Abstract
The ability to maintain human fungiform papillae cells in culture for multiple cell cycles would be of considerable utility for characterizing the molecular, regenerative, and functional properties of these unique sensory cells. Here we describe a method for enzymatically isolating human cells from fungiform papillae obtained by biopsy and maintaining them in culture for more than 7 passages (7 months) without loss of viability and while retaining many of the functional properties of acutely isolated taste cells. Cells in these cultures exhibited increases in intracellular calcium when stimulated with perceptually appropriate concentrations of several taste stimuli, indicating that at least some of the native signaling pathways were present. This system can provide a useful model for molecular studies of the proliferation, differentiation, and physiological function of human fungiform papillae cells.
Keywords: culture, fungiform, gustducin, regeneration, sweet, taste receptor
Introduction
Taste receptor cells are specialized epithelial cells with unique histological, molecular, and physiological characteristics that permit detection of a wide range of both simple and structurally complex molecules. In mammals, taste buds are associated with fungiform papillae on the anterior two-thirds of the tongue and circumvallate and foliate papillae are on the posterior third of the tongue. Taste buds are also present in the epithelium of the soft palate and pharynx (Travers and Nicklas 1990). Taste receptor molecules and their downstream signaling components have also been found in enteroendocrine cells throughout the lining of the gastrointestinal tract (Bezencon et al. 2007; Margolskee et al. 2007; Kokrashvili et al. 2009). Human fungiform papillae contain from zero to over 25 taste buds, with over half having no taste buds and the rest having an average of 3 or 4 buds (Arvidson 1979). Each taste bud contains 50–100 cells of 4 morphologically and functionally distinct types, which exhibit properties of both neuronal and epithelial cells (Finger 2005). About half of the cells in the taste bud are spindle-shaped type I (dark) cells, which appear to have a glial-like function because they surround other celltypes and express molecules involved in neurotransmitter inactivation (Paran et al. 1975). An additional 25% of the taste bud cells are type II (light) cells, which express several proteins including the G-protein alpha-gustducin, phospholipase C-β2 (PLC-β2), inositol 1,4,5-trisphosphate receptor type 3, and the transient receptor potential channel M5 (TRPM5), which have been implicated in transduction of sweet, umami, and bitter taste responses (McLaughlin et al. 1992; Takami et al. 1994; Rossler et al. 1998; Asano- Miyoshi et al. 2000; Gilbertson et al. 2000; Miyoshi et al. 2001; Clapp et al. 2001; Perez et al. 2002). Current evidence indicates that G-protein–coupled receptors implicated in sweet and umami taste (taste receptor family 1 [T1Rs]) and bitter taste (taste receptor family 2 [T2Rs]) are expressed in nonoverlapping subsets of type II cells (Zhao et al. 2003; Chandrashekar et al. 2006). Cells mediating sour (acid) taste are likely to be a subset of type III cells (Huang et al. 2008), which comprise an additional 15% of the taste cells. The cells mediating salty taste have not yet been identified. Importantly, taste buds are one of the very few truly regenerative organs in the human body, with taste cells having an average life span of about 12 days (Beidler and Smallman 1965). A small number of type IV or basal cells in each bud are generally considered to be the stem cells giving rise to the other cell types (Stone, Tan, et al. 2002; Nakayama et al. 2008; Thirumangalathu et al. 2009; Sullivan et al. 2010).
Properties of taste cells often vary among species, and it is presently not known how maturational stage or lineage relates to the stimulus–response properties of these cells (Barlow and Northcutt 1994). Functional studies of taste cells have been done using freshly isolated cells in primary culture, explant cultures from rodents, or semi-intact taste buds in tissue slices (Mbiene et al. 1997; Miyamoto et al. 1999; Caicedo et al. 2000; Qin et al. 2008). Although each of these preparations has advantages, the development of long-term cultures would have provided significant benefits, particularly for studies of taste cell proliferation and differentiation. Most attempts to culture taste cells have reported limited viability, with cells typically not lasting beyond 3–5 days (Spielman et al. 1989; Caicedo et al. 2000; Kishi et al. 2001; Ruiz et al. 2001; Stone, Wilcox, and Kinnamon 2002). We recently reported on a method for the extended culture of rodent taste cells (Ozdener et al. 2006). Here, we describe the establishment of long-term primary cultures of cells from human fungiform papillae that have molecular and physiological properties consistent with both developing and mature taste cells. In addition, these cultures were amenable to the use of moderate throughput screening (MTS) to examine responses to individual or combinations of taste stimuli. These cultures provide the basis for the development of a variety of taste cell models with which to study intracellular signaling, the effects of trophic or toxic agents on taste cell growth and function, and the assessment of potential taste stimuli and taste modifiers.
Materials and methods
Criteria for selection of sample donors
Healthy, 28- to 56-year-old men (n = 7) and women (n = 6) were screened as potential donors of lingual fungiform papillae. Subjects read, understood, and signed a consent form. The protocol (#0934) was approved by Schulman Associates Institutional Review Board Inc., Cincinnati, OH. Ethical standards as set forth by the Helsinki Declaration of 1973, as revised in 1983, were adhered to. Subject exclusion criteria on initial screening included those reporting a medical history of or current manifestation of systemic, chronic disease; those reporting the regular use of prescription medications; those reporting dry mouth or oral disease; and those reporting adverse reaction to lidocaine anesthesia in a dental setting. Subjects meeting these criteria were invited to participate in the study. Exclusion criteria at the time of biopsy included a heart rate >100 or <60, systolic blood pressure >145 or <100, diastolic blood pressure >90 or <60, and the oral surgeon's opinion that the subject was not appropriate for the study. The oral cavity was inspected for general tissue health. All data collected were recorded by subject identity code and maintained confidential.
Human fungiform biopsy and culture
Fungiform papilla were removed from the dorsal surface of the anterior portion of the tongue using curved spring microscissors and immediately placed into an isolation solution (in millimolar, NaHCO3, 26; NaH2PO4, 2.5; glucose, 20; NaCl, 65; KCl, 20; and Ethylenediaminetetraacetic acid [EDTA], 1). Six to 8 papillae are taken from each subject. The isolation solution was removed by pipette and approximately 1 mL of isolation buffer mixed with 1.5 mg/mL pronase E (Sigma), and 1 mg/mL elastase (Sigma) was added. After a 30-min incubation period at room temperature, the papillae were then transferred to Iscove's Modified Dulbecco's medium (Gibco BRL) containing 10% fetal bovine serum, 1:5 ratio of MCDB 153 (Sigma), and a triple cocktail of antibiotics (100 U/mL/100 μg/mL, penicillin/streptomycin, 2.5 μg/mL gentamycine, and 0.5 μg/mL fungizone). The papillae were gently minced with a razor blade in a glass dish and the pieces seeded into cloning cylinders onto 18 mm round glass coverslips (Fisher) (using tissue culture cylinders) coated with rat tail collagen type 1 (3.96 mg/mL diluted 1:4 in distilled nuclease-free water, BD Sciences) and incubated at 36 °C in a humidified environment containing 5% CO2. Alternatively, we incubated fungiform papillae with collagenase (550 U/mL), elastase (10 U/mL), and soy bean trypsin inhibitor (0.9 mg/mL) in calcium-free Ringer solution (145 g NaCl, 0.373 g KCl, 0.203 g MgCl2, 0.147 g CaCl2, 0.110 g Na-pyruvate, and 4.76 g (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-Na in 1 L water and adjust pH to 7.1–7.2 and osmolarity to 300–310). For calcium-free solution, calcium chloride was replaced with 2 mM EDTA in 35 °C water bath with circulation and gentle oxygenation with 95% O2/5% CO2 for 30 min. After incubation, cells were washed with Ringer solution, triturated with a fire-polished glass Pasteur pipette 10 times, and centrifuged for 3 min at 2500 revolutions per minute (rpm) at room temperature. Isolation solution was replaced with 1 mL of taste cell culture medium. Digested fungiform papillae were then gently minced with a surgical razor in a glass dish and transferred into cloning cylinders on rat tail collagen type-1 coated coverslips. Culture medium was replaced after 48 h, and then every 5–7 days, one-third of culture medium was replaced.
