Abstract
Yeast prions are atypical genetic elements that are transmitted as heritable protein conformations. [PSI+], [URE3], and [PIN+] are three well-studied prions in the budding yeast, Saccharomyces cerevisiae. In the last three years, several additional prions have been reported in yeast, including [SWI+], [OCT+], [MCA], [GAR+], [MOT3+], [ISP+], and [NSI+]. The growing number of yeast prions suggests that protein-based inheritance might be a widespread biological phenomenon. In this review, we summarize the characteristics of each prion element, and discuss their potential functional roles in yeast biology.
Keywords: prion, yeast, protein aggregation, epigenetics, amyloid
1. Introduction
Yeast prions are self-perpetuating protein conformations which manifest as dominant, cytoplasmically transmitted phenotypes. A prion is formed when a native protein adopts an alternative conformation (known as the prion fold), which is able to recruit soluble protein isomers and induce them to form the same prion fold. Almost all prion proteins form insoluble amyloid aggregates, apparently sequestering the protein from its native function. The prion phenotype is generally metastable, meaning that [PRION+] cells can revert to [prion−] cells at a low frequency.
[PSI+] and [URE3] were the first two yeast prions to be identified. The [PSI+] and [URE3] phenotypes were described in 1965 [1] and 1971 [2], respectively, but it was not until 1994 that Wickner proposed that they represented a prion-like phenomenon [3]. [PIN+], the third identified yeast prion, was also described as a phenotype first [4], before its protein determinant was identified [5–7]. Since that time, multiple new prions have been identified, with widely varying phenotypes, and arising from proteins across the functional spectrum (Table 1) (for a recent review, also see [8]). In this article, we will briefly describe the three well-characterized yeast prions ([PSI+], [URE3], and [PIN+]), provide a more detailed review of the newly identified yeast prions, and speculate on possible functions for yeast prions in nature.
Table 1.
Summary of the characteristics of known yeast prions.
| Prion | Protein Determinant |
Native function | Amyloid structure |
Curability | Ref | ||
|---|---|---|---|---|---|---|---|
| hsp104Δ | HSP104↑ | GdnHCl | |||||
| [PSI+] | Sup35 | translational termination | yes | yes | yes | yes | [9,16,73,74] |
| [URE3] | Ure2 | Gln3 repressor | yes | yes | no | yes | [2,20,75] |
| [PIN+]/[RNQ+] | Rnq1 | Unknown | yes | yes | no | yes | [7,5,76] |
| [SWI+] | Swi1 | transcriptional regulation | yes | yes | no | yes | [39,42,45] |
| [OCT+] | Cyc8 | transcriptional regulation | N/D | N/D | no | yes | [43] |
| [MCA] | Mca1 | metacaspase homologue | N/D | N/D | yes | yes | [47] |
| [MOT3] | Mot3 | transcriptional regulation | yes | yes | N/D | yes | [45] |
| [GAR+] | Pma1/Std1* | proton pump, glucose signaling | no | no | no | no | [49] |
| [ISP+] | Sfp1 | transcription factor | N/D | no | no | yes | [56] |
| [NSI+] | unknown | unknown | N/D | yes | no | yes | [57] |
[GAR+] appears to be propagated by the interaction between two proteins
N/D: not determined
Δ: deletion; ↑: overexpression
2. Known yeast prions
2.1. The classic yeast prions
2.1.1. [PSI+]
[PSI+] is the prion form of Sup35, a component of the translational termination complex in yeast [9]. [PSI+] is the most extensively studied yeast prion, and has been reviewed in multiple publications (for example: [10–13]). [PSI+] aggregates sequester soluble Sup35, resulting in defects in faithful translation termination. The prion therefore acts as a nonsense suppressor, allowing read-through of premature stop codons [14]. The [PSI+] prion can be cured by treatment with millimolar concentrations of guanidine hydrochloride (GdnHCl) [15], and can re-occur in cured cells, reflecting the fact that prions are a protein-only phenomenon [3]. Overexpression or deletion of HSP104 also cures [PSI+][16], suggesting that an intermediate level of Hsp104 expression is required for [PSI+] propagation.
2.1.2. [URE3]
[URE3] is the prion form of Ure2, a protein involved in the regulation of nitrogen metabolism [3]. Ure2 represses the activity of Gln3, a transcriptional activator which upregulates genes controlled by nitrogen catabolite repression [17, 18]. Sequestration of Ure2 in [URE3] aggregates results in Gln3 activation to allow cells to assimilate poor nitrogen sources, such as ureidosuccinate (USA), in the presence of good nitrogen sources, such as ammonia [19]. Like [PSI+], [URE3] exhibits dominant transmission, and is reversibly curable by GdnHCl [3]. [URE3] can be cured by HSP104 deletion, but not HSP104 overexpression [20].
