Abstract
Green fluorescent protein (GFP) has been widely used in several molecular and cellular biology applications, since it is remarkably stable in vitro and in vivo. Interestingly, native GFP is resistant to the most common chemical denaturants; however, a low fluorescence signal has been observed after acid-induced denaturation. Furthermore, this acid-denatured GFP has been used as substrate in studies of the folding activity of some bacterial chaperones and other chaperone-like molecules. Protein disulfide isomerase enzymes, a family of eukaryotic oxidoreductases that catalyze the oxidation and isomerization of disulfide bonds in nascent polypeptides, play a key role in protein folding and it could display chaperone activity. However, contrasting results have been reported using different proteins as model substrates. Here, we report the further application of GFP as a model substrate to study the chaperone activity of protein disulfide isomerase (PDI) enzymes. Since refolding of acid-denatured GFP can be easily and directly monitored, a simple micro-assay was used to study the effect of the molecular participants in protein refolding assisted by PDI. Additionally, the effect of a well-known inhibitor of PDI chaperone activity was also analyzed. Because of the diversity their functional activities, PDI enzymes are potentially interesting drug targets. Since PDI may be implicated in the protection of cells against ER stress, including cancer cells, inhibitors of PDI might be able to enhance the efficacy of cancer chemotherapy; furthermore, it has been demonstrated that blocking the reductive cleavage of disulfide bonds of proteins associated with the cell surface markedly reduces the infectivity of the human immunodeficiency virus. Although several high-throughput screening (HTS) assays to test PDI reductase activity have been described, we report here a novel and simple micro-assay to test the chaperone activity of PDI enzymes, which is amenable for HTS of PDI inhibitors.
Keywords: green fluorescent protein, protein disulfide isomerase, folding, chaperone
1. Introduction
Green fluorescent protein (GFP) is an autofluorescent protein that was first identified and isolated from the jellyfish, Aequorea victoria [1]. GFP is a soluble protein that contains a fluorophore consisting of three post-translationally modified amino acid residues (Ser65–Tyr66–Gly67) [2]. Because its fluorescence is remarkably stable, in vitro and in vivo, GFP has been widely used in several molecular and cellular biology applications [3,4]. Interestingly, GFP is resistant to denaturation by 8 M urea, 6 M guanidine hydrochloride, and 1% sodium dodecyl sulfate (SDS) [5]; moreover, it is unusually thermo-stable at different pH values and after chemical denaturation [6,7]. However, a low fluorescence signal has been observed after chemical-induced denaturation at pH 6.5 [8] and after acid-induced denaturation at pH 1.5 [9]. Acid-denatured GFP has been used as model substrate to study the protein folding activity of bacterial chaperones [10–13] as well as the chaperone-like activity of imidazole [14], and CPH nanogel [15].
Protein disulfide isomerase enzymes (PDI, EC 5.3.4.1) are eukaryotic oxidoreductases that catalyze the oxidation and isomerization of disulfide bonds in nascent polypeptides. PDI enzymes play a key role in the folding of proteins delivered to the secretory pathway; moreover, they are multifunctional proteins that display chaperone activity [16,17]. Interestingly, their function as oxidoreductase or chaperone is substrate-dependent [18–20]. Moreover, it seems that the chaperone activity of PDI might be independent of its enzymatic activity [21].
One function of chaperones is to suppress aggregation of non-native forms of proteins during the refolding or unfolding processes; however, bona fide chaperones are proteins that promote the complete folding of denatured substrates to their native conformation and are different from chaperone-like proteins which, on the contrary, only retain partially that ability (e.g., prevent aggregation) [22]. Here, we report the further application of GFP as a model substrate to study the chaperone activity of yeast and human PDI homologues (YPDI and HuPDI, respectively). Since refolding of acid-denatured GFP can be easily and directly monitored by real-time fluorescence, a simple micro-assay was used to study the effect of the chaperone or substrate concentration on protein folding kinetics. Moreover, this micro-assay format was used to evaluate the effect of a well-known inhibitor of PDI enzymes.
