Abstract
Alphavirus nonstructural protein nsP1 possesses distinct methyltransferase (MTase) and guanylyltransferase (GTase) activities involved in the capping of viral RNAs. In alphaviruses, the methylation of GTP occurs before RNA transguanylation and nsP1 forms a covalent complex with m7GMP unlike the host mRNA guanylyltransferase which forms GMP-enzyme complex. In this study, full length SINV nsP1 was expressed in a soluble form with an N-terminal histidine tag in Escherichia coli and purified to homegeneity. The purified protein is enzymatically active and contains both MTase and GTase activity indicating that SINV nsP1 does not require membrane association for its enzymatic function. Biochemical analysis shows that detergents abolish nsP1 GTase activity, whereas nonionic detergents do not affect MTase activity. Furthermore, SINV nsP1 contains the metal-ion dependent GTase whereas MTase does not require a metal ion. Circular dichroism spectroscopic analyses of purified protein indicate that nsP1 has a mixed α/β structure and is in the folded native conformation.
Keywords: Alphavirus, SINV, nsP1, methyltransferase, guanylyltransferase, purification
1. Introduction
The alphavirus genus of the Togaviridae family of plus-strand RNA viruses contains a single RNA genome of ~11.8 kb with a 5’ cap structure and a 3’ polyA tail. Sindbis virus (SINV) is the prototype virus of the genus. Members of the genus alphavirus, including Chikungunya virus (CHIKV) and Venezuelan equine encephalitis virus (VEEV) are important pathogens that can cause fatal disease in humans and animals. Recently, CHIKV and VEEV caused serious outbreaks that were reported from the Indian-sub continent, Indian Ocean islands, the south east coast of Africa and Latin America [1, 2]. However, currently there is no human vaccine or drug treatment available against alphavirus infection.
Alphaviruses replicate in the cytoplasm of infected cells. After virus entry, the viral genome is released into the cytoplasm and serves directly as mRNA for translation of the four nonstructural (ns) proteins, nsP1 to nsP4, encoded by the 5’ two-thirds of the viral genome. These nonstructural proteins along with unidentified host factors form the membrane-associated replication complex involved in replication of the viral genome. During infection, the replication complexes synthesize a 26S subgenomic RNA and the 42S viral genome [3]. Both the viral genome and the subgenomic RNA contain m7GpppA cap structure at their 5’ ends [4]. The alphavirus genome encodes a virus-specific capping enzyme required for its replication in the cytoplasm of the host cell, whereas the host capping reactions occur in the nucleus. In general, the cap structure, which is present at the 5’end of eukaryotic and most viral mRNAs, plays an important role in mRNA stability and efficient translation [5].
Nonstructural protein nsP1 catalyzes methyltransferase (MTase) and guanylyltransferase (GTase) enzymatic reactions responsible for the formation of the cap structure at the 5’ end of the viral RNAs. The mechanism of cap structure formation in alphaviruses is unique. In alphaviruses, the methylation of GTP occurs before the transfer of guanine to the 5’ end of mRNA, whereas in host cells the transguanylation reaction occurs before the guanosine moiety is methylated [6, 7]. A series of three reactions occur during cap structure formation. The first reaction in capping the alphavirus genome is the removal of the 5’ γ-phosphate of the nascent RNA molecule catalyzed by nsP2 RNA triphosphatase. The second reaction is the transfer of a methyl group from S-adenosylmethionine (AdoMet) to GTP to form m7GTP and this reaction is catalyzed by the AdoMet-dependent nsP1 guanosine-7-methyltransferase (MTase). In the third reaction, the nsP1 gunaylyltransferase (GTase) forms an intermediate m7GMP-nsP1 covalent complex that is derived by hydrolyzing m7GTP, and finally the m7GMP moiety is transferred to the 5’ end of RNA by forming a 5’-5’ triphosphate linkage. The MTase and GTase capping reactions catalyzed by nsP1 are distinct from the host capping mechanism, which is commonly found in eukaryotes and several viruses including vaccinia virus. In the host cell, the first reaction is similar to the one used by alphaviruses in which 5’ RNA triphosphatase removes the 5’ γ-phosphate of the nascent RNA molecule. Thereafter, host GTase forms a covalent intermediate with GMP and then transfers GMP to the 5’ end of RNA. In the last step, an AdoMet-dependent MTase methylates the terminal guanosine at position 7 by transferring a methyl group from AdoMet to the guanosine nucleotide [8, 9].
The alphavirus nsP1 N-terminal domain is predicted to be structurally very similar to the S-adenosylmethionine-dependent Rossmann fold methyltransferase enzymes (SAM_MTases) [6]. Despite the very low sequence identity among SAM-MTases, these enzymes share a highly conserved structural motif for binding the cofactor. The residues involved in binding AdoMet are not well conserved but are localized at the same position in the fold [10]. Some residues of the AdoMet binding motif were identified in Semliki Forest virus (SFV) nsP1 by mutational and crosslinking studies. Asp64 and Asp90, the two conserved acidic residues of SFV nsP1, were shown to be involved in AdoMet binding and are present in the cofactor-binding G-loop motif of nsP1 [6]. Based on amino acid sequence alignment, these conserved residues are present at Asp65 and Asp91 in SINV nsP1.
The nsP1 enzymatic activities are indispensable for alphavirus replication since it has been demonstrated that point mutations that abolish nsP1 enzymatic activity render the virus non-infectious [11]. Because capping of the alphavirus genome is an essential step in viral replication, nsP1 is considered to be a potential antiviral target. However, drug development has been hampered due to the lack of availability of pure nsP1 required for enzyme inhibitor screening and structure-function studies that would facilitate rational drug design studies.
In this study, we expressed SINV nsP1 with six histidine residues fused at its N-terminus in Escherichia coli in soluble form and purified the recombinant protein to homogeneity. To our knowledge this is the first report of purification of an enzymatically active alphavirus nsP1 capping enzyme. Furthermore, biochemical studies performed using purified protein indicates that SINV nsP1 MTase activity is independent of a divalent metal ion but GTase activity requires a divalent metal ion. The effects of various detergents on the MTase and GTase were also investigated and it was demonstrated that nonionic detergents have little or no effect on nsP1 MTase activity but abolished the GTase activity of SINV nsP1. However, both enzymatic activities of the purified protein were eliminated by the ionic detergents used in this study. Circular dichroism analysis and secondary structure predictions indicate SINV nsP1 to be a mixed α/β protein.
2. Materials and Methods
2.1. Strains, plasmids, biochemicals and chemicals
Oligonucleotides for PCR and site-directed mutagenesis were ordered from Integrated DNA Technologies Inc. (Coralville, IA). QIAprep spin miniprep kit for plasmid isolation, PCR purification kit and DNA Gel Extraction kit were from Qiagen Inc. Restriction enzymes NcoI, NdeI and XhoI were purchased from New England BioLabs, Inc. Escherichia coli strain XL1-Blue used for transformation and amplification of plasmids and QuikChange mutagenesis kit were purchased from Stratagene, La Jolla, CA. E. coli BL21(DE3), BL21(DE3)pLysS and Rosetta (DE3) expression strains used as a host for protein overexpression and pET-28a vector were obtained from Novagen Inc. Madison, WI, USA. HiTrap Chelating HP column was purchased from GE Healthcare. Imidazole (low absorbance at 280) was obtained from Acros. AKTA Explorer (Model 100 AIR) system from GE Healthcare was used for protein purification. Amicon Ultra protein concentrators were purchased from Millipore. Tween 20, Triton X-100, octyl beta-D-glucopyranoside and sodium deoxycholate were obtained from Sigma. Protein estimation assay kit was obtained from Bio-Rad (Hercules, CA). S-adenosyl[methyl-3H]methionine (75 Ci/mmol) and [α-32P]-GTP (400 Ci/mmol), were purchased from GE Healthcare. AdoMet and guanosine 5’-(β–γ-imido) triphosphate (guanylylimidodiphosphate or GIDP) were obtained from Sigma.
