Abstract
Plant disease resistance proteins commonly belong to the nucleotide binding-leucine rich repeat (NB-LRR) protein family. These specialized immune proteins mediate recognition of diverse pathogen-derived effector proteins and initiate potent defense responses. NB-LRRs exhibit a multidomain architecture and each domain appears to have discrete functions depending on the stage of NB-LRR signaling. Novel proteins that were found to interact with the core HSP90 chaperone complex regulate accumulation and activation of NB-LRR immune receptors. Recent studies have also advanced our understanding of how accessory proteins contribute to NB-LRR activation. The dynamic nature of NB-LRR localization to different subcellular compartments before and after activation suggests that NB-LRRs may activate immune responses in multiple parts of the cell. In this review we highlight recent advances in understanding NB-LRR function.
INTRODUCTION
Plants use a sophisticated innate immune surveillance system to recognize pathogens (reviewed in [1]). One part of the system uses integral plasma membrane proteins with extracellular receptor domains to perceive conserved pathogen associated molecular patterns (PAMPs) that are presented by pathogens during infection. Another branch of the immune monitoring system uses primarily intracellular Resistance (R) proteins to recognize the presence of specific pathogen effector proteins in host cells. In susceptible plant genotypes, pathogen effectors promote virulence by targeting plant proteins or processes to manipulate host cell physiology to be more amenable to pathogen growth and replication [2]. Recognition of effectors by R proteins in resistant plant genotypes results in activation of effector-triggered immunity (ETI) which is often associated with programmed cell death at sites of infection, termed the hypersensitive response (HR) [3].
Most R genes cloned to date encode nucleotide-binding leucine-rich repeat (NB-LRR) proteins which mediate recognition of diverse effectors from all classes of plant pathogens. It is accepted that two general modes of effector recognition exist: (1) direct physical interaction between the immune receptor and effector and (2) indirect interaction mediated by additional plant proteins that the immune receptor associates with and monitors for effector-induced modifications. These indirect recognition “accessory” proteins may be either genuine virulence targets of the effector (the guard model) or decoy proteins that the plant has evolved to mimic bona fide effector targets (the decoy model) [1,4]. A third model for effector recognition is a hybrid of direct and indirect modes [5]. In this model, the accessory protein serves as “bait” which the effector associates with prior to direct interaction with the NB-LRR and this two-step process activates immune signaling [5]. Although R proteins were originally thought to recognize effectors in a “gene-for-gene” manner, emerging data suggest that some NB-LRRs can function in pairs, at least genetically and perhaps biochemically, to establish a full immune response (reviewed in [6]). Activation of ETI signaling results in massive reprogramming of the cell to defense responses, and therefore must be tightly regulated. Many factors control NB-LRR signaling upstream and downstream of activation. In this review we highlight the latest advances in understanding NB-LRR biology.
MAIN TEXT OF REVIEW
Multi-talented Domain Architecture
NB-LRRs belong to a subfamily of proteins within the STAND (signal transduction ATPase with numerous domains) superfamily which also contains regulators of immunity, inflammation, and apoptosis in animals [7,8]. Plant NB-LRRs contain a variable N terminus, a central nucleotide binding pocket (NB-ARC domain), and a C-terminal LRR domain. The N-terminal domain usually exhibits either a toll/interleukin-1 receptor (TIR) domain or a coiled coil domain (CC), which influences the requirement for distinct downstream signaling components [9]. TIR-NB-LRRs require Enhanced Disease Susceptibility 1 (EDS1), while most CC-NB-LRRs require Non-race specific Disease Resistance 1 (NDR1) for activation of immune responses. Furthermore, the CC domain of many NB-LRRs is required for interaction with accessory proteins [7], while the TIR domain has been implicated in accessory protein binding, effector recognition specificity, and initiation of the HR [7,10]. The NB-ARC domain mediates nucleotide binding and exchange/hydrolysis and might also serve additional downstream signaling functions [5,7]. For instance, the NB domain of the CC-NB-LRR Rx has recently been shown to be sufficient for HR induction [11]. The C-terminal LRR domain is predicted to form a curved intra- and/or inter-molecular protein interaction surface and evidence exists for positive (i.e. recognition specificity) and negative (i.e. autoinhibition) roles in signaling [5,7]. This domain likely provides a platform that keeps NB-LRRs in a primed, signaling competent state in the absence of effector stimulus, yet is labile enough to allow conformational changes upon effector-induced perturbations [5,7].