After approximately a month, cells on the coverslips were harvested using EDTA/trypsin and reseeded into a T25 culture flask (Corning). One-third of the medium was replaced each week. Cells were maintained in T25 flasks until reaching confluence (approximately 1 month) and then reseeded into a T75 culture flask (passage 1). Cells were maintained for additional 1 month until reaching full confluence. Then cells were either frozen at −80 °C until their next use or were maintained in culture for experimental purposes. Cultured human fungiform cells continued to passage in T75 flasks for up to 7 passages. When cells reached confluence, cells were replated for additional 4–6 days and experiments performed as described. Assessment of viability was done by staining with 0.04% Trypan Blue (Sigma).
Prior to use, coverslips were treated with 2 M NaOH for 1 h and left overnight in 70% nitric acid. After 1 h in an HCl acid wash, the coverslips were autoclaved in water, rinsed with 70% ethanol and 100% ethanol, and then air-dried.
Adenosine triphosphatase staining of human fungiform taste bud
Fungiform papillae were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 2 h at room temperature. Specimens were then cryoprotected with sequential immersions in 10%, 20%, and 30% sucrose in PBS for 24 h each. Sagittal sections of the entire papilla were cut at 8–10 μm on a Microm HM 500 OM cryostat. The tissue sections were thaw mounted onto Superfrost Plus slides (Fisher). We employed the lead (Pb) method for membrane-associated calcium adenosine triphosphatase (ATPase) detection (Barry 1992). Slides containing tissue sections were incubated for 10 min at room temperature in a solution of 20 mL of 0.84 mmol ATP, 20 mL of 80 mmol Tris-maleate buffer (pH 7.2), 3 mL of 2% Pb(NO3)2, 5 mL of 0.1 mol/L Mg(NO3)2, and 2 mL of deionized water. After 6–10 min, the slides were dipped 3 times in deionized water, developed in 1% yellow ammonium sulfide for 1 min, and then dipped 3 times again in deionized water. The slides were air-dried and observed using light microscopy. Brownish-black deposits indicated the presence of ATPase in the stained tissue sections.
Immunoprecipitation
Human fungiform cells were maintained for many passages, reseeded into a dish, and cultured for additional 4–7 days, and then cells were lysed into 1 mL of cold RIPA buffer (150 mM NaCl, 10 mM Tris, pH 7.2, 0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 1% deoxycholate, 5 mM EDTA) supplemented with protease inhibitor cocktail. The cell lysate was centrifuged at 2500 rpm (Z233 LabNet) for 2 min, and a 10-μL aliquot of the supernatant was used to determine protein concentration using the Bio-Rad Dc Protein estimation kit (Bio-Rad Laboratories) according to the manufacturer's protocol. Supernatant with 1 mg total protein was incubated on ice for 30 min with 25 μL protein A-sepharose beads (Invitrogen) to remove those lysate proteins showing nonspecific binding. After centrifugation at 3000 rpm for 3 min, the supernatant was incubated with 2 μg of rabbit immunoglobulin G (IgG) (Vector Laboratories) on ice for 30 min. To this was added 40 μL of a 0.50% suspension of protein A-sepharose beads (Invitrogen), and the mixture was incubated with constant rotation at 4 °C for 45 min. The beads were centrifuged, and this precleared lysate was incubated with either 4 μg of rabbit anti-gustducin antibody (I-20, Santa Cruz, sc-395) or 4 μg of rabbit anti-PLC-β2 antibody (Q-15, Santa Cruz; sc-206) overnight with rotation at 4 °C. The mixture was then incubated with 50 μL of a 50% protein A-sepharose suspension at 4 °C with rotation for 4 h. The beads were washed 3 times with 1 mL of cold RIPA buffer containing protease inhibitors after being centrifuged at 3500 rpm for 3 min. Washed beads were resuspended in 60 μL of SDS gel-loading buffer (50 mM Tris–HCl [pH 6.8], 5% 2-mercaptoethanol, 2% SDS, 0.1% bromophenol blue, 10% glycerol), heated at 100 °C for 5 min, and kept on ice for additional 5 min. Samples were briefly centrifuged at 3000 rpm for 2 min, and the supernatants were separated by SDS–polyacrylamide gel electrophoresis as described below.
Western blot
The products of immunoprecipitation were separated by SDS–polyacrylamide (5–15%) gradient gel (Bio-Rad Laboratories) electrophoresis and transferred to a Polyvinylidene Fluoride membrane (Bio-Rad Laboratories) that was incubated at room temperature for 1 h with 1% nonfat dry milk. Polyclonal rabbit anti-gustducin (Gαgust, I-20; sc-395, dilution 1:500–1:1000, Santa Cruz Biotechnology) and polyclonal rabbit anti-PLC-β2 (Q-15; sc-206, dilution 1:500–1:1000, Santa Cruz Biotechnology) were used. After overnight incubation at 4 °C with primary antibodies, the membrane was washed 3 times with PBS/T (0.1 M phosphate buffer solution with 0.05% Tween-20) for 10 min. Membranes were reprobed with donkey anti-rabbit antibody conjugated with horseradish peroxidase (NA934, dilution 1:1 0000, Amersham Biosciences) for 30–45 min at room temperature followed by three 15-min washes with PBS/T. Signal was detected with the enhanced chemiluminescence plus immunoblot detection system (Amersham Biosciences) following the manufacturer's instructions. X-ray films were later scanned for documentation and analysis.
To determine specificity of gustducin and PLC-β2 detection, both antibodies were preincubated with 5-fold (by weight) of their specific peptide (Santa Cruz, sc-395 P, sc-206 P, respectively) for 4 h at room temperature and then used to replace the primary antibody for western blot, as described above.
Reverse transcription–polymerase chain reaction
Total RNA from cultured human fungiform cells was extracted with Versagene kit (Gentra systems) following the manufacturer's instructions. RNA (0.5 μg) was reverse transcribed for 50 min at 42 °C using the Superscript First Strand Synthesis System for reverse transcription–polymerase chain reaction (RT–PCR) (Invitrogen). As a control for genomic DNA contamination and nonspecific amplification, samples of RNA were treated in parallel in the presence and absence of reverse transcriptase and used for PCR by amplifying with primers designed for detection of β-actin, gustducin, PLC-β2, T2R5, T1R3, and TRPM5 (Table 1). Primers were chosen to span one or more introns to exclude confusion with amplified fragments from genomic DNA. PCR amplification of cDNA for each RT reaction was performed in a final volume of 25 μL containing 1 μL of cDNA, 1× AmpliTaq Gold PCR buffer, 2.5 mM MgCl2, 1 mM deoxynucleoside triphosphates, 0.4 μM of each primer, and 0.25 U/μL of AmpliTaq Gold polymerases (Applied Biosystems). PCR amplification was determined individually for each gene (Table 1). PCR products were separated on 2% agarose gels and stained with 0.2 μg/mL of ethidium bromide to verify their expected size. Amplified product on gel was excised with razor. DNA was extracted from gel using QIAquick Gel Extraction Kit (Qiagen). The PCR product obtained from gel extraction was sequenced using both reverse and forward amplification primers in University of Pennsylvania Sequencing facilities.
