulate the levels of endogenous kinase without affecting our ability to express different recombinant forms of Nek9 (Figure 4B; Supplementary Figure S4C). Cotransfection of Nek9 wild type partially rescued the effect of the Nek9 UTR siRNAs, significantly reducing the number of cells with unseparated centrosomes while increasing the percentage of cells with fully separated centrosomes. In contrast, Nek9[Ser869Ala], which is unable to bind Plk1 PBD (see Figure 1C and Supplementary Figure S2C), although expressed at similar levels that wild-type Nek9, was not able to significantly rescue the observed effects of endogenous Nek9 downregulation, thus further stressing the relationship between Plk1 and Nek9 functions.
Active Nek9 and Nek6 induce centrosome separation in an Eg5-dependent manner
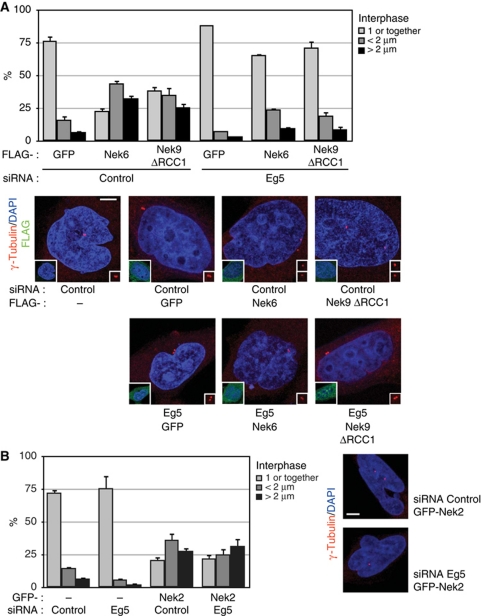
We next sought to determine whether the activation of the Nek9/Nek6/7 module could be sufficient to induce centrosome separation. For this, we artificially increased the cellular activity of either Nek9 or Nek6 by expressing Nek9[Δ346–732], a constitutively active form of the kinase that lacks the autoinhibitory RCC1 domain (Roig et al, 2002), or wild-type Nek6 (partially active when expressed above endogenous levels; Belham et al, 2003). To test whether active Nek9 and Nek6 exerted their effect through the regulation of Eg5, we simultaneously transfected the cells with control or Eg5 siRNAs. Figure 5A shows that expression of Nek6 or Nek9[Δ346–732] significantly increased the number of cells with separated centrosomes. The effect was cell-cycle independent, as the expression of Nek6 or Nek9[Δ346–732] did not change the cell-cycle profile of the cells (as assessed by FACS; Supplementary Figure S5) and consequently most of the transfected cells that contained separated centrosomes were in interphase (Figure 5A, see example cells). Centrosome separation was not induced by wild-type Nek9 or Nek7, which are not active under the expression conditions used, by neither kinase-deficient Nek9 [K81M,Δ346–732] or Nek6[K74,75M] (data not shown).
Figure 5.
Active Nek9 and Nek6 induce centrosome separation in an Eg5-dependent manner. (A) HeLa cells were transfected with either control or Eg5 siRNAs, after 16 h retransfected with expression plasmids for the indicated FLAG-tagged proteins (Nek9ΔRCC1, Nek9[Δ346–732]) and 24 h latter fixed and stained with anti-γ-tubulin (red) and anti-FLAG (green) antibodies plus DAPI (blue). The percentage of FLAG-positive cells showing 1 or 2 unseparated centrosomes (1 or together), two centrosomes separated <2 μm (<2 μm) or fully separated centrosomes (>2 μm) is shown in the upper graphic (mean±s.e.m. of three independent experiments; ∼50 cells counted in each experiment). Representative examples of the observed phenotypes (anti-γ-tubulin plus DAPI) are shown below (bar, 5 μm). Insets show the same field stained with anti-FLAG plus DAPI. The effect of the different treatments on the levels of Eg5 can be seen in Figure 6, upper right panel. (B) As in (A), cells transfected with either control or Eg5 siRNAs and expression plasmids for GFP or GFP-Nek2 (mean±s.e.m. of three independent experiments; ∼50 cells counted in each experiment).
Remarkably, downregulation of Eg5 almost totally abrogated Nek6 or Nek9-induced centrosome separation, suggesting that Nek6 and Nek9 induce centrosome separation through the regulation of the kinesin. The dependence on Eg5 additionally distinguished Nek9/Nek6-induced centrosome separation from Nek2-induced centrosome disjunction, the dissolution of the physical link that keeps together duplicated centrosomes, precedes separation and is controlled by this NIMA-family kinase (Fry et al, 1998; Faragher and Fry, 2003). Nek2 effects on centrosomes were totally independent of Eg5 (Figure 5B) and can be attributed to non-directed drifting of the disjointed centrosomes after Nek2 ectopic activation in interphase.