2.1.3. [PIN+]
[PIN+], also known as [RNQ+], is the prion form of Rnq1 [5–7], a protein of unknown function. [PIN+] was first described as a non-Mendelian [PSI+]-inducing factor, which promotes [PSI+] de novo formation upon Sup35 overproduction [4]. Subsequent research has shown that [PIN+] also enhances the aggregation of polyglutamine (polyQ) [6] and polyasparagine (polyN) [21], and that overproduction of several Q/N-rich polypeptides can substitute for [PIN+] to induce [PSI+] de novo formation [5, 22]. Like [PSI+] and [URE3], [PIN+] is a dominant and reversibly curable epigenetic trait [4]. Aggregation and promoting [PSI+] formation are the only assayable [PIN+] functions, since the native function of Rnq1 is unknown.
2.1.4 Common features of the classic yeast prions
[PSI+], [URE3], and [PIN+] share several common characteristics that have come to be thought of as identifying features of yeast prions. For example, all three of these prion proteins contain Q/N-rich regions (known as prion domains, or PrDs), which are largely dispensable for normal function of the protein, but are necessary for maintenance and propagation of the prion [7, 23, 24]. These prions also exhibit reversible curability, meaning that after a cell is cured of a prion (for instance, by treatment with GdnHCl), the prion can arise again in the same cell [25]. Deletion of the gene encoding the prion protein abolishes the prion completely, since maintenance of a prion requires a constant supply of the prion protein [26]. Overproduction of prion proteins increases the rate of de novo appearance of the associated prion [3, 7, 23, 24, 27], presumably due to increased opportunities for the soluble protein to misfold. Finally, the prion phenotype mimics a partial knockout of the associated protein, due to the sequestration of prion protein isomers in insoluble prion aggregates [28]. Due to the aggregation of prion conformers, formation of the prion can often be detected by a shift from the soluble to the insoluble fraction of the cell [7, 29, 30].
Another common feature of [PSI+], [URE3], and [PIN+] is their dependence on the molecular chaperone network. Most notable is their strict requirement for Hsp104. Inhibition of Hsp104 function by HSP104 deletion, treatment with GdnHCl, or expression of a dominant negative allele of HSP104, leads to curing of [PSI+], [PIN+], and [URE3] [3, 7, 15, 16, 20, 29], because of the central role it plays in creating prion “seeds”. Hsp104 is responsible for breaking prion aggregates into smaller fragments, which then seed the formation of new aggregates [31]. In the absence of Hsp104, individual prion aggregates grow so large that they are unable to be transmitted to daughter cells during cell division; thus, the mother cell retains all of the prion aggregates, and the daughter cell is prion-free [32, 33]. Introduction of a dominant negative allele of HSP104 exerts a similar effect, since Hsp104 functions as a hexamer [34, 35]; thus, a dominant negative allele prevents the formation of a functional Hsp104 complex. GdnHCl treatment inhibits the activity of Hsp104, thus leading to prion curing [36, 37]. Hsp104 overproduction can also cure [PSI+] [16], but not [PIN+] or [URE3] [4, 7, 20], although the mechanism by which this occurs is unknown. In addition to Hsp104, the Hsp70/Ssa1 and Hsp40/Sis1 system is central to the propagation of most yeast prions [38–43]. While the exact mechanism of prion curing induced by modulating chaperone activity has been difficult to elucidate, it is clear that molecular chaperones play a crucial role in the maintenance and propagation of yeast prions.
3.1. Newly identified yeast prions
Since the discovery and characterization of [PSI+], [URE3], and [PIN+], multiple new yeast prion proteins have been identified. While many of these novel prions exhibit traits that are characteristic of the three classic, or well-characterized, prions, others behave in new and unexpected ways. In the following section we will discuss these new prions, their similarities to [PSI+], [URE3], and [PIN+], and unique features that distinguish them from other prions.
3.1.1. [SWI+]
[SWI+], which was initially identified by our laboratory and later confirmed by Alberti and colleagues, is the prion form of Swi1, a component of the SWI/SNF chromatin remodeling complex [44, 45]. Swi1 was identified as a prion candidate due to its high Q/N content [46], and its ability to promote [PSI+] formation [5]. The [SWI+] phenotype manifests as reduced growth on carbon sources other than glucose, reminiscent of Swi1 knockdown [47]. [SWI+] can be cured by HSP104 deletion but not HSP104 overexpression [44]. [SWI+] is a dominant phenotype, which can be transmitted by mating or by cytoduction [44]. Swi1 forms aggregates in [SWI+] cells, and deletion of SWI1 cures the prion phenotypes [44]. One unique feature of [SWI+] that sets it apart from the three previously identified yeast prions, is that the soluble form of Swi1 is a nuclear protein. Interestingly, [SWI+] aggregates appear to be cytoplasmic [44, 48], although further work is required to determine whether [SWI+] aggregates also appear in the nucleus.
