SUMMARY
SSB proteins bind to and control the accessibility of single stranded (ss) DNA, likely facilitated by their ability to diffuse on ssDNA. Using a hybrid single-molecule method combining fluorescence and force, we probed how proteins with large binding site sizes can migrate rapidly on DNA and how protein-protein interactions and tension may modulate the motion. We observed force-induced progressive unraveling of ssDNA from the SSB surface between 1 and 6 pN, followed by SSB dissociation at ~10 pN, and obtained experimental evidence of a reptation mechanism for protein movement along DNA wherein a protein slides via DNA bulge formation and propagation. SSB diffusion persists even when bound with RecO and at forces under which the fully wrapped state is perturbed, suggesting that even in crowded cellular conditions SSB can act as a sliding platform to recruit and carry its interacting proteins for use in DNA replication, recombination and repair.
INTRODUCTION
In the cell, a multitude of proteins compete for access to ssDNA but it is unclear how these proteins coordinate with each other. ssDNA binding (SSB) proteins bind selectively to ssDNA with high affinity and little sequence specificity, protecting ssDNA from degradation and recruiting other proteins necessary for DNA replication, recombination and repair (Richard et al., 2009; Shereda et al., 2008). E. coli (Eco)SSB, a representative homotetrameric SSB, wraps ~ 65 nt of ssDNA around it ((SSB)65 mode) at moderately high salt conditions (≥ 200 mM Na+ or ≥ 2 mM Mg2+) (Bujalowski and Lohman, 1986; Chrysogelos and Griffith, 1982; Lohman and Overman, 1985; Roy et al., 2007) such that the two ssDNA ends enter and exit the protein in close proximity (Raghunathan et al., 2000). At lower salt concentrations and higher SSB to DNA ratios, a cooperative SSB binding mode is favored in which only two subunits of the tetramer interact with ~35 nucleotides of ssDNA (Lohman and Ferrari, 1994). In order for subsequent DNA metabolic processes to occur, EcoSSB plays a second major role in DNA metabolism in that it interacts directly with at least 14 other proteins that we term SIPs (SSB Interacting Proteins), including DNA Polymerase II, III and V, primase, RecQ, RecO, RecJ, RecG, PriA, PriB, Exonuclease I and IX, Uracil DNA Glycosylase and phage N4 RNA polymerase (Shereda et al., 2008), bringing them to their sites of function. How SSB permits access of SIPs to SSB-bound DNA is unclear.
The process by which protein diffusion along DNA facilitates location of specific target sites or its repositioning has been studied almost exclusively on double-stranded DNA (Gorman and Greene, 2008). Recently, we reported the direct observation of a protein diffusing on ssDNA (Roy et al., 2009). Using single molecule two- and three-color FRET (fluorescence resonance energy transfer (Ha et al., 1996)), we found that EcoSSB can diffuse on ssDNA (diffusion coefficient ~300 nt2/s at 37 °C) with a mean step size of 3 nt, and that this SSB activity transiently melts DNA secondary structures and stimulates RecA filament elongation (Roy et al., 2009). The underlying mechanism for diffusion is fundamental to understanding cellular functions but how a protein with such a large binding site size (~65 nt for SSB) and high affinity can diffuse rapidly on DNA remains unknown. In addition, how the application of force or the binding of SIPs might modulate these SSB dynamics has not been investigated.
Here, we apply a recently developed optomechanical tool combining single molecule fluorescence detection and force (Hohng et al., 2007) to monitor the tension-dependent conformational transitions of DNA/protein complexes with nanometer resolution at the single protein level. Our earlier study on SSB (Roy et al., 2009) was based on only fluorescence measurements. Here, with tension applied to the DNA and our capability to measure temporal changes of arbitrary coordinates (i.e., not just the end-to-end distance of a biopolymer) at low forces, we were able to obtain information that is unattainable by mechanical manipulations or fluorescence techniques alone. The fluorescence probes can be positioned at different desired locations on DNA and/or protein to probe the dynamics along various vectors, which maximizes the information content. This approach allowed us to probe the dissociation mechanism of an EcoSSB bound to ssDNA and directly observe the mechanical regulation of an individual SSB tetramer diffusing along ssDNA.
RESULTS
Near-Equilibrium DNA Unwrapping and Rewrapping at Low Forces
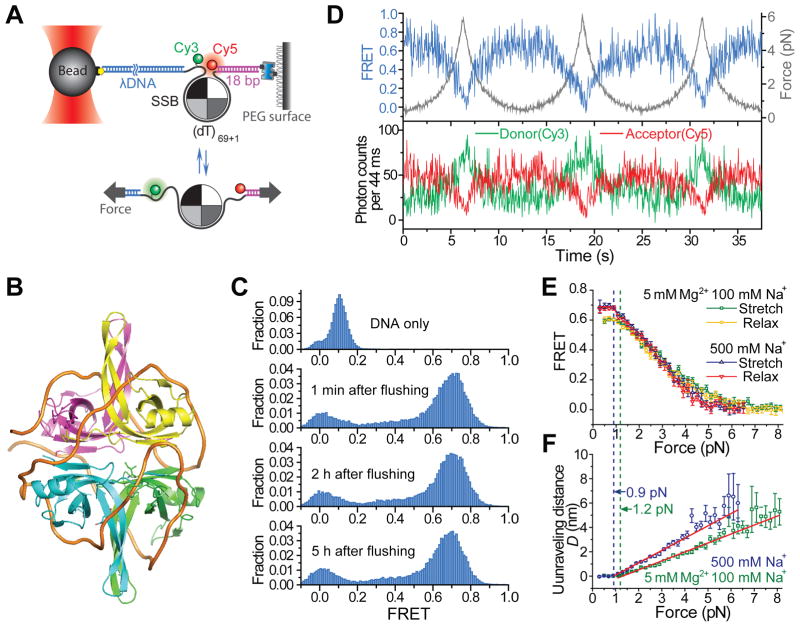
To investigate the removal of tightly wrapped ssDNA from SSB in its fully wrapped (SSB)65 mode, we first examined the effect of applying an unwrapping force to SSB-bound DNA in 500 mM NaCl using fluorescence-force spectroscopy that combines confocal fluorescence microscopy-based single molecule FRET and optical tweezers (Hohng et al., 2007) (See also Experimental Procedures). At zero force, high FRET (~ 0.7) is observed between a donor (Cy3) and an acceptor (Cy5) attached near the two ends of a 70-mer ssDNA wrapped around a single SSB tetramer (Figures 1A and 1B). For simplicity, we depict the complex as a protein disc surrounded by a line, whereas in the 3D structural model of the SSB/DNA complex in its (SSB)65 mode, the path of the DNA around SSB resembles the seam of a tennis ball (Figure 1B). SSB remains bound to the surface-tethered DNA even 5 hours after removing free SSB from solution (Figure 1C). For each DNA stretching cycle, the sample stage was moved from a low force position (~0.5 pN) at a constant speed v (455 nm s−1) until the force reached a predetermined value (~6 pN), followed by returning the stage at the same speed to the initial low force position (at 20 nM SSB tetramer concentration). The FRET efficiency E decreased and increased gradually (between ~ 0.7 and ~ 0) as the force was increased and then decreased respectively, demonstrating force-induced, progressive DNA unravelling from SSB (Figure 1D). Superimposed on the curve are the anti-correlated fluctuations of donor signal ID and acceptor signal IA reflecting SSB diffusion as we discuss below. In contrast, the E vs. force curve of ssDNA alone without SSB showed E values below 0.2 for the entire force range (Figure S1).
Figure 1. Force-induced Unraveling of ssDNA from SSB Measured by Fluorescence-Force Spectroscopy.
