Abstract
A marine Verrucosispora sp. isolated from the sponge Chondrilla caribensis f. caribensis was found to produce thiocoraline, a potent cytotoxic compound. Five new analogs of thiocoraline were isolated and represent the first analogs of thiocoraline. 22'-Deoxythiocoraline (2), thiochondrilline C (5), and 12'-sulfoxythiocoraline (6) demonstrated significant cytotoxicity against the A549 human cancer cell line with EC50 values of 0.13, 2.86, and 1.26 µM, respectively. The analogs provide insight into the SAR and biosynthesis of thiocoraline. The DP4 probability method was used to analyze ab initio NMR calculations to confirm stereochemical assignments.
INTRODUCTION
Natural products isolated from terrestrial bacteria have historically contributed to the development of therapeutics.1 However, the high rate of rediscovery2 (99.5%) from terrestrial bacteria has necessitated a change in focus for drug discovery sources. The marine environment, which harbors over twenty million microbes,3 has provided several microbial-derived compounds, such as salinosporamide A,4 TZT-1027,5 and ILX-651,6 that are currently in clinical trials.7 Among the list of microbial-derived marine natural products with therapeutic relevance is thiocoraline (1), a potential candidate for clinical trials.8 First isolated in 1997 from the mycelia of Micromonospora marina,9,10 thiocoraline, a bisintercalator, has shown potent cytotoxicity in lung, breast, colon, renal, and melanoma cancer cells,10–12 and in vivo efficacy against human carcinoma xenografts.8 As a result of the in vivo efficacy, thiocoraline has been the subject of several synthetic13–17 and biosynthetic18–22 studies. The 3-OH-quinaldic system, which has been proposed to stabilize the complex with DNA,12 provides thiocoraline with a unique mechanism of action and sequence specificity over other bisintercalators, such as echinomycin and triostin A, which contain a quinoxaline ring system.23 While bisintercalators containing the quinoxaline ring result in DNA damage and inhibition of topoisomerase II, thiocoraline inhibits DNA elongation by DNA polymerase α.11 The synthesis of several thiocoraline analogs,14–17 has provided insight into the SAR of thiocoraline. Synthetic analogs from the Boger group14,15 demonstrated that the 3-OH-quinaldic system is a key contributor to the bioactivity of thiocoraline, and the synthesis of N-Me-azathiocoraline17 demonstrated an increase in potency over thiocoraline.
We report the first isolation of thiocoraline analogs. Five analogs, including three monomers we named the thiochondrillines, were isolated from a marine Verrucosispora sp. (Strain WMMA107), cultivated from the sponge Chondrilla caribensis f. caribensis (Rützler, Duran & Piantoni, 2007; order Chondrosida, family Chondrillidae). Verrucosispora is a gram-positive, spore-forming actinomycete genus,24 and only three classes of compounds, the antibiotic abyssomicins,25 proximicins,26 and gifhornenolones27 are known to be produced by Verrucosispora. Several Verrucosispora spp. have previously been cultivated from sponges.28,29 No natural products have been previously reported from the sponge Chondrilla caribensis f. caribensis, but natural products have been reported from other Chondrilla spp.30,31 We report the first isolation of thiocoraline from a Verrucosispora sp. Spectroscopic methods, supported by mass spectrometry and molecular modeling, led to the elucidation of the structures. Cytotoxicity against the A549 human cancer cell line was determined for each compound. The new analogs provide insight into the biosynthesis of thiocoraline and a better understanding of the SAR of thiocoraline (1).
RESULTS AND DISCUSSION
Extracts from five Verrucosispora strains (WMMA102, WMMA105, WMMA107, WMMA110, and WMMA111) were screened in an antibacterial disc diffusion assay, and despite antibacterial activity from the extracts, no abyssomicins, proximicins, or gifhornenolones were identified. The source of the antibacterial activity was identified as thiocoraline (1), produced by strain WMMA107. Five analogs of thiocoraline were also isolated; NMR, MS, and molecular modeling provided the necessary data for determining the structures.
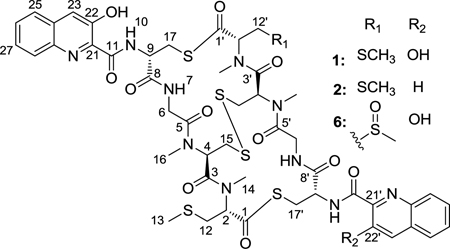
HRMS supported the molecular formula of C48H56O11N10S6 for 22'-deoxythiocoraline (2). Integration of the 1H spectrum and coupling between H-22' and H-23' (Table 1) indicated a difference of one phenol group. HMBC and COSY data confirmed that the rest of the aromatic ring moiety remained the same as thiocoraline (1). 22'-Deoxythiocoraline (2) is assumed to have the same biosynthetic machinery as thiocoraline (1) and consequently, will have the same absolute configuration as thiocoraline (1). Therefore, the absolute configuration of 22'-deoxythiocoraline (2) was assumed to be the same as thiocoraline (1) on the basis of biosynthetic precedent,18 as well as a comparison of NMR shifts of thiocoraline.9
TABLE 1.