2.2. Construction of bacterial expression plasmid
The SINV genomic cDNA, pToto64 [12], was used as the template for polymerase chain reaction (PCR) amplification of a DNA fragment encoding nsP1. Two oligonucleotide primers containing NcoI and XhoI restriction endonuclease sites designed to anneal to the 5’ and 3’ ends of the gene, respectively, were synthesized for nsP1 gene amplification. The NcoI and XhoI restriction sites introduced in the S1 sense and A1 antisense primers (Table 1) allowed cloning of the nsP1 gene into the corresponding sites of the pET-28a vector. The SINV nsP1 gene was amplified by PCR using these primers, and the amplified DNA fragment was purified using a PCR purification kit according to the manufacturer's instructions to remove unincorporated nucleotides and primers. The purified PCR fragment and pET-28a vector were restriction digested with NcoI and XhoI restriction enzymes. The digested DNA fragments were separated on 1% agarose gel electrophoresis, purified using a DNA gel extraction kit and ligated with T4 DNA ligase. E. coli XL1-Blue competent cells were transformed by the heat shock method [13] using the ligation mixture. Some of the colonies obtained by plating the transformed cells on Luria-Bertani (LB) agar plates containing 50 µg of kanamycin/ml were picked and grown overnight. Plasmids were isolated using a MiniPrep plasmid isolation kit and screened for the presence of the nsP1 gene by restriction enzyme digestion. The integrity of the resulting plasmid, pETnsP1, was confirmed by sequencing in both directions using T7 promoter and T7 terminator primers. The internal NdeI site of the SINV nsP1 gene in pETnsP1 was removed by site-directed mutagenesis. The pETnsP1-NdeI plasmid with the mutated Nde I site was then used as a template for amplification of the nsP1 gene using sense primer S2 (Table 1) containing an NdeI site and antisense primer A1 containing a stop codon and XhoI site. The resultant PCR product was subcloned into the NdeI and XhoI sites of pET-28a vector to generate pETHis6-nsP1, the plasmid that allowed expression from the T7 promoter of full-length SINV nsP1 with an N-terminal 6xHis-tag. The ligation product was used to transform E. coli XL1-Blue cells. Colonies were screened by restriction analysis with plasmids isolated from overnight cultures and the construct containing the nsP1 gene insert was sequenced for verification. Further expression and solubility optimization experiments were done using the pETHis6-nsP1 plasmid.
2.3. Site-directed mutagenesis
To substitute Ala for Asp65, the putative AdoMet binding residue in the N-terminal MTase domain of SINV nsP1, the sense S4 and antisense A3 mutagenic primers (Table 1) were designed and used to construct the pETHis6-nsP1D65A mutant expression plasmid using the QuikChange mutagenesis kit. The pETHis6-nsP1 plasmid was used as a template in the primer extension reaction for constructing the mutant. PCR was performed using Pfu DNA polymerase and samples were subjected to 20 cycles of 0.5 min denaturation at 95 °C, 1 min annealing at 58 °C, and 5 min elongation at 72 °C. The parent template plasmid was digested with the DpnI restriction enzyme and the digested product was directly used to transform XL-1 Blue competent cells. The presence of the D65A mutation in pETHis6-nsP1D65A plasmid was confirmed by DNA sequencing.
2.4. Production of SINV nsP1 in Escherichia coli
Plasmid pETHis6-nsP1 containing the SINV nsP1 gene was used for transforming various E. coli expression strains such as BL21(DE3), BL21(DE3)pLysS and Rosetta(DE3). The expression and solubility of His-tagged SINV nsP1 were tested by growing 5 ml bacterial cultures in Luria broth (LB) supplemented with the appropriate antibiotic. The effect of temperature was tested to optimize the expression and solubility of recombinant nsP1. Cells were initially grown at 37 °C to an optical density of 0.4 at 600 nm (OD600). Growth was continued at 37 °C, 25 °C or 19 °C until OD600 reached 0.8 and expression was induced with 0.4 mM isopropyl-ß-thiogalactopyranoside (IPTG). Cultures were grown for ~4 h at 37 °C, ~6 h at 25 °C and ~16 h at 19 °C. Cells were harvested by centrifugation at 5,000 × g at 4 °C, and cell pellets were re-suspended in 500 µl of 50 mM Tris, pH 8.0. Cells were lysed by sonication on ice (30% amplitude and a pulse of 20 sec). The cell lysates were centrifuged at 15,000 × g at 4 °C for 2.5 min to separate soluble and insoluble fractions. Samples were analyzed on 13% SDS-PAGE gels and visualized by Coomassie Blue staining. The identified optimal conditions and E. coli strain that produced soluble protein were further used for large-scale expression and purification.
For the production of His-tagged recombinant nsP1 for purification, Rosetta(DE3) cells transformed with pETHis6-nsP1 were grown in LB supplemented with 50 µg/ml kanamycin and 35 µg/ml chloramphenicol at 37 °C to an optical density of 0.4 at 600 nm (OD600). The temperature was then reduced to 19 °C and growth was continued until OD600 reached 0.8. Then expression was induced with 0.4 mM IPTG and the culture was grown overnight at 19 °C. Finally, cells were harvested by centrifugation at 5,000 × g at 4 °C and pellets were stored at −20 °C until further use.
2.5. Purification of recombinant nsP1
Purification of His-tagged nsP1 was performed at low temperature (~ 4 °C). Frozen cell pellets from a 1-liter culture were thawed on ice, resuspended in 20 ml of buffer A [20 mM Tris-HCl (pH 7.5) 1 mM DTT, 100 mM NaCl, 2 mM EDTA (pH 8.0)], and disrupted using a French press. The cell lysate was clarified by centrifugation at 18,000 × g for 40 min at 4 °C. The resulting supernatant was fractionated further by ammonium sulfate precipitation. The lysate was gently stirred during slow addition of solid (NH4)2SO4 to 10% (w/v) and then incubated for 30 min at 4 °C. The precipitate was collected by centrifugation (15,000 × g, 10 min) and discarded. This step was repeated and protein was precipitated again with addition of (NH4)2SO4 to 20% (w/v) to the supernatant. After incubation for 30 min at 4 °C, the precipitate was collected by centrifugation (15,000 × g, 10 min) and the supernatant was discarded. The pellet was dissolved in 30 ml of buffer B [20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 40 mM imidazole, 5% glycerol] and applied to a 5 ml HiTrap chelating column charged with NiSO4 and pre-equilibrated with buffer B. Elution was performed using a linear gradient of 40–500 mM imidazole of 60 ml at a flow rate of 1 ml/min and the fractions were analyzed by SDS-PAGE. Fractions containing pure nsP1 were pooled and first dialyzed for 3 h against 1L of dialysis buffer [20 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5% glycerol, 0.5 mM DTT] containing 2 mM EDTA. Dialysis was continued [(1L for 3 h) × 2] against dialysis buffer containing 5 mM MgCl2. Purified protein was concentrated using an Amicon Ultra-10 kDa concentrator and stored at −70 °C. The yield and concentration of purified His-tagged nsP1 was measured using a Bio-Rad protein-assay kit with bovine serum albumin as a standard. The SINV nsP1D65A mutant was expressed and purified using the same protocol.