Active Nek9 and Nek6 can rescue Plk1 but not Eg5 downregulation in prophase centrosome separation
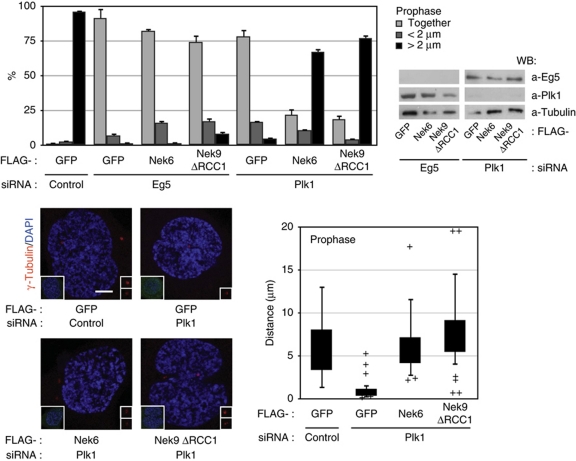
We inquired whether expression of active Nek9 or Nek6 could compensate for either Eg5 or Plk1 downregulation during prophase centrosome separation. For this we transfected cells with control, Eg5 and Plk1 siRNAs in combination with expression plasmids for GFP (control), active Nek9[Δ346–732] or Nek6 and determined the distance between centrosomes in transfected prophase cells (Figure 6). As expected, most of control cells contained fully separated centrosomes; expression of either Nek9[Δ346–732] or Nek6 did not change this significantly (not shown). In accordance with our previous results, depletion of Eg5 abrogated prophase centrosome separation and active Nek9 or Nek6 expression was not able to rescue this effect. In cells depleted of Plk1 and expressing GFP as a control protein, centrosome separation in prophase was almost completely abolished, but in contrast, expression of active Nek9[Δ346–732] or Nek6 in Plk1-depleted cells was able to restore the percentage of cells with separated centrosomes to levels similar to control cells (see also box plot indicating the distribution of intercentrosomal distances in individual cells).
Figure 6.
Active Nek9 and Nek6 can rescue Plk1 but not Eg5 downregulation in prophase centrosome separation. HeLa cells were transfected with control, Eg5 or Plk1 siRNAs plus the indicated plasmids and processed as in Figure 5 (Nek9ΔRCC1, Nek9[Δ346–732]). The percentage of FLAG-positive prophase cells showing two unseparated centrosomes (together), two centrosomes separated <2 μm (<2 μm) or fully separated centrosomes (>2 μm) is shown in the upper graphic (mean±s.e.m. of three independent experiments; ∼40 cells counted in each experiment). The effect of the different transfections on the levels of Plk1 and Eg5 is shown (right). Lower panels show representative examples of the observed phenotypes (anti-γ-tubulin plus DAPI staining, bar, 5 μm; insets show the same field stained with anti-FLAG plus DAPI) and a box plot of the distribution of distances from the centre of the duplicated centrosomes in FLAG-positive cells (as in Figure 4; 30 cells counted for each experimental condition).
Our results show that Nek9, Nek6 and Nek7 act downstream of Plk1 and upstream of Eg5 during early centrosome separation and suggest that Plk1 inhibition precludes centrosome separation as a result of the failure to activate the Nek9/Nek6/7 module.
Plk1 controls Eg5 phosphorylation at the Nek6 site Ser1033, that together with the CDK1 site Thr926 is necessary for prophase centrosome separation and Eg5 recruitment
Eg5 is phosphorylated during mitosis at Ser1033, a site that we have previously shown is modified by Nek6/7 (Rapley et al, 2008). Eg5[Ser1033-P] accumulates at centrosomes in prophase, and we therefore speculated that Plk1 and Nek9 might control prophase centrosome separation through Nek6/7 phosphorylation of this residue. Using an antibody that specifically recognizes Eg5[Ser1033-P] (Rapley et al, 2008), we first confirmed by RNAi that mitotic levels of Eg5[Ser1033-P] depend on Nek6, but also Nek7 and their upstream kinase Nek9 (Supplementary Figure S6A). Next, we sought to determine whether Eg5[Ser1033] phosphorylation also depends on Plk1. For this, we arrested cells in mitosis with nocodazole or by depleting either Plk1 or Eg5 by RNAi. Mitotic arrest was confirmed by FACS (not shown) and the phosphorylation state of Cdc27, and the levels of Eg5[Ser1033-P] were compared with those present in exponentially growing cells. Figure 7A shows that Eg5[Ser1033-P] was detected in nocodazole-arrested cells but not in exponentially growing cells. Plk1 downregulation by RNAi resulted in the abrogation of Eg5[Ser1033-P] from mitotic cells. Additionally, Eg5 depletion had a similar effect, thus confirming the specificity of the antibody. Similar results were obtained by using the Plk1 inhibitor BI2536 that resulted in mitotic cells without any observable Eg5[Ser1033-P] accumulation (Supplementary Figure S6B).
Figure 7.
Plk1 controls Eg5 phosphorylation at the Nek6 site Ser1033. Both Ser1033 and the CDK1 site Thr926 phosphorylation are necessary for prophase centrosome separation and Eg5 recruitment. (A) HeLa cells were arrested in mitosis by either nocodazole (ND) treatment or RNAi against Plk1 or Eg5 (24 h transfection), collected after mitotic shake off and cell extracts were analysed by western blot (WB) using the indicated antibodies. Mitotic arrest was confirmed by FACS (not shown) and the phosphorylatin state of Cdc27. Untreated cells (Exp) are shown in the first lane as a control. Asterisks mark protein bands with altered mobility due to phosphorylation. (B) HeLa cells were transfected with either control or Eg5 siRNAs, after 16 h retransfected with expression plasmids for the indicated Myc-tagged proteins (cDNAs rendered resistant to the siRNA by several silent point mutations), fixed and stained with antibodies against Myc, γ-tubulin and DAPI. The percentage of Myc-positive prophase cells showing two unseparated centrosomes (together), two centrosomes separated <2 μm (<2 μm) or fully separated centrosomes (>2 μm) is shown in the upper graphic (mean±s.e.m. of three independent experiments; ∼40 cells counted in each experiment). Levels of endogenous and recombinant Eg5 as determined by WB are shown in Supplementary Figure S6C. (C) Cells transfected and processed as in (B). Representative examples of the observed phenotypes (Myc–Eg5, green) are shown below (bar, 5 μm). Insets show the same field stained with γ-tubulin (red) plus DAPI (blue). Centrosomal accumulation of Eg5 is noted with arrows.