Table 1.
Primers and conditions used for detecting taste-specific molecules
| Gene | Sequence | PCR condition | Expected size (bp) | Reference | |
| β-Actin | GGACTTCGAGCAAGAGATGG | 95 | 7 min | 234 | NM_001101.3 |
| 94 | 45 s | ||||
| 53 | 45 s | ||||
| AGCACTGTGTTGGCGTACAG | 72 | 45 s | |||
| 72 | 7 min | ||||
| Gustducin | TCTGGGTATGTGCCAAATGA | 95 | 7 min | 386 | NM_001102386 |
| 94 | 45 s | ||||
| 53 | 45 s | ||||
| GGCCCAGTGTATTCTGGAAA | 72 | 45 s | |||
| 72 | 7 min | ||||
| PLC-β2 | GTCACCTGAAGGCATGGTCT | 95 | 3 min | 333 | NM_004573 |
| 94 | 30 s | ||||
| 53 | 30 s | ||||
| TTAAAGGCGCTTTCTGCAAT | 72 | 60 s | |||
| 72 | 7 min | ||||
| T2R5 | TATGGTTTGCCACCTTCCTC | 95 | 7 min | 394 | NM_01898 |
| 94 | 45 s | ||||
| 53 | 45 s | ||||
| AAGGACTTCAGCGCAGTGAT | 72 | 45 s | |||
| 72 | 7 min | ||||
| T1R3 | CTTTTGTGGCCAGGATGAGT | 95 | 4 min | 345 | NM_152228 |
| 94 | 45 s | ||||
| 56 | 45 s | ||||
| TGCAGGAAGAGTGTGCTCAG | 72 | 50 s | |||
| 72 | 10 min | ||||
| TRPM5 | TGGTAGAGCGCATGATGAAG | 95 | 10 min | 301 | NM_001101 |
| 94 | 20 s | ||||
| 63 | 30 s | ||||
| ACCAACAGGAAGGTGACCAG | 72 | 45 s | |||
| 72 | 7 min | ||||
Immunohistochemistry and immunocytochemistry
Human fungiform papillae were fixed in 4% PFA in PBS (pH 7.2) for 1 h at room temperature and then cryoprotected by sequential immersions in 10%, 20%, and 30% sucrose in PBS for 24 h each. Sagittal sections of each papilla were cut at 10 μm on a Microm HM 500 OM cryostat. Tissue sections were thaw mounted onto Superfrost Plus slides and stained as described below.
Cultured human fungiform cells were maintained for many passages and reseeded for 2–6 days followed by fixing with 4% PFA in PBS for 10 min at room temperature. Cells were blocked with 3% normal goat serum, 3% bovine serum albumin, and 0.3% Triton X-100 in PBS for 60 min and then incubated with primary antibodies (Table 2) overnight at 4 °C followed by incubation with secondary antibodies (Table 2) diluted in blocking buffer for 30 min at room temperature. After washing in PBS and water, coverslips were mounted using Vectashield with 4',6-diamidino-2-phenylindole (Vector Laboratories). Controls for immunofluorescence consisted of omitting the primary antibody or substituting the primary antibody with the host IgG from which the antibody was generated. In all cases, the controls revealed no nonspecific labeling. Immunoreactive cells were counted in at least 3 fields at ×20 magnification. Images were acquired with a Leica TCS-SP2 Spectral Confocal Microscope (Leica Microsystems Inc.)
Table 2.
Antibodies used for detecting expression of specific molecules
| Primary antibody | Source | Host | Dilution | Secondary antibody | Source | Host | Dilution |
| Gustducin | Santa Cruz | Rabbit | 1:500 | Anti-rabbit IgG Alexa 633 | Molecular Probes | Goat | 1:500 |
| PLC-β2 | Santa Cruz | Rabbit | 1:500 | Anti-rabbit IgG Alexa 633 | Molecular Probes | Goat | 1:500 |
| NCAM | Sigma | Mouse | 1:500 | Anti-mouse IgG Alexa 488 | Molecular Probes | Goat | 1:500 |
| GLAST | Chemicon | Guinea pig | 1:2500 | Anti-guinea pig-FITC | Sigma | Donkey | 1:500 |
FITC, Fluorescein isothiocyanate.
Confocal imaging
Fluorescent images were captured with a Leica TCS-SP2 Spectral Confocal Microscope (Leica Microsystems Inc.) using UV, Argon, and HeNe lasers and an HC PL APO CS ×20.0 (0.070 NA) objective. Excitation wavelengths used were at 405 nm for DAPI, 488 nm for Alexa Fluor 488, and 633 nm for Alexa Fluor 633 with emissions detected at appropriate wavelengths. The pinhole diameter was set at the first minimum diameter of the Airy disc for the objective used, giving acceptable resolution of the z axis for the fluorescent focal plane. Laser power and photomultiplier gain were adjusted for optimal signal/noise ratio and held constant for comparison of antibody-labeled and control slides. Sequential acquisition of each wavelength was used for some double-labeling experiments to prevent cross talk or bleed-through between fluorophores. Leica Scanware software was used to acquire confocal images scanning unidirectionally at a 1024 × 1024 pixel format with 3 lines and 2 frames averaging. Computer-controlled digital zoom was used to increase magnification to a maximum of ×2.3 with the ×20 objective. Images were arranged and minimally adjusted for contrast and brightness using LCS software (Leica Microsystems Inc.) and Adobe Photoshop elements 2.0 (Adobe Systems Inc.).
MTS and data analysis
Cultured human fungiform papillae cells (from 7 independent ultures) were grown for 2–4 days after replating in 96-well plates. Cells were loaded with 1 mM Fura-2 AM (Molecular Probes Inc.) in10 mg/mL Pluronic F127 (Molecular Probes Inc.) in modified Ringer solution for 1 h at 36 °C. Changes in intracellular calcium levels ([Ca2+]i) in response to taste stimuli were measured in 96-well format using a Discovery-1 imaging station and Metamorph software (Molecular Devices). Stimuli were dissolved in Ringer and adjusted for pH and osmolarity if necessary (Table 3). The cells in each well were visualized with an inverted fluorescence microscope using excitation wavelengths of 340 and 380 nm and an emission wavelength set by a band-pass filter centered at 510 nm. The stimulus delivery head was fitted onto the plate and allowed for constant flow of fluid into and out of the well, providing a constant level of fluid over the cells. Stimulus delivery and removal was controlled by a 2 channel peristaltic pump (Spetec). The delivery head was moved from well to well by a robotic plate handler (Hudson Controls). Cells were first superfused with bath solution and then exposed to taste stimuli via superfusion, with each stimulus applied for 15 s. A series of image pairs was acquired for each well at ×20 with a CCD camera (Photometrics) as follows: 4 baseline, 12 poststimulus, and 4 baseline for each of 3 stimuli. A 2-min washout period followed each poststimulus block of images. Stimulus selection and delivery, focusing, and image acquisition, as well as plate movement were controlled by custom-designed Discovery-1 software (Molecular Devices). Following acquisition, Metamorph software (Molecular Devices) was used to define regions of interest (ROIs), calculate fluorescence intensity values, and compute average fluorescence ratios for individual cells. Ratio data for the cells were subsequently analyzed with custom software to determine which cells had responded with a significant change in intracellular calcium. The analysis yields raw baseline ratio and noise levels, identified peaks, and calculated the magnitude and latency for each cell's response to each stimulus. Data from cells that failed to return to baseline were not included, nor were apparent responses that occurred later than 150 s following stimulation. Autofluorescence was negligible and, with exposure times of 100–200·ms, we did not find appreciable photo bleaching.