2. Results and Discussion
2.1. Acid-Denatured GFP as Model Substrate
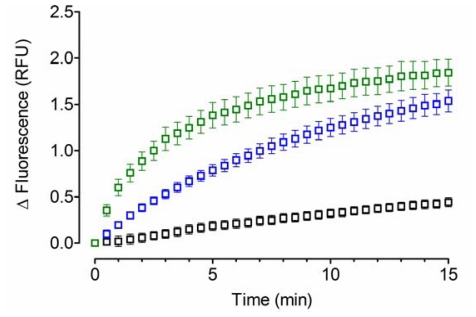
To demonstrate the feasibility of applying the acid-denatured GFP as substrate to study the chaperone activity of YPDI and HuPDI, a preliminary assay was performed (Figure 1). As expected, spontaneous refolding was observed in the absence of a molecular chaperone, with kinetics that followed an exponential one-phase association model with a rate constant (k) of 0.037 ± 0.021 min−1 and fluorescence at infinite time (Plateau) of 1.04 ± 0.48 RFU. Significant improvement of refolding was obtained with the assistance of YPDI or HuPDI (p < 0.05), indicating the participation of their chaperone activity. Furthermore, the refolding kinetics also followed an exponential one-phase association model with k values of 0.107 ± 0.012 min−1 for YPDI and 0.311 ± 0.027 min−1 for HuPDI, but with similar Plateau values (1.91 ± 0.12 and 1.78 ± 0.04 RFU, respectively). Interestingly, the k value exhibited by HuPDI was three-fold higher than that for YPDI. This dissimilarity could be likely related to the molecular differences in their substrate binding sites [23–25] and the multifunctional behavior of HuPDI [16,17].
Figure 1.
Green fluorescent protein (GFP) refolding kinetics. 0.05 μM of acid-denatured GFP was refolded in renaturing buffer containing 1 μM of a protein disulfide isomerase (PDI as chaperone): YPDI [blue] and HuPDI [green]; or in the absence of any chaperone [black]. Data represent mean ± S.E.M. (bars) of three independent experiments.
2.2. Effect of PDI Concentration
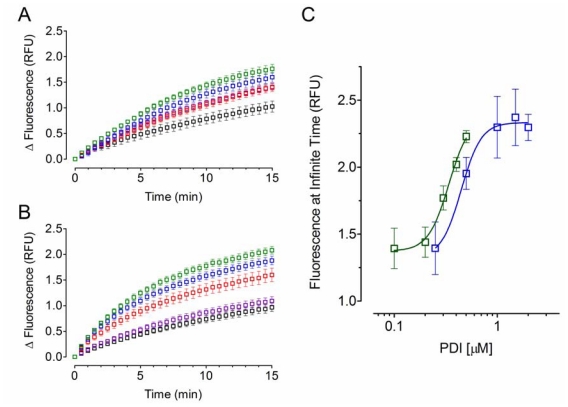
Since acid-denatured GFP was a feasible substrate model and considering that refolding assisted by PDI proteins followed an exponential one-phase association model, the effect of PDI concentration on their chaperone activity was then analyzed. As observed in Figure 2, an amplification of the fluorescent signal over time was detected as result of increasing the PDI concentration (Figure 2A and 2B). Although, no significant change was observed in the k values (p > 0.05), the Plateau values were used to estimate the half maximal effective concentration (EC50) for each PDI protein (Figure 2C). The concentration/Plateau data were fitted to a four-parameter dose-response variable slope model and the apparent EC50 values of 0.45 ± 0.09 μM for YPDI and 0.34 ± 0.04 μM for HuPDI were obtained. Considering that there might be differences in the substrate affinity and specificity, the observed EC50 values suggest that both PDI proteins have similar chaperone activity.
Figure 2.
Effect of PDI concentration on its chaperone activity. 0.05 of acid-denatured GFP was refolded in renaturing buffer containing different μM concentrations of YPDI: 0 [black], 0.5 [purple], 1.0 [red], 1.5 [blue], and 2.0 [green] (A); or HuPDI: 0.1 [black], 0.2 [purple], 0.3 [red], 0.4 [blue], and 0.5 [green] (B); dependence of the refolding kinetics (fluorescence at infinite time, Plateau) on PDI concentration: YPDI [blue] or HuPDI [green] (C). Data represent mean ± S.E.M. (bars) of three independent experiments.