(A) Experimental scheme for force-induced unravelling of ssDNA, (dT)69+1, from SSB measured via FRET. One end of the construct was immobilized on a PEG surface via biotin-neutravidin interaction and the other end was linked to a bead held in an optical trap via a Digoxigenin-Anti-digoxigenin interaction.
(B) Structural model for an SSB tetramer bound to a 70nt ssDNA (thick orange line) in the fully wrapped (SSB)65 binding mode, based on an X-ray crystallographic structure of a C-terminal truncated SSB tetramer (SSBΔC) bound to two (dC)35 oligonucleotides (Raghunathan et al., 2000).
(C) FRET histograms of the DNA construct at zero force with and without SSB bound. The peak at zero FRET corresponds to DNA molecules with active Cy3 only, and the second major peak corresponds to DNA molecules with both active Cy3 and Cy5. Excess SSB proteins were removed from the solution after incubating at 500 mM NaCl and 1 nM SSB tetramer concentration. The FRET histograms were obtained 1 minute, 2 hours, and 5 hours after the removal of free SSB in solution.
(D) Fluorescence-force traces obtained while stretching and relaxing the DNA at the stage-moving speed v of 455 nm s−1 (20 nM SSB in solution) when the maximum force achieved was set to ~ 6 pN (Averaged among ~ 50 cycles from 10 molecules with a bin size of 0.2 pN).
(E) The averaged FRET vs. force curves for stretching and relaxing the DNA when the maximum force achieved was set to ~ 6 pN (in 500 mM Na+) or ~8 pN (in 5 mM Mg2+ and 100 mM Na+).
(F) Unraveling distance vs. force curves in two ionic conditions fit to straight lines (red lines), D = α·(F-β) (when the force F ≥ β), where α = 1.0 ± 0.03 nm/pN, β = 0.9 ± 0.2 pN for 500 mM Na+, and α = 0.7± 0.02 nm/pN, β = 1.2 ± 0.3 pN for 5 mM Mg2+ and 100 mM Na+, determined from the fit Error bars are the standard errors.
See also Figure S1.
In 500 mM Na+, ssDNA unravelling begins once the force goes above a threshold of β = 0.9 ± 0.2 pN and the averaged stretching and relaxation curves coincide (Figure 1E), indicating that the initial peeling off of ssDNA from the SSB surface below 6 pN of force is reversible. The change in the distance between Cy3 and Cy5 (estimated from FRET efficiency, see Extended Experimental Procedures), D, scales linearly with force within the FRET detectable range with a slope of α = 1.0 ± 0.03 pN/nm (Figure 1F). The unravelling experiment performed at a different ionic condition (5 mM Mg2+ and 100 mM Na+) gave a similar result except that α = 0.7± 0.02 nm/pN and β = 1.2 ± 0.3 pN (Figure 1F). The mechanical work to unravel ssDNA is given by ( ) pN nm (D is in nm), yielding the interaction energy density of (0.22 ± 0.05) kBT/nm or (0.13 ± 0.03) kBT/nt for the SSB-DNA complex in 500 mM Na+, and (0.29 ± 0.07) kBT/nm or (0.17 ± 0.04) kBT/nt in 5 mM Mg2+ and 100 mM Na+ (see Figures S2 and Extended Experimental Procedures). This interaction energy density is smaller than that between nucleosomal DNA and a histone octamer (0.5 –1.0 kBT/nm) (Kulic and Schiessel, 2003b; Polach and Widom, 1995), potentially explaining the more rapid diffusion for SSB.
SSB Dissociation Events at Higher Forces
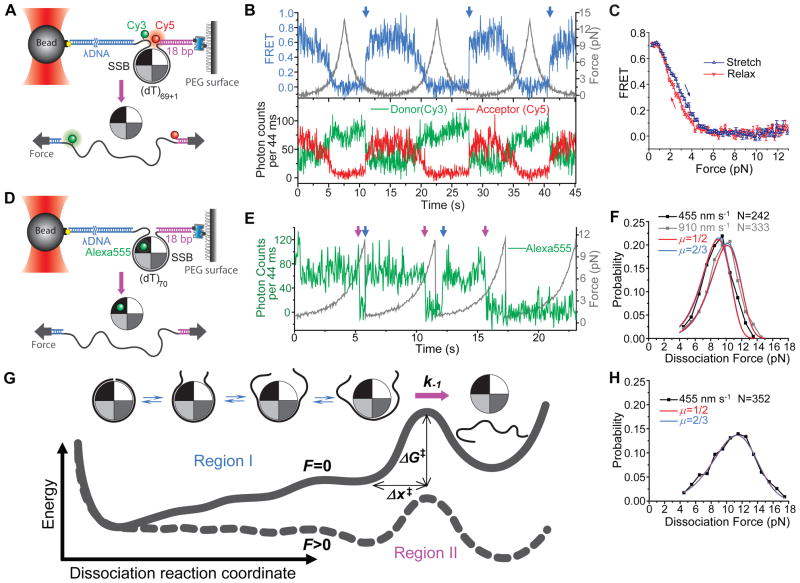
If a maximum force of ~13 pN was reached, hysteresis was often observed (Figures 2A and 2B) where the initial return segment resembled that observed for ssDNA alone (Figure S1B), indicating that the SSB had dissociated fully at this higher force and did not immediately rebind. The averaged stretching and relaxation curves did not overlap and displayed hysteresis due to suppression of rebinding at high forces (Figure 2C), further indicating that full SSB dissociation occurs mainly at forces between 6 and 13 pN.
Figure 2. Direct Observation of Force-Induced Individual SSB Dissociation Events.
(A) Experimental scheme for force-induced unravelling of ssDNA, (dT)69+1, from SSB measured via FRET as in Figure 1A.
(B) Fluorescence-force traces obtained while stretching and relaxing the DNA at the stage-moving speed v of 455 nm s−1 (20 nM SSB in solution) when the maximum force achieved was set to ~ 13 pN (in 500 mM Na+). Blue arrows indicate SSB binding events.
(C) The averaged FRET vs. force curves for stretching and relaxing the DNA when the maximum force achieved was set to ~ 13 pN. Error bars are ± s.e.m. (Averaged among 45 cycles from 12 molecules with a bin size of 0.2 pN).
(D) Experimental scheme for direct observation of individual SSB dissociation events.
(E) Fluorescence-force curves that indicate the binding (blue arrows) and dissociation (magenta arrows) of individual SSBf at v = 455 nm s−1.
(F) Dissociation force distributions obtained in 500 mM Na+ at the two stage-moving speeds. The small population assigned to fluorophore photobleaching has been removed. The solid lines are the global fits to the Dudko model with the parameter (μ) that controls the shape of the energy barrier set to 1/2 (blue) or 2/3 (red) (see Experimental Procedures).
(G) Energy landscape along the SSB dissociation reaction coordinate with two distinct regions.
(H) Dissociation force distribution obtained in 5 mM Mg2+ and 100 mM Na+ at the stage-moving speed of 455 nm s−1.
See also Figure S3.