We have previously shown that Eg5[Ser1033] phosphorylation is necessary for normal mitotic progression (Rapley et al, 2008). We now tested whether phosphorylation of Ser1033 was necessary for centrosome separation during prophase. We depleted endogenous Eg5 by RNAi and concomitantly expressed different Myc-tagged Eg5 variants, which were rendered resistant to the siRNA by several silent point mutations and were all expressed at similar levels (Supplementary Figure S6C). Figure 7B shows that in cells depleted of endogenous Eg5, wild-type Eg5 (but not the CDK1 site mutant Eg5[Thr926Ala]) was capable of supporting centrosome separation in prophase cells. Remarkably, Eg5[Ser1033Ala] was substantially less effective in rescuing endogenous Eg5 depletion. An additional mutant, Eg5[Ser1033Asp], showed a tendency to be more efficient in sustaining centrosome separation that Myc–Eg5[Ser1033Ala], although the differences between these two forms were statistically not significant, leading us to conclude that Eg5[Ser1033Asp] only partially mimicked Eg5[Ser1033-P] (data not shown).
During mitosis, Eg5 binding to centrosomes and microtubules depends on CDK1 phosphorylation of Thr926 (Blangy et al, 1995). We explored the possibility that Ser1033 phosphorylation could be in addition necessary for Eg5 centrosomal recruitment during prophase, thus explaining the requirement of this site for normal centrosome separation during early mitosis. Prophase localization of different Eg5 recombinant forms in transfected cells is shown in Figure 7C. As expected (Blangy et al, 1995; Sawin and Mitchison, 1995), in prophase cells, wild-type Eg5 (but not Eg5[Thr926Ala]) accumulated on centrosomes and the proximal ends of microtubules. Strikingly, Eg5[Ser1033Ala] was not present at prophase centrosomes in cells that had failed to separate them. Reduced centrosomal amounts of this mutant were observed in cells with separated centrosomes. In addition to these observations, it is worth noting that except Eg5[Thr926Ala], all Eg5 forms showed a cytoplasmatic distribution that was compatible with that of microtubules.
Thus, our results show that, like Nek9 and Nek6/7, Plk1 is necessary for mitotic Eg5[Ser1033] phosphorylation, and suggest that this modification together with Thr926 phosphorylation by CDK1, is required for normal Eg5 recruitment to centrosomes and subsequent centrosome separation during prophase.
Plk1, Nek9, Nek6, Nek7 are necessary for centrosome recruitment of Eg5 during prophase; active Nek9 and Nek6 are able to rescue Plk1 downregulation in prophase Eg5 recruitment
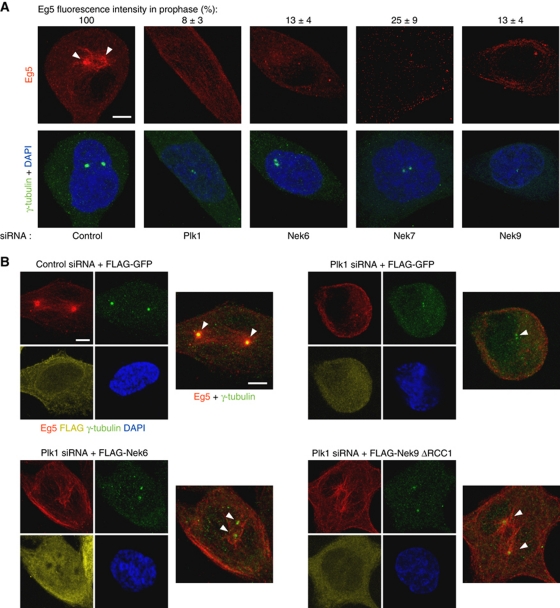
We reasoned that Plk1, Nek9, Nek6 and Nek7 should be necessary for Eg5 centrosomal recruitment, and that cells that failed to separate the centrosomes during prophase as a result of interfering with the different protein kinases should present diminished centrosomal levels of Eg5. Downregulation of the different kinases by RNAi confirmed this (Figure 8A). While prophase cells transfected with control siRNA showed separated centrosomes with an evident accumulation of Eg5 in the vicinity of centrosomes, transfection with Plk1, Nek6, Nek7 or Nek9 siRNAs resulted in prophase cells with unseparated centrosomes and without any apparent recruitment of Eg5 to these organelles. We next sought to determine whether the observed ability of active Nek9 and Nek6 to rescue Plk1 downregulation during centrosome separation is concomitant with a recovery in the amount of centrosomal Eg5. Figure 8B shows that active Nek9[Δ346–732] or Nek6 cotransfection at least partially restores Eg5 pericentrosomal accumulation in cells transfected with Plk1 siRNA (see Supplementary Figure S7 for additional examples of Eg5 localization under these conditions).
Figure 8.