2.3. Effect of Acid-Denatured GFP Concentration
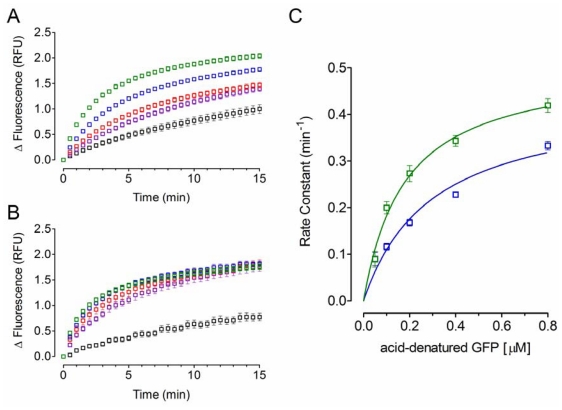
Additionally, the effect of the substrate concentration on the chaperone activity of PDI was also determined. As shown in Figure 3, a higher fluorescence signal over time was detected as a consequence of increasing the concentration acid-denatured GFP (Figures 3A and 3B); thus, increasing k values were observed (Figure 3C), demonstrating that the chaperone activity PDI was dependent on the substrate concentration. Furthermore, the displayed increase on the k values was statistically significant (p < 0.05). Moreover, analysis of the concentration/k data with the Hill equation yields a Hill coefficient of 0.89 ± 0.11 and 1.06 ± 0.08 for YPDI and HuPDI, respectively, indicating that both chaperones bind one molecule substrate per monomer with no cooperativity. Thus, by fitting data to a specific one-binding site model, apparent Kd values of 0.30 ± 0.09 μM for YPDI and 0.18 ± 0.03 μM for HuPDI were observed. These results add further evidence to the notion that differences in the substrate affinity and specificity exist [23–25]. Table 1 summarizes the kinetic parameters exhibited by the chaperone activity of YPDI and HuPDI.
Figure 3.
The effect of acid-denatured GFP concentration on PDI chaperone activity. Different μM concentrations of acid-denatured GFP were refolded in renaturing buffer containing 0.25 μM of YPDI (A) or 0.20 μM of HuPDI (B): 0.05 [black], 0.1 [purple], 0.2 [red], 0.4 [blue], and 0.8 [green]; dependence of the refolding kinetics (rate constant, k) on the substrate concentration: YPDI [blue] or HuPDI [green] (C). Data represent mean ± S.E.M. (bars) of three independent experiments.
Table 1.
Kinetic parameters of the chaperone activity exhibited by YPDI and HuPDI proteins using acid-denatured GFP as substrate*.
| Spontaneous | YPDI | HuPDI | |
|---|---|---|---|
| k ( min−1) | 0.037 ± 0.021 | 0.107 ± 0.012 | 0.311 ± 0.027 |
| Plateau (RFU) | 1.04 ± 0.48 | 1.91 ± 0.12 | 1.78 ± 0.04 |
| EC50 (μM) | n.d. | 0.45 ± 0.09 | 0.34 ± 0.04 |
| Hill coefficient | n.d. | 0.89 ± 0.11 | 1.06 ± 0.08 |
| Kd (μM) | n.d. | 0.30 ± 0.09 | 0.18 ± 0.03 |
Data represent mean ± S.E.M. of three independent experiments; n.d., not determined.
2.4. Bacitracin Inhibits the Chaperone Activity of PDI
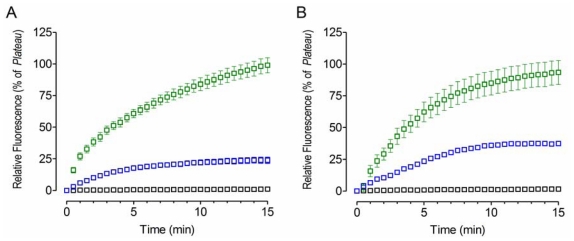
The antibiotic bacitracin, a well-known inhibitor of PDI proteins [26], was then used to test the effect of an inhibitor on the chaperone activity of PDI (Figure 4). Typical inhibition kinetics was displayed, i.e., diminished refolding was observed as the outcome of bacitracin binding to YPDI or HuPDI (Figures 4A and 4B), indicating the inhibition of its chaperone activity (p < 0.05). Interestingly, full inhibition was observed at 1 mM of bacitracin. Although, the apparent k values determined from the residual chaperone activity at 0.1 mM of bacitracin remain practically unchanged (p < 0.05), a reduction of the apparent Plateau was detected, 75% for YPDI and 56% for HuPDI. These results support the notion that bacitracin inhibits PDI apparently through competition for the substrate binding site [27].