A universal paradigm for the respective roles of individual protein domains during activation of NB-LRR signaling has not been established. It is likely that single domains have multiple, discrete functions depending on stage of signaling (i.e. pre- or post- activation). Furthermore, functions of similar domains may differ depending on the context of the NB-LRR to which they belong, its interacting protein partners, and mode of effector recognition. In-depth reviews of NB-LRR domain function, molecular dynamics, and models of activation have been published recently [5,7,12,13].
Upstreams: Accumulation and Activation– New Regulators Revealed
Plant NB-LRRs require a conserved chaperone complex for proper folding, accumulation and regulation. Major components of this complex include Heat shock protein 90kDa (HSP90) and its co-chaperones Required for MLA12 Resistance 1 (RAR1) and Suppressor of the G2 allele of SKP1 (SGT1) (reviewed in [14]). Each component interacts pairwise with the other two, forming a stable complex [14,15*]. HSP90 functions as a homo-dimer and it has been proposed that RAR1 bridges the N-termini of each HSP90 monomer to regulate “open” and “closed” conformational states that regulate NB-LRR stabilization [15*,16*]. HSP90 and RAR1 generally function as positive regulators of NB-LRR accumulation [14]. By contrast, SGT1 proteins can influence NB-LRR protein levels both positively and negatively [14,17,18]. The seemingly dual functions of SGT1 and the finding that it can associate with components of the SCF (Skp1-Cullin-F-box) ubiquitin ligase complex has led to the hypothesis that SGT1 can also regulate NB-LRR protein turnover via the proteasome [18,19].
SRFR1 – another player in NB-LRR accumulation
Recent studies have identified SRFR1 (Suppressor of rps4-RLD 1) as an additional negative regulator of NB-LRR accumulation [20,21**,22**]. SRFR1 mutants in the Col-0 background exhibit constitutive defense responses mediated by overaccumulation and ectopic activation of the SNC1 (Suppressor of npr1, Constitutive 1) NB-LRR [21**,22**]. However, in srfr1/snc1 double knockout mutants, defense-associated genes are up-regulated and other NB-LRRs are detected at increased levels, suggesting that SRFR1 can also regulate defenses independent of SNC1 [21**,22**]. SRFR1 is a highly conserved eukaryotic protein containing N-terminal tetratricopeptide repeats (TPR) and a conserved C-terminal domain of unknown function [20]. Interestingly, the SRFR1 TPR domain interacts with the TPR domains of SGT1a and SGT1b [22**]. Similar to srfr1 mutants, increased amounts of several NB-LRR proteins were observed in sgt1b mutants [18,22**], suggesting that both proteins may share related function(s). SRFR1 can associate in complex with the SNC1 and/or RPS4 NB-LRRs [21**]. Presumably, SGT1 also resides in this complex.
At what level(s) of NB-LRR accumulation does SRFR1 operate? Due to their mutual interaction and related phenotypes, it is possible that SRFR1 functions with SGT1 in proteasome-mediated NB-LRR turnover [22**]. Thus, it will be important to establish whether SRFR1 interaction with SGT1 is necessary for its regulation of NB-LRR accumulation. Notably, both SGT1 and SRFR1 localize to the cytoplasm and nucleus [20,21**,23]. SRFR1 exhibits similarity with eukaryotic transcriptional repressors and has been hypothesized to function as a negative regulator of defense gene expression [20,21**]. Enhanced defense-associated gene expression in the srfr1/snc1 double knockout might also be explained by weak activation of additional over-accumulating NB-LRRs. It will be interesting to determine if SRFR1 protein levels change upon NB-LRR activation. SRFR1 degradation might cause deregulation of additional NB-LRRs (e.g. SNC1) that could result in an amplification of immune responses.