Table 3.
Response frequency of cultured human fungiform taste cells to different chemical stimuli
| Stimuli | Response frequency (%) | Positive ROI/total ROI |
| 2 mM Denatonium | 15 | 2220/1 4971 |
| 1 mM Sucralose | 2 | 292/1 4971 |
| 250 ppm Acesulfame-K | 2 | 316/1 4971 |
| 3 mM Monopotassium glutamate | 1 | 96/1 4971 |
| 10 μM ATP | 22 | 1297/5825 |
Results
The isolation and characterization of cells from human fungiform papillae
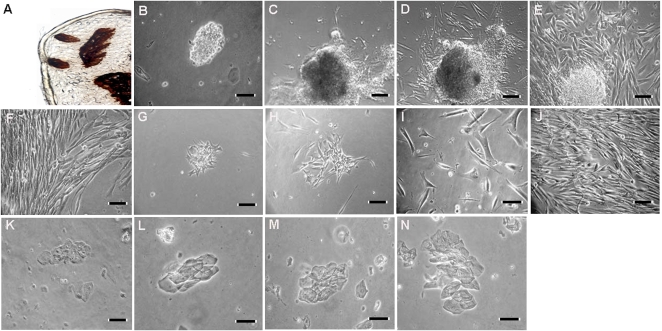
Our goal was to develop a simple and efficient method for culturing human fungiform cells through several cell cycles. The presence of taste buds in excised human fungiform papillae was demonstrated using a mammalian taste bud marker, calcium-dependent ecto-ATPase (Barry 1992) (Figure 1A) . After treatment of the fungiform papillae with the enzyme mixture, they were minced and the resulting cell suspension plated on either cell culture plate or on glass coverslips coated with rat tail collagen type 1. Utilizing both isolation methods worked equally effective to obtain human fungiform cells in culture. After 24–48 h in culture, small pieces of tissue and individual cells began to adhere and grow on the bottom of the culture flask (Figure 1B). Attachment efficiency varied from 30% to 50%. No cell attachment was observed on uncoated glass culture plates or coverslips. Cells typically began to grow outward from the pieces of fungiform tissue that remained after the dissociation procedure (Figure 1C,D). Cells grew for 2–4 weeks both under attached cell clusters and within small colonies of cells (Figure 1D,E). It does appear that clusters of proliferating cells grow out from underneath the older layer of cells. The older layer of cells may provide trophic or mechanical support for the proliferating cells. After 2–4 weeks, the cell clusters began to detach (Figure 1F). The cells typically grew to confluence within 4 weeks (Figure 1F), at which time they were reseeded into T25 flasks for 3–4 weeks and then transferred into T75 flasks where they continued to grow for more than 7 passages (Figure 1G–J). Cells with epithelial cell morphology did not appear to grow under these conditions over the same period of time (Figure 1K–N). During the first 15–30 days, most of the cells retained their original compact cell bodies, sometimes with one or more processes. After reaching confluence, most cells had an polarized elongated, migratory-like, and often multipolar appearance. The morphology of the expanding populations of cells was similar to that observed for cultured taste bud cells and other chemosensory cells in culture (Gomez et al. 2000; Kishi et al. 2001, 2002; Murata et al. 2006).
Figure 1.
Attachment and morphology of cultured human fungiform taste cells. ATPase staining of papillae showing several taste buds (A). Primary cell cultures grown on collagen type 1–coated plates were imaged after 2 days (B), 4 days (C), and 6 days (D). Cells from human fungiform papillae grew for up to 2–4 weeks under attached cell clusters (E). After 2–4 weeks, the clusters detached and the cells typically reached confluence within 4 weeks (F). (G–J) Represents day 2, day 4, day 10, and 4 weeks after harvesting and reseeding, respectively. Cultured cells continued to grow for at least 8 months. During this period, we did not observe growth of cells with the appearance of nontaste epithelial cells (K–N). Scale bars = 50 (C, F–H) and 100 μm (A, B, D, E, I, and J).
The cultures remained viable for several passages after the biopsy. In addition, the cells withstood cryopreservation; however, renewed growth of the cryopreserved cells after thawing and subsequent culture was considerably quicker than growth of cultures directly from biopsy by 3–4 weeks.
Staining with Trypan Blue indicated that cells maintained 98–99% viability for over 7 passages. We did not observe any bacterial or fungal contamination problems throughout the study, indicating the elimination of any contamination that may have been present in the initial tissue sample.
Expression of specific taste marker proteins and mRNA in cultured human fungiform cells
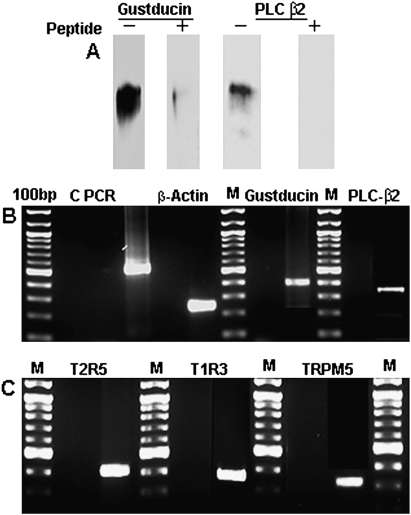
Homogenates of cultured fungiform cells were subjected to immunoprecipitation with polyclonal antisera against α-gustducin and PLC-β2. Both gustducin and PLC-β2 proteins were detected by western blot (Figure 2A), and the single bands for each corresponded to those previously reported (Ozdener et al. 2006). We also examined mRNA expression of taste cell markers, including gustducin, PLC-β2, T2R5, T1R3, and TRPM5. Human β-actin was used as a positive control housekeeping marker (Figure 2B). RT–PCR results demonstrated that both taste receptor subunits (T2R5 and T1R3), as well as all 3 taste signaling components (gustducin, PLC-β2, and TRPM5), were expressed in the cultures, with amplification products of the expected size (Figure 2B,C). As a control, PCR reactions carried out with RNA samples amplified in the absence of reverse transcriptase enzyme yielded no products using all sets of primers, indicating the lack of genomic DNA contamination. All amplified PCR products purified from agarose gels were sequenced and showed 100% homology to expected sequences.
Figure 2.
Immunoprecipitation and western blot analysis showed that both gustducin and PLC-β2 were expressed in cultured cells maintained for many passages. Preincubation of gustducin and PLC-β2 antibodies with their specific peptides removed their respective bands completely, supporting the specificity of reactions (A). RT–PCR results demonstrated the expression of β-actin, gustducin, PLC-β2, (B) T1R3, T2R5, and TRPM5 (C) mRNA in cultured human fungiform taste cells. The cDNA transcribed from total RNA was amplified with intron-spanning specific primers. PCR products were found at expected size and confirmed by sequencing. Specific mRNA was not detected in control experiments without reverse transcriptase, indicating no genomic DNA contamination. M, marker (100-bp division).