Figure 4.
Effect of bacitracin on the PDI chaperone activity. 0.05 μM of acid-denatured GFP was refolded in renaturing buffer containing 2 μM of YPDI (A); or HuPDI (B), inhibited with 0 [green], 0.1 [blue], and 1 mM [black] of the antibiotic bacitracin. Data represent mean ± S.E.M. (bars) of three independent experiments.
3. Experimental Section
3.1. Vectors, Bacterial Strains, Enzymes and Chemicals
The E. coli XL1-Blue MRF’ strain and the plasmid pBluescript SK(-) were from Stratagene. The plasmid pQE30 and the Ni-NTA-agarose resin were from Qiagen. The plasmids pQBI25 and pCMV6-XL4/NM_000918.2 were from Qbiogene and Origene, respectively. The plasmid pCT38 was kindly provided by Christine Tachibana (University of Washington, USA) [28]. The routine molecular biology enzymes were from New England Biolabs. The biochemicals and other reagents were from Sigma-Aldrich.
3.2. Construction of Recombinant Plasmids
The jellyfish GFP gene (sgGFP, that is an optimized variant of the A. victoria GFP: F64L, S65C, I167T, in which solubility and protein folding has been improved for more rapid and efficient fluorophore formation) was obtained by endonuclease cleavage from plasmid pQBI25. A KspI-XbaI GFP-containing fragment was subcloned into the plasmid pBluescript SK(-). After that, a stretch of the multiple cloning sites, from BamHI to EcoRI, was removed by endonuclease digestion, overhangs filling and autoligation. Then, a SacI-HindIII GFP-containing fragment was subcloned into the plasmid pQE30. To get the recombinant plasmid pQHGF301, a segment from BssHII to BamHI was removed as previously. The PDI genes from yeast (YPDI) and human (HuPDI) were amplified by PCR using the plasmids pCT38 and pCMV6-XL4/NM_000918.2 as templates, respectively, and the corresponding synthetic oligonucleotides as primers (Table 2). Then, the amplified YPDI and HuPDI products were digested with specific endonucleases and subcloned into the plasmid pQE30, to get the recombinant plasmids pQYPDI and pQHuPDI. All recombinant plasmids were isolated from stable transformants and confirmed by endonuclease restriction analyses and DNA sequencing.
Table 2.
Primers used for the amplification and cloning of YPDI and HuPDI genes.
| Primer | Sequence | Endonuclease |
|---|---|---|
| ScPDIF | acactcggatccCAACAAGAGGCTGTGGCC | BamHI |
| ScPDIR | acactcctgcagTTACAATTCATCGTGAATGGC | PstI |
| BamPDIH | cgggatccGACGCCCCCGAGGAGGAGGAC | BamHI |
| HsPDIR2 | gcccaagcttACAGTTCATCTTTCACAGCTTTCTG | HindIII |
3.3. Expression and Purification of GFP, YPDI and HuPDI
The expression and purification of recombinant GFP, YPDI, and HuPDI proteins from E. coli XL1-Blue MRF’ cell cultures (harboring the plasmid pQHGF301, pQYPDI or pQHuPDI) was performed according to standard protocols.
GFP, YPDI and HuPDI expression
A single clone of each transformant was selected and grown overnight at 37 °C with agitation; the culture was diluted 100-fold in fresh media and grown at 37 °C for 2 h with agitation (pre-induction); then, IPTG was added to a final concentration of 0.1 mM and grown at 37 °C for another 4 h with agitation (protein induction). The cells were harvested by centrifuging at 9,500 rpm for 5 min.
GFP purification
The cell pellet was resuspended in 5 volumes of lysis buffer (8 M urea, 50 mM Tris-HCl, pH 8.0) and disrupted by vortexing (1–2 min). The total cell lysate was clarified by centrifugation at 9,500 rpm for 15 min (4 °C). Recombinant GFP was purified by Ni-chelating affinity chromatography following the native conditions protocol, described in the QIAexpressionist® manual (Qiagen), to eliminate the denaturant and allow the on-column refolding. Finally, refolded GFP was collected from Sephadex™ G-25 columns (PD-10, Amersham Biosciences) using 20 mM Tris-HCl (pH 8.0) as elution buffer. Protein concentration was determined by UV spectrophotometry at 280 nm, using the calculated molar absorptivity (21,890 M−1 cm−1) [29]. Additionally, the maximum absorbance peak was identified (474 nm) and the ratio of absorbance 474/280 was calculated (1.49) as control of purity.