3.1.2. [OCT+]
[OCT+] is the prion form of Cyc8, a component of the Cyc8/Tup1 corepressor complex [49]. Cyc8 was identified as a prion candidate based on its ability to promote [PSI+] formation when overproduced [5]. [OCT+] exhibits many standard features of yeast prions: its ability to use lactate as the sole carbon source in cyc1 mutant cells (Lac+) with a high, constitutive invertase activity is a milder version of CYC8 deletion [50]; overproduction of Cyc8 increases its appearance; and the prion phenotype can be masked by expression of the functional domain alone of Cyc8 [49]. Moreover, the [OCT+] phenotype is dominant, and can be transmitted to wild-type cells by cytoduction or extract transformation. Cyc8 forms aggregates in the cytoplasm of [OCT+] cells, and deletion of CYC8 abolishes the prion. [OCT+] is cured when Hsp104 is inhibited by treatment with GdnHCl, or by expression of a dominant negative HSP104 allele [49]; however, overexpression of HSP104 did not cure the [OCT+] phenotype. Since the non-prion form of Cyc8 is associated with chromatin, it precipitates in the insoluble fraction of wild-type cells; thus it was difficult to establish a change in solubility in [OCT+] cells. The fact that both Swi1 and Cyc8 are normally involved in transcriptional regulation is an unexpected coincidence, and suggests a possible link between yeast prions and transcriptional regulation.
3.1.3. [MOT3+]
[MOT3+], the prion form of Mot3, was identified by screening for PrD-like regions in the yeast proteome using a computational algorithm based on the PrDs of [PSI+], [URE3], and [PIN+] [45]. Based on this algorithm and assays including fluorescence aggregation, nonsense suppression of Sup35C fusion, and thioflavin-T binding, the authors identified multiple candidate PrDs with a high probability of prionogenicity. To validate their algorithm and experimental methodology, they chose one of their candidates, Mot3, to analyze in depth and demonstrated that it is in fact a prion. Mot3 is a transcriptional regulator that affects many cellular processes, including the repression of anaerobic genes during aerobic growth [51]. Taking advantage of the fact that Mot3 inhibits the expression of DAN1, the authors established a dan1::URA3 based reporter system to positively select for [MOT3+] [45]. Using this system, the authors showed that [MOT3+] cells exhibit a loss-of-function phenotype for Mot3. Additionally, transient overproduction of Mot3 enhances the spontaneous rate of [MOT3+] prion formation, and purified Mot3 fibers can transmit the [MOT3+] phenotype. Finally, [MOT3+] is curable by GdnHCl-mediated Hsp104 inhibition, and this curability is reversible. Mot3 is thus the fourth known yeast prion protein that is also a transcriptional regulator.
3.1.4. [MCA]
[MCA], the prion form of Mca1, was discovered by fusing random segments of yeast chromosomal DNA to SUP35MC, and assaying for nonsense suppression [52]. Sequestration of Sup35MC resulting from aggregation of the N-terminal fusion peptide resulted in read-through of a reporter nonsense codon, and the ability of this strain to grow on adenine deficient media. Three independent clones containing fragments of MCA1 induced aggregation of Sup35MC, suggesting that Mca1 may be a prion protein. The Ade+ phenotype conferred by the Mca1-Sup35MC fusion protein is a dominant and cytoplasmic trait, and Mca11–117-GFP (representing the putative Q/N-rich prion domain) formed fluorescent foci in [MCA] cells. Furthermore, full-length Mca1 protein was insoluble in [MCA] cells, and deletion of MCA1 resulted in the loss of [MCA]. Finally, cytoduction of Mca1 or Mca1N aggregates into [mca-o] (non-prion) cells induced [MCA] phenotypes in the recipient cells. Importantly, the authors showed that [MCA] can re-occur in the cured cells, demonstrating that the phenotype is reversibly curable. Unfortunately, Mca1 has few readily assayable phenotypes, so a knockdown of Mca1 function was not shown [52]. Interestingly, GdnHCl treatment appeared to have no effect on [MCA], but overexpression of HSP104 did cure the prion. [MCA] and [PSI+] are the only known yeast prions that can be cured by HSP104 overexpression.
Shortly after the identification of [MCA], another group showed that Mca1 has two alternative translation start sites, and can possibly give rise to three different isoforms. The two longer isoforms contain an N-terminal extension upstream of the Q/-N-rich putative PrD. The authors explored differences in the aggregate-forming ability of the different Mca1 isoforms expressed from these alternative start sites, and found that a longer Mca1 isoform (Mca1454) forms insoluble aggregates, whereas the shorter isoform (Mca1432) does not [53]. However, since these experiments were conducted in cells that were presumably [mca-o] (prion status was not assessed), it is unclear how relevant their findings are to the behavior of the different Mca1 isoforms in [MCA] cells. While the potential impact of alternative translational start sites on the strength or appearance of a prion phenotype is intriguing, its significance remains to be fully explored.
A more recent study by Lee, et al. implicates Mca1 in clearing insoluble protein aggregates from the cell [54]. The authors propose that the Q/N-rich region of Mca1 may mediate interactions with aggregated proteins. If this is the case, it might be expected that prion formation would affect the association of Mca1 with insoluble protein aggregates, and decrease clearance of these aggregates. Future studies of the impact of [MCA] formation on this newly identified Mca1 function should yield interesting insights into the regulation of protein solubility in yeast.