Plk1, Nek9, Nek6, Nek7 are necessary for centrosome recruitment of Eg5 during prophase. Active Nek9 and Nek6 can rescue Plk1 downregulation in prophase Eg5 recruitment. (A) HeLa cells were transfected with the indicated siRNAs, and after 24 (Plk1) or 48 (control, Nek6, Nek7, Nek9) hours, fixed and stained with antibodies against Eg5, γ-tubulin and DAPI. Representative examples of Eg5 (red) distribution in prophase cells are shown. Insets show the same field stained with γ-tubulin (green) and DAPI (blue). Centrosomal accumulation of Eg5 is noted with arrowheads. Bar, 5 μm. The efficiency of the different RNAi treatments can be seen in Figure 4C. Centrosomal Eg5 fluorescence intensity was quantified with ImageJ software on images acquired under constant exposure, using a circular area of 2 μm diameter surrounding a single centrosome (identified by γ-tubulin staining; an adjacent area of the same dimensions within each cell was quantified and subtracted as background). Results are expressed as a percentage of the intensities measured in control cells±s.e.m. (three independent experiments; >20 centrosomes counted in each experiment). (B) HeLa cells were cotransfected with either control or Plk1 siRNAs and expression plasmids for the indicated proteins and after 24 h fixed and stained with antibodies against Eg5, γ-tubulin and DAPI (Nek9ΔRCC1, Nek9[Δ346–732]). After incubation with labelled secondary antibodies, FLAG was detected with Fab-prelabelled anti-FLAG (see Materials and methods). Representative examples of Eg5 distribution in prophase cells are shown. Images show the same field stained with Eg5 (red), γ-tubulin (green), FLAG (yellow) and DAPI (blue), and a composite of Eg5 (red) plus γ-tubulin (green). Centrosomal accumulation of Eg5 is noted with arrowheads. Bar, 5 μm.
Additional experiments showed that in fact, transfection of active Nek9[Δ346–732] or Nek6 is able to induce ectopic Eg5 accumulation around centrosomes in parallel to centrosome separation even in interphase (Supplementary Figure S8A) and that this is accompanied with Eg5[S1033] phosphorylation (Supplementary Figure S8B; Rapley et al, 2008). Thus, a physiological correlation exists between Eg5 recruitment and centrosome separation in prophase cells, and activation of the Nek9/Nek6 module is both necessary and sufficient to induce both phenomena in a cell-cycle-independent manner.
Failure to phosphorylate Eg5[Ser1033] results in a delay in prometaphase
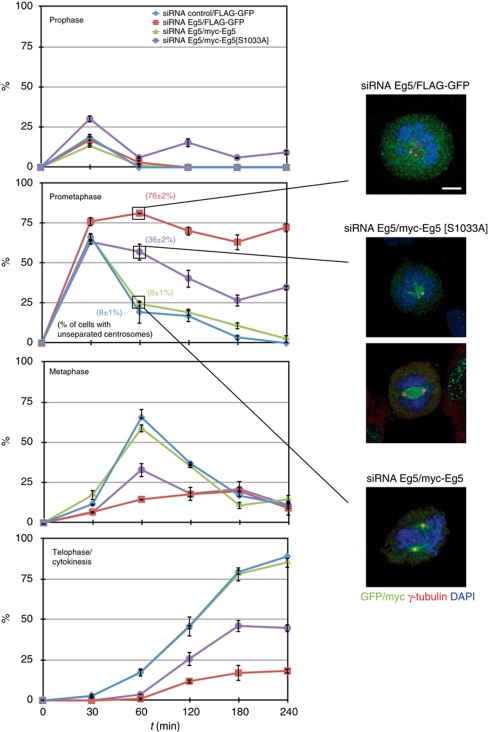
We finally wished to determine how mitotic progression would be affected by substituting endogenous Eg5 by the non-phosphorylable form Eg5[Ser1033Ala] (and thus by interfering with normal centrosome separation during prophase). For this, HeLa cells were transfected with control siRNA plus a control protein (FLAG-GFP) or Eg5 siRNA plus either a FLAG-GFP, Eg5 wild type or Eg5[Ser1033Ala], and arrested at the G2/M border using the Cdk1 inhibitor RO-3306. Cells were released by repeated washes, fixed at different time points and mitotic cells categorized according to mitotic phase. Centrosome separation was assessed in prometaphase cells by γ-tubulin staining (Figure 9). Note that, similarly to other synchronization methods (Gavet and Pines, 2010), RO-3306 treatment results in premature centrosome splitting/separation in a significative amount of G2 cells (see Supplementary Figure S9); this effect is Eg5 independent and may result in an underestimation of the effects on centrosome separation of the different forms of Eg5 used. In all conditions, ∼50% of cells immediately entered mitosis upon removal of the CDK1 inhibitor, thus confirming both synchronization and the reversibility of the treatment. Of these, most were in prometaphase 30 min postrelease. Control cells and cells transfected with Eg5 siRNAs plus wild-type Eg5 progressed into metaphase (∼60% of cells at 60 min) and then into telophase/cytokinesis (∼80% at 240 min). As expected, most of the cells transfected with Eg5 siRNAs plus a control protein remained in prometaphase with unseparated centrosomes for the length of the experiment (76% at 240 min). Cells transfected with Eg5 siRNAs plus Eg5[Ser1033Ala] entered prometaphase at a similar rate than control cells (63% at 30 min), but only a small percentage of them progressed into metaphase and later into telophase. Remarkably, at 60 min, >57% of the cells expressing Eg5[Ser1033Ala] were still in prometaphase (as compared with 25% for cells expressing wild-type Eg5) and a significative part of the cells showed unseparated centrosomes (36±2% of prometaphase cells with centrosomes separated <2 μm, as compared with 8±1% in cells expressing wild-type Eg5). This percentage slowly diminished with time in parallel with the apparition of metaphase and telophase/cytokinetic cells at longer time points. After 240 min, 35% of cells expressing Eg5[Ser1033Ala] remained in prometaphase, while 45% of cells having progressed to telophase/cytokinesis. We conclude that failure to phosphorylate Eg5[Ser1033], and thus to recruit the kinesin to centrosomes and properly separate these organelles during early mitosis results in prometaphase delay (and in some cases possibly in prometaphase arrest). This highlights the importance for normal mitotic progression of the mechanism responsible for the phosphorylation of Eg5[Ser1033].