2.6. Far-UV Circular Dichroism spectrum
For estimation of secondary structure content, purified SINV nsP1 was subjected to circular dichroism (CD) analysis using Chirascan Circular Dichroism Spectrometer (Applied photophysics Ltd, Surrey KT22 7PB, United Kingdom). CD spectra were collected using a 1 mm quartz cell under constant nitrogen purge between 190 to 260 nm in 0.5 nm wavelength steps and an average time of 3.0 sec at 25 °C. Protein samples at concentrations 0.15, 0.25 and 0.35 mg/ml were analyzed in 20 mM sodium phosphate, pH 7.4 buffer containing 100 mM NaCl, 0.5 mM DTT, 5% glycerol, and 5 mM MgCl2. For each sample, three scans were collected and averaged, and the baseline, corresponding to the buffer, was subtracted to obtain the final values. The averaged, background-subtracted spectra analyzed using the software CDPro with the SELCON analysis program (http://lamar.colostate.edu/sreeram/CDPro/) [14, 15]. Effects of chemical denaturants urea and guanidine hydrochloride (GdnHCl) on the conformation of SINV nsP1 were determined by incubating the protein with 8 M urea or 5 M GdnHCl for 4 h at 4 °C followed by the far UV-CD measurements.
2.7. MTase assay
MTase activity was assayed in a reaction volume of 30 µl containing 50 mM Tris-HCl (pH 7.8), 10 mM KCl, 5 mM MgCl2, 5 mM DTT, 20 mM GIDP (a nonhydrolyzable analog of GTP) and 4 µCi of S-adenosyl[methyl-3H]Methionine [3H-AdoMet] (75 Ci/mmol). The reaction was initiated by the addition of ~25 pmol of nsP1 and incubated for 30 min at 30 °C. The reaction was terminated by the addition of an equal (30 µl) volume of 2% SDS [16]. The samples were either analyzed immediately by phenol extraction of [3H-methyl]7 GIDP or stored at −20 °C until further use.
The nsP1 MTase product [3H-methyl]7GIDP was separated from S-adenosyl [methyl-3H] methionine (used as methyl donor) by phenol extraction. Phenol extraction was performed using a published protocol [16]. The extraction is based on the fact that when phenol is mixed with the reaction mixture, AdoMet partitions into the phenol phase, and the methylated acceptor, [3H-methyl]7GIDP, partitions into the aqueous phase. Briefly, the reaction was quenched by addition of SDS to a final concentration of 2% and NH4Cl2 to a final concentration of 1 M (60 µl of 2M NH4Cl2). The reaction mixture was extracted three times with a double volume (240 µl) of phenol buffered with 100 mM Tris-HCl (pH 8.0). Finally, the aqueous phase containing the methylated [3H-methyl]7 GIDP was removed carefully and added to 5 ml of liquid scintillation cocktail. To quantify the transfer of 3H-labeled methyl from AdoMet to GIDP by nsP1, the counts of radioactivity in the extracted aqueous phase containing methylated GIDP were measured with a Beckman Coulter scintillation counter.
2.8. GTase assay
The GTase activity of nsP1 was assayed in a 30 µl final reaction volume containing 50 mM Tris-HCl (pH 7.8), 10 mM KCl, 5 mM MgCl2, 5 mM DTT, 100 µM AdoMet, and 5 µCi of [α-32P] GTP (400 Ci/mmol) [7]. The reaction was initiated by the addition of purified protein and incubated for 30 min at 30 °C. The reaction was terminated by addition of 7 µl of 6x sample buffer (Laemmli buffer) and incubated at 95 °C for 2 min. For the positive control, the GTase activity of vaccinia virus capping enzyme (VVCE) was assayed at pH 8.0 by incubating the reaction mixture at 37 °C for 15 min without the addition of AdoMet [17]. The samples were analyzed by loading 18 µl of the boiled reaction mixture on 10% SDS-PAGE gel. The gel was fixed for 30 min in a fixing solution containing methanol/acetic acid/water (40:10:50 by volume). To remove unincorporated or free [α-32P] GTP, the gel was washed three times with distilled water for 10 min each. The gel was then soaked for 10 min with amplify reagent (GE healthcare) by shaking in the dark. The gel was dried and exposed to an X-ray film and the 32P-labeled intermediate covalent complexes, m7GMP-nsP1 or GMP-VVCE, were visualized by autoradiography. The negative control for the nsP1 GTase assay was the reaction in the absence of AdoMet because GTP methylation of nsP1 MTase is a prerequisite for the formation of the m7GMP-nsP1 covalent intermediate [6].
2.9. Effects of detergents, EDTA and NaCl on nsP1 activity
The influence of ionic and nonionic detergents on nsP1 enzymatic activities was investigated. Purified nsP1 was pre-incubated with detergent (1% w/v) for 1 hr at room temperature and the enzyme reactions were initiated by the addition of AdoMet. The detergents used for studying the effect on nsP1 activities were Tween 20, Triton X-100, octyl beta-D-glucopyranoside (OGP), sodium deoxycholate (DOC) and sodium dodecyl sulfate (SDS). The requirement of a divalent metal ion for both enzymatic activities of SINV nsP1 was tested by performing the MTase and GTase assays in the presence of 20 mM EDTA. The effect of NaCl on the MTase and GTase activities of SINV nsP1 was examined by incubating the reaction mixtures with increasing concentrations of NaCl (0 mM to 500 mM), followed by the measurement of nsP1 activities as described above.