To determine precisely the force at which SSB dissociates, Fd, we repeated the same experiment, but using 1 nM SSBf, an A122C SSB mutant labeled with ~ one Alexa555 fluorophore per SSB tetramer (Roy et al., 2009) and after Cy3 and Cy5 on the DNA have been photobleached (Figure 2D). Alexa555 fluorescence increases abruptly (Figures 2E and S3, green trace) upon SSBf binding to the DNA and disappears later due to either SSBf dissociation or photobleaching. The probability distribution p(Fd) was obtained (Figure 2F) after removing a population at low force (~ 1 pN) that we attribute to photobleaching (Figures S3F and S3G). p(Fd) is broad and asymmetric, and the mean Fd shifted from 8.8±0.2 to 9.5±0.2 pN upon doubling the pulling rate v (Figures 2F and S3B), indicating that the final SSB dissociation from a partially wrapped intermediate (Kozlov and Lohman, 2002) is a non-equilibrium process. We used the theory of Dudko et al (Dudko et al., 2006; Dudko et al., 2008; Greenleaf et al., 2008) to obtain the rate of SSB dissociation from the partially wrapped intermediate at zero force, k−1 = 0.010 ± 0.006 s−1; the distance to the transition state from the intermediate, Δx‡ = 3.2 ± 0.5 nm; and the height of the free energy barrier between the intermediate and unbound state ΔG‡ = (11 ± 2) kBT (Experimental Procedures). Similar results were obtained in 5 mM Mg2+ and 100 mM Na+ (Figure 2H) with mean Fd = 10.7±0.3 pN, k−1 = 0.010 ± 0.005 s−1, Δx‡ = 2.4 ± 0.3 nm, and ΔG‡ = (8.0 ± 0.3) kB T, Combining our data both at low (< 6 pN) and high force ranges (> 6 pN), the overall energy landscape can be stitched together with two major regions along the dissociation reaction coordinate (Figure 2G).
Rolling vs. Sliding Mechanisms for SSB Diffusion on DNA
Next, we investigated the mechanism of SSB diffusion on ssDNA. How a protein with such extensive interactions (~ 65 nt) with DNA and high affinity can spontaneously and rapidly migrate on the DNA has been a mystery. At present, there is only one proposed mechanism in the literature on how SSB may achieve this feat (Kuznetsov et al., 2006; Romer et al., 1984). In this rolling mechanism, a partial unwrapping of one end segment of ssDNA from an SSB tetramer is followed by rewrapping of the other end of the ssDNA in its place, resulting in a one-dimensional random walk of SSB along DNA (Figure 3A, Movie S1). The rolling mechanism utilizes the closed wrapping of ssDNA on the SSB surface and allows SSB diffusion while maintaining most of its contacts with ssDNA (with relatively low energetic cost), and is therefore an attractive mechanism. An alternative scenario that has not been previously considered for SSB is that the whole ssDNA ‘slides’ relative to the protein surface (Figure 3B).
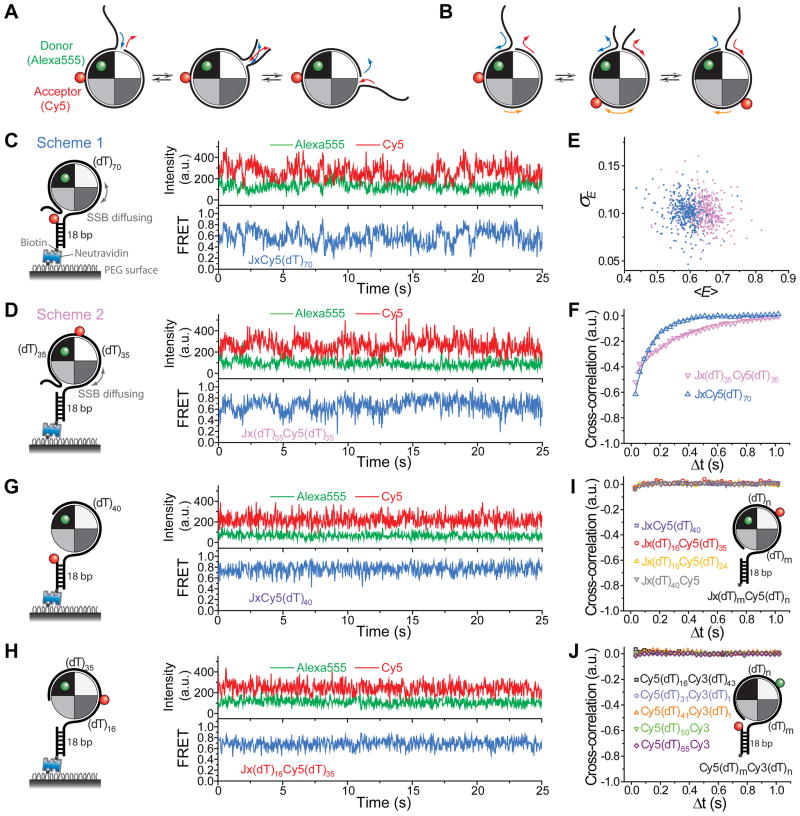
Figure 3. Evidence Favoring the Sliding Mechanism Over the Rolling Mechanism.
(A) Rolling mechanism for SSB diffusion. One end of the wrapped DNA could partially dissociate from the SSB while the other end of the DNA binds to the same newly open DNA binding site. This mechanism is facilitated by the ‘closed wrapping’ topology of the DNA around the SSB tetramer and there is no relative ‘sliding’ motion between ssDNA and the SSB surface in this model. Only the ends but not the mid-section of the bound DNA slide/move relative to SSB during diffusion.
(B) Sliding mechanism for SSB diffusion. In this mechanism, the whole SSB-bound DNA (65 nt) slides relative to protein surface during diffusion.
(C and D) Representative single-molecule time traces of donor(Alexa555) and acceptor(Cy5) intensities and corresponding FRET efficiency show fluctuations induced by SSBf diffusion along the ssDNA if Cy5 is attached near one end of (dT)70 (Scheme 1) or to the middle of (dT)70 (Scheme 2; 30 ms time resolution). a.u., arbitrary units.
(E) A scatter plot of <E> versus σE for individual FRET time trajectories obtained from the two Cy5 labelling schemes.
(F), Cross-correlation analysis of single-molecule intensity-time traces fit to single exponential function for data obtained with the two labeling schemes in (C) and (D) (averaged over > 300 molecules each).
(G and H) Representative single-molecule intensity-time traces (30 ms time resolution) suggest the FRET fluctuations are inhibited if SSBf binds to a Cy5-labeled ssDNA that is shorter than the SSB tetramer binding site size ((dT)40 and (dT)51).
(I), Cross-correlation analysis of single-molecule intensity-time traces for data obtained with the shorter ssDNA in (G) and (H) (averaged over > 100 molecules each).
(J), Cross-correlation analysis of single-molecule intensity-time traces for data obtained with five Cy3–Cy5 labeled DNA constructs whose ssDNA tail lengths are equal to or shorter than the SSB tetramer binding site size (averaged over > 100 molecules each).
See also Figure S5.
To distinguish between ‘rolling’ and ‘sliding’, we performed smFRET experiments with SSBf and DNA constructs with Cy5 attached to either the end (Figure 3C, Scheme 1) or the mid-section (Figure 3D, Scheme 2) of (dT)70 ssDNA. In rolling, only the end segments of the ssDNA would display motion relative to SSB while the mid-section of bound ssDNA would not because the 70 nt ssDNA is only slightly longer than the SSB binding site. Therefore, Scheme 2 should show FRET fluctuations for sliding due to the change in the distance between Alexa555 and Cy5 (Figure 3B), but not for rolling (Figure 3A). FRET time traces for both schemes show fluctuations of similar amplitudes (Figures 3C and 3D), strongly supporting the sliding model in which the whole SSB-bound ssDNA moves relative to the SSB surface during diffusion. Plotting the mean FRET efficiency of each molecule, <E>, versus its standard deviation over time, σE, (Figure 3E) revealed no significant differences in the amplitude of FRET fluctuations between the two labelling schemes, further indicating that sliding is likely the dominant mechanism and that contributions from rolling, if any, must be much smaller. The diffusion-induced fluctuation time scales were 117 ± 3 ms for Scheme 1 and 301 ± 22 ms for Scheme 2 obtained from single-exponential fits to the cross-correlation of ID and IA (Figure 3F and Experimental Procedures). These time scales were larger than that obtained from labelled (dT)70 bound with unlabeled SSB likely due to a different degree of degeneracy in the FRET states (Figures S4 and S5).