Figure 9.
Effects of Eg5[Ser1033Ala] on cell-cycle progression. HeLa cells were transfected with either control or Eg5 siRNAs, after 16 h retransfected with expression plasmids for the indicated Myc-tagged proteins (cDNAs rendered resistant to the siRNA by several silent point mutations). After 24 h, cells were incubated 20 h with 9 μM RO-3306. Synchronization in G2 was confirmed by FACS. Cells were released in fresh media after repeated washes, and at the indicated times fixed and stained with antibodies against myc or GFP, γ-tubulin and DAPI. Mitotic myc- or GFP-positive cells were categorized according to mitotic phase (mean±s.e.m. of three independent experiments; ∼40 mitotic cells counted in each experiment). Representative examples of prometaphase cells as well as the percentage of cells in this phase of the cell cycle with unseparated centrosomes (distance <2 μm) at 60 min postrelease is shown, see Supplementary Figure S9 (mean±s.e.m. of three independent experiments; ∼30 cells counted in each experiment; bar, 5 μm).
Altogether, our results show that Plk1 controls Eg5[Ser1033] phosphorylation through the activation of Nek9, Nek6 and Nek7, and that this modification is required for the recruitment of Eg5 to centrosomes, early centrosome separation and normal progression through mitosis.
Discussion
The NIMA-family kinases Nek9, Nek6 and Nek7 form a signalling module required for normal spindle assembly and function during mitosis (Roig et al, 2002, 2005; Belham et al, 2003; O’Regan and Fry, 2009). While Nek6 and Nek7 are both directly activated by Nek9, the mechanism of activation of Nek9 has remained elusive. Based on the observations presented in this work, we put forward CDK1 and Plk1 as Nek9 physiologic activators. We propose a two-step activation mechanism for this NIMA-family kinase in which CDK1 (together with cyclin B1 and perhaps cyclin A) phosphorylates Nek9 at Ser869, inducing Plk1 binding and subsequent Plk1 phosphorylation and activation of Nek9. Nek9 activation could directly result from Plk1 phosphorylation of Nek9[Thr210], although conceivably Plk1 phosphorylation of additional sites outside the Nek9 activation loop may also contribute to activation by releasing Nek9 autoinhibition thus triggering Thr210 autophosphorylation (Roig et al, 2005). Our model provides a basis to understand the temporal and spatial pattern of Nek9 activation, occurring at prophase centrosomes, where it colocalizes with active CDK1 and Plk1. Additionally, it integrates signalling through the NIMA family with that of CDK1 and Plk1, suggesting new ways through which these two major mitotic kinases could control the organization and function of the mitotic machinery.
Regarding the conservation of the proposed Nek9 activation mechanism, an S[S/T]P site in a similar position to that of human Ser869 is lacking in mouse and rat Nek9, although in these and other organisms Ser750 is conserved. We thus suggest that this residue (that in human cells is also phosphorylated in vivo but only marginally affects Plk1 binding) could act as the main Plk1-binding site when the homologue of Ser869 is not present. A relationship between the NIMA, CDK and Polo families may have been long conserved through evolution and may even be bidirectional, as Aspergillus NIMA is activated in mitosis through a mechanism that involves NIMXCDC2 (Ye et al, 1995) and Schizosaccharomyces pombe Fin1, like NIMA and Nek9 involved in the regulation of spindle formation, has been described to be necessary for Polo (Plo1) association to the spindle pole body (Grallert and Hagan, 2002).
Early functional reports of Drosophila Polo (Llamazares et al, 1991) or mammalian Plk1 (Lane and Nigg, 1996) described the failure to separate centrosomes in mitosis (associated to the appearance of monopolar spindles) as one of the major results of interfering with these kinases. Since then it has been well established that Plk1 has diverse functions during early, mid and late mitosis (Petronczki et al, 2008; Archambault and Glover, 2009), among them the regulation of centrosome separation and maturation, two Plk1 roles the molecular basis of which still remains to be fully understood. We herein propose that Plk1 controls centrosome separation in prophase through Nek9 and Nek6/7 signalling to the kinesin Eg5. Accordingly, and without discarding the existence of additional Nek9/Nek6/7-independent roles of Plk1 during centrosome separation, we suggest that the main cause for the failure of this process in cells with diminished Plk1 activity is the absence of activation of the Nek9/Nek6/7 module. This results in lack of Eg5 phosphorylation at Ser1033, a previously described modification (Rapley et al, 2008) that we now show is necessary for Eg5 recruitment to centrosomes and prophase centrosome separation.