3. Results
3.1. Expression optimization and purification of SINV nsP1
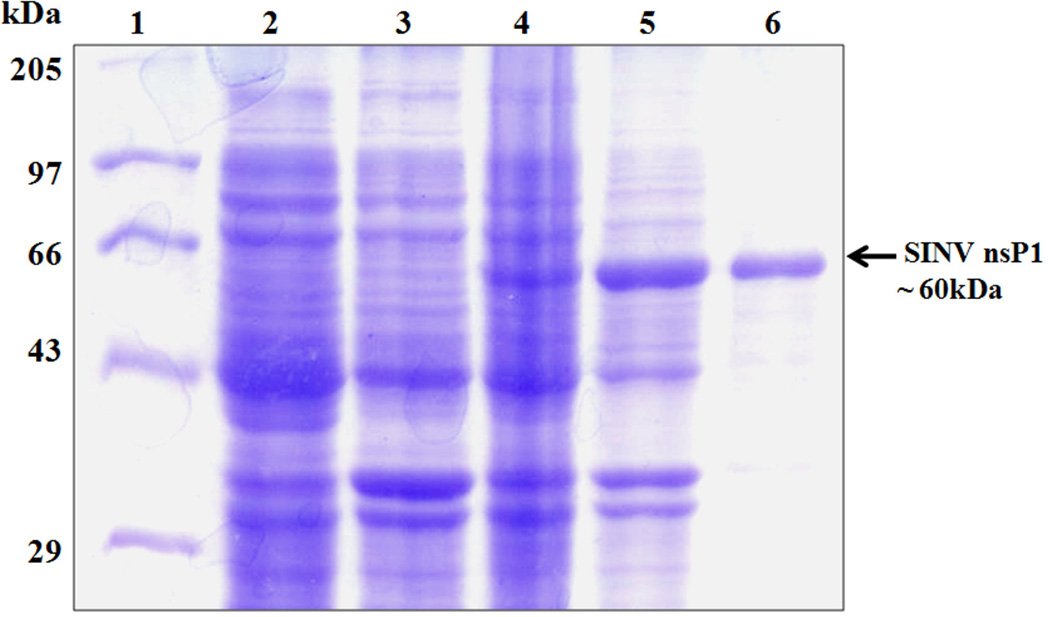
For production of recombinant SINV nsP1 replication protein in a bacterial expression system, the PCR amplified nsP1 gene was cloned into expression vector pET-28a generating a construct (pETHis6-nsP1) expressing nsP1 with an N-terminal 6xHistag. Small scale SINV nsP1 expression and solubility tests were performed using BL21(DE3), BL21(DE3)pLysS and Rosetta(DE3) E. coli expression strains. For solubility optimization, expression was carried out at several temperatures (37 °C, 25 °C and 19 °C), using different IPTG concentrations for induction of expression (0.05, 0.1, 0.2, 0.4 and 1.0 mM), and by varying induction times (3, 4, 6 and 16 h). Unfortunately, BL21(DE3) cells did not show any expression of nsP1, whereas nsP1 was produced in BL21(DE3)pLysS but was in an insoluble form. Primary sequence analysis revealed the presence of 27 rare tRNA codons in the SINV nsP1 sequence. Therefore, heterologous expression to increase the solubility levels of recombinant nsP1 was carried out in the Rosetta(DE3) E. coli expression strain. It harbors the pRARE plasmid encoding tRNA genes that are expressed at a low level in E. coli. SDS-PAGE analysis showed that expression of recombinant SINV nsP1 protein in soluble form with the expected molecular mass (~60 kDa) could be achieved using Rosetta(DE3) cells transformed with pETHis6-nsP1 and grown at 19 °C for ~ 16 h after induction with 0.4 mM IPTG (Fig. 1). Solubility and expression of SINV nsP1 in Rosetta(DE3) cells was also confirmed by western blot analysis using antisera containing anti-SINV nsP1 antibody (a gift from James Strauss, California Institute of Technology) (data not shown). Expression conditions were optimized for large-scale (1L) production of recombinant nsP1 as described in the Materials and Methods section.
Fig. 1.
Expression and purification of Sindbis virus nsP1 from E. coli. SDS-PAGE gel stained with Coomassie Blue. Lanes: 1, molecular mass standards (kDa); 2, Uninduced insoluble fraction of E. coli Rosetta (DE3); 3, Uninduced soluble fraction of lysed Rosetta(DE3); 4, Induced insoluble fraction of lysed Rosetta(DE3); 5, Induced soluble fraction of Rosetta(DE3) containing nsP1; 6, Purified full length (~ 60 kDa) SINV nsP1.
Full length SINV nsP1 with an N-terminal His tag was purified from the soluble fraction of an E. coli lysate using a simple two-step procedure consisting of ammonium sulfate precipitation followed by metal-affinity chromatography as described in the Materials and Methods. In the first step, ammonium sulfate precipitation of the soluble fraction of the nsP1 lysate assisted in removing some of bacterial proteins. The second step of purification was performed using a 5 ml HiTrap Ni2+ chelating column, where an imidazole gradient was used for eluting bound proteins. The bound His-tagged SINV nsP1 eluted from the column at an imidazole concentration of ~150 mM. Homogeneity of the purified sample was assessed by SDS-PAGE, which exhibited a single band of ~60 kDa corresponding to the molecular weight of SINV nsP1 (Fig. 1). The estimated yield was ~1.5 mg of purified protein per liter of culture. Purified protein was concentrated to about 1 mg/ml. The first 25 amino acid residues of purified protein were determined by automated N-terminal amino acid sequencing at the Purdue Proteomics Facility to confirm the identity of purified protein.
3.2. Secondary structure analysis of SINV nsP1
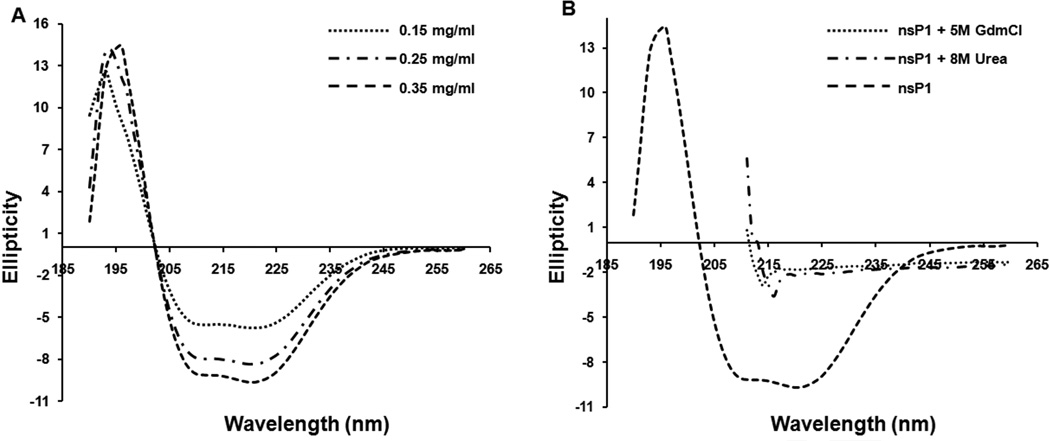
Alphavirus non-structural protein nsP1 does not share significant sequence identity with any protein in the NCBI data bank. Thus, it was difficult to predict the secondary and tertiary structure of the protein. In this study, an attempt was made to study the secondary structure of nsP1 using far-UV circular dichroism (CD). The CD spectral data were analyzed and deconvoluted by the CD spectra SELCON algorithm available through the CDPro software. SELCON yields a structural content of 32.8% α-helix, 19.5% β-sheet, 19% β-turn, and 32.5% random coil (Fig. 2). Prediction of secondary structure elements from the sequence of SINV nsP1 using SOPMA indicates the presence of 38% helix, 22.6 % β-sheet, 6.85% β-turn and 32.6% random coil [18]. Results from the deconvolution of the CD spectra of nsP1 using SELCON were comparable with those obtained using the secondary structure prediction method SOPMA (Table 2). The far-UV CD spectra obtained for SINV nsP1 after incubating the protein with high concentrations of denaturants (urea and GdnHCl) resulted both in loss of ellipiticity values at 222 nm and change in the shape of the CD spectra, indicating the loss of secondary structure and protein folding (Fig. 2). Taken together, the results of CD experiments and the predictions indicate that the purified nsP1 is in the folded native conformation and contains an α/β tertiary structure.
Fig. 2.