Immunocytochemical detection of taste cell markers in cultured human fungiform cells
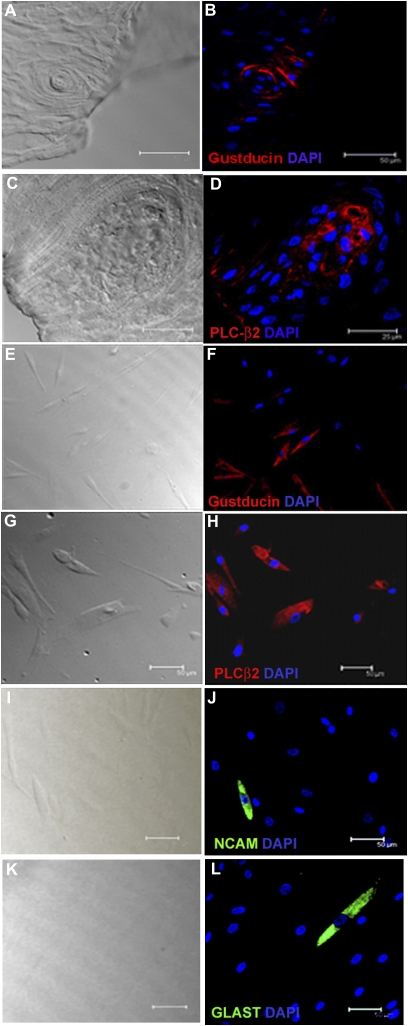
Human fungiform papilla tissue and cell cultures derived from them were examined for the presence of specific taste cell markers using standard immunocytochemical techniques. Staining fungiform papillae tissue with gustducin and PLC-β2 antibodies indicated presence of both proteins (Figure 3A–D, respectively). Approximately 60% of cultured cells expressed gustducin (Figure 3E,F) and about 25% expressed PLC-β2 (Figure 3G,H), which are taste signaling molecules found in type II taste cells. We also observed immunoreactivity to NCAM (Figure 3I,J) and GLAST (Figure 3K,L) antibodies, each in about 10% of the cells. These are markers for type III and type I taste cells, respectively. Some cultured cells were immunoreactive to only a single biomarker, and many cells were not immunolabeled with any of these antibodies. No reactivity was observed when primary antibodies were substituted with Ig generated from the same animal. Peptide inhibition of gustducin antibody showed complete removal of gustducin immunostaining, indicating antibody specificity (data not shown). Thus, our results demonstrate that cells at different stages of maturation, which have persisted and divided in culture for several months, continue to express proteins found in mature taste cells in vivo.
Figure 3.
Immunostaining of human fungiform tissue and cultured cells showed presence of taste cell–specific biomarkers. Images were acquired with a Leica TCS-SP2 confocal laser scanning microscope. Cells were maintained in culture plate for 4–5 days before fixation. Transmission images of corresponding fields are shown on the left. Immunoreactivity was observed for gustducin in papillae (red in B) and in about 60% of cultured cells (F). Immunoreactivity for PLC-β2 (red) was also observed in papillae (D) and about 25% of cultured cells (H). Immunoreactivity to NCAM (green in J) and GLAST (green in L) antibodies was observed in some cells. Nuclei of cells were stained as blue with DAPI. For controls, immunostaining with antibody-specific Ig demonstrated the absence of nonspecific immunoreactivity (data not shown). Scale bars = 25 (A–D) and 50 μm (E–L).
Chemical stimuli induce intracellular Ca2+ responses
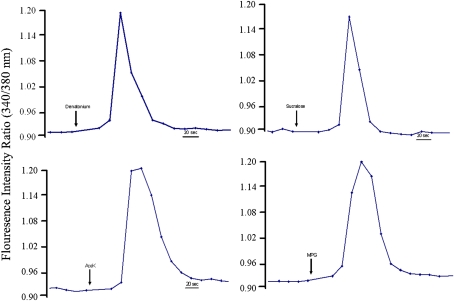
The responses of cultured human fungiform cells representing 7 independent cultures from different passage numbers (1–7) maintained for 2–10 days in plates were tested with representative stimuli of bitter, sweet, and umami qualities using the ratio metric Ca2+-sensitive dye Fura-2. Approximately 20% of the tested ROI of cultured cells exhibited increased [Ca+2]i levels in response to one of the stimuli, which is consistent with previous report (Ozdener et al. 2006). Overall response frequency was similar for all independent culture preparations. Highest frequency of responses was elicited by 2 mM denatonium (15%). Figure 4A shows a representative tracing of the transient increase in [Ca+2]i to denatonium. The artificial sweeteners 1 mM sucralose (Figure 4B) and 250 ppm acesulfame-K (Figure 4C) elicited an increase in [Ca+2]i in about 2% of ROI tested independently. Only a few cells responded to both artificial sweeteners. We also observed that monopotassium glutamate (Figure 4D) elicited an increased [Ca2+]i responses by 1% of tested cells. Note the return of [Ca+2]i to baseline levels over time after washing with buffer. We also tested whether ATP (10 μM) could elicit [Ca2+]i responses (data not shown). Similar to frequency of total responsive cells, 22% of ROI of tested cells responded to ATP and cells that responded to ATP also responded to bitter or sweet stimuli.
Figure 4.
Cultured taste cells responded to sweet, umami, and bitter stimuli. Changes in intracellular calcium levels ([Ca2+]i) in cultured human fungiform taste cell were measured in 96-well plates using Fura-2, a medium throughput system. The ratio refers to fluorescence intensity recorded at excitation wavelengths of 340 and 380.Stimuli were dissolved in modified Ringer solution and adjusted for pH and osmolarity. Graphs illustrate representative changes in [Ca+2]i levels in individual cells during exposure to (A) denatonium benzoate (2 mM), (B) sucralose (1 mM), (C) acesulfame-K (AceK, 250 ppm), (D) monopotassium glutamate (MPG, 3 mM). This figure appears in color in the online version of Chemical Senses.
Discussion
We have maintained cells obtained from human fungiform papillae for over 7 passages, spanning a period of 7 months. These cultures generate new cells, many of which mature in vitro to the stage where they express several markers of mature taste bud cell types. Previous studies indicated the importance of specific microenvironmental features controlling the survival and regeneration of cells obtained from mammalian taste tissues (Spielman et al. 1989; Kishi et al. 2001; Ruiz et al. 2001; Stone, Wilcox, and Kinnamon 2002; Ozdener et al. 2006). Studies indicated that the precursor cell fate was sensitive to culture conditions and that cellular phenotype is highly influenced by factors within the media that may be secreted or exogenously added (Raff et al. 1983; Tang et al. 2000; Babu et al. 2007). In a culture environment, taste cells lose a variety of components that are present in intact taste buds, such as nerve fiber connections, growth factors, neurotrophins, surrounding taste and epithelial cells, and saliva. This makes selecting an appropriate medium critical to successful multipassage cell culture. Our results indicate that the medium we employed the coating of slides with rat collagen type 1 and perhaps the persistence of supportive cells in the heterogeneous cell culture were sufficient to allow fungiform cells to grow and at least partially mature (Ozdener et al. 2006). However, which specific factors influence proliferation and differentiation of human fungiform cells in culture is not known. Both the similarities and differences between the cultured cells and their in vivo counterparts provide an opportunity to examine the presence and regulation of these characteristics at a molecular and cellular level, under conditions which can be manipulated and monitored.