3.1.5. [GAR+]
[GAR+], an atypical yeast prion, was identified by screening the literature for dominant phenotypes that could not be explained by DNA-based genetic inheritance [55]. The [GAR+] phenotype consists of growth in glycerol in the presence of glucosamine, a nonmetabolizable glucose analog, and is a dominant cytoplasmic trait [56]. Interestingly, [GAR+] is not curable by GdnHCl treatment, Hsp104 deletion, or Hsp104 overexpression. This is strikingly different from all previously described yeast prions, which are all cured by either Hsp104 inhibition or overexpression (or both, in the case of [PSI+]) [7, 16, 20, 44, 45, 49, 52]. Moreover, [GAR+] does not appear to be affected by the presence of other prions, which is unusual, since yeast prions often have a profound effect on the formation and/or propagation of other yeast prions [5, 43, 44]. Perhaps the most unique feature of this prion is that it appears to be propagated, not by a misfolded form of a single protein, but by the interaction between two proteins: Std1 and Pma1. In [GAR+] cells, Std1 co-immunoprecipitates with Pma1, a transmembrane protein which normally associates with Mth1 (an Std1 paralog) when the prion is not present. Overexpression of STD1, a component of the Snf3/Rgt2 regulatory pathway, increases the rate of [GAR+] appearance, which is a characteristic of prion protein determinants. However, deletion of STD1 does not cure the prion, nor do stdΔ cells exhibit resistance to glucose-associated repression (the prion phenotype). Deleting PMA1 does not cure [GAR+] either; but deleting PMA1 and the coding sequence for the putative Std1 prion domain does. Finally, [GAR+] does not appear to be amyloid in nature, as there is no change in localization or SDS resistance of Std1 and Pma1 in [GAR+] cells as compared to [gar−] cells. This fact, and the fact that [GAR+] appears to be propagated by a complex of at least two proteins, make this prion strikingly different from all other known yeast prion proteins.
3.1.6. [ISP+]
[ISP+] is the prion form of Sfp1, a protein was first identified as a split finger protein that binds to DNA and regulates cell growth and budding [57]. Later, Sfp1 was shown to be a global transcriptional regulator that influences the expression of ~10% of total yeast genes, including ribosomal protein genes [58]and genes that regulate G2/M transitions during mitotic cell cycle and DNA-damage response [59], and modulate cell size [60]. The [ISP+] phenotype manifests as a suppressor of nonsense codon read-through (or “anti-suppression”) in strains with mutations in SUP35 and SUP45. Intriguingly, [ISP+] can be cured by treatment with GdnHCl, but not by deletion or overexpression of HSP104 [61]. This raises the interesting possibility that GdnHCl treatment may have other effects in addition to Hsp104 inactivation. [ISP+] forms nuclear aggregates, which has not been previously reported for a yeast prion. This nuclear localization may explain the low efficiency of [ISP+] transfer by cytoduction. Another unique feature of [ISP+] is that its phenotype differs markedly from the sfp1Δ phenotype. Deletion of SFP1 leads to a decrease in cell size and increased sensitivity to drugs targeting translation. Based on the behavior of other prions, it would be expected that the presence of the prion would lead to a partial loss-of-function of SFP1, i.e. slightly reduced cell size and enhanced drug sensitivity. However, the opposite appears to be true: in strains containing the prion, cell size is significantly larger than wild-type cells, and drug resistance is increased [62]. These atypical features of [ISP+] could lead to a better understanding of how prions behave within the cellular environment.
3.1.7. [NSI+]
The most recently discovered yeast prion is [NSI+] [63]. The [NSI+] phenotype manifests itself as nonsense suppression in strains expressing Sup35 with a mutated or deleted N-terminus. Any contribution of Sup35 to these phenotypes was ruled out by the demonstration that maintenance of [NSI+] does not require Sup35N or Sup35NM, and the fact that Sup35 is soluble in cells containing the [NSI+] prion. [NSI+] can be cured by GdnHCl treatment and by HSP104 deletion, but not by HSP104 overexpression [63]. The protein that forms the [NSI+] prion has not yet been identified. Deleting any one of several known prion-encoding genes had no effect on the propagation of [NSI+] [63], which strongly suggests that the protein determinant of [NSI+] is a novel prion protein.
4. Hypotheses on the function of prions
The growing number and variety of yeast prions that have been reported are beginning to yield clues to potential roles that prions may play in yeast biology. Whether prions represent a disease in yeast [64, 65], or are actually advantageous to cells [66–68], is a topic of considerable debate. On the one hand, prions can confer adaptability in highly specialized or stressed conditions [68, 69] and increase evolvability [67, 70]. Furthermore, many Q/N-rich regions have resisted evolutionary pressure, suggesting that their function has been conserved for some reason [71]. One the other hand, prion phenotypes tend to be detrimental to the cell, and [PIN+] is the only prion that has been found even in a very small percentage of yeast strains in nature [64]. The [Het-s] prion of Podospora anserina shows a clear example that functional prions can exist in nature. This prion regulates heterokaryon incompatibility, and is transmitted in a dominant, protein-only manner [72, 73]. [Het-s] can be propagated in yeast [74], implying that similar chaperone systems, and possibly other cellular factors, may contribute to the propagation of yeast and non-yeast prions. If prions do represent a disease of yeast, then it is to be expected that further investigation will yield only more negative and random phenotypes. If, however, prions are a functional part of yeast biology, then the behavior and characteristics of the known yeast prions can offer clues as to what this role may be, and how it has been conserved over evolution. In the following section we will explore possible functional roles for prions in yeast.