In order to further test that the FRET fluctuations observed are due to SSB diffusion on DNA, we first obtained FRET time traces using the two labelling schemes but using (dT)40 and (dT)51, which are shorter than an SSB tetramer binding site size and therefore are not expected to allow SSB diffusion. Indeed, FRET fluctuations beyond measurement noise were eliminated (Figures 3G, 3H and S5D) and the cross-correlation of ID and IA averaged over >100 molecules showed no significant anti-correlation (Figure 3I). Next, we performed a systematic experiment using unlabeled SSB and DNA constructs with ssDNA equal to or shorter than the SSB tetramer binding site size and labelled with Cy3 and Cy5 separated by 16, 31, 41, 50 and 60 nt (Figures 3J and S5E). We did not observe any FRET fluctuations beyond measurement noise, indicating that the FRET fluctuations observed when ssDNA is longer than the binding site size are not due to conformational changes of SSB-bound ssDNA.
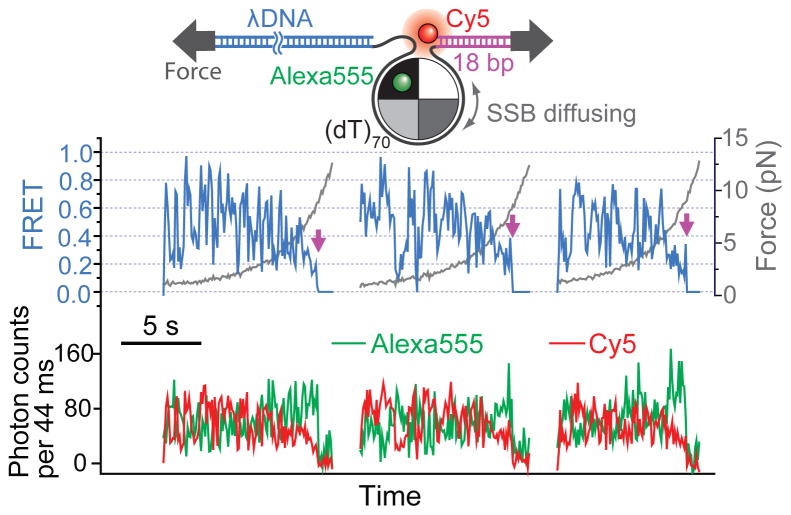
As a further test, we applied tension to SSB-bound DNA to disrupt the closed wrapping which is a prerequisite for rolling (Kozlov and Lohman, 2002; Roy et al., 2009). The diffusion-induced FRET fluctuations persisted even at forces up to ~ 5 pN (Figure 4), a force regime where the ssDNA unravelling, as measured by FRET, is essentially complete (Figure 1C). If diffusion on ssDNA indeed does not require closed wrapping, the ability to diffuse on ssDNA may be shared by other ssDNA binding proteins that do not display closed wrapping. This result also supports the sliding mechanism.
Figure 4. SSB Diffusion Persists Under Tension.
FRET trajectories of SSBf continue to show diffusion-induced fluctuations with increasing force up to ~ 5 pN. Magenta arrows indicate SSBf dissociation events.
Reptation (Sliding-With-Bulge) Mechanism for SSB diffusion
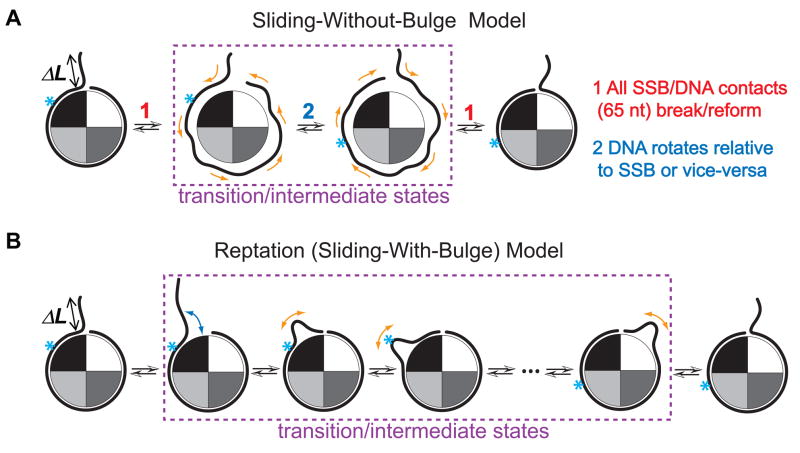
Having ruled out rolling as a dominant mechanism, how might the sliding of 65 nt of SSB-bound DNA be achieved every time SSB takes a step? There are two general classes of model with different transition or intermediate states between diffusional steps. In one, all of the contacts are broken simultaneously before the protein can slide relative to the DNA to arrive at the adjacent position (Class A, Figure 5A). In the other, only a few contacts between the protein and the DNA are broken and then reformed at a time, i.e., all contacts are broken in piecemeal (Class B). An attractive possibility for a Class B model is ‘reptation’ (Figure 5B, Movie S2) where propagation of a defect (or a loop/bulge) in a polymer chain gives rise to an overall translation of the chain (de Gennes, 1971; Perkins et al., 1994; Sukhishvili et al., 2000). A DNA bulge is formed via thermal fluctuations with an excess length ΔL, equivalent to the diffusion step size, which then propagates via a random walk: if the bulge happens to be annihilated at the position where it formed, there will be no net motion, but if it reaches the other end of the SSB-bound DNA, the protein would be repositioned by ΔL (Schiessel et al., 2001). The bulge can form spontaneously if an unwrapped DNA segment is rewrapped but with an offset of size ΔL. Because this would reduce the overall end-to-end length of the DNA tether, an applied force would make such an event less likely to occur, slowing down the reaction. Therefore, the reptation model predicts that increased tension on the DNA will slow down SSB diffusion. One would expect just the opposite for Class A models where the DNA needs to be transiently detached from the protein surface for each step of diffusion because higher forces will make it easier to achieve such a transient state (hence faster diffusion) by reducing the number of contacts between the protein and the DNA via DNA unraveling.
Figure 5. Schemes for Two Possible SSB Sliding Mechanisms.
(A and B) For the sliding mechanism, the whole bound DNA sliding would occur through different transition or intermediate states. The sliding-without-bulge model (or ‘hopping’) would require the simultaneous rupture of all of the binding interactions between ~ 65 nts of DNA and the SSB protein surface as the transition state (A). Alternatively, a sliding-with-bulge model, namely the reptation mechanism for SSB diffusion, allows the sliding of the whole bound DNA relative to SSB surface to occur little by little. As the transition states in reptation, the ssDNA at the ‘edge’ of the SSB partially dissociates from the protein surface and distortion of this unwrapped segment of DNA can form a loop-bulge with an extra length of three nucleotides, and this ‘defect in stored length’ propagates back and forth over the entire wrapped portion until it emerges on the other side, leading to one step of SSB diffusion along the ssDNA (B). The arrows represent the DNA movements and the cyan asterisk represents a single nucleotide position on the DNA. The asterisk-marked position on ssDNA will slide along the protein surface by the end of the diffusion cycle.