The production of long-term proliferating, primary cell cultures that are capable of generating cells exhibiting many features characteristic of the mature, differentiated phenotype have been reported for other regenerative tissues such as the liver and olfactory epithelium. Under certain culture conditions, early passages of primary rat liver epithelial cells showed true diploid karyotype for up to 6 months. During this time, the growth characteristics of the cells also followed a normal pattern (Herrinig et al. 1983). Similarly, primary cultures derived from human olfactory epithelial biopsies can be maintained over many passages and support production of neuroepithelial cells exhibiting diverse olfactory and neuronal characteristics (Borgmann-Winter et al. 2009). Although we did not perform any specific experiments regarding senescence of cultured human fungiform cells, each passage and preparation of cultured fungiform cells demonstrated similar physiological and molecular and immunological properties. In particular, we did not see any differences in response to major tastant between early (e.g., less than 4) and late (e.g., 5–7) passages.
Mammalian taste receptor cells are distributed throughout the oral, laryngeal, and gastrointestinal epithelium. Several types of cells have been described in mammalian taste buds. Although there is some disagreement over their classification, most authors agree that there are 4 major types, referred to as type I, II, III, and basal cells, which can be distinguished on the basis of their ultrastructural and molecular characteristics (Farbman 1965a, 1965b; Delay et al. 1986). In our culture system, we demonstrated the presence of cells exhibiting features of the 3 types of taste cells. Although we did not utilize any test for basal cells, the proliferation and differentiation of cultured cells suggest the presence of at least some basal cells in the primary culture. We observed that approximately 50–60% of cells showed immunoreactivity to gustducin; 40% of these cells were also immunoreactive to PLC-β2 antibodies demonstrating the presence of type II cells. We also observed approximately 5–10% of cells that were immunoreactive to NCAM and GLAST antibodies, indicating presence of type III and type I taste cells, respectively. The relationship between molecular expression profiles and functional and morphological classification of taste cell type is not clearly established. The nonselective expression of molecular markers among cell types remains a possibility (Yee et al. 2003; Miura et al. 2006). The prevalence and relative expression patterns reflecting cell signaling pathways and cell types do not precisely reflect that seen in vivo. The factors governing the relative distribution of different cell types within a taste bud are unknown and may not be replicated in the culture conditions. Similarly, relative expression quantities and prevalence of transduction factors may depend on a variety of intrinsic and extrinsic factors. Cellular differentiation and lineage commitment are considered to be robust and irreversible processes during development. However, recent studies have independently demonstrated that transcription factors and environmental factors such as changes in culture medium composition and/or exogenous growth factors may induce pluripotency in primary cells. These conditions may induce cells to fully differentiate into various types of cells and/or undergo cell fate changes (Rieske et al. 2007, 2009; Vierbuchen et al. 2010). These findings suggest that cultured human fungiform cells may differentiate from immature or basal cells or may cross-differentiate from one mature cell type into another under the influence of particular environmental conditions such as cell culture medium and the unique cellular milieu retained within the primary culture (Luo et al. 2009; Nakamura et al. 2010).
An important criterion for a primary cell culture system is the presence of relevant functional properties. Although the cells in our cultures did not have the characteristic spindle-shaped morphology of in situ taste cells, they did display several key molecular and physiological properties of taste receptor cells. Most importantly, some cells responded to applications of sweet and bitter taste stimuli with transient increases in intracellular calcium, which is a feature of dissociated taste cells (Zhao et al. 2002), cells in taste bud slices (Caicedo et al. 2000; Zhao et al. 2002), and cultured rat taste cells (Ozdener et al. 2006). This indicates not only that some of the cells in culture expressed molecular receptors for sweet and bitter but also that they had enough of the G-protein–coupled transduction cascade to generate calcium responses (San Gabriel et al. 2009). The presence of gustducin- and PLC-β2 immunoreactivity in subsets of cultured cells is consistent with this conclusion.
Although the details of the transduction pathways are not definitively established for all taste stimuli, the increase in intracellular calcium elicited by bitter, sweet, and umami stimuli is thought to be dependent upon activation of phospholipase C, generation of second messengers followed by release of calcium from internal stores, and activation of the calcium-dependent TRPM5 channel, which is a monovalent-selective cation channel. The expression pattern of TRPM5 is highly selective, with expression in lingual tissue localized to type II taste cells where it coexists with T1Rs and T2Rs, gustducin, and PLC-β2. TRPM5 channel activity is strongly regulated by voltage, phosphoinositides, and temperatureand is blocked by acid pH (Liman 2007). At the same time, the G-protein α-gustducin activates a phosphodiesterase that metabolizes accumulated cyclic nucleotides (Yan et al. 2001). However, cyclic guanosine monophosphate and cyclic adenosine monophosphate are known to rise in response to sweet taste–induced activation of adenylyl cyclase (Naim et al. 1991). The pathways responsible for calcium responses in these cultured fungiform cells remains to be established, but our data support the presence of several key transduction elements and thus the use of this model system for further studies of these pathways.
The art of primary cell culture aims to provide an environment similar to the physiological conditions that a cell would experience in vivo. Primary cultures of many cell types have facilitated efforts to understand the signals involved in proliferation, differentiation, and yielded tools to rapidly assay new molecules targeting specific receptor pathways. Thus, they may be useful not only for examining cellular function in a more representative system than heterologously expressed receptors. Although these applications amply justify work with primary cultures, a number of limitations must be mentioned. Foremost among these is that each batch of primary cultured cells can vary due to differences in the initial population of cells used to start the culture. Primary cell cultures often miss the original tissue organization and structure and factors that play a role in the physiological function of cells. Cells are often not always and entirely in contact with other cells, as cultures are never grown to 100% confluence. Although the identification of a specific marker or set of markers and/or a control stimulus that can be used to compare cell profiles among batches is helpful in interpreting the results of functional or molecular assays, unfortunately there is no specific marker and single stimulus that can be used to identify a “mature taste cell.” On a more subtle level, primary cells are also subject to dedifferentiation, and chromosomal instability, characterized by losses or gains of chromosomes during cell replication in continuous cell lines, may be another problem. The comparison of cultured human fungiform cells with cultured nontaste cells of tongue needs to be explored. These experiments will give further insight into the regulation of taste cell development, proliferation, and differentiation.
In summary, the protocol described here enables the primary culture of cells derived from human fungiform buds for at least 7 passages (7 months) with the persistence of relevant cell physiological and molecular characteristics. The further development and application of this long-term primary culture will provide a model system for studies of proliferation and differentiation, stimulus specificity, cross talk, and adaptation properties of human taste receptor cells, as well as tissue regeneration. Great progress has been made toward understanding the development of taste receptor cells and the mechanisms by which they respond to chemical stimuli; yet, many open questions remain. These questions will require the use of many experimental approaches and model systems before our understanding is complete. This model system will also provide a tool to examine, in a controlled environment, the effects of conditions that impair taste cell function such as infection, medications, radiation, toxic, or chemical exposures.
Funding
This work was supported in part by grants (P50-DC006760 and NIH DC02995 to P.B.); National Science Foundation (0216310); and Givaudan Inc. grant.