4.1. Prions as transcriptional regulators
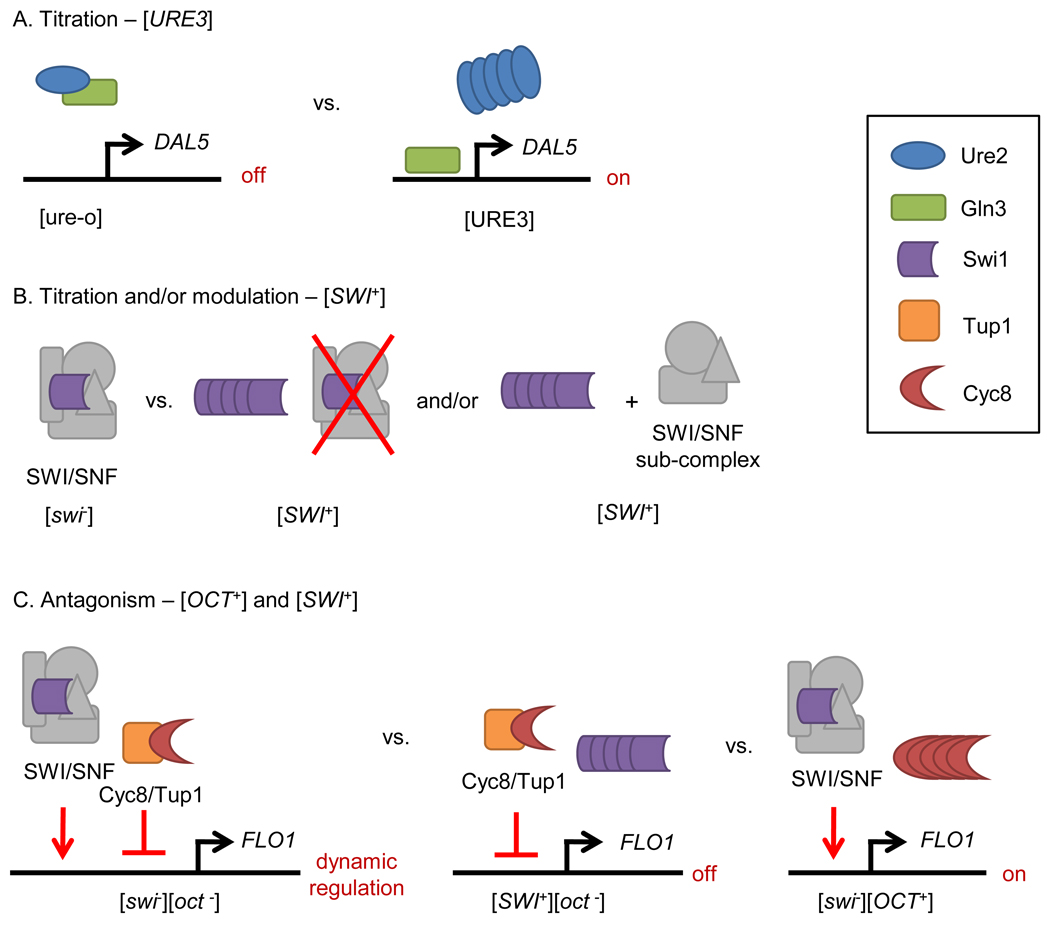
The relative prevalence of transcriptional regulators among yeast prion proteins is striking, and has provocative implications for the function of prions. In addition to Swi1 and Cyc8, Sfp1, Mot3 and Ure2 are also transcriptional regulators. Furthermore, the list of strong prion candidates identified by Alberti, et al. also showed a marked enrichment for transcriptional regulators [45]. There are several potential models for how prionization of a transcription regulator may affect the expression of its target genes (Figure 1). The simplest titration model predicts that prion aggregates sequester functional transcription regulators so that [PRION+] cells exhibit complete or partial loss-of-function phenotypes. Amyloid prions such as [URE3] and [SWI+] likely fall into this category. In the case of Swi1 a titration/modulation model can be postulated, in which [SWI+] formation leads to a titration of the functional SWI/SNF complex, or assembly of Swi1-free sub-complexes that might modulate different target genes [75]. It is noteworthy that the SWI/SNF and Cyc8/Tup1 complexes work together to regulate the transcription of a set of yeast genes [76]; for instance, mutations in SWI1 and CYC8 have profound (though opposite) effects on the utilization of alternative carbon sources and invasive growth [50, 77, 78]. Thus, an antagonism model for [SWI+] and [OCT+] is proposed, using FLO1 locus regulation as an example (Figure 1). The fact that an overlapping set of genes is prone to “regulation” by two separate prions suggests that there may be an adaptive advantage to this sort of global regulation, possibly enabling yeast cells to survive extremely stressful environmental conditions. Given the large number of known and potential yeast prion proteins that are involved in global or gene-specific transcriptional regulation, it is possible that the prion phenomenon, if functional, may be closely tied to transcription.
Figure 1.