Far-UV Circular Dichroism analysis of nsP1 at 25 °C. A. CD spectra of the protein at concentrations 0.15 mg/ml, 0.25 mg/ml, and 0.35 mg/ml in 20 mM Sodium phosphate, pH 7.4 buffer containing 100 mM NaCl, 0.5 mM DTT, 5% glycerol and 5 mM MgCl2. Each spectrum represents an average of three scans. The calculated secondary structure of SINV nsP1 is 32.8% α-helix, 19.5% β-sheets, 19% β-turns and 32.5% random coil. B. CD spectra of the protein in the same buffer, buffer containing 5M GdnHCl, and 8M urea.
3.3. Transfer of methyl group from AdoMet to GIDP
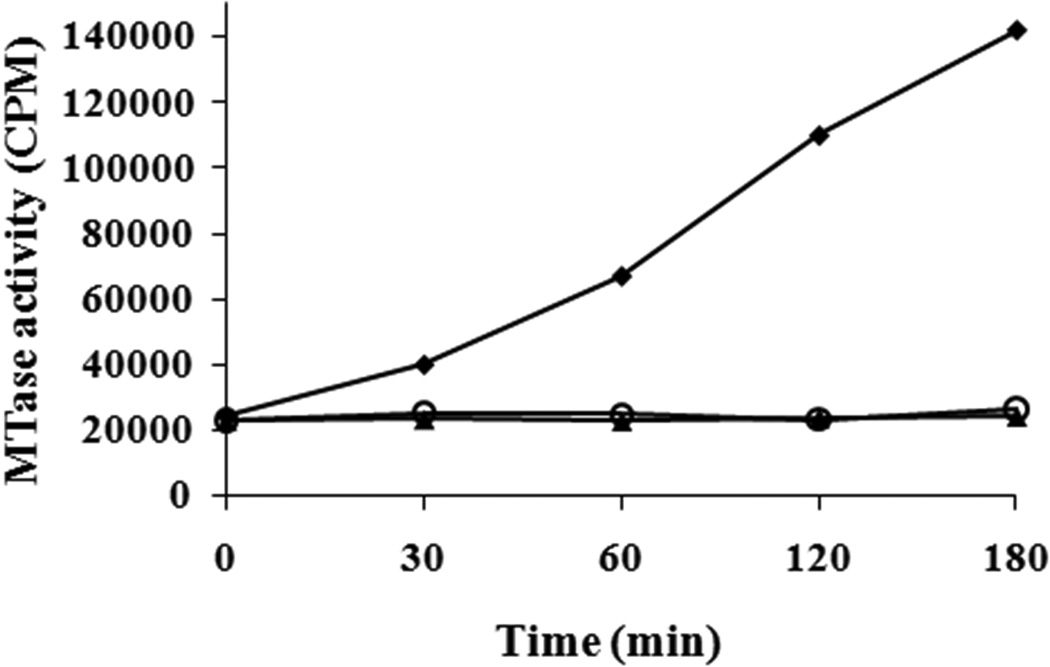
The AdoMet-dependent MTase activity of purified SINV nsP1 with a His-tag at the N-terminus was assayed by monitoring the transfer of a 3H-labeled methyl group from [3H]-AdoMet to the methyl acceptor, GIDP (guanylylimidodiphosphate). The incorporation of 3H-methyl was measured over a period of time (0, 5, 10, 15, 30, 60, 90, and 120 min). The time course of the MTase shows incorporation of 3H-methyl over the period of time (Fig. 3). Negative controls for the MTase assay were performed using the purified nsP1D65A mutant and also in the absence of methyl acceptor GIDP. As expected, no incorporation of 3H-methyl was observed for either negative control reaction. The requirement of divalent cations for the MTase activity was also tested by carrying out an MTase reaction in the presence of 20 mM EDTA. No effect on nsP1 MTase activity was observed in the presence of EDTA (data not shown). This indicates that the transfer of the methyl group from AdoMet to GTP by the Sindbis virus nsP1 MTase does not require divalent cations.
Fig. 3.
Time course of [3H]-methyl incorporation by SINV nsP1 methyltransferase. The methylation reaction mixture containing 20 mM guanylylimidodiphosphate (GIDP), 4 µCi of S-adenosyl [methyl-3H] Methionine, 10 µM AdoMet, and 25 pmol nsP1 was incubated at 30 °C. Reaction samples at the indicated time intervals were collected and analyzed by experimental procedure given in Materials and Methods. Representations in graph: SINV nsP1 MTase reaction (♦); SINV nsP1 D65A MTase negative mutant (○); and negative control containing no GIDP (▲). Each value in the graph represents an average of three measurements.
3.4. Formation of the covalent m7GMP-nsP1 intermediate
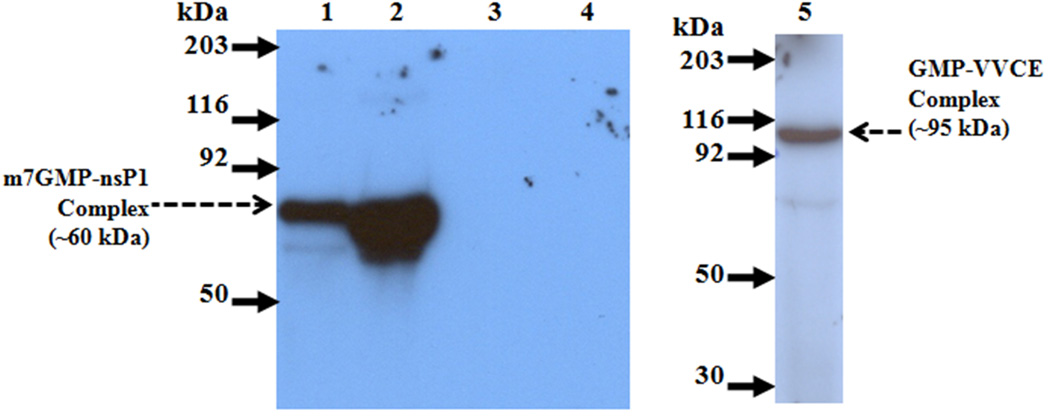
The GTase activity of recombinant SINV nsP1 was assayed by analyzing the formation of the 32P-labeled m7GMP-nsP1 covalent intermediate by SDS-PAGE. The formation of m7GMP-nsP1 was examined by incubating the purified N-terminal His-tagged SINV nsP1 with [α-32P]GTP in a reaction mixture containing AdoMet, the methyl donor. A 32P-labeled m7GMP-nsP1 complex of ~60 kDa corresponding to the molecular weight of nsP1 was detected by autoradiography (Fig. 4). As a positive control in the GTase assay, vaccinia virus capping enzyme (VVCE), which forms an intermediate covalent complex with the GMP moiety, was used. The transfer of the methyl group to GTP is not a prerequisite for GMP-VVCE complex formation. Therefore, AdoMet was not included in the VVCE capping reaction. As a negative control, AdoMet was excluded from the nsP1 GTase reaction because the methylation of GTP by nsP1 MTase is a prerequisite for nsP1 GTase activity [7]. As expected, a 32P-labeled m7GMP-nsP1 band was not detected for the negative control. The presence of 20 mM EDTA in the GTase reaction resulted in no 32P-labeled m7GMP-nsP1 band by SDS-PAGE autoradiography, demonstrating an absolute requirement of divalent metal ion for SINV nsP1 GTase activity (data not shown).