Acknowledgments
We acknowledge the support and helpful discussions of Jay Slack, Ping Zhong, Karen Yee, and Bruce Bryant and the technical assistance of Esi Quayson, Aimee Myers, Linda Wysocki, and Valerie Audige. Author contributions: M.H.O. and N.E.R. designed the study and wrote the manuscript. M.H.O. performed the majority of the experiments. A.S. performed the biopsies of human fungiform papillae. P.B., J.H.T., F.W.L., and J.G.B. contributed to preparation and editing of the manuscript.
References
- Arvidson K. Location and variation in number of taste buds in human fungiform papillae. Scand J Dent Res. 1979;87:435–442. doi: 10.1111/j.1600-0722.1979.tb00705.x. [DOI] [PubMed] [Google Scholar]
- Asano-Miyoshi M, Abe K, Emori Y. Co-expression of calcium signaling components in vertebrate taste bud cells. Neurosci Lett. 2000;283:61–64. doi: 10.1016/s0304-3940(00)00911-3. [DOI] [PubMed] [Google Scholar]
- Babu H, Cheung G, Kettenmann H, Palmer TD, Kempermann G. Enriched monolayer precursor cell cultures from micro-dissected adult mouse dentate gyrus yield functional granule cell-like neurons. PLoS One. 2007;2:e388. doi: 10.1371/journal.pone.0000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow LA, Northcutt RG. Analysis of the embryonic lineage of vertebrate taste buds. Chem Senses. 1994;19:715–724. doi: 10.1093/chemse/19.6.715. [DOI] [PubMed] [Google Scholar]
- Barry MA. Ecto-calcium-dependent ATPase activity of mammalian taste bud cells. J Histochem Cytochem. 1992;40:1919–1928. doi: 10.1177/40.12.1453008. [DOI] [PubMed] [Google Scholar]
- Beidler LM, Smallman RL. Renewal of cells within taste buds. J Cell Biol. 1965;27:263–272. doi: 10.1083/jcb.27.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezencon C, Le Coutre J, Damak S. Taste-signaling proteins are coexpressed in solitary intestinal epithelial cells. Chem Senses. 2007;32:41–49. doi: 10.1093/chemse/bjl034. [DOI] [PubMed] [Google Scholar]
- Borgmann-Winter KE, Rawson NE, Wang HY, Wang H, Macdonald ML, Ozdener MH, Yee KK, Gomez G, Xu J, Bryant B, et al. Human olfactory epithelial cells generated in vitro express diverse neuronal characteristics. Neuroscience. 2009;158:642–653. doi: 10.1016/j.neuroscience.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo A, Jafri MS, Roper SD. In situ Ca2+ imaging reveals neurotransmitter receptors for glutamate in taste receptor cells. J Neurosci. 2000;20:7978–7985. doi: 10.1523/JNEUROSCI.20-21-07978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- Clapp TR, Stone LM, Margolskee RF, Kinnamon SC. Immunocytochemical evidence for co-expression of Type III IP3 receptor with signaling components of bitter taste transduction. BMC Neurosci. 2001;2:6. doi: 10.1186/1471-2202-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay RJ, Kinnamon JC, Roper SD. Ultrastructure of mouse vallate taste buds: II. Cell types and cell lineage. J Comp Neurol. 1986;253:242–252. doi: 10.1002/cne.902530210. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Electron microscope study of the developing taste bud in rat fungiform papilla. Dev Biol. 1965a;11:110–135. doi: 10.1016/0012-1606(65)90040-0. [DOI] [PubMed] [Google Scholar]
- Farbman AI. Fine structure of the taste bud. J Ultrastruct Res. 1965b;12:328–350. doi: 10.1016/s0022-5320(65)80103-4. [DOI] [PubMed] [Google Scholar]
- Finger TE. Cell types and lineages in taste buds. Chem Senses. 2005;30(Suppl 1):i54–i55. doi: 10.1093/chemse/bjh110. [DOI] [PubMed] [Google Scholar]
- Gilbertson TA, Damak S, Margolskee RF. The molecular physiology of taste transduction. Curr Opin Neurobiol. 2000;10:519–527. doi: 10.1016/s0959-4388(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Gomez G, Rawson NE, Hahn CG, Michaels R, Restrepo D. Characteristics of odorant elicited calcium changes in cultured human olfactory neurons. J Neurosci Res. 2000;62:737–749. doi: 10.1002/1097-4547(20001201)62:5<737::AID-JNR14>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Herrinig AS, Raychaudhuri R, Kelley SP, Iype PT. Repeated establishment of diploid epithelial cell cultures from normal and partially hepatectomized rats. In Vitro. 1983;19:576–588. doi: 10.1007/BF02619606. [DOI] [PubMed] [Google Scholar]
- Huang YA, Maruyama Y, Stimac R, Roper SD. Presynaptic (Type III) cells in mouse taste buds sense sour (acid) taste. J Physiol. 2008;586:2903–2912. doi: 10.1113/jphysiol.2008.151233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi M, Emori Y, Tsukamoto Y, Abe K. Primary culture of rat taste bud cells that retain molecular markers for taste buds and permit functional expression of foreign genes. Neuroscience. 2001;106:217–225. doi: 10.1016/s0306-4522(01)00184-1. [DOI] [PubMed] [Google Scholar]
- Kishi M, Emori Y, Tsukamoto Y, Abe K. Changes in cell morphology and cell-to-cell adhesion induced by extracellular Ca2+ in cultured taste bud cells. Biosci Biotechnol Biochem. 2002;66:484–487. doi: 10.1271/bbb.66.484. [DOI] [PubMed] [Google Scholar]
- Kokrashvili Z, Mosinger B, Margolskee RF. Taste signaling elements expressed in gut enteroendocrine cells regulate nutrient-responsive secretion of gut hormones. Am J Clin Nutr. 2009;90:822S–825S. doi: 10.3945/ajcn.2009.27462T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER. TRPM5 and taste transduction. Handb Exp Pharmacol. 2007;179:287–298. doi: 10.1007/978-3-540-34891-7_17. [DOI] [PubMed] [Google Scholar]
- Luo X, Okubo T, Randell S, Hogan BL. Culture of endodermal stem/progenitor cells of the mouse tongue. In Vitro Cell Dev Biol Anim. 2009;45:44–54. doi: 10.1007/s11626-008-9149-2. [DOI] [PubMed] [Google Scholar]
- Margolskee RF, Dyer J, Kokrashvili Z, Salmon KS, Ilegems E, Daly K, Maillet EL, Ninomiya Y, Mosinger B, Shirazi-Beechey SP. T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc Natl Acad Sci U S A. 2007;104:15075–15080. doi: 10.1073/pnas.0706678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbiene JP, Maccallum DK, Mistretta CM. Organ cultures of embryonic rat tongue support tongue and gustatory papilla morphogenesis in vitro without intact sensory ganglia. J Comp Neurol. 1997;377:324–340. doi: 10.1002/(sici)1096-9861(19970120)377:3<324::aid-cne2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- McLaughlin SK, McKinnon PJ, Margolskee RF. Gustducin is a taste-cell-specific G protein closely related to the transducins. Nature. 1992;357:563–569. doi: 10.1038/357563a0. [DOI] [PubMed] [Google Scholar]