Postulated mechanisms for prion-mediated transcriptional regulation. A, A titration model shows that [URE3] formation leads to DAL5 activation due to Ure2 insufficiency. In [ureo] cells, Ure2 binds to the DAL5 activator Gln3 and as a consequence, DAL5 promoter is off. B, [SWI+] formation can lead to the destruction of SWI/SNF complex assembly and/or formation of Swi1-free SWI/SNF sub-complexes to further modulate gene expression. C, An antagonism model shows that formation of [SWI+] or [OCT+] can have opposite effects on FLO1 expression.
4.2. Prion proteins as regulators of translational fidelity
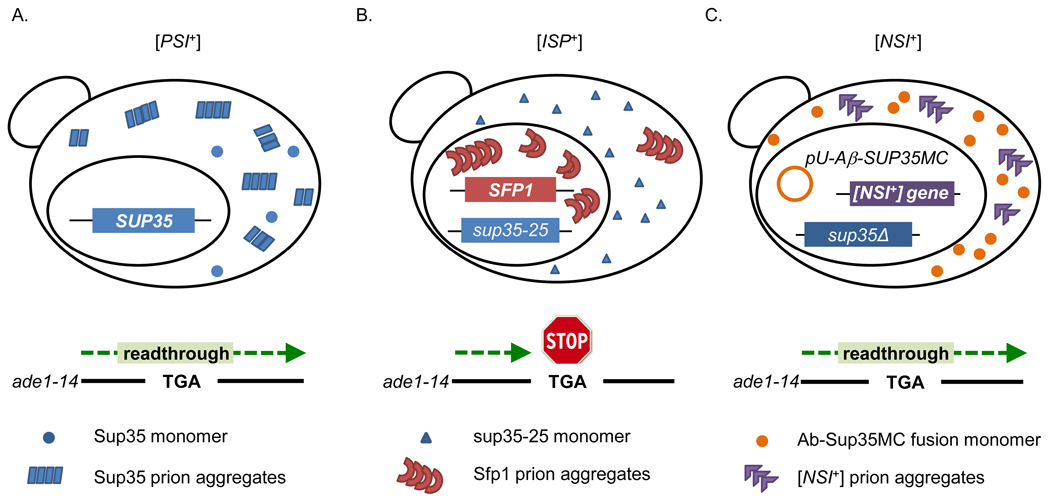
Another intriguing example of multiple prion phenotypes affecting a single cellular function, is the case of [PSI+], [ISP+], and [NSI+]. All three of these prions are global regulators of translation, meaning that they affect translational fidelity in a non-specific manner (Figure 2). [PSI+] acts as a suppressor of nonsense codons, allowing read-through of stop codons. [ISP+] is an anti-suppressor, antagonizing read-through of stop codons, which essentially results in faithful translational termination. [NSI+] exhibits a similar phenotype to [PSI+], suppressing and reading through nonsense codons. The fact that at least three prions regulate the same process in yeast strongly suggests that there is a functional role for prions in translational regulation. [NSI+] was detected in strains containing a deleted or modified Sup35 N-terminus [63]. Since deletion or modification of Sup35N prohibits the formation of [PSI+], it is intriguing to find that another prion which exerts a similar effect can appear when [PSI+] is unable to form. The [ISP+] antisuppressor phenotype is detectable when certain nonsense mutations of SUP35 and cryptic SUP45 mutations are present [61, 62]. Since [PSI+] also confers nonsense suppression, like these mutations, it is possible that [ISP+] could antagonize the effects of [PSI+] within the same cell. Alternatively, de novo formation of [ISP+] might be a mechanism for yeast to cope with the harmful mutations. Future investigation of their mutual interactions will likely yield interesting insights into their regulatory role in translation fidelity.
Figure 2.
Three prions, [PSI+], [ISP+], and [NSI+], affect translation fidelity. A, In [PSI+] cells, the majority of Sup35 is sequestered in prion aggregates, resulting in partial read-through of nonsense translation codons, such as TGA in ade1-14. B, In [ISP+] cells, Sfp1 forms nuclear prion aggregates that result in suppression of nonsense read-through in some sup35 and sup45 mutant strains. C, In [NSI+] cells, an unidentified prion protein forms prion aggregates, resulting in nonsense codon read-through, a phenotype similar to that of [PSI+]. Aggregated Sup35 is not implicated in this phenotype.
4.3. Atypcial yeast prions
[GAR+] is an extremely unusual prion that is difficult to characterize. It seems to share some important characteristics with typical prion proteins: namely, that it is a protein-based phenomenon that is transmitted in a dominant and self-propagating manner. However, it is not affected by Hsp104 levels, does not form amyloid aggregates, and its infectious or heritable form appears to be an interaction between two proteins. The yeast prion [β] is also very different from the classic yeast prions. It differs from typical yeast prions in that it does not represent an aggregated, inactive form of a normally functional protein; instead, [β] is “infectious” because it activates its own enzyme activity [79]. The mature yeast protease B (PrB) enzyme is derived from pro-PrB, which must be cleaved by a series of enzymes including PrB itself in order to become catalytically active [80, 81]. As such, it can be technically classified as a prion, due to its self-propagating nature, but does not belong in the same category of the yeast prions described here. While many outstanding questions remain as to the nature of [GAR+], it is safe to say that it is a highly atypical prion, and perhaps exists in a sub-class by itself, like [β].