CRT1 – an early component of NB-LRR protein activation
CRT1 (Compromised Recognition of TCV), a member of the GHKL (Gyrase, Hsp90, histidine kinase, MutL) ATPase/kinase superfamily, was originally identified in a genetic screen as being required for HRT-mediated defense against turnip crinkle virus [24]. However, CRT1 function is not limited to the HRT NB-LRR. Genetic analyses demonstrate that CRT1 and its close homologs are required for immune responses mediated by both TIR-NB-LRRs and CC-NB-LRRs. Furthermore, CRT1 can bind the NB domains of multiple NB-LRRs, indicating that CRT1 may generally facilitate R protein function [24,25*]. CRT1 also interacts with HSP90, but not RAR1 or SGT1. Unlike HSP90 and its co-chaperones (and SRFR1 above), CRT1 does not affect NB-LRR steady-state levels [25*]. Interestingly, CRT1 cannot be co-immunoprecipitated with activated forms of several NB-LRRs. This finding suggests that CRT1 likely functions early in NB-LRR folding and/or activation and then dissociates from actively signaling NB-LRRs [25*]. As CRT1 localizes to endosomal compartments in the cytoplasm [25*], it remains to be determined how it might regulate diverse NB-LRRs with different localization patterns.
Midstream: The Recognition Event
Effector recognition is hypothesized to induce conformational changes in NB-LRRs, releasing inhibition, and freeing the NB-LRR to activate downstream signaling [5,7,12,13]. At least two general strategies for effector recognition by NB-LRRs exist. Direct physical interactions between the fungal effectors AvrPita, AvrL567, and AvrM with the corresponding NB-LRRs Pi-ta, L, and M, respectively, are likely to occur, and in these cases activation of ETI correlates absolutely with effector/NB-LRR interactions in yeast two-hybrid assays [26–28]. Likewise, co-immunoprecipitation experiments in planta between alleles of Arabidopsis RPP1 and its cognate Hyaloperonospora arabidopsidis effector ATR1 demonstrate that (1) association of the RPP1 LRR with ATR1 is necessary for activation of ETI and (2) mutations in the TIR or NB domains abolish ETI but not ATR1 binding [29*]. These observations are consistent with the idea that during direct recognition events, the LRR confers recognition specificity, while the TIR and NB domains function in activation and downstream signaling [5,7,29*]. In contrast to the above examples of direct binding to eukaryotic effectors, recognition of most bacterial effectors is mediated indirectly through an accessory host protein that interacts with both the effector and NB-LRR. It remains to be established how NB-LRRs function in combination with their accessory proteins to establish a functional immune recognition complex and whether this complex changes dynamically during recognition events.
Indirect Recognition - RIPK modifies RIN4 to trigger RPM1 activation
Immune responses regulated by the conserved plant protein RIN4 have been instrumental in understanding how NB-LRRs indirectly perceive effectors. Arabidopsis RIN4 is targeted by multiple bacterial effectors (AvrRpt2, AvrRpm1, AvrB, and HopF2) and is monitored for effector-induced modification by at least two NB-LRRs (RPS2 and RPM1) [30–33]. AvrRpt2 is a protease that directly cleaves RIN4, a modification which activates RPS2-ETI [31,32,34]. HopF2 is an ADP-ribosyltransferase that can modify RIN4 and RIN4 is required for HopF2 virulence activity [33,35]. Although AvrB and AvrRpm1 exhibit no sequence homology to known kinases, both effectors associate with and induce phosphorylation of RIN4 [30]. It is hypothesized that RPM1 recognizes RIN4 phosphorylation and initiates ETI. However, the kinase(s) involved remained elusive and direct evidence for this model has been lacking.