YPDI and HuPDI purification
The pellet was resuspended in 5 volumes of lysis buffer (CelLytic® B, as recommended by the manufacturer) and disrupted by rocking (15 min). Total cell lysate was clarified by centrifugation at 9,500 rpm for 15 min (4 °C). Recombinant YPDI or HuPDI were purified by Ni-chelating affinity chromatography following the native conditions protocol described in the QIAexpressionist® manual (Qiagen). Finally, active PDI proteins were collected from Sephadex™ G-25 columns (PD-10, Amersham Biosciences) using 20 mM Tris-HCl (pH 8.0) as elution buffer. Protein concentration was determined by UV spectrophotometry at 280 nm, using the calculated molar absorptivity (M−1 cm−1: YPDI = 49,655; HuPDI = 45,755) [29]. The enzyme activity of PDI proteins was determined by an insulin-reduction turbidimetric assay reported elsewhere.
3.4. Acid-Denaturation of GFP
A 5 μM acid-denatured GFP solution (pH 1.5) was prepared by mixing a 10 μM GFP solution in denauring buffer (0.3 mM EDTA, 1 mM DTT, 50 mM Tris-HCl, pH 7.5) with an equal volume of 125 mM HCl solution and incubated at room temperature for 1 min [13]. After that, protein denaturation (low fluorescence signal) was confirmed by fluorescence measurements. Fluorescence was expressed in relative fluorescence units (RFU, i.e., arbitrary florescence units per pmol of GFP).
3.5. Refolding of GFP by PDI Proteins
GFP refolding was started by diluting the 5 μM acid-denatured GFP in the renaturing buffer (25 mM MgCl2, 100 mM KCl, 50 mM Tris-HCl, pH 7.5) containing PDI enzymes. Refolding reactions were carried out in final volumes of 200 μL. All experiments were performed at room temperature and the fluorescence was continuously determined for 15 min. Fluorescence measurements were carried out using a Fluoroskan Ascent® FL microplate fluorometer and luminometer (Thermo Scientific) at 485 nm excitation and 538 nm emission wavelengths.
3.6. Inhibition of the PDI Chaperone Activity by Bacitracin
The antibiotic bacitracin was used as inhibitor of the chaperone activity of PDI proteins. Different concentrations of bacitracin (0, 0.1, or 1 mM) were added to a renaturing buffer containing 2 μM of YPDI or HuPDI. After 30 min of the PDI/bacitracin interaction, the residual chaperone activity was measured as previously described, using 0.05 μM of acid-denatured GFP as substrate.
3.7. Statistical and Data Analysis
All statistical and data analysis were performed using Prism® v.5 (GraphPad Software, San Diego, CA, USA). Mean, standard error of the mean (S.E.M.) and one-way ANOVA were calculated from data of three independent experiments. At a statistical value p < 0.05, the ANOVA was followed by either Tukey’s or Dunnett’s multiple comparison test, as appropriate. All acid-denatured GFP refolding data were fitted using the standard least square regression method. Association kinetics data were fitted to an exponential one-phase association model using the following Equation (1):
| (1) |
where Y0 and Plateau represent the fluorescence signal at time (t) equal to zero and at infinite time, respectively, while k is the observed constant rate of association. The chaperone concentration/Plateau data were fitted to a four-parameter logistic (4PL) dose-response variable slope model using the following Equation (2):
| (2) |
where H describes the steepness of the curves, Ym and YM are the apparent plateaus of response, minimal and maximal respectively, and EC50 is the concentration of chaperone (i.e., YPDI or HuPDI) that gives a response half way between Ym and YM. The acid-denatured GFP concentration/k data were fitted to a specific one-binding site model using the following Equation
| (3) |
where Bmax represents the maximum specific binding and Kd is the equilibrium binding constant.