Fig. 4.
Autoradiograph of SDS-PAGE gel showing the formation of α-32P labeled m7GMP-nsP1 covalent complex. The reaction mixture included the standard components as described in Materials and Methods with 5 µCi of [α-32P] GTP, and ~25 pmol/100 pmol of nsP1. Incubation was at 30 °C for 30 min. A. Arrows indicate position of prestained molecular weight markers; Lane 1, 25 pmol nsP1; 2, 100 pmol of nsP1; 3, no protein; 4, no AdoMet; 5, Positive control vaccinia virus capping enzyme.
3.5. Effect of detergents on nsP1 enzyme activities
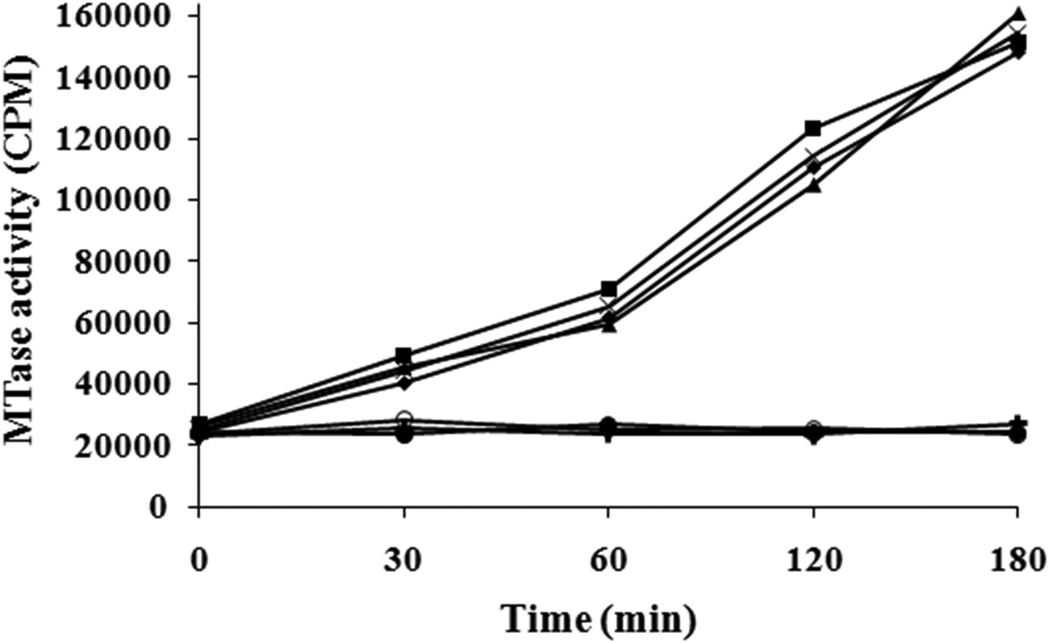
The replication of alphaviruses takes place in membrane associated replication complexes composed of four nonstructural proteins (nsP1- nsP4) and unidentified host factors on cytoplasmic membranes [19–21]. Studies on SFV nsP1 suggested that nsP1 contains an amphipathic helix through which it associates with the membrane and targets the alphavirus replication complex to the plasma membranes [20, 22–25]. Based on enzymatic studies that were carried out in the presence of various detergents and lipids using E. coli-expressed and detergent-extracted SFV nsP1, it was proposed that SFV nsP1 requires membrane association for its enzyme activity [26]. Our results, the first with an alphavirus nsP1 purified to homogeneity, show both MTase and GTase activities in absence of a membrane (Figs. 3 & 4). Reasoning that detergent extraction may have caused the activity loss in the earlier study, we examined the effects of added detergents on enzyme activity of purified SINV nsP1. Therefore, experiments were performed using purified SINV nsP1 to investigate the effects of various detergents on in vitro enzymatic activity. When nsP1 enzyme activities were measured in the presence of two ionic detergents, deoxycholate and sodium dodecyl sulfate, both enzymatic activities of nsP1 were abolished. Interestingly, three nonionic detergents, Tween 20, Triton X-100 and octyl beta-D-glucopyranoside (OGP), eliminate all detectable nsP1 GTase activity but had little, if any, effect on the MTase activity of SINV nsP1 (Fig. 5).
Fig. 5.
Time course of [3H]-methyl incorporation by SINV nsP1 methyltransferase in the presence of ionic and nonionic detergents. The methylation reaction mixture containing 20 mM GIDP, 4 µCi of S-adenosyl [methyl-3H] Methionine, and 25 pmol nsP1 was preincubated with detergents at room temperature for 1 hr. Reactions were initiated by addition of 10 µM AdoMet and by incubating the reaction mixtures at 30 °C for various time intervals. Representations in graph: reaction in the absence of detergent (♦); Tween 20 (■); Triton X-100 (▲); OGP (×); SDS (●); Deoxycholate (○); negative control containing no AdoMet (+). Each value in the graph represents an average of three measurements.
3.6. Effect of NaCl on nsP1 enzyme activities
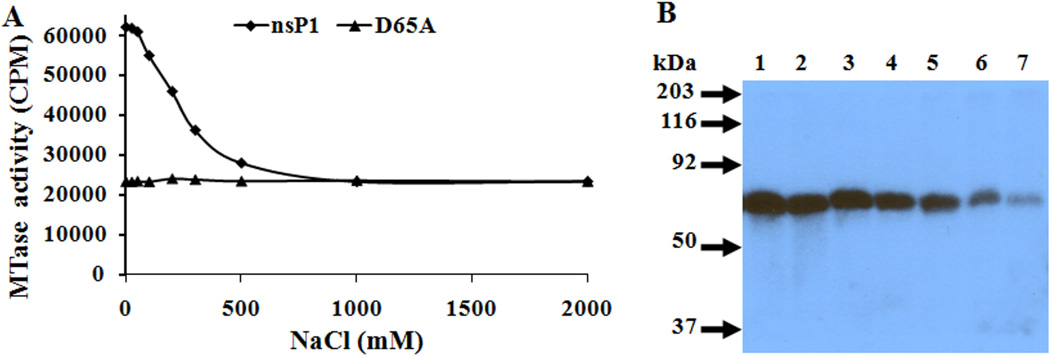
Purified protein was used to study the effect of NaCl on the enzymatic activities of SINV nsP1. Alphavirus nsP1 has been proposed to be a membrane-associated protein, and NaCl and various detergents have been used to disrupt nsP1 membrane association [27, 28]. Therefore, the effect of NaCl on the enzymatic activities of nsP1 was studied. The results could be important for future biochemical and structural analysis of nsP1. The effect of different NaCl concentrations (25, 50, 100, 200, 300 and 500 mM) on both the MTase as well as GTase activity of SINV nsP1 was determined. Changes in the ionic strength over the range of 0.025 to 0.1 M NaCl had little, if any, effect on either enzymatic activity of nsP1. However, at higher ionic strength (200–500 mM), a gradual decrease in MTase and GTase activity of SINV nsP1 was observed (Fig. 6).
Fig. 6.