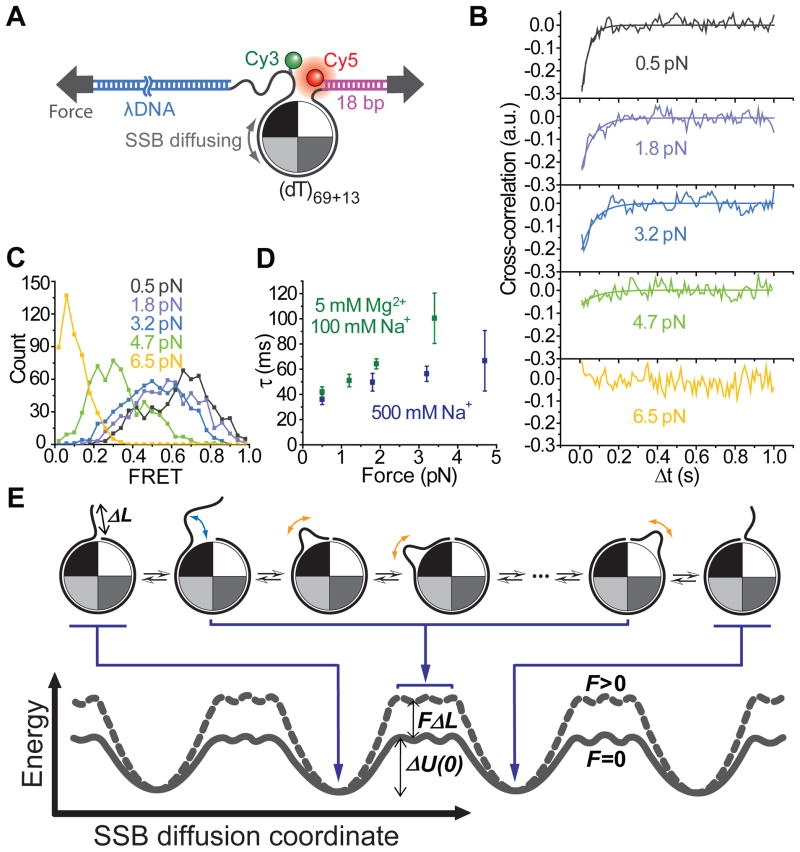
In order to probe if diffusion becomes faster or slower with increasing force, we extended the ssDNA region by 13 nt beyond the 69 nt that separate Cy3 and Cy5 (Figure 6A) and measured FRET at constant forces and 1 nM SSB. The anti-correlated fluctuations in ID and IA confirm that SSB can diffuse along the ssDNA even under tensions up to ~ 5 pN (Figures 6B and S6). The peak of the FRET histogram shifted to a value near zero at the higher (6.5 pN) force (Figure 6C) due to ssDNA unwrapping from the protein surface. Importantly, the characteristic time scale for diffusion τ calculated from the cross-correlation curve increased with force F under both ionic conditions that we have investigated (Figure 6D). The slower diffusion at higher forces favors the reptation model (‘sliding-with-bulge’), over the Class A models (‘sliding-without-bulge’) as a mechanism for sliding. In addition, the data in Figures 1 and 2 indicate that the energy cost of breaking multitudes of bonds simultaneously between ~65 nt of DNA and the protein surface is (11 + 0.13·N) kBT (N is the number of nucleotides unraveled before reaching the partially wrapped intermediate) and that an SSB tetramer remains bound to ssDNA in the absence of free SSB for several hours (Figure 1B). Therefore, it is highly unlikely that a complete/global dissociation of SSB occurs every time SSB diffuses on ssDNA by each step, further discounting Class B models in favour of the reptation model.
Figure 6. Mechanical Control of SSB Diffusion Along DNA and the Reptation (Sliding-With-Bulge) Mechanism.
(A) Experimental scheme with extended ssDNA region.
(B and C) Cross-correlations of donor and acceptor intensities and exponential decay fits in the presence of 1 nM SSB (B) and FRET efficiency histograms (C) at five different constant forces (10 ms time resolution). Fluorophores are conjugated to the 82 nt long ssDNA as shown.
(D) The characteristic time scale τ of SSB diffusion determined from the exponential fit of cross-correlation vs force obtained under two ionic conditions. Error bars are s.e.m.
(E) Energy landscape along the SSB diffusion coordinate and the proposed reptation model for SSB diffusion on DNA. ΔL is the reduction in the overall DNA length when the thermally activated DNA bulge is formed. Solid line for force F = 0; dashed line for F > 0.
See also Figure S6.
In reptation, to step from one site to the other at zero force, SSB needs to overcome an energy barrier, ΔU(0) (Figure 6E), associated with the extra curvature energy for the DNA loop-bulge formation and the adsorption energy of the protein surface and DNA (Schiessel et al., 2001). The ragged plateaus in the energy landscape represent the intermediates when a DNA loop-bulge of about 3 nt in extra length is formed, and in this model, we envision that the bulge propagates to either of the two ends rapidly and is then annihilated. The tension, F, applied to the ends of the ssDNA adds an extra mechanical energy penalty (~FΔL) to loop formation, and increases the energy barrier by the same amount, resulting in a force-dependent diffusion time scale.
Why is the force dependence of the diffusion time scale steeper in 5 mM Mg2+ and 100 mM Na+ than in 500 mM Na+? We obtained FRET histograms of naked ssDNA with Cy3 and Cy5 separated by either (dT)31 or (dT)50 (Figures S6F and S6G). We observed lower FRET values in 5 mM Mg2+ and 100 mM Na+, suggesting that ssDNA is more extended and the persistence length of ssDNA is larger due to the lower salt concentration (Murphy et al., 2004). If the minimum bulge size during reptation is limited by the persistence length of ssDNA, one would expect a larger ΔL at lower salt concentrations, resulting in a stronger force dependence as observed.