4. Conclusions
Eukaryotic PDI enzymes have been shown to be up-regulated during ER stress, suggesting that they might be involved in cellular protection, including protection against induced apoptosis [30,31]. Additionally, several studies have shown that PDI-mediated reductive cleavage of disulfide bonds at the cellular surface is essential for the infectivity of some pathogens, such as HIV, Chlamydia, and Leishmania [32–34]. Because of the diversity of their functional activities, PDI enzymes are potentially interesting drug targets. Given that PDI may be implicated in the protection of cells against ER stress, including cancer cells, inhibitors of PDI might be able to enhance the efficacy of chemotherapy in some cancers [35,36]; furthermore, it has been demonstrated that blocking the reductive cleavage of disulfide bonds of proteins associated with the cell surface markedly reduced the infectivity of HIV [37]. Although several high-throughput screening (HTS) assays to test PDI reductase activity have been reported [38,39], we described here a novel and simple micro-assay to test the chaperone activity of PDI enzymes that is amenable for HTS of PDI inhibitors.
Acknowledgements
We thank Christine Tachibana (University of Washington) for providing the plasmid pCT38; and Rodrigo Lameda-Díaz, Analleli E. Malváez and Blanca C. Saucedo for the construction of plasmids harboring PDI genes. This work was supported in part by CONACYT (SEP-2004-C01-47554) and UABC (CPI-C15a-3868) grants. Authors are National Researchers (SNI-CONACYT) and members of the Biological-Pharmaceutical Academic Group (Health Sciences, UABC).
References
- 1.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 2.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bizzarri R, Serresi M, Luin S, Beltram F. Green fluorescent protein based pH indicators for in vivo use: A review. Anal. Bioanal. Chem. 2009;393:1107–1122. doi: 10.1007/s00216-008-2515-9. [DOI] [PubMed] [Google Scholar]
- 4.Morris MC. Fluorescent biosensors of intracellular targets from genetically encoded reporters to modular polypeptide probes. Cell. Biochem. Biophys. 2010;56:19–37. doi: 10.1007/s12013-009-9070-7. [DOI] [PubMed] [Google Scholar]
- 5.Ward WW, Bokman SH. Reversible denaturation of Aequorea green-fluorescent protein: physical separation and characterization of the renatured protein. Biochemistry. 1982;21:4535–4540. doi: 10.1021/bi00262a003. [DOI] [PubMed] [Google Scholar]
- 6.Penna TC, Ishii M, Junior AP, Cholewa O. Thermal stability of recombinant green fluorescent protein (GFPuv) at various pH values. Appl. Biochem. Biotechnol. 2004;114:469–483. doi: 10.1385/abab:114:1-3:469. [DOI] [PubMed] [Google Scholar]
- 7.Nagy A, Malnasi-Csizmadia A, Somogyi B, Lorinczy D. Thermal stability of chemically denatured green fluorescent protein (GFP)—A preliminary study. Thermochim. Acta. 2004;410:161–163. [Google Scholar]
- 8.Alkaabi KM, Yafea A, Ashraf SS. Effect of pH on thermal- and chemical-induced denaturation of GFP. Appl. Biochem. Biotechnol. 2005;126:149–156. doi: 10.1385/abab:126:2:149. [DOI] [PubMed] [Google Scholar]
- 9.Enoki S, Saeki K, Maki K, Kuwajima K. Acid denaturation and refolding of green fluorescent protein. Biochemistry. 2004;43:14238–14248. doi: 10.1021/bi048733+. [DOI] [PubMed] [Google Scholar]
- 10.Makino Y, Amada K, Taguchi H, Yoshida M. Chaperonin-mediated folding of green fluorescent protein. J. Biol. Chem. 1997;272:12468–12474. doi: 10.1074/jbc.272.19.12468. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida T, Kawaguchi R, Taguchi H, Yoshida M, Yasunaga T, Wakabayashi T, Yohda M, Maruyama T. Archaeal group II chaperonin mediates protein folding in the cis-cavity without a detachable GroES-like co-chaperonin. J. Mol. Biol. 2002;315:73–85. doi: 10.1006/jmbi.2001.5220. [DOI] [PubMed] [Google Scholar]