Effect of NaCl on enzymatic activities of SINV nsP1. A. Effect of NaCl on MTase activity. Transfer of [3H]-methyl from AdoMet to GIDP in the presence of increasing concentrations of NaCl. B. Autoradiograph of SDS-PAGE gel showing the formation of α-32P labeled m7GMP-nsP1 covalent complex in the presence of varying concentrations of NaCl. Lanes: 1, 0 mM NaCl; 2, 25 mM NaCl; 3, 50 mM NaCl; 4, 100 mM NaCl; 5, 200 mM NaCl; 6, 300 mM NaCl; 7, 500 mM NaCl.
4. Discussion
Alphaviruses replicate in the cytoplasm of infected cells and have developed virus-specific transcription and capping mechanisms for replication of their genomes [3]. Although the cap structures of eukaryotic mRNA and viral RNAs are similar, RNA viruses have diverse pathways for cap formation such as cap-snatching or virus-specific capping mechanisms [29–31]. Some viruses have developed a cap-independent mechanism by using the 5’UTR internal ribosomal entry site (IRES) for efficient translation initiation [32]. On the other hand, alphaviruses and other members of the alphavirus-like superfamily encode for virus-specific capping enzymes and have developed a novel capping mechanism for the formation of a cap structure at the 5’ end of viral RNAs [7, 26]. Therefore, the alphavirus nsP1 replication protein, possessing virus-specific capping enzyme activities, is a potential antiviral drug target. The expression and purification of nsP1 in an active form is essential for biochemical and structural studies that could lead to the development of a pharmacologically active inhibitor. In this study, details of heterologous production and purification of SINV nsP1 are reported. To demonstrate that the purified protein is active and in the native conformation, enzymatic assays and CD analysis were carried out.
Using purified nsP1, we performed the first conformational analysis of an alphavirus replication protein nsP1 using CD spectroscopy. Secondary structural elements of SINV nsP1 were also predicted by statistical methods using the primary structure of the protein and compared with the estimated secondary structure obtained from CD data. CD experiments were validated by the reproducibility of the spectra at different protein concentrations and by the loss of protein folding in the presence of high concentrations of denaturants (urea and GdnHCl). The secondary structure of SINV nsP1 evaluated experimentally by CD measurements indicates that purified SINV nsP1 contains ~33% α-helical and ~20% β-sheet structure. In addition, the secondary structure predictions for SINV nsP1 protein using the primary sequence were in good agreement with the CD measurements (Table 2). Another interesting observation made from CD analysis and secondary structure prediction is the high percentage (~40–50%) of residues not in regular secondary structure. Bioinformatics analysis was consistent with the observed low regular secondary structure content as a disordered region (amino acids 374–429) was predicted in the nsP1 sequence using the Fold-Index server [33]. Structurally disordered regions have been predicted at the N-termini of the alphavirus nonstructural protein nsP4 and the capsid protein [34–37]. Recently, the disordered region of nsP4 has been proposed to be involved in protein-protein interactions with other alphavirus replication proteins [38]. If nsP1 contains intrinsically disordered regions, they may carry out important functions, for example to participate in specific interactions with other proteins, nucleic acids or membranes [39].
Purified N-terminal His-tagged SINV nsP1 was used to determine MTase activity. The transfer of a 3H-labeled methyl group from [3H]-AdoMet to the methyl acceptor GIDP and a gradual increase in incorporation of 3H-methyl over time were detected, demonstrating that the recombinant, purified protein contains MTase activity. Mutation of conserved Asp65 of the AdoMet binding G-loop motif in SINV nsP1 to alanine completely abolished the MTase activity. This confirmed that the transfer of a 3H-labeled methyl to GIDP was catalyzed by the purified SINV nsP1 and that Asp65 is essential for MTase activity as deduced from studies with SFV nsP1 [6]. Divalent metal ion is not required for nsP1 MTase activity as revealed by the unchanged enzyme activity in presence of EDTA.
We also performed a GTase assay with purified SINV nsP1 and observed the formation of a ~60 kDa [32P]-labeled m7GMP-nsP1 covalent intermediate by SDS-PAGE and autoradiography. The possibility that a bacterial protein impurity catalyzed formation [32P]-labeled-GMP-nsP1 was ruled out when nsP1-guanylate was not detected in a negative control reaction in which AdoMet was not included in the reaction mix. These experiments demonstrate that the purified, recombinant SINV nsP1 protein catalyzed the formation of the expected ~60 kDa [32P]-labeled m7GMP-nsP1 covalent intermediate.
It has been proposed that the membrane association of alphavirus nsP1 is essential for its enzymatic activities, based on studies in which the use of detergents abolished both the MTase and GTase enzyme activities of SFV nsP1 [27, 28]. However, in our studies, purified Sindbis virus nsP1 possessed both MTase and guanyltransfer activities in the absence of any membrane. The effect of detergents on purified SINV nsP1 activity revealed a detergent effect. The SINV MTase was active in the presence of nonionic detergents (Triton X-100, Tween 20 and OGP), while MTase activity was abrogated in the presence of ionic detergents (DOC and SDS) (Fig. 5). In general, nonionic detergents are less likely to cause protein denaturation than are ionic detergents [40]. Therefore, ionic detergents may perturb the native conformation of the nsP1 protein, leading to inactivation of nsP1 MTase.
In contrast to the MTase, the nsP1 GTase was inactive in the presence of either ionic or nonionic detergents, suggesting that the GTase is quite sensitive to detergent-stimulated structural perturbations. The effect of solution environment on nsP1 activity was also apparent in experiments that varied NaCl concentration as both the GTase and MTase activities of nsP1 decreased as ionic strength was increased.
This is the first report of purification of an enzymatically active alphavirus capping enzyme, nsP1. The availability of purified recombinant SINV nsP1 enzyme will not only accelerate the structural investigations for understanding the detailed enzyme mechanisms but will also facilitate development of a high throughput enzyme assay for screening alphavirus specific capping enzyme inhibitors.
Highlights.
> We have purified enzymatically active alphavirus capping enzyme, nsP1. > Purified recombinant Sindbis virus (SINV) nsP1 possess the methyltransferase (MTase) and guanylyltransferase (GTase) activities. > This indicates that alphavirus nsP1 does not require membrane association for its enzymatic function. > SINV nsP1 GTase activity is metal-ion dependent whereas MTase does not require a metal ion. > Circular dichroism studies show that nsP1 has a mixed α/β structure with a structural content of 32.8% α-helix, 19.5% β-sheet, 19% β-turn, and 32.5% random coil.
Acknowledgements
The authors thank Steven S. Broyles, Purdue University, for the kind gift of vaccinia virus capping enzyme, and James Strauss, California Institute of Technology, for the kind gift of anti-SINV nsP1 antibody, and Anita Robinson for assistance with the manuscript. We also thank Macromolecular Crystallographic Unit (MCU) at the Institute Instrumentation Centre (IIC), Indian Institute of Technology (IIT), Roorkee for providing protein production and purification facilities. This work was supported by Department of Science and Technology, India (ST) and grant P01 AI-055672 from the US National Institutes of Health (R.J.K. and J.L.S.).
Appendix
Table 1.
List and sequences of oligonucleotides used
| Primer | Sequence |
|---|---|
| S1 | 5′-atataCCATGGagaagccagtagtaaacgtagac-3′ |
| A1 | 5′-atatcCTCGAGTCAtgctccgatgtccgcctggag-3′ |
| S2 | 5′-atataCATATGgagaagccagtagta aacgtagac-3′ |
| S3 | 5′-catcccggccaccatTtgcgatcagatgactg-3′ |
| A2 | 5′-cagtcatctgatcgcaAatggtggccgggatg-3′ |
| S4 | 5′-acagcgacgatcttggCAataggcagcgca-3′ |
| A3 | 5′-tgcgctgcctatTGccaagatcgtcgctgt-3′ |
Table 2.