- Miura H, Kusakabe Y, Harada S. Cell lineage and differentiation in taste buds. Arch Histol Cytol. 2006;69:209–225. doi: 10.1679/aohc.69.209. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Fujiyama R, Okada Y, Sato T. Strain difference in amiloride-sensitivity of salt-induced responses in mouse non-dissociated taste cells. Neurosci Lett. 1999;277:13–16. doi: 10.1016/s0304-3940(99)00828-9. [DOI] [PubMed] [Google Scholar]
- Miyoshi MA, Abe K, Emori Y. IP(3) receptor type 3 and PLCbeta2 are co-expressed with taste receptors T1R and T2R in rat taste bud cells. Chem Senses. 2001;26:259–265. doi: 10.1093/chemse/26.3.259. [DOI] [PubMed] [Google Scholar]
- Murata Y, Ozaki M, Nakamura T. Primary culture of gustatory receptor neurons from the blowfly, Phormia regina. Chem Senses. 2006;31:497–504. doi: 10.1093/chemse/bjj052. [DOI] [PubMed] [Google Scholar]
- Naim M, Ronen T, Striem BJ, Levinson M, Zehavi U. Adenylate cyclase responses to sucrose stimulation in membranes of pig circumvallate taste papillae. Comp Biochem Physiol B. 1991;100:455–458. doi: 10.1016/0305-0491(91)90203-p. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Kawai T, Kamakura T, Ookura T. TGF-beta3 is expressed in taste buds and inhibits proliferation of primary cultured taste epithelial cells. In Vitro Cell Dev Biol Anim. 2010;46:36–44. doi: 10.1007/s11626-009-9239-9. [DOI] [PubMed] [Google Scholar]
- Nakayama A, Miura H, Shindo Y, Kusakabe Y, Tomonari H, Harada S. Expression of the basal cell markers of taste buds in the anterior tongue and soft palate of the mouse embryo. J Comp Neurol. 2008;509:211–224. doi: 10.1002/cne.21738. [DOI] [PubMed] [Google Scholar]
- Ozdener H, Yee KK, Cao J, Brand JG, Teeter JH, Rawson NE. Characterization and long-term maintenance of rat taste cells in culture. Chem Senses. 2006;31:279–290. doi: 10.1093/chemse/bjj030. [DOI] [PubMed] [Google Scholar]
- Paran N, Mattern CF, Henkin RI. Ultrastructure of the taste bud of the human fungiform papilla. Cell Tissue Res. 1975;161:1–10. doi: 10.1007/BF00222109. [DOI] [PubMed] [Google Scholar]
- Perez CA, Huang L, Rong M, Kozak JA, Preuss AK, Zhang H, Max M, Margolskee RF. A transient receptor potential channel expressed in taste receptor cells. Nat Neurosci. 2002;5:1169–1176. doi: 10.1038/nn952. [DOI] [PubMed] [Google Scholar]
- Qin YM, Shi JQ, Zhang GH, Deng SP, Wang TH. A reliable method to obtain cells of taste buds from fungiform papillae of mice. Acta Histochem. 2010;112:107–112. doi: 10.1016/j.acthis.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303:390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- Rieske P, Azizi SA, Augelli B, Gaughan J, Krynska B. A population of human brain parenchymal cells express markers of glial, neuronal and early neural cells and differentiate into cells of neuronal and glial lineages. Eur J Neurosci. 2007;25:31–37. doi: 10.1111/j.1460-9568.2006.05254.x. [DOI] [PubMed] [Google Scholar]
- Rieske P, Golanska E, Zakrzewska M, Piaskowski S, Hulas-Bigoszewska K, Wolanczyk M, Szybka M, Witusik-Perkowska M, Jaskolski DJ, Zakrzewski K, et al. Arrested neural and advanced mesenchymal differentiation of glioblastoma cells-comparative study with neural progenitors. BMC Cancer. 2009;9:54. doi: 10.1186/1471-2407-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossler P, Kroner C, Freitag J, Noe J, Breer H. Identification of a phospholipase C beta subtype in rat taste cells. Eur J Cell Biol. 1998;77:253–261. doi: 10.1016/s0171-9335(98)80114-3. [DOI] [PubMed] [Google Scholar]
- Ruiz CJ, Stone LM, McPheeters M, Ogura T, Bottger B, Lasher RS, Finger TE, Kinnamon SC. Maintenance of rat taste buds in primary culture. Chem Senses. 2001;26:861–873. doi: 10.1093/chemse/26.7.861. [DOI] [PubMed] [Google Scholar]
- San Gabriel A, Uneyama H, Maekawa T, Torii K. The calcium-sensing receptor in taste tissue. Biochem Biophys Res Commun. 2009;378:414–418. doi: 10.1016/j.bbrc.2008.11.060. [DOI] [PubMed] [Google Scholar]
- Spielman AI, Mody I, Brand JG, Whitney G, MacDonald JF, Salter MW. A method for isolating and patch-clamping single mammalian taste receptor cells. Brain Res. 1989;503:326–329. doi: 10.1016/0006-8993(89)91684-3. [DOI] [PubMed] [Google Scholar]
- Stone LM, Tan SS, Tam PP, Finger TE. Analysis of cell lineage relationships in taste buds. J Neurosci. 2002;22:4522–4529. doi: 10.1523/JNEUROSCI.22-11-04522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone LM, Wilcox CL, Kinnamon SC. Virus-mediated transfer of foreign DNA into taste receptor cells. Chem Senses. 2002;27:779–787. doi: 10.1093/chemse/27.9.779. [DOI] [PubMed] [Google Scholar]
- Sullivan JM, Borecki AA, Oleskevich S. Stem and progenitor cell compartments within adult mouse taste buds. Eur J Neurosci. 2010;31:1549–1560. doi: 10.1111/j.1460-9568.2010.07184.x. [DOI] [PubMed] [Google Scholar]
- Takami S, Getchell TV, McLaughlin SK, Margolskee RF, Getchell ML. Human taste cells express the G protein alpha-gustducin and neuron-specific enolase. Brain Res Mol Brain Res. 1994;22:193–203. doi: 10.1016/0169-328x(94)90047-7. [DOI] [PubMed] [Google Scholar]
- Tang DG, Tokumoto YM, Raff MC. Long-term culture of purified postnatal oligodendrocyte precursor cells. Evidence for an intrinsic maturation program that plays out over months. J Cell Biol. 2000;148:971–984. doi: 10.1083/jcb.148.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumangalathu S, Harlow DE, Driskell AL, Krimm RF, Barlow LA. Fate mapping of mammalian embryonic taste bud progenitors. Development. 2009;136:1519–1528. doi: 10.1242/dev.029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers SP, Nicklas K. Taste bud distribution in the rat pharynx and larynx. Anat Rec. 1990;227:373–379. doi: 10.1002/ar.1092270313. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Sunavala G, Rosenzweig S, Dasso M, Brand JG, Spielman AI. Bitter taste transduced by PLC-beta(2)-dependent rise in IP(3) and alpha-gustducin-dependent fall in cyclic nucleotides. Am J Physiol Cell Physiol. 2001;280:C742–C751. doi: 10.1152/ajpcell.2001.280.4.C742. [DOI] [PubMed] [Google Scholar]
- Yee CL, Jones KR, Finger TE. Brain-derived neurotrophic factor is present in adult mouse taste cells with synapses. J Comp Neurol. 2003;459:15–24. doi: 10.1002/cne.10589. [DOI] [PubMed] [Google Scholar]
- Zhao FL, Lu SG, Herness S. Dual actions of caffeine on voltage-dependent currents and intracellular calcium in taste receptor cells. Am J Physiol Regul Integr Comp Physiol. 2002;283:R115–R129. doi: 10.1152/ajpregu.00410.2001. [DOI] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]