Previous studies have suggested that vertebrate cells can separate centrosomes through two distinct and partially redundant pathways: a prophase pathway that relies on microtubule-based motors, the nuclear membrane and possibly microtubule pushing forces, and a prometaphase pathway that is based in interactions between the two microtubule asters, astral microtubule pulling forces and kinetochore-generated pushing forces (see Rosenblatt, 2005 and Tanenbaum and Medema, 2010 for reviews). The prophase pathway is, at least in mammalian cells, strongly dependent on Eg5 (our data and Whitehead and Rattner, 1998; Tanenbaum et al, 2008; Woodcock et al, 2010) and our results indicate that phosphorylation of Eg5[Ser1033] controls this pathway by allowing the recruitment of the kinesin to the vicinity of the centrosomes during prophase. Attesting to the redundancy of the two centrosome separation pathways and the robustness of the mechanisms that results in spindle bipolarity, cells that fail to phosphorylate Eg5[Ser1033] remain longer in prometaphase, but for the most part reach metaphase (although with a marked delay) and progress to later mitotic phases. Thus, Ser1033 phosphorylation is not necessary for prometaphase centrosome separation or the non-prophase functions of Eg5 during spindle assembly (a process that depends on Eg5 unless it initiates with well-separated centrosomes; Ferenz et al, 2009). Nevertheless, our results suggest that Eg5[Ser1033] phosphorylation and thus prophase centrosome separation promote and accelerate the building of the bipolar spindle, probably allowing prometaphase mechanisms to work more efficiently on already separated centrosomes. It remains to be determined whether, in addition of the timely formation of a bipolar spindle, Eg5[Ser1033] phosphorylation influence the accuracy of chromosome segregation as well.
How does Ser1033 phosphorylation induce Eg5 pericentrosomal localization and control prophase centrosome separation? Mutation of this residue to a non-phosphorylable alanine results in a form of the kinesin that has a greatly impaired centrosomal localization but is still able to bind microtubules and to localize to the metaphase spindle (Rapley et al, 2008). Conversely, mutation of the CDK1 phosphorylation site Thr926 (required for Eg5 microtubule binding during mitosis; Blangy et al, 1995; Sawin and Mitchison, 1995) results in abrogation of Eg5 recruitment to centrosomes and of centrosome separation. Consequently, Ser1033 phosphorylation does not control the ability of Eg5 to bind microtubules but it relies on it to concentrate the kinesin at the vicinity of centrosomes and separate them (Thr926 phosphorylation may not be required in conditions in which high Nek9/Nek6/7 activity results in a significant increase in the levels of Eg5[Ser1033-P], see Figure 5 and Supplementary Figure S8). We can hypothesize that phosphorylation of Ser1033 allows a pool of Eg5 to preferentially bind microtubules proximal or even anchored to the centrosome, either directly or through the interaction with a yet to be identified centrosomal protein (in turn this may be directly modulated by Plk1 or other yet to be described Plk1 targets, accounting for the only partial ability of Nek9 and Nek6 to rescue Eg5 recruitment to the centrosomes in cells with diminished levels of Plk1, see Figure 8 and Supplementary Figure S7). Accumulation of enough Eg5 at the vicinity of a prophase centrosome will allow separation from the opposing centrosome by exerting forces on the relatively few microtubules emanating from it. Centrosomal localization of Eg5 may not be necessary during prometaphase, as in this latter stage of mitosis the increased number and length of microtubules (with Eg5 bound through their lengths) would ensure sufficient overlap as to produce force in collaboration with other separation mechanisms and without the need of concentrating Eg5 at centrosomes. Whether this hypothesis is correct, as well as how it is related to novel functions of Eg5 C-terminal domain (Weinger et al, 2011), the action of other motor systems like dynein/dynactin (Blangy et al, 1997; Uteng et al, 2008; Ferenz et al, 2009) or the positioning of centrosomes in respect to the nucleus (Splinter et al, 2010) remains to be determined.
In summary, our data identify two major mitotic regulators, Plk1 and CDK1 as upstream activators of the Nek9/Nek6/7 module, firmly positioning the NIMA-family kinases Nek9, Nek6 and Nek7 at the centre of mitotic signalling. A first example of the roles that these kinases can perform downstream of CDK1 and Plk1 is described, shedding light on one of the most conspicuous but less understood roles of Plk1 during early mitosis, centrosome separation, and defining the elements that in mammalian cells control this process as well as its importance during mitotic progression. It is now clear that NIMA-family kinases control different but consecutive steps of the centrosomal cycle, namely centrosome disjunction (regulated by Nek2 in an Eg5-independent manner and essential when Eg5 function is partially compromised, see Mardin et al, 2010) and separation (regulated by Nek9 and Nek6/7 and executed by Eg5). Whether this is the result of sharing the diverse functions of an ancestral NIMA and whether Nek9, directly or through Nek6 and Nek7, is responsible for additional mitotic roles downstream of Plk1 as the phenotypes that result from interfering with these kinases suggest (Roig et al, 2002, 2005; O’Regan and Fry, 2009), will surely be the subject of future investigations.
Materials and methods
Plasmids and reagents
Different Nek9 and Nek6 expression plasmids have been described elsewhere (Roig et al, 2002; Belham et al, 2003). Additional Nek9 mutants were constructed using the QuickChange Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's instructions, using specific primers (S869A 5′-CAAGTAGAAGCCTCGGCACCTCGGCTGAATCCTGC-3′, S29A 5′-GGTTGCGGGGACTCGGCTCCGGGGCCTAGCGCC-3′, S750A 5′-ACTGTGTTTCAGAGCTCTGCCCCGGGAGGAGGCGGCGG-3′- with the appropriate reverse complements). pCMV5-FLAG-GFP was constructed by cloning eGFP into pCMV5-FLAG. For construction of PGEX-Plk1[345–603], a PCR fragment corresponding to Plk1 PBD was cloned into a modified PGEX vector (Pharmacia Biotech). All constructs were sequenced after generation. RNAi-resistant forms of Eg5 have been described in Rapley et al (2008).