Interaction with RecO via SSB-Ct Slows SSB Diffusion on DNA
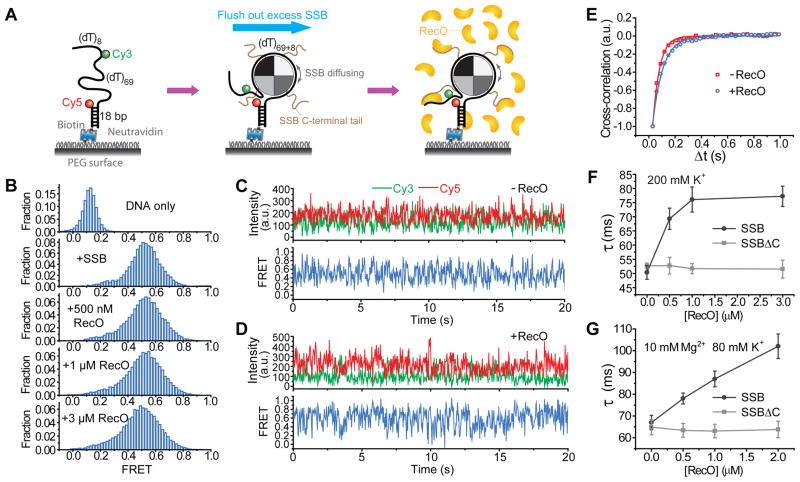
Our data thus far show that SSB diffusion on DNA is robust against moderate tension. Would SSB diffusion persist even when bound to SIPs? We examined the effect of RecO, one of the 14 SIPs in E. coli (Shereda et al., 2008). RecO promotes annealing between SSB-coated DNA strands (Kantake et al., 2002) and stimulates RecA loading onto SSB-coated ssDNA (Cox, 2007; Hobbs et al., 2007; Manfredi et al., 2008; Sakai and Cox, 2009; Umezu and Kolodner, 1994). RecO is monomeric in solution under our conditions (Experimental Procedures) and binds with 1:1 stoichiometry to each of the four SSB-Cterminal tail (SSB-Ct) but not to SSB without the Ct (Figure S7)(Hobbs et al., 2007; Ryzhikov et al., 2011). After forming a stable complex of a single SSB tetramer with (dT)69+8, we added RecO to the solution (Figure 7A). These FRET histograms (Figure 7B) differed from those observed for DNA/RecO interactions (Figure S6E). FRET fluctuations persisted even with RecO present in solution (Figures 7C and 7D), indicating that SSB diffusion occurs even with RecO bound to the DNA/SSB complex. However, cross-correlation analysis (Figure 7E) showed that RecO binding does slow SSB diffusion and the characteristic time scale of SSB diffusion increased from 50 ± 2 to 77 ± 4 ms at the highest RecO concentration tested. This effect is not merely due to RecO binding to ssDNA (Luisi-DeLuca and Kolodner, 1994; Ryzhikov et al., 2011; Sakai and Cox, 2009) because it is abolished if the SSB/RecO interaction is disrupted by deleting the last 42 amino acid residues from the SSB-Ct (Raghunathan et al., 2000) (termed SSBΔC; Figure 7F). SSBΔC or an 8 amino acid C-terminal truncation of SSB (termed SSBΔC8) displayed the same ssDNA binding features as wild type SSB under these conditions (Kozlov et al., 2010; Roy et al., 2009), but SSBΔC8 does not bind RecO (Figure S7B). Because RecO-promoted ssDNA annealing requires Mg2+(Luisi-DeLuca and Kolodner, 1994), we repeated our experiment in buffer containing 10 mM Mg2+ and 80 mM K+. Similar diffusion-induced FRET fluctuations were observed in the absence and presence of RecO (Figures S7F and S7G). Slowing of diffusion was also observed for wild type SSB but not for SSBΔC8 (Figures 7G).
Figure 7. SSB Diffusion Along ssDNA Persists but Slows Down when SSB Interacts with RecO via SSB-Ct.
(A) Schematic of reaction steps.
(B) FRET efficiency histograms for (dT)69+8 DNA only, and DNA/SSB complexes in the absence and presence of RecO.
(C) Representative single-molecule time traces of DNA/SSB in the absence of free SSB and RecO (in 200 mM K+).
(D) Representative single-molecule time traces of DNA/SSB/RecO complex in 3 μM RecO and in the absence of free SSB (in 200 mM K+).
(E), Normalized cross-correlations of donor and acceptor intensity time traces as shown in (C) and (D) averaged over more than 300 molecules each with and without 3 μM RecO. Single exponential fits are also shown.
(F) The characteristic time scale τ of SSB diffusion determined from the exponential fits of cross correlations as shown in (E) as a function of RecO concentrations for wild type SSB and SSBΔC in 200 mM K+. τ of SSB diffusion as a function of RecO concentrations for wild type SSB and SSBΔC in 10 mM Mg2+ and 80 mM K+.
See also Figure S7.
DISCUSSION
Two Stages of SSB Dissociation from DNA
Our data suggest that DNA unravels from SSB in two distinct stages (Figure 2G). Under moderate tension DNA is peeled off from SSB gradually at near equilibrium with a uniform SSB/DNA interaction energy density of 0.1–0.2 kBT per nt, followed by complete dissociation at higher tension (~10 pN) that involves a large energy barrier (11 kBT for 500 mM Na+, 8 kBT for 5 mM Mg2+ and 100 mM Na+). The partially wrapped intermediate that separates the two regions represents a state where SSB stays bound to the last short stretch of ssDNA before final dissociation. All SIPs tested so far bind SSB via the last 8–10 amino acids in the unstructured SSB-Ct (Shereda et al., 2008). After the initial binding to an SSB-Ct, the resulting high local concentration of a SIP and the unwrapping of ssDNA at moderate tension may allow progressive ssDNA transfer from SSB to the SIP while avoiding the exposure of the ssDNA region to nucleases.
Reptation as a Diffusion Mechanism
Our probing of SSB diffusion along ssDNA has provided new insights into the fundamental mechanism of the one-dimensional random walk of proteins on ssDNA. Our data ruled out ‘rolling’ as a dominant mechanism for SSB diffusion on DNA and suggest that the SSB-bound DNA would ‘slide’ all together relative to the protein surface during diffusion. Reptation may also offer an explanation for why SSB diffusion may occur with a step size larger than 1 nt (Roy et al., 2009). The minimum step size in reptation is constrained by the minimum size of the DNA bulge. Because the persistence length ranges from 1 to 3 nm between 25 mM and 2 M Na+ (Murphy et al., 2004), a bulge of the minimal size, 1 nt, could be too energetically costly to form. A reptation step size of ~3 nt may also be rationalized by the crystal structure of SSB bound by ssDNA which showed clusters of 2–4 nt in size that bind to specific sites on the protein (Raghunathan et al., 2000).
The force-dependence data on SSB diffusion (Figure 6) provides direct experimental evidence for a ‘reptation’ model of protein motion on DNA. Although we have not directly observed bulge formation and propagation, which is presumably too fast to detect, and we have not technically ruled out all other Class B models, reptation (or sliding-with-bulge) is the only model we are aware of that is consistent with all of the data reported here.
Functional Role of SSB Diffusion on DNA
The fact that SSB diffusion along ssDNA was detected with up to 5 pN of tension, even when the SSB-ssDNA structure is not fully wrapped, suggests that SSB diffusion may persist during its cellular functioning even in the crowded conditions in vivo where the DNA is likely to experience tension of various magnitudes, and that the ability to diffuse on ssDNA may be shared by other ssDNA binding proteins that do not display closed wrapping, as suggested for phage T4 gene 32 protein (Lohman, 1984). SSB appears to diffuse continually as long as there is an available extension of ssDNA beyond its binding site size. This small-scale (tens of nucleotides) SSB diffusion along DNA should be important in the redistribution of SSB on ssDNA after its initial binding to a random location because for proteins with such high affinities, redistribution would be difficult if it required complete dissociation and reassociation. SSB diffusion over short lengths would be important for protecting these small DNA gaps and allowing access of SIPs to the ssDNA and hairpin removal by SSB. In addition, single SSB tetramers can be moved by the action of a directed motion as we have shown for RecA filament formation (Roy et al., 2009). Our data also suggest RecO and other SIPs that bind to SSB via the SSB-Ct would not prevent but only moderately slow down SSB diffusion along ssDNA. The slowing of diffusion may be due to the weak interaction of RecO with ssDNA facilitated by SSB-Ct binding (Ryzhikov et al., 2011), and/or the increased radius of the SSB-RecO complex. Our data overall suggest that SSB diffusion may occur even when a SIP interacts simultaneously with both ssDNA and SSB and that SSB may serve as a dynamic platform to recruit SIPs for use in DNA replication, recombination and repair.
Implications for Nucleosomes
The closed wrapping of ssDNA around SSB bears some resemblance to the wrapping of ~ 147 bp of dsDNA around the histone core in a nucleosome (Chrysogelos and Griffith, 1982; Luger et al., 1997; Raghunathan et al., 2000). The mechanistic insights that we obtained for SSB diffusion and dissociation processes parallel those observed for nucleosomes (Beard, 1978; Brower-Toland et al., 2002; Li et al., 2005; Mihardja et al., 2006; Ranjith et al., 2007). Nucleosomes can also be repositioned along duplex DNA (Beard, 1978), likely through the spontaneous unwrapping of the DNA ends (Li et al., 2005), and RNA polymerase can rectify this thermal motion to move through chromatin (Hodges et al., 2009). A similar mechanism allows a growing RecA filament to rectify the diffusion of SSB into a directed movement (Roy et al., 2009). Two models for nucleosome sliding were proposed, based on the reptation of defects in polymer chains: through 10 bp bulge defects (Kulic and Schiessel, 2003b; Schiessel et al., 2001) and through 1 bp twist defects (Kulic and Schiessel, 2003a) but no experimental support is yet available for either. Our study provides direct experimental evidence for a reptation model of protein motion on ssDNA.