Recent studies confirm that effector-induced phosphorylation of specific residues of RIN4 can activate the RPM1 NB-LRR [36**,37**]. Phosphomimetic mutations demonstrate that phosphorylation of a single amino acid residue, RIN4 T166, is sufficient to activate RPM1-mediated HR when expressed in N. benthamiana and Arabidopsis [36**]. Phosphorylation of T166 increases when AvrB and AvrRpm1 are delivered into plant cells by bacteria [36**,37**]. Furthermore, Arabidopsis RIPK (RPM1-induced protein kinase) interacts with and phosphorylates RIN4 at multiple residues (T21, S160, and T166) [37**]. RIPK also interacts with AvrB and ripk knockout plants exhibit reduced AvrB-induced RIN4 phosphorylation and defects in RPM1-mediated immune responses, illustrating the importance of RIPK in RPM1-mediated immune responses [37**]. RIPK is a member of the large receptor-like cytoplasmic kinase (RLCK) family and RPM1-mediated ETI is not completely abolished in the ripk mutant. It is likely that other related kinases can also interact with AvrB to induce RIN4 phosphorylation and contribute to RPM1-ETI. Taken together, these data support a model in which AvrB induces RIN4 phosphorylation via host kinases and modification of a specific RIN4 residue activates RPM1 (Fig 1). Despite the attempts of multiple laboratories, no kinase activity has been detected for AvrB. However, it is also possible that AvrB itself mimics host kinase activity to phosphorylate RIN4, or AvrB may work with host kinases to induce target protein phosphorylation.
Figure 1.
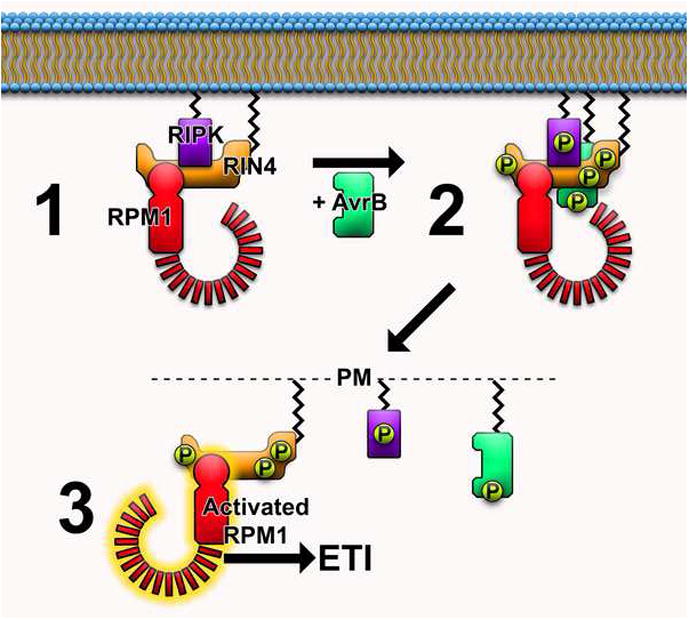
Model for RPM1 activation. 1) At steady state, RIPK, RIN4, and RPM1 form a complex. Additional proteins (not shown) may associate with this complex. 2) When delivered into the cell, AvrB associates with both RIN4 and RIPK, inducing RIPK-mediated phosphorylation of both AvrB and RIN4. RIN4 is phosphorylated at amino acid residues T21, S160, and T166. 3) RIN4 phosphorylation alters interaction dynamics of complex members. AvrB and RIPK dissociate from phosphorylated RIN4. RPM1 perceives RIN4 phosphorylation at amino acid residue T166 and initiates signal cascades culminating in effector triggered immunity.
Downstreams: Events Post NB-LRR Activation
NB-LRRs in the nucleus
A clear model of the signaling events that link NB-LRR activation to downstream immune responses remains elusive. Several recent studies suggest that nuclear activity of some NB-LRRs is necessary to trigger defense responses (Table 1). The CC-NB-LRR MLA10 (barley) and TIR-NB-LRRs RRS1-R (Arabidopsis), N (tobacco), RPS4 (Arabidopsis), and SNC1 (Arabidopsis) require nuclear localization and accumulation for full activation of immunity [38–42] (Table 1). Genetic screens looking for modifiers of the snc1-1 autoactive NB-LRR allele (mos mutants) have revealed several nuclear processes that are important for NB-LRR-mediated immune responses including nuclear protein export and import, mRNA export, and regulation of transcription [38,43,44]. Moreover, the presence of a WRKY DNA-binding domain in RRS1-R and the association of MLA10 and N with plant WRKY and SPL transcription factors, respectively, have led to the hypothesis that NB-LRRs have a direct role in regulating immune related transcriptional changes [12]. This idea is supported by the demonstration that upon recognition of the effector AvrA10, MLA10 associates with WRKY transcription factors to relieve repression of defense genes [39]. Thus, a major recent focus of R protein study has been to understand the specific functions of other nuclear-localized NB-LRRs.