- 12.Okochi M, Matsuzaki H, Nomura T, Ishii N, Yohda M. Molecular characterization of the group II chaperonin from the hyperthermophilic archaeum Pyrococcus horikoshii OT3. Extremophiles. 2005;9:127–134. doi: 10.1007/s00792-004-0427-y. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Hu ZJ, Luo YM, Huo YW, Ma Q, He YZ, Zhang YY, Sun F, Dong ZY. Distinct symmetry and limited peptide refolding activity of the thermosomes from the acidothermophilic archaea Acidianus tengchongensis S5(T) Biochem. Biophys. Res. Commun. 2010;393:228–234. doi: 10.1016/j.bbrc.2010.01.106. [DOI] [PubMed] [Google Scholar]
- 14.Shi R, Pan Q, Guan Y, Hua Z, Huang Y, Zhao M, Li Y. Imidazole as a catalyst for in vitro refolding of enhanced green fluorescent protein. Arch. Biochem. Biophys. 2007;459:122–128. doi: 10.1016/j.abb.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Asayama W, Sawada S, Taguchi H, Akiyoshi K. Comparison of refolding activities between nanogel artificial chaperone and GroEL systems. Int. J. Biol. Macromol. 2008;42:241–246. doi: 10.1016/j.ijbiomac.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Chen S, Wang X, Wang L, Wallis AK, Freedman RB, Wang CC. Plasticity of human protein disulfide isomerase: evidence for mobility around the X-linker region and its functional significance. J. Biol. Chem. 2010;285:26788–26797. doi: 10.1074/jbc.M110.107839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim. Biophys. Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Katiyar S, Till EA, Lennarz WJ. Studies on the function of yeast protein disulfide isomerase in renaturation of proteins. Biochim. Biophys. Acta. 2001;1548:47–56. doi: 10.1016/s0167-4838(01)00214-x. [DOI] [PubMed] [Google Scholar]
- 19.Hatahet F, Ruddock LW. Substrate recognition by the protein disulfide isomerases. FEBS J. 2007;274:5223–5234. doi: 10.1111/j.1742-4658.2007.06058.x. [DOI] [PubMed] [Google Scholar]
- 20.Kozlov G, Määttänen P, Thomas DY, Gehring K. A structural overview of the PDI family of proteins. FEBS J. 2010;277:3924–3936. doi: 10.1111/j.1742-4658.2010.07793.x. [DOI] [PubMed] [Google Scholar]
- 21.Fu XM, Zhu BT. Human pancreas-specific protein disulfide-isomerase (PDIp) can function as a chaperone independently of its enzymatic activity by forming stable complexes with denatured substrate proteins. Biochem. J. 2010;429:157–169. doi: 10.1042/BJ20091954. [DOI] [PubMed] [Google Scholar]
- 22.Spinozzi F, Mariani P, Rustichelli F, Amenitsch H, Bennardini F, Mura GM, Coi A, Ganadu ML. Temperature dependence of chaperone-like activity and oligomeric state of alphaB-crystallin. Biochim. Biophys. Acta. 2006;1764:677–687. doi: 10.1016/j.bbapap.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Pirneskoski A, Klappa P, Lobell M, Williamson RA, Byrne L, Alanen HI, Salo KE, Kivirikko KI, Freedman RB, Ruddock LW. Molecular characterization of the principal substrate binding site of the ubiquitous folding catalyst protein disulfide isomerase. J. Biol. Chem. 2004;279:10374–10381. doi: 10.1074/jbc.M312193200. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen VD, Wallis K, Howard MJ, Haapalainen AM, Salo KE, Saaranen MJ, Sidhu A, Wierenga RK, Freedman RB, Ruddock LW, Williamson RA. Alternative conformations of the x region of human protein disulphide-isomerase modulate exposure of the substrate binding b’ domain. J. Mol. Biol. 2008;383:1144–1155. doi: 10.1016/j.jmb.2008.08.085. [DOI] [PubMed] [Google Scholar]
- 25.Byrne LJ, Sidhu A, Wallis AK, Ruddock LW, Freedman RB, Howard MJ, Williamson RA. Mapping of the ligand-binding site on the b’ domain of human PDI: Interaction with peptide ligands and the x-linker region. Biochem. J. 2009;423:209–217. doi: 10.1042/BJ20090565. [DOI] [PubMed] [Google Scholar]