FLAG-Nek9 and FLAG-Nek9[K81M] were expressed in 293T cells and purified by immunoprecipitation with anti-FLAG antibody (Sigma), followed by repeated washes and elution using FLAG peptide (Sigma). Purified Plk1 and CDK1/cyclin B were purchased from Invitrogen. Histone H3 was from Roche. Nocodazole, MG132 and Purvalanol A were from Sigma. Bi2536 was from Axon Medchem. RO-3306 was from Enzo Life Sciences.
Cell culture and transfection
HeLa, U2OS and HEK 293T cells were cultured as described (Roig et al, 2002). Cells in mitosis were obtained by mitotic shake off of nocodazole-arrested (0.25 mM, 16 h) cultures. HEK 293T cells were transfected using different expression plasmids with Lipofectamine (Invitrogen) according to the manufacturer's instructions. HeLa cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNAs were transfected using siPORT NeoFX Transfection Agent (Ambion) according to the manufacturer's instructions. siRNA and DNA cotransfection was performed using Lipofectamine 2000.
siRNA duplexes were as following: Eg5, 5′-CUGAAGACCUGAAGACAAUdTdT-3′ (Ambion) (Weil et al, 2002); Plk1, 5′-CGAGCTGCTTAATGACGAGTT-3′ (Dharmacon) (Oshimori et al, 2006); Nek6, 5′-AAUAGCAGCUGUGUGAGUCUUGCCU-3′ (Ambion) (O’Regan and Fry, 2009); Nek7, 5′-AAUAGUGAUCUGAAGGAAGAGGUGG-3′ (Invitrogen); Nek9, 5′-AAUAGCAGCUGUGUGAGUCUUGCCU-3′ (Invitrogen); Nek9 UTR, 5′-GCTGCCTTGGGAATTCAGTdTdT-3′ and 5′-GCAGCCAAACTTTGATTAAdTdT-3′ (Ambion).
Immunoprecipitation and western blot analysis
Immunoprecipitations and western blotting were performed as described in Roig et al (2002). Anti-Nek9, anti-Nek9[Thr210-P], anti-Nek6 and anti-Eg5[Ser1033-P] polyclonal antibodies have been described in Roig et al (2002, 2005); Belham et al (2003) and Rapley et al (2008). Other antibodies used are anti-Nek7 (Cell Signaling), anti-cdc27, anti-cyclin B1 (Santa Cruz Biotechnology), anti-Plk1 (Calbiochem), anti-Eg5 (BD Bioscience), anti-GFP (Roche and Invitrogen), anti-FLAG and anti-β-tubulin (Sigma). Secondary antibodies were from Jackson ImmunoResearch Laboratories and were detected by ECL chemiluminescence (Thermo Scientific).
MS analysis
For phosphopeptide identification, Coomassie-stained protein bands were excised and in situ digested with trypsin and LC/MS/MS analysis of phosphorylation sites was performed at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School, Boston, MA) as described previously (Roig et al, 2005).
Two-hybrid analysis
cDNAs coding for the human Plk1 and Nek9 fragments indicated in Figure 1 was subcloned into pGBKT7 and pGADT7, respectively, and yeast two-hybrid analysis was performed as described in Rapley et al (2008).
Kinase assays
Protein kinase assays were carried out as described previously (Roig et al, 2002) using 100 μM ATP.
Immunofluorescence
Cells were grown on coverslips fixed with methanol and permeabilized as described earlier (Rapley et al, 2008). Primary antibodies used were mouse anti-γ-tubulin (Sigma), mouse anti-FLAG (Sigma), mouse anti-GFP (Invitrogen), rabbit anti-centrin (Groen et al, 2004), rabbit anti-histone H3[Ser10-P] (Cell Signaling), rabbit anti-Myc (Sigma) and mouse anti-Eg5 (BD). Primary antibodies were detected with Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 555 goat anti-mouse IgG (Invitrogen). When needed, anti-FLAG antibodies were detected with Alexa Fluor 647-Fab fragments using the Zenon mouse IgG labelling kit (Invitrogen). DNA was stained with DAPI (Sigma).
Images were taken using a Leica TCS SPE confocal system with a DM2500 CSQ upright microscopy and a × 63 1.30 ACS Apo lens, and edited using Leica LAS AF software (Leica Microsystems) and Photoshop (Adobe).
Supplementary Material
Acknowledgments
We thank C Vila and other members of the extended Cell Signaling group as well as the staff of the IRB Barcelona Advanced Digital Microscopy Core Facility for their help. We also thank I Vernos (Centre de Regulació Genòmica, Barcelona, Spain) and J Lüders (IRB Barcelona) for comments, A Fry (University of Leicester, Leicester, UK) for GFP-Nek2 and comments and A Groen, R Ohi and the Mitchison Laboratory (Harvard University, Boston, USA) for anti-centrin antibodies. JA acknowledges support from NIH grant DK17776 and institutional funds. JR acknowledges support from the Ramón y Cajal Program and the Plan Nacional I+D Grant BFU2008-03441/BMC (MICINN, Spain), and institutional funds.