Table 1.
Importance of nucleocytoplasmic partitioning in NB-LRR function
| NB-LRR (Localization) | Effector (Localization) | Subcellular compartment required for defense | Interactors (Localization) | References |
|---|---|---|---|---|
| RPS4 (C, N) | AvrRps4 (C) | N | Unknown | [40] |
| N (C, N) | p50 helicase (C, N) | N (HR) | NRIP1 (CP, C, N) | [42,51] |
| SNC1 (C, N) | N/A | N | TPR1 (N) | [38,45*] |
| RRS1-R (N) | PopP2 (N) | N/A | PopP2 (N) | [41] |
| MLA10 (C, N) | AvrA10 (N/A) | N | WRKY1 and WRKY2 (N) | [39] |
| Rx (C, N) | PVX coat protien (C, N) | C + N | RanGAP1 and RanGAP2 (C) | [48**,49**] |
Abbreviations: N=nucleus, C=cytoplasm, CP=chloroplast, N/A=data not available, HR=hypersensitive response
SNC1 interacts with transcriptional regulators to initiate defenses
Recently, SNC1 was found to associate with the transcriptional corepressor TPR1 (Topless Related 1) to control gene expression during immune responses. Null mutations in TPR1 partially suppress the snc1-1 autoactive immune phenotype [45*]. The TIR domain of SNC1 interacts directly with TPR1 to repress expression of Defense no Death 1 (DND1) and Defense no Death 2 (DND2), two cyclic nucleotide-gated ion channels that are known negative regulators of plant immunity [45*–47]. Genetic analysis demonstrated that TPR1 and its close homologs are required for defenses mediated by other TIR-NB-LRRs, but not immunity controlled by the CC-NB-LRR RPS2 [45*]. These findings suggest that some TIR-NB-LRRs interact with transcriptional co-repressors to inhibit the expression of negative immune regulators to trigger defense responses. As expression of only the TIR domain from several NB-LRRs can elicit the HR [10], it will be important to determine if interaction with TPR-like proteins is required for this response.
Importance of nucleocytoplasmic distribution of immune regulators: the case of Rx
Emerging evidence is revealing that NB-LRRs can activate defense mechanisms in multiple subcellular compartments. The CC-NB-LRR Rx, which recognizes the coat protein of Potato virus X (PVX), localizes to the cytoplasm and nucleus and both pools contribute to full immunity [48**,49**] (Table 1). Experiments using either a nuclear export signal or a nuclear localization signal fused to Rx, the PVX coat protein, or RanGAP2 (a Rx cytoplasmic retention factor necessary for Rx function) demonstrate that the PVX coat protein activates Rx in the cytoplasm, and forced nuclear hyperaccumulation of Rx suppresses immune responses [48**,49**]. However, nuclear export signal-mediated expulsion of Rx from the nucleus moderately reduced resistance, indicating that the nuclear pool of Rx also functions in immunity [48**]. This is the first demonstration of both nuclear and cytoplasmic pools of a NB-LRR being required for resistance.
Intriguingly, stabilization of Rx in the cytoplasm by over-expressing RanGAP2 actually increased resistance [49**], suggesting that cytoplasmic Rx is predominately responsible for limiting PVX replication. The cytoplasmic pool of Rx likely activates several immune signaling cascades. These could stimulate antiviral mechanisms in the cytoplasm such as translational inhibition of viral RNAs [50]. The nuclear pool of Rx could serve additional functions such as activation of defense gene expression. It is certainly conceivable that cytoplasmic Rx activates signaling cascades that are transmitted to the nucleus, and nuclear Rx contributes redundantly to these immune signals. The likelihood of Rx activating cytoplasm-localized antiviral defenses also raises questions regarding commonalities in resistance mechanisms downstream of NB-LRR activation. Thus, it appears possible that NB-LRRs conferring resistance to different pathogen types may utilize fundamentally different mechanisms. The nature of NB-LRR-mediated cytoplasmic and nuclear immune signaling and their relative importance is not clear. Future experiments addressing the importance of these findings in Rx and other NB-LRRs will be an important area of research.