4.4. Prions in the nucleus
The unique localization of [ISP+] aggregates in the nucleus opens up new avenues of understanding prion behavior and function within the cell. There are several interesting questions raised by the presence of prion aggregates in the nucleus: first, do these aggregates rely on the same chaperone network as cytoplasmic aggregates? Cytoplasmic chaperones play an important role in the formation, propagation, and transmission of most yeast prions. The presence of prion aggregates in the nucleus suggests the involvement of nuclear chaperones, whether alone or in combination with cytoplasmic chaperones. Second, what is the effect of nuclear localization on propagation of the prion? [ISP+] can be transmitted by cytoplasmic mixing, but only at a very low rate [62], most likely due to the fact that the majority of the prion aggregates are not exposed to the cytoplasm. Perhaps a mechanism exists to shuttle prion conformers in and out of the nucleus, exposing the prion protein to an environment where it can encounter cytoplasmic chaperones and be passed on to other cells by cytoplasmic transfer. Third, does nuclear aggregation affect the phenotype of the prion? The presence of the [SWI+] or [OCT+] prion noticeably affects the transcriptional regulation of SWI/SNF and Cyc8/Tup1 targets, respectively, despite the fact that aggregates of these two prions are located primarily in the cytoplasm. It seems likely that aggregates of a transcription factor within the nucleus itself have the potential to affect transcription in different, possibly more pronounced, ways. For instance, aggregated Sfp1 may be able to bind to DNA and block binding sites for monomeric protein. Alternatively, the prion form of Sfp1 may be able to more effectively sequester binding partners in the nucleus than in the cytoplasm. The presence of [ISP+] aggregates in the nucleus and its unknown effects on nuclear processes may explain why the prion phenotype differs so greatly from the sfp1Δ phenotype. This first demonstration of nuclear localization of a yeast prion provides a unique opportunity to explore diverse features of yeast prion proteins and their behavior within the cell.
5. Conclusions
The yeast prion field has come a long way since the [PSI+] phenotype was first described in 1965. It was almost thirty years until this strange dominant phenotype was ascribed to a prion-like phenomenon, and since then more and more proteins have been identified that behave in this unusual manner. We anticipate that the continuing characterization of known prions and the identification of novel prions will bring to light the role of these entities in nature. Do they play a part in the regulation and adaptation of the yeast genome, or are they a deleterious by-product of evolution? The answer to this question and others are part of the exciting future of prion research.
Acknowledgements
This work was supported by a grant from the U.S. National Institutes of Health (R01NS056086) to L.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Emily T. Crow, Email: e-crow@northwestern.edu.
Liming Li, Email: limingli@northwestern.edu.
References
- 1.Cox B. [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity. 1965;20:505–521. [Google Scholar]
- 2.Lacroute F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J Bacteriol. 1971;206:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wickner RB. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 4.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 6.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 7.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 8.Tuite MF. Cell biology: the strain of being a prion. 2010 doi: 10.1038/428265a. [comment] [DOI] [PubMed] [Google Scholar]
- 9.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, et al. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindquist S. Mad cows meet psi-chotic yeast: the expansion of the prion hypothesis. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 11.Liebman SW, Derkatch IL. The yeast [PSI+] prion: making sense of nonsense. J Biol Chem. 1999;274:1181–1184. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- 12.Serio TR, Lindquist SL. [PSI+]: an epigenetic modulator of translation termination efficiency. Annu Rev Cell Dev Biol. 1999;15:661–703. doi: 10.1146/annurev.cellbio.15.1.661. [DOI] [PubMed] [Google Scholar]
- 13.Tuite MF, Cox BS. The [PSI(+)] prion of yeast: A problem of inheritance. Methods. 2006 doi: 10.1016/j.ymeth.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Wickner RB, Masison DC, Edskes HK. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 15.Tuite MF, Mundy CR, Cox BS. Agents that cause a high frequency of genetic change from [PSI+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 17.Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coschigano PW, Magasanik B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol Cell Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edskes HK. Protein-based inheritance in Saccharomyces cerevisiae: [URE3] as a prion form of the nitrogen regulatory protein Ure2. Res Microbiol. 2001;152:605–612. doi: 10.1016/s0923-2508(01)01239-6. [DOI] [PubMed] [Google Scholar]
- 20.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters TW, Huang M. Protein aggregation and polyasparagine-mediated cellular toxicity in Saccharomyces cerevisiae. Prion. 2007;1:144–153. doi: 10.4161/pri.1.2.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 25.Wickner RB, Edskes HK, Maddelein ML, Taylor KL, Moriyama H. Prions of yeast and fungi. Proteins as genetic material. J Biol Chem. 1999;274:555–558. doi: 10.1074/jbc.274.2.555. [DOI] [PubMed] [Google Scholar]
- 26.Masison DC, Edskes HK, Maddelein ML, Taylor KL, Wickner RB. [URE3] and [PSI] are prions of yeast and evidence for new fungal prions. Curr Issues Mol Biol. 2000;2:51–59. [PubMed] [Google Scholar]
- 27.Chernoff YO, Derkach IL, Inge-Vechtomov SG. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 28.Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol. 2002;56:703–741. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- 29.Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 30.Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci U S A. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 32.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 33.Kushnirov VV, Ter-Avanesyan MD. Structure and replication of yeast prions. Cell. 1998;94:13–16. doi: 10.1016/s0092-8674(00)81216-7. [DOI] [PubMed] [Google Scholar]