Secondary structure contents of SINV nsP1 obtained by Circular Dichorism and Protein secondary structure prediction methods.
| CDPro Secondary structure content | ||||
|---|---|---|---|---|
| Algorithm | α-Helix | β-sheets | β-turn | Random coil |
| SELCON | 32.8 | 19.5 | 19 | 32.5 |
| Secondary structure prediction | ||||
| SOPMA | 37.96 | 22.59 | 6.85 | 32.59 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chevillon C, Briant L, Renaud F, Devaux C. The Chikungunya threat: an ecological and evolutionary perspective. Trends Microbiol. 2008;16:80–88. doi: 10.1016/j.tim.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Weaver SC, Ferro C, Barrera R, Boshell J, Navarro JC. Venezuelan equine encephalitis. Annu. Rev. Entomol. 2004;49:141–174. doi: 10.1146/annurev.ento.49.061802.123422. [DOI] [PubMed] [Google Scholar]
- 3.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pettersson RF, Söderlund H, Kääriäinen L. The nucleotide sequences of 5′-terminal T1 oligonucleotides of Semliki Forest virus 42-S and 26-S RNAs are different. Eur. J. Biochem. 1980;105:435–443. doi: 10.1111/j.1432-1033.1980.tb04518.x. [DOI] [PubMed] [Google Scholar]
- 5.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahola T, Laakkonen P, Vihinen H, Kääriäinen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 1997;71:392–397. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahola T, Kääriäinen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA. 1995;92:507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizumoto K, Kaziro Y. Messenger RNA capping enzymes from eukaryotic cells. Prog Nucleic Acid Res Mol Biol. 1987;34:1–28. doi: 10.1016/s0079-6603(08)60491-2. [DOI] [PubMed] [Google Scholar]
- 9.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 10.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:83–93. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 11.Wang HL, O’Rear J, Stollar V. Mutagenesis of the Sindbis virus nsP1 protein: effects on methyltransferase activity and viral infectivity. Virology. 1996;217:527–531. doi: 10.1006/viro.1996.0147. [DOI] [PubMed] [Google Scholar]
- 12.Owen KE, Kuhn RJ. Identification of a Region in the Sindbis Virus Nucleocapsid Protein That Is Involved in Specificity of RNA Encapsidation. J. Virol. 1996;70:2757–2763. doi: 10.1128/jvi.70.5.2757-2763.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue H, Nojima H, Okayama H. E. coli competent cells and transformation. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 14.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 15.Sreerama N, Venyaminov SY, Woody RW. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheidel LM, Durbin RK, Stollar V. SVLM21, a Sindbis virus mutant resistant to methionine deprivation, encodes an altered methyltransferase. Virology. 1989;173:408–414. doi: 10.1016/0042-6822(89)90553-9. [DOI] [PubMed] [Google Scholar]
- 17.Shuman S, Hurwitz J. Mechanism of mRNA capping by vaccinia virus guanylyltransferase : characterization of an enzyme--guanylate intermediate. Proc. Natl. Acad. Sci. U.S.A. 1981;78:187–191. doi: 10.1073/pnas.78.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geourjon C, Deléage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 19.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1996;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kujala P, Ikäheimonen A, Ehsani N, Vihinen H, Auvinen P, Kääriäinen L. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 2001;75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 2010;84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spuul P, Salonen A, Merits A, Jokitalo E, Kääriäinen L, Ahola T. Role of the amphipathic peptide of Semliki forest virus replicase protein nsP1 in membrane association and virus replication. J. Virol. 2007;81:872–883. doi: 10.1128/JVI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peränen J, Laakkonen P, Hyvönen M, Kääriäinen L. The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles. Virology. 1995;208:610–620. doi: 10.1006/viro.1995.1192. [DOI] [PubMed] [Google Scholar]
- 24.Salonen A, Vasiljeva L, Merits A, Magden J, Jokitalo E, Kääriäinen L. Properly folded nonstructural polyprotein directs the semliki forest virus replication complex to the endosomal compartment. J. Virol. 2003;77:1691–1702. doi: 10.1128/JVI.77.3.1691-1702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laakkonen P, Ahola T, Kääriäinen L. The effects of palmitoylation on membrane association of Semliki Forest virus RNA capping enzyme. J. Biol. Chem. 1996;271:28567–28571. doi: 10.1074/jbc.271.45.28567. [DOI] [PubMed] [Google Scholar]
- 26.Ahola T, Ahlquist P. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 1999;73:10061–10069. doi: 10.1128/jvi.73.12.10061-10069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahola T, Lampio A, Auvinen P, Kääriäinen L. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 1999;18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lampio A, Kilpelainen I, Pesonen S, Karhi K, Auvinen P, Somerharju P, Kaariainen L. Membrane binding mechanism of an RNA virus-capping enzyme. J. Biol. Chem. 2000;275:37853–37859. doi: 10.1074/jbc.M004865200. [DOI] [PubMed] [Google Scholar]
- 29.Duijsings D, Kormelink R, Goldbach R. In vivo analysis of the TSWV cap-snatching mechanism: single base complementarity and primer length requirements. EMBO J. 2001;20:2545–2552. doi: 10.1093/emboj/20.10.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao P, Yuan W, Kru RM. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 2003;22:1188–1198. doi: 10.1093/emboj/cdg109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih SR, Krug RM. Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology. 1996;226:430–435. doi: 10.1006/viro.1996.0673. [DOI] [PubMed] [Google Scholar]
- 32.Ehrenfeld E, Semler BL. Anatomy of the poliovirus internal ribosome entry site. Curr. Top. Microbiol. Immunol. 1995;203:65–83. doi: 10.1007/978-3-642-79663-0_3. [DOI] [PubMed] [Google Scholar]
- 33.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann J, Silman I, Sussman JL. Foldindex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 34.Tomar S, Hardy RW, Smith JL, Kuhn RJ. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J. Virol. 2006;80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi HK, Tong L, Minor W, Dumas P, Boege U, Rossmann MG, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 36.Tong L, Wengler G, Rossmann MG. Refined structure of Sindbis virus core protein and comparison with other chymotrypsin-like serine proteinase structures. J. Mol. Biol. 1993;230:228–247. doi: 10.1006/jmbi.1993.1139. [DOI] [PubMed] [Google Scholar]
- 37.Choi H, Lee S, Zhang Y, McKinney B, Wengler G, Rossmann MG, Kuhn RJ. Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis. J. Mol. Biol. 1996;262:151–167. doi: 10.1006/jmbi.1996.0505. [DOI] [PubMed] [Google Scholar]
- 38.Rupp JC, Jundt N, Hardy RW. Requirement for the amino-terminal domain of sindbis virus nsP4 during virus infection. J. Virol. 2011;85:3449–3460. doi: 10.1128/JVI.02058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 40.Goddard ED, Anathapadmanabhan KP. Interactions of Surfactants with Polymers and Proteins. Boca Raton, FL: CRC Press, Inc.; 1993. pp. 319–365. [Google Scholar]