CONCLUSIONS
Plant NB-LRR R proteins are highly diverse in terms of pathogen proteins recognized and interacting plant protein partners. NB-LRRs are subject to multiple levels of regulation in order to prevent erroneous activation of potent and metabolically costly immune responses. Newly discovered proteins that regulate NB-LRR accumulation and activation appear to associate with the core HSP90-SGT1-RAR1 chaperone complex. Knowledge of how these components are integrated into the complex may lead to the elucidation of specific biochemical mechanisms of NB-LRR stabilization, activation, and turnover. Furthermore, identification of all components of these macromolecular complexes in planta coupled to structural analyses will be required to gain a firm understanding of how these critical protein complexes regulate plant immunity.
Many NB-LRRs exhibit complex patterns of subcellular location pre- and post- activation. Recent studies demonstrating contributions of both cytoplasmic and nuclear localizations of the Rx NB-LRR to immune signaling suggests that a subset of NB-LRRs can activate defenses in multiple subcellular compartments. It is plausible that cytoplasmic defense signaling could be sufficient for effective immunity in certain situations, and that NB-LRR nuclear activity is a way for the plant to “hedge its bet” against some pathogens. Being able to measure the exact nature of NB-LRR signaling in the cytoplasm and nucleus is the first step to understanding the relative contribution of each to disease resistance. The complexity of NB-LRR function and associated signaling can only by matched by their importance in plant-microbe interactions.
Acknowledgments
We thank members of the Coaker laboratory for critical reading of the manuscript. GC is supported by a grant from NIH (1R01GM092772-01) and USDA (2010-65108-20527). JME is supported by an NSF IGERT graduate research training grant (DGE-0653984). We apologize to our colleagues whose work we were unable to cite due to page limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 2.Cui H, Xiang T, Zhou JM. Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol. 2009;11:1453–1461. doi: 10.1111/j.1462-5822.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- 3.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 4.van der Hoorn RAL, Kamoun S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier SM, Moffett P. NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 2009;14:521–529. doi: 10.1016/j.tplants.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukasik E, Takken FL. STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol. 2009;12:427–436. doi: 10.1016/j.pbi.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Ye Z, Ting JP-Y. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr Opin in Immun. 2008;20:3–9. doi: 10.1016/j.coi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swiderski MR, Birker D, Jones JDG. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. MPMI. 2009;22:157–165. doi: 10.1094/MPMI-22-2-0157. [DOI] [PubMed] [Google Scholar]
- 11.Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P. The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell. 2008;20:739–751. doi: 10.1105/tpc.107.056036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caplan J, Padmanabhan M, Dinesh-Kumar SP. Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe. 2008;3:126–135. doi: 10.1016/j.chom.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Takken FL, Tameling WI. To nibble at plant resistance proteins. Science. 2009;324:744–746. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- 14.Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- *15.Zhang M, Kadota Y, Prodromou C, Shirasu K, Pearl LH. Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: implications for chaperoning of NLR innate immunity receptors. Mol Cell. 2010;39:269–281. doi: 10.1016/j.molcel.2010.05.010. This paper describes the crystal structure of the interacting domains of the HSP90(N domain)-SGT1(CS domain)-RAR1(CHORDII domain) chaperone complex. The authors present in vitro interaction data supporting the proposition that the two CHORD domains of a single RAR1 molecule bridge the N-termini of the HSP90 monomers, thus regulating the “open” and “closed” state of the HSP90 dimer that coordinates NB-LRR stabilization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Hubert DA, He Y, McNulty BC, Tornero P, Dangl JL. Specific Arabidopsis HSP90.2 alleles recapitulate RAR1 cochaperone function in plant NB-LRR disease resistance protein regulation. Proc Natl Acad Sci USA. 2009;106:9556–9563. doi: 10.1073/pnas.0904877106. Genetic suppressor screens in the rar1 background identify HSP90.2 alleles that bypass the requirement for RAR1 interaction in regulating NB-LRR accumulation and function. The authors propose that RAR1 directly regulates the open and closed state of the HSP90 dimer, thereby influencing NB-LRR stabilization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noel L, Sadanandom A, Casais C, Parker J, Shirasu K. Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J. 2006;25:2007–2016. doi: 10.1038/sj.emboj.7601084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt BF, 3rd, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 19.Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 20.Kwon SI, Kim SH, Bhattacharjee S, Noh JJ, Gassmann W. SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J. 2009;57:109–119. doi: 10.1111/j.1365-313X.2008.03669.x. [DOI] [PubMed] [Google Scholar]
- **21.Kim SH, Gao F, Bhattacharjee S, Adiasor JA, Nam JC, Gassmann W. The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 2010;6:e1001172. doi: 10.1371/journal.ppat.1001172. Together with Li, et al., SRFR1 is identifed as a negative regulator of SNC1 accumulation and activation. SRFR1 also regulates NB-LRR defenses independent of SNC1, indicating that it is a general factor controlling plant defense. Furthemore, SRFR1 can associate in complex with the NB-LRRs RPS4 and SNC1. Intriguingly, the authors show that SNC1 contributes to immunity in response to AvrRps4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **22.Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y. SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 2010;6:e1001111. doi: 10.1371/journal.ppat.1001111. Together with Kim, et al., this study identifes SRFR1 as a negative regulator of SNC1-mediated immune accumulation and activation. The authors show that the TPR domains of SRFR1 interact with TPR domains of SGT1 proteins. Similar to SGT1, SRFR1 appears to negatively regulate the accumulation of multiple NB-LRR proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noel LD, Cagna G, Stuttmann J, Wirthmuller L, Betsuyaku S, Witte C-P, Bhat R, Pochon N, Colby T, Parker JE. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell. 2007;19:4061–4076. doi: 10.1105/tpc.107.051896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang HG, Kuhl JC, Kachroo P, Klessig DF. CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to turnip crinkle virus. Cell Host Microbe. 2008;3:48–57. doi: 10.1016/j.chom.2007.11.006. [DOI] [PubMed] [Google Scholar]
- *25.Kang HG, Oh CS, Sato M, Katagiri F, Glazebrook J, Takahashi H, Kachroo P, Martin GB, Klessig DF. Endosome-associated CRT1 functions early in resistance gene-mediated defense signaling in Arabidopsis and tobacco. Plant Cell. 2010;22:918–936. doi: 10.1105/tpc.109.071662. This study identifies CRT1 and its homologs as being required for immune responses mediated by diverse NB-LRRs. CRT1 associates with HSP90 and the NB domain of NB-LRRS and appears to regulate an early step of NB-LRR maturation and/or activation. This hypothesis is supported by the observation that CRT1 does not associate with activated NB-LRRs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catanzariti A-M, Dodds PN, Ve T, Kobe B, Ellis JG, Staskawicz BJ. The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. MPMI. 2009;23:49–57. doi: 10.1094/MPMI-23-1-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. Embo J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:2444–2458. doi: 10.1105/tpc.110.075358. This recent study characterizes an example of likely direct recognition between NB-LRR and an oomycete effector. Mutational analysis separates effector binding to the LRR from downstream signaling mediated by the TIR and NB-ARC domains, indicating that LRR-effector association is a necessary first step of NB-LRR activation in this case. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 31.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 32.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 33.Wilton M, Subramaniam R, Elmore J, Felsensteiner C, Coaker G, Desveaux D. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc Natl Acad Sci USA. 2010;107:2349–2354. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coaker G, Falick A, Staskawicz B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science. 2005;308:548–550. doi: 10.1126/science.1108633. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou J-M. A Pseudomonas syringae ADP-Ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell. 2010;22:2033–2044. doi: 10.1105/tpc.110.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **36.Chung E-H, da Cunha L, Wu A-J, Gao Z, Cherkis K, Afzal A, Mackey D, Dangl J. Specific threonine phosphorylation of a host target mediated by two unrelated type III effector proteins results in activation of a host innate immune receptor. Cell Host Microbe. 