- 34.Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 35.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 36.Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- 37.Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- 38.Kushnirov VV, Kochneva-Pervukhova NV, Chechenova MB, Frolova NS, Ter-Avanesyan MD. Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J. 2000;19:324–331. doi: 10.1093/emboj/19.3.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci U S A. 2008;105:16596–16601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathur V, Hong JY, Liebman SW. Ssa1 overexpression and [PIN(+)] variants cure [PSI(+)] by dilution of aggregates. J Mol Biol. 2009;390:155–167. doi: 10.1016/j.jmb.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci U S A. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 48.Du Z, Crow ET, Kang HS, Li L. Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Mol Cell Biol. 2010;30:4644–4655. doi: 10.1128/MCB.00225-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlson M, Osmond BC, Neigeborn L, Botstein D. A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics. 1984;107:19–32. doi: 10.1093/genetics/107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grishin AV, Rothenberg M, Downs MA, Blumer KJ. Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signaling in Saccharomyces cerevisiae. Genetics. 1998;149:879–892. doi: 10.1093/genetics/149.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nemecek J, Nakayashiki T, Wickner RB. A prion of yeast metacaspase homolog (Mca1p) detected by a genetic screen. Proc Natl Acad Sci U S A. 2009;106:1892–1896. doi: 10.1073/pnas.0812470106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Erhardt M, Wegrzyn RD, Deuerling E. Extra N-terminal residues have a profound effect on the aggregation properties of the potential yeast prion protein Mca1. PLoS One. 2010;5:e9929. doi: 10.1371/journal.pone.0009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee RE, Brunette S, Puente LG, Megeney LA. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc Natl Acad Sci U S A. 2010;107:13348–13353. doi: 10.1073/pnas.1006610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown JC, Lindquist S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 2009;23:2320–2332. doi: 10.1101/gad.1839109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ball AJ, Wong DK, Elliott JJ. Glucosamine resistance in yeast. I. A preliminary genetic analysis. Genetics. 1976;84:311–317. doi: 10.1093/genetics/84.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blumberg H, Silver P. A split zinc-finger protein is required for normal yeast growth. Gene. 1991;107:101–110. doi: 10.1016/0378-1119(91)90302-r. [DOI] [PubMed] [Google Scholar]
- 58.Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci U S A. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Z, Norris D. The SFP1 gene product of Saccharomyces cerevisiae regulates G2/M transitions during the mitotic cell cycle and DNA-damage response. Genetics. 1998;150:1419–1428. doi: 10.1093/genetics/150.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volkov KV, Aksenova AY, Soom MJ, Osipov KV, Svitin AV, Kurischko C, et al. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics. 2002;160:25–36. doi: 10.1093/genetics/160.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, et al. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci U S A. 2010;107:10573–10577. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saifitdinova AF, Nizhnikov AA, Lada AG, Rubel AA, Magomedova ZM, Ignatova VV, et al. [NSI (+)]: a novel non-Mendelian nonsense suppressor determinant in Saccharomyces cerevisiae. Curr Genet. 2010;56:467–478. doi: 10.1007/s00294-010-0314-2. [DOI] [PubMed] [Google Scholar]
- 64.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci U S A. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T, Engel A, McCann L, et al. Yeast prions: evolution of the prion concept. Prion. 2007;1:94–100. doi: 10.4161/pri.1.2.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 67.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 68.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 69.Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harrison LB, Yu Z, Stajich JE, Dietrich FS, Harrison PM. Evolution of budding yeast prion-determinant sequences across diverse fungi. J Mol Biol. 2007;368:273–282. doi: 10.1016/j.jmb.2007.01.070. [DOI] [PubMed] [Google Scholar]
- 72.Glass NL, Kaneko I. Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryot Cell. 2003;2:1–8. doi: 10.1128/EC.2.1.1-8.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saupe SJ. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev. 2000;64:489–502. doi: 10.1128/mmbr.64.3.489-502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taneja V, Maddelein ML, Talarek N, Saupe SJ, Liebman SW. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol Cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat Struct Mol Biol. 2007;14:540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
- 76.Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 77.Fleming AB, Pennings S. Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J. 2001;20:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peterson CL, Dingwall A, Scott MP. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci U S A. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roberts BT, Wickner RB. Heritable activity: a prion that propagates by covalent autoactivation. Genes Dev. 2003;17:2083–2087. doi: 10.1101/gad.1115803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moehle CM, Tizard R, Lemmon SK, Smart J, Jones EW. Protease B of the lysosomelike vacuole of the yeast Saccharomyces cerevisiae is homologous to the subtilisin family of serine proteases. Mol Cell Biol. 1987;7:4390–4399. doi: 10.1128/mcb.7.12.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nebes VL, Jones EW. Activation of the proteinase B precursor of the yeast Saccharomyces cerevisiae by autocatalysis and by an internal sequence. J Biol Chem. 1991;266:22851–22857. [PubMed] [Google Scholar]