EXPERIMENTAL PROCEDURES
DNA Sequences and Annealing Procedures
Details of DNA sequences with modifications and annealing indicated in the text are reported in Extended Experimental Procedures.
Protein Purification, Characterization and Labeling
E. coli SSB, SSB-C proteins (>99% homogeneity), SSBΔC (Lohman et al., 1986), SSBΔC8 (Kozlov et al., 2010) and SSB mutant (A122C labelled with ~ one Alexa555 per SSB tetramer) (Roy et al., 2009) were purified as described. E. coli RecO protein was expressed and purified as described (Makharashvili et al., 2004; Makharashvili et al., 2009). The assembly state and stability of RecO protein was verified using sedimentation equilibrium at two concentrations (3 and 4 μM) and three rotor speeds (20, 25 and 30 thousands RPM) as described (Kumaran et al., 2006). All sedimentation profiles (not shown) obtained either under conditions of single molecule assays (Figures 7 and S7E) or ITC binding experiments (Figure S7A–C) fit well to a model for a single ideal species with molecular masses 26.9 ± 0.3 kD and 25.9 ± 0.4, respectively, similar to that expected for a RecO monomer (27.3 kD).
Fluorescence-Force Spectroscopy Instrument
The combined optical trapping and single-molecule confocal fluorescence instrument was built as previously described (Hohng et al., 2007). Briefly, the trapping laser beam (1064 nm, 350 mW, CLAS-106-STF02-02, Blue Sky Research) was coupled through the back port of the microscope, while the fluorescence excitation laser beam (532 nm, 30 mW, World StarTech) was directionally controlled by a two-dimensional piezo-controlled steering mirror (S-334K.2SL, Physik Instrument) and coupled through the right side port. The fluorescence emission was isolated from the reflected infrared light (F3: HNPF-1064.0-1.0, Kaiser) and was band-pass filtered (F1: HQ580/60m, F2: HQ680/60m, Chroma) before being imaged onto two avalanche photodiodes. The bright-field image of the trapped bead was obtained using a CCD camera (GW-902H, Genwac). Two dimensional calibration of the QPD (UDT SPOT/9DMI) over the full detector range and trap stiffness determination were performed as described (Hohng et al., 2007; Lang et al., 2002).
Sample Assembly
For fluorescence-force measurements, about 10–50 pM of the complete DNA templates were immobilized on a coverslip surface which is coated with polyethyleneglycol (mPEG-SC, Laysan Bio) in order to eliminate nonspecific surface adsorption of proteins and reduce the surface interactions with DNA and beads(Ha et al., 2002; Roy et al., 2008). The immobilization was mediated by biotin-Neutravidin binding between biotinylated DNA, Neutravidin (Pierce), and biotinylated polymer (Bio-PEG-SC, Laysan Bio). Next anti-digoxigenin-coated 1 μm polystyrene beads (Polysciences) were added so that one bead can attach to the free end of each tethered DNA. Finally, 1 nM or 20 nM of SSB protein was added in an imaging buffer containing 500 mM NaCl (or 5 mM MgCl2, 100 mM NaCl), 20 mM Tris:HCl (pH8.0), 0.1mM EDTA, 0.5mg/ml BSA(New England Biolabs), 0.01 mg/ml anti-digoxigenin, 0.5 % (wt/vol) D-glucose (Sigma), 165 U/ml glucose oxidase (Sigma), 2170 U/ml catalase (Roche), 3 mM Trolox (Sigma), and 0.1% (vol/vol) Tween 20 (Sigma).
For the diffusion measurement of SSB at zero force, the partial duplex DNA was surface immobilized as described above but the beads were not added afterwards. Instead, 1 nM of SSB was directly added with the aforementioned imaging buffer and then incubated for 1 min to form the SSB-ssDNA complexes before flushing with the same imaging buffer (but with no SSB) to remove the excess SSB from solution. All the experiments with RecO were performed with a total internal reflection (TIR) microscope described previously (Roy et al., 2008). After SSB/DNA complexes were formed and excess SSB proteins were removed, RecO was added at varying concentrations with buffer: 200 mM KCl, 0.2% DMSO (or 10 mM MgCl2, 80 mM KCl, 0.8% DMSO), 20 mM Hepes:NaOH (pH 7.5), 0.5mM TCEP, 1 mM EDTA, 0.1 mg/ml BSA, 5% (vol/vol) glycerol, 0.5 % (wt/vol) D-glucose, 165 U/ml glucose oxidase, 2170 U/ml catalase, 3 mM Trolox, and RecO at concentrations as stated.
Single-Molecule Data Acquisition
All single molecule measurements were performed at 22 ± 1°C. For fluorescence-force measurements, once a tethered bead was trapped, the coverslip was moved back and forth with the piezo-stage to roughly determine the tethered position by finding the central position of the stretching curves in two orthogonal directions in the sample plane. The origin of the piezo stage was then reset to this central position. Next a more accurate position of the fluorescently labeled molecule was determined by displacing the molecule by 13 μm from the trap center and taking a confocal image around the tethered position. For the SSB dissociation experiment, the piezo-stage was then moved back and forth between a starting position (typically 13–14 μm separation between the tethered point and the trap center) to an end position (16.5 –16.8 μm separation between the tethered point and the trap center) at a constant stage-moving speed (455 or 910 nm s−1) for several force cycles. The confocal excitation beam was programmed to follow the motion of the molecule so that in the meantime we were able to record the donor and acceptor fluorescence intensities with 44ms time resolution as the applied force ramped up. To obtain the averaged FRET vs. force curve, averaging was done over 30–50 cycles from 10–20 molecules using a force bin size of 0.2 pN. To test the force dependence of the SSB diffusion rates, the stage was sequentially moved to five different positions. At different constant forces, single-molecule fluorescence signals were collected for 6 s with 10 ms time resolution. For force-free smFRET experiments, the confocal microscope in the combined setup or a TIR microscope was used and single-molecule FRET histograms were generated by averaging for 300 ms.
FRET Efficiency Calculation
Apparent FRET efficiency was calculated from the fluorescence intensities of the donor (ID) and acceptor (IA) using the formula E = IA/(IA + ID). The background and the cross-talk between the donor and acceptor were considered as previously described (Ha et al., 2002; Hohng et al., 2007; Roy et al., 2008).