Author contributions: MTB designed and performed the experiments in Figures 1, 2 and 3 and 7, and Supplementary Figures S1–S3 and S6; SS designed and performed the experiments in Figures 4, 5, 6, 7, 8 and 9 and Supplementary Figures S4–S9; LR performed the experiments in Figure 1 and Supplementary Figure S2; JA was instrumental for the initial stages of the study including the reported MS analysis; CC was instrumental for the realization of the study, discussed the data and commented and contributed to the paper; JR conceived the study, designed experiments and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Archambault V, Glover DM (2009) Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol 10: 265–275 [DOI] [PubMed] [Google Scholar]
- Belham C, Roig J, Caldwell JA, Aoyama Y, Kemp BE, Comb M, Avruch J (2003) A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J Biol Chem 278: 34897–34909 [DOI] [PubMed] [Google Scholar]
- Blangy A, Arnaud L, Nigg EA (1997) Phosphorylation by p34cdc2 protein kinase regulates binding of the kinesin-related motor HsEg5 to the dynactin subunit p150. J Biol Chem 272: 19418–19424 [DOI] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d’Hérin P, Harper M, Kress M, Nigg EA (1995) Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83: 1159–1169 [DOI] [PubMed] [Google Scholar]
- Elia AE, Cantley LC, Yaffe MB (2003) Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299: 1228–1231 [DOI] [PubMed] [Google Scholar]
- Faragher AJ, Fry AM (2003) Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell 14: 2876–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P (2009) Dynein antagonizes Eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol 19: 1833–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, Meraldi P, Nigg EA (1998) A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J 17: 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J (2010) Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell 18: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Hagan IM (2002) Schizosaccharomyces pombe NIMA-related kinase, Fin1, regulates spindle formation and an affinity of Polo for the SPB. EMBO J 21: 3096–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, Kim SH, Lockhart DJ, Schultz PG (1998) Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science 281: 533–538 [DOI] [PubMed] [Google Scholar]
- Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R (2004) XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr Biol 14: 1801–1811 [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435: 114–118 [DOI] [PubMed] [Google Scholar]
- Lane HA, Nigg EA (1996) Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol 135: 1701–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM (2007) The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol 17: 304–315 [DOI] [PubMed] [Google Scholar]
- Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE (1991) polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev 5: 2153–2165 [DOI] [PubMed] [Google Scholar]
- Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E (2010) Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol 12: 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA (2001) Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol 2: 21–32 [DOI] [PubMed] [Google Scholar]
- O’Connell MJ, Krien MJ, Hunter T (2003) Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol 13: 221–228 [DOI] [PubMed] [Google Scholar]
- O’Regan L, Blot J, Fry AM (2007) Mitotic regulation by NIMA-related kinases. Cell Div 2: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan L, Fry AM (2009) The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol Cell Biol 29: 3975–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Ohsugi M, Yamamoto T (2006) The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol 8: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Petronczki M, Lénárt P, Peters JM (2008) Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev Cell 14: 646–659 [DOI] [PubMed] [Google Scholar]
- Quarmby LM, Mahjoub MR (2005) Caught Nek-ing: cilia and centrioles. J Cell Sci 118: 5161–5169 [DOI] [PubMed] [Google Scholar]
- Rapley J, Nicolàs M, Groen A, Regué L, Bertran MT, Caelles C, Avruch J, Roig J (2008) The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci 121: 3912–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig J, Groen A, Caldwell J, Avruch J (2005) Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol Biol Cell 16: 4827–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig J, Mikhailov A, Belham C, Avruch J (2002) Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev 16: 1640–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J (2005) Spindle assembly: asters part their separate ways. Nat Cell Biol 7: 219–222 [DOI] [PubMed] [Google Scholar]
- Salem H, Rachmin I, Yissachar N, Cohen S, Amiel A, Haffner R, Lavi L, Motro B (2010) Nek7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene 29: 4046–4057 [DOI] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ (1992) Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359: 540–543 [DOI] [PubMed] [Google Scholar]
- Sawin KE, Mitchison TJ (1995) Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci USA 92: 4289–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, Demmers J, Fornerod M, Melchior F, Hoogenraad CC, Medema RH, Akhmanova A (2010) Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 8: e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macu̇rek L, Galjart N, Medema RH (2008) Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J 27: 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH (2010) Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell 19: 797–806 [DOI] [PubMed] [Google Scholar]
- Uteng M, Hentrich C, Miura K, Bieling P, Surrey T (2008) Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J Cell Biol 182: 715–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt MA, van de Weerdt BC, Vader G, Janssen H, Calafat J, Klompmaker R, Wolthuis RM, Medema RH (2004) Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J Biol Chem 279: 36841–36854 [DOI] [PubMed] [Google Scholar]
- Weil D, Garçon L, Harper M, Duménil D, Dautry F, Kress M (2002) Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. Biotechniques 33: 1244–1248 [DOI] [PubMed] [Google Scholar]
- Weinger JS, Qiu M, Yang G, Kapoor TM (2011) A nonmotor microtubule binding site in Kinesin-5 is required for filament crosslinking and sliding. Curr Biol 21: 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead CM, Rattner JB (1998) Expanding the role of HsEg5 within the mitotic and post-mitotic phases of the cell cycle. J Cell Sci 111(Part 17): 2551–2561 [DOI] [PubMed] [Google Scholar]
- Woodcock SA, Rushton HJ, Castañeda-Saucedo E, Myant K, White GR, Blyth K, Sansom OJ, Malliri A (2010) Tiam1-Rac signaling counteracts Eg5 during bipolar spindle assembly to facilitate chromosome congression. Curr Biol 20: 669–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye XS, Xu G, Pu RT, Fincher RR, McGuire SL, Osmani AH, Osmani SA (1995) The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO J 14: 986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yissachar N, Salem H, Tennenbaum T, Motro B (2006) Nek7 kinase is enriched at the centrosome, and is required for proper spindle assembly and mitotic progression. FEBS Lett 580: 6489–6495 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.