2011;9:125–136. doi: 10.1016/j.chom.2011.01.009. In-depth mutational analysis identified phosphorylation of RIN4 T166 as being essential for RPM1 activation. The T166 residue is absolutely critical for AvrB- mediated RPM1 activation and contributes to AvrRpm1-induced RPM1 activation. These findings suggest that while AvrB and AvrRpm1 induce phosphorylation of this residue, additional factors might be involved during AvrRpm1-triggered RPM1 activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **37.Liu J, Elmore JM, Lin ZD, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe. 2011;9:137–146. doi: 10.1016/j.chom.2011.01.010. This study used an affinity purification strategy of phosphoylated RIN4 to identify the kinase RIPK as an important component of RPM1-mediated immunity. RIPK phosphorylates RIN4 at multiple residues (T21, S160, T166) which results in RPM1 activation. Along with Chung, et al., phosphorylation of T166 was identified as upregulated after delivery of AvrB and AvrRpm1 and important for RPM1 activation. Thus, RPM1 monitors phosphorylation of its accessory protein RIN4 and AvrB-induced phosphorylation of RIN4 by a plant kinase intiates ETI signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng YT, Germain H, Wiermer M, Bi D, Xu F, Garcia AV, Wirthmueller L, Despres C, Parker JE, Zhang Y, et al. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell. 2009;21:2503–2516. doi: 10.1105/tpc.108.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- 40.Wirthmueller L, Zhang Y, Jones JD, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 41.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiermer M, Palma K, Zhang Y, Li X. Should i stay or should i go? Nucleocytoplasmic trafficking in plant innate immunity. Cell Microbiol. 2007;9:1880–1890. doi: 10.1111/j.1462-5822.2007.00962.x. [DOI] [PubMed] [Google Scholar]
- 44.Germain H, Qu N, Cheng YT, Lee E, Huang Y, Dong OX, Gannon P, Huang S, Ding P, Li Y, et al. MOS11: a new component in the mRNA export pathway. PLoS Genet. 2010;6:e1001250. doi: 10.1371/journal.pgen.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci USA. 2010;107:13960–13965. doi: 10.1073/pnas.1002828107. This paper describes the second published example (the first being Mla10) of a NB-LRR interacting directly with a transcriptional regulator to induce defense responses. SNC1 associates with TPR1 to repress expression of known negative regulators of immune signaling. TPR1 and its homologs are required for defense induced by TIR-NB-LRRs, but not CC-NB-LRRs. Furthermore, the TIR domain of SNC1 interacts directly with TPR1, suggesting that other TIR-NB-LRRs might signal via a similar mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RK, Jr, Bent AF. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA. 2000;97:9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jurkowski GI, Smith RK, Yu Ic, Ham JH, Sharma SB, Klessig DF, Fengler KA, Bent AF. Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. MPMI. 2004;17:511–520. doi: 10.1094/MPMI.2004.17.5.511. [DOI] [PubMed] [Google Scholar]
- **48.Slootweg E, Roosien J, Spiridon LN, Petrescu A-J, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell. 2010;22:4195–4215. doi: 10.1105/tpc.110.077537. The authors show that NB-LRR Rx1 is localized to both the nucleus and cytoplasm. Additionally, both nuclear and cytoplasmic pools of NB-LRR Rx1 are necessary for full immune responses to Potato virus X. Potato virus X coat protein activates Rx in the cytoplasm. This is the first demonstration of both nuclear and cytoplasmic pools of a NB-LRR being required for resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **49.Tameling WIL, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MHAJ. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell. 2010;22:4176–4194. doi: 10.1105/tpc.110.077461. This study shows that RanGAP2 functions to inhibit Rx1 nuclear localization and stabilizes the cytoplasmic Rx1 pool. Forced nuclear hyperaccumulation of Rx1 is demonstrated to greatly compromise resistance. Co-expression of Rx1 with RanGAP2 is shown to enhance defense signaling and resistance, suggesting that cytoplasmic Rx is predominately responsible for limiting PVX replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhattacharjee S, Zamora A, Azhar MT, Sacco MA, Lambert LH, Moffett P. Virus resistance induced by NB LRR proteins involves Argonaute4-dependent translational control. Plant J. 2009;58:940–951. doi: 10.1111/j.1365-313X.2009.03832.x. [DOI] [PubMed] [Google Scholar]
- 51.Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell. 2008;132:449–462. doi: 10.1016/j.cell.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]