- 26.Mandel R, Ryser HJ, Ghani F, Wu M, Peak D. Inhibition of a reductive function of the plasma membrane by bacitracin and antibodies against protein disulfide-isomerase. Proc. Natl. Acad. Sci. USA. 1993;90:4112–4116. doi: 10.1073/pnas.90.9.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Versteeg HH, Ruf W. Tissue factor coagulant function is enhanced by protein-disulfide isomerase independent of oxidoreductase activity. J. Biol. Chem. 2007;282:25416–25424. doi: 10.1074/jbc.M702410200. [DOI] [PubMed] [Google Scholar]
- 28.Tachibana C, Stevens TH. The yeast EUG1 gene encodes an endoplasmic reticulum protein that is functionally related to protein disulfide isomerase. Mol. Cell. Biol. 1992;12:4601–4611. doi: 10.1128/mcb.12.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka S, Uehara T, Nomura Y. Up-regulation of protein-disulfide isomerase in response to hypoxia/brain ischemia and its protective effect against apoptotic cell death. J. Biol. Chem. 2000;275:10388–10293. doi: 10.1074/jbc.275.14.10388. [DOI] [PubMed] [Google Scholar]
- 31.Rao RV, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program. Cell Death Differ. 2004;11:372–380. doi: 10.1038/sj.cdd.4401378. [DOI] [PubMed] [Google Scholar]
- 32.Barbouche R, Miquelis R, Jones IM, Fenouillet E. Protein-disulfide isomerase-mediated reduction of two disulfide bonds of HIV envelope glycoprotein 120 occurs post-CXCR4 binding and is required for fusion. J. Biol. Chem. 2003;278:3131–3136. doi: 10.1074/jbc.M205467200. [DOI] [PubMed] [Google Scholar]
- 33.Abromaitis S, Stephens RS. Attachment and entry of Chlamydia have distinct requirements for host protein disulfide isomerase. PLoS Pathog. 2009;5:e1000357. doi: 10.1371/journal.ppat.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos CX, Stolf BS, Takemoto PV, Amanso AM, Lopes LR, Souza EB, Goto H, Laurindo FR. Protein disulfide isomerase (PDI) associates with NADPH oxidase and is required for phagocytosis of Leishmania chagasi promastigotes by macrophages. J. Leukoc. Biol. 2009;86:989–998. doi: 10.1189/jlb.0608354. [DOI] [PubMed] [Google Scholar]
- 35.Corazzari M, Lovat PE, Armstrong JL, Fimia GM, Hill DS, Birch-Machin M, Redfern CP, Piacentini M. Targeting homeostatic mechanisms of endoplasmic reticulum stress to increase susceptibility of cancer cells to fenretinide-induced apoptosis: The role of stress proteins ERdj5 and ERp57. Br. J. Cancer. 2007;96:1062–1071. doi: 10.1038/sj.bjc.6603672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovat PE, Corazzari M, Armstrong JL, Martin S, Pagliarini V, Hill D, Brown AM, Piacentini M, Birch-Machin MA, Redfern CP. Increasing melanoma cell death using inhibitors of protein disulfide isomerases to abrogate survival responses to endoplasmic reticulum stress. Cancer Res. 2008;68:5363–5369. doi: 10.1158/0008-5472.CAN-08-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallina A, Hanley TM, Mandel R, Trahey M, Broder CC, Viglianti GA, Ryser HJ. Inhibitors of protein-disulfide isomerase prevent cleavage of disulfide bonds in receptor-bound glycoprotein 120 and prevent HIV-1 entry. J. Biol. Chem. 2002;277:50579–50588. doi: 10.1074/jbc.M204547200. [DOI] [PubMed] [Google Scholar]
- 38.Smith AM, Chan J, Oksenberg D, Urfer R, Wexler DS, Ow A, Gao L, McAlorum A, Huang SG. A high-throughput turbidometric assay for screening inhibitors of protein disulfide isomerase. J. Biomol. Screen. 2004;9:614–620. doi: 10.1177/1087057104265292. [DOI] [PubMed] [Google Scholar]
- 39.Khalaf NB, De Muylder G, Ratnam J, Ang KK-H, Arkin M, McKerrow J, Chenik M. A high-throughput turbidometric assay for screening inhibitors of Leishmania major protein disulfide isomerase. J. Biomol. Screen. 2011;16:545–551. doi: 10.1177/1087057111401026. [DOI] [PubMed] [Google Scholar]