Dissociation Force Distributions
Unfolding force distributions were created by reading out the corresponding force value for the SSB dissociation event indicated by fluorescence. The two distributions obtained in 500 mM Na+ with different pulling speeds were fit to the non-equilibrium model of Dudko et al (Dudko et al., 2006).
where
r is the loading rate, k−1 is the SSB dissociation rate from the partially wrapped intermediate at zero force, Δx‡ is the distance to the transition state from the intermediate; and ΔG‡ is the height of the energy barrier between the intermediate and unwrapped state, and μ is a parameter characterizing the shape of the energy barrier. We found the fitting results to be insensitive to the absolute values of the two loading rates but sensitive to the ratio between the two. Shifting the loading rate values by 10 % (the ratio maintains at 1:2) caused a shift of less than 1% in the fitted values of the three parameters. Considering that the majority of the dissociation events happened in a short span between 5 and 13 pN and the contour length of the DNA tether is very long, the loading rates can be treated as constant to a good approximation (Dudko et al., 2008). We therefore performed a linear fit to the force-time curve in the range of 5–13 pN to determine the approximate loading rates for the two pulling speeds. We used two values of v to fit the force distributions and the fitting was performed globally between the two dissociation force distributions with three shared parameters k−1, Δx‡ and ΔG‡. We obtained k−1 = 0.010 ± 0.007 s−1, Δx‡=3.4 ± 0.7 nm., and ΔG‡ = (12 ± 3) kBT for a sharp, cusp-like energy barrier (μ = 1/2), whereas k−1 = 0.010 ± 0.006 s−1, Δx‡ = 3.2 ± 0.5 nm, and ΔG‡ = (11 ± 2) kBT for a softer, cubic potential (μ = 2/3).
Cross-correlation Analysis
The cross-correlation analysis was performed as previously described (Dijk et al., 2004; Kim et al., 2002). The calculated cross-correlation functions were calculated between donor and acceptor time traces for a given molecule. By fitting the calculated cross-correlation functions to a single exponential function, one obtains two parameters (the characteristic time of the exponential, τ, and the amplitude of the exponential at τ = 0).
Supplementary Material
Acknowledgments
We thank all the members of Ha laboratory for experimental help and discussion. These studies were supported by grants from the National Institutes of Health (TML, SK and TH) and the National Science Foundation (TH). TH is an employee of the Howard Hughes Medical Institute.
Footnotes
Supplementary Information includes Extended Experimental Procedures and seven figures and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beard P. Mobility of histones on the chromosome of simian virus 40. Cell. 1978;15:955–967. doi: 10.1016/0092-8674(78)90279-9. [DOI] [PubMed] [Google Scholar]
- Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc Natl Acad Sci U S A. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM. Regulation of bacterial RecA protein function. Crit Rev Biochem Mol Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- de Gennes PG. Reptation of a Polymer Chain in the Presence of Fixed Obstacles. The Journal of Chemical Physics. 1971;55:572–579. [Google Scholar]
- Dijk MA, Kapitein LC, Mameren J, Schmidt CF, Peterman EJ. Combining optical trapping and single-molecule fluorescence spectroscopy: enhanced photobleaching of fluorophores. J Phys Chem B. 2004;108:6479–6484. doi: 10.1021/jp049805+. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, Szabo A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys Rev Lett. 2006;96:108101. doi: 10.1103/PhysRevLett.96.108101. [DOI] [PubMed] [Google Scholar]
- Dudko OK, Hummer G, Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc Natl Acad Sci U S A. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nat Struct Mol Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- Hobbs MD, Sakai A, Cox MM. SSB protein limits RecOR binding onto single-stranded DNA. J Biol Chem. 2007;282:11058–11067. doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DM, Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc Natl Acad Sci U S A. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AG, Cox MM, Lohman TM. Regulation of Single-stranded DNA Binding by the C Termini of Escherichia coli Single-stranded DNA-binding (SSB) Protein. Journal of Biological Chemistry. 2010;285:17246–17252. doi: 10.1074/jbc.M110.118273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002;41:6032–6044. doi: 10.1021/bi020122z. [DOI] [PubMed] [Google Scholar]
- Kulic IM, Schiessel H. Chromatin dynamics: nucleosomes go mobile through twist defects. Phys Rev Lett. 2003a;91:148103. doi: 10.1103/PhysRevLett.91.148103. [DOI] [PubMed] [Google Scholar]
- Kulic IM, Schiessel H. Nucleosome repositioning via loop formation. Biophys J. 2003b;84:3197–3211. doi: 10.1016/S0006-3495(03)70044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–11973. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SV, Kozlov AG, Lohman TM, Ansari A. Microsecond dynamics of protein-DNA interactions: direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J Mol Biol. 2006;359:55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Lang MJ, Asbury CL, Shaevitz JW, Block SM. An automated two-dimensional optical force clamp for single molecule studies. Biophys J. 2002;83:491–501. doi: 10.1016/S0006-3495(02)75185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- Lohman TM. Kinetics and mechanism of dissociation of cooperatively bound T4 gene 32 protein-single-stranded nucleic acid complexes. 1. Irreversible dissociation induced by sodium chloride concentration jumps. Biochemistry. 1984;23:4656–4665. doi: 10.1021/bi00315a022. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luisi-DeLuca C, Kolodner R. Purification and characterization of the Escherichia coli RecO protein. Renaturation of complementary single-stranded DNA molecules catalyzed by the RecO protein. J Mol Biol. 1994;236:124–138. doi: 10.1006/jmbi.1994.1123. [DOI] [PubMed] [Google Scholar]
- Makharashvili N, Koroleva O, Bera S, Grandgenett DP, Korolev S. A novel structure of DNA repair protein RecO from Deinococcus radiodurans. Structure. 2004;12:1881–1889. doi: 10.1016/j.str.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Makharashvili N, Mi T, Koroleva O, Korolev S. RecR-mediated modulation of RecF dimer specificity for single- and double-stranded DNA. J Biol Chem. 2009;284:1425–1434. doi: 10.1074/jbc.M806378200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi C, Carrasco B, Ayora S, Alonso JC. Bacillus subtilis RecO nucleates RecA onto SsbA-coated single-stranded DNA. J Biol Chem. 2008;283:24837–24847. doi: 10.1074/jbc.M802002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihardja S, Spakowitz AJ, Zhang Y, Bustamante C. Effect of force on mononucleosomal dynamics. Proc Natl Acad Sci U S A. 2006;103:15871–15876. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins TT, Smith DE, Chu S. Direct observation of tube-like motion of a single polymer chain. Science. 1994;264:819–822. doi: 10.1126/science.8171335. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- Ranjith P, Yan J, Marko JF. Nucleosome hopping and sliding kinetics determined from dynamics of single chromatin fibers in Xenopus egg extracts. Proc Natl Acad Sci U S A. 2007;104:13649–13654. doi: 10.1073/pnas.0701459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DJ, Bolderson E, Khanna KK. Multiple human single-stranded DNA binding proteins function in genome maintenance: structural, biochemical and functional analysis. Crit Rev Biochem Mol Biol. 2009:1–19. doi: 10.1080/10409230902849180. [DOI] [PubMed] [Google Scholar]
- Romer R, Schomburg U, Krauss G, Maass G. Escherichia coli single-stranded DNA binding protein is mobile on DNA: 1H NMR study of its interaction with oligo- and polynucleotides. Biochemistry. 1984;23:6132–6137. doi: 10.1021/bi00320a036. [DOI] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J Mol Biol. 2007;369:1244–1257. doi: 10.1016/j.jmb.2007.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryzhikov M, Koroleva O, Postnov D, Tran A, Korolev S. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Cox MM. RecFOR and RecOR as distinct RecA loading pathways. J Biol Chem. 2009;284:3264–3272. doi: 10.1074/jbc.M807220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessel H, Widom J, Bruinsma RF, Gelbart WM. Polymer reptation and nucleosome repositioning. Phys Rev Lett. 2001;86:4414–4417. doi: 10.1103/PhysRevLett.86.4414. [DOI] [PubMed] [Google Scholar]
- Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhishvili SA, Chen Y, Muller JD, Gratton E, Schweizer KS, Granick S. Materials science. Diffusion of a polymer ‘pancake’. Nature. 2000;406:146. doi: 10.1038/35018166. [DOI] [PubMed] [Google Scholar]
- Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J Biol Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.