1H and 13C NMR data for 2, 6 (600 MHz, CDCl3)
| 2 | 6 | |||
|---|---|---|---|---|
| Position | δC, mult. | δH (J in Hz) | δC, mult. | δH (J in Hz) |
| 1 | 199.6, C | 199.7, C | ||
| 2 | 61.0, CH | 5.79, dd (3.7, 11.5) | 61.1, CH | 5.78, dd (11.4, 4.0) |
| 3 | 170.1, C | 170.4, C | ||
| 4 | 56.6, CH | 6.39, m (6.0) | 56.0, CH | 6.41, br t (6.0) |
| 5 | 168.3, C | 168.9, C | ||
| 6a | 40.3, CH2 | 3.62, m | 40.5, CH2 | 3.63, m (3.2) |
| 6b | 4.59, m (8.0) | 4.55, m (6.0) | ||
| 7-NH | 6.77, m (10.5) | 6.76, m | ||
| 8 | 169.5, C | 169.5, C | ||
| 9 | 54.2, CH | 4.89, m | 54.0, CH | 4.91, m |
| 10-NH | 8.80, d (5.5) | 8.80, d (6.3) | ||
| 11 | 169.1, C | 169.4, C | ||
| 12a | 32.1, CH2 | 2.84, m | 32.1, CH2 | 2.82, m |
| 12b | 3.23, m (4.1) | 3.18, m | ||
| 13 | 15.1, CH3 | 2.12, s | 15.3, CH3 | 2.11, s |
| 14 | 30.7, CH3 | 3.04, s | 31.0, CH3 | 3.07, s |
| 15a | 41.6, CH2 | 2.79, m | 40.6, CH2 | 2.88, m |
| 15b | 3.54, m | 3.42, m | ||
| 16 | 30.6, CH3 | 2.99, s | 30.8, CH3 | 2.96, s |
| 17a | 30.2, CH2 | 3.50, m | 30.6, CH2 | 3.50, dd (3.1, 14.4) |
| 17b | 3.73, m (5.4) | 3.68, m (2.8) | ||
| 21 | 133.6, C | 134.0, C | ||
| 22 | 153.5, C | 153.9, C | ||
| 22-OH | 11.26, s | 11.27, s | ||
| 23 | 120.5, CH | 7.59, s | 121.2, CH | 7.60, s |
| 24 | 132.1, C | 132.4, C | ||
| 25 | 128.6, CH | 7.74, m | 131.1, CH | 7.76, m |
| 26 | 127.7, CH | 7.48, m | 128.1, CH | 7.47, m |
| 27 | 129.7, CH | 7.47, m | 129.1, CH | 7.46, m |
| 28 | 126.2, CH | 7.65, m (7.7) | 126.9,CH | 7.67, m |
| 29 | 141.3, C | 141.5, C | ||
| 1' | 199.6, C | 198.7, C | ||
| 2' | 61.0, CH | 5.80, dd (3.7, 11.5) | 60.1, CH | 6.06, dd (3.2, 11.5) |
| 3' | 169.8, C | 170.3, C | ||
| 4' | 56.6, CH | 6.40, m (6.0) | 56.0, CH | 6.41, br t (6.0) |
| 5' | 168.3, C | 168.6, C | ||
| 6a' | 40.3, CH2 | 3.58, m | 40.5, CH2 | 3.65, m (3.2) |
| 6b' | 4.62, m (8.0) | 4.58, m (6.0) | ||
| 7'-NH | 6.77, m (10.5) | 6.76, m | ||
| 8' | 169.5, C | 169.7, C | ||
| 9' | 54.3, CH | 4.95, m | 53.6, CH | 4.98, m |
| 10'-NH | 8.66, d (5.5) | 8.76, d (6.3) | ||
| 11' | 170.1, C | 169.4, C | ||
| 12a' | 32.1, CH2 | 2.84, m | 53.4, CH2 | 3.15, m |
| 12b' | 3.23, m (4.1) | 3.33, dd (3.7, 13.8) | ||
| 13' | 15.1, CH3 | 2.12, s | 40.2, CH3 | 2.64, s |
| 14' | 30.7, CH3 | 3.09, s | 32.1, CH3 | 3.21, s |
| 15a' | 41.6, CH2 | 2.79, m | 40.6, CH2 | 2.90, m |
| 15b' | 3.54, m | 3.46, m | ||
| 16' | 30.6, CH3 | 2.99, s | 30.8, CH3 | 3.00, s |
| 17a' | 30.2, CH2 | 3.50, m | 30.6, CH2 | 3.55, dd (3.1, 14.4) |
| 17b' | 3.73, m (5.4) | 3.70, m | ||
| 21' | 148.4, C | 134.0, C | ||
| 22' | 119.2, CH | 8.20, d (8.5) | 153.9, CH | |
| 22' -OH | 11.27, s | |||
| 23' | 138.4, CH | 8.27, d (8.5) | 121.3, CH | 7.59, s |
| 24' | 129.5, C | 132.4, C | ||
| 25' | 128.9, CH | 7.84, m (8.4) | 131.1, CH | 7.72, m |
| 26' | 128.9, CH | 7.59, m | 127.9, CH | 7.54, m |
| 27' | 131.2, CH | 7.73, m | 129.2, CH | 7.52, m |
| 28' | 129.7, CH | 7.87, m (8.4) | 126.9, CH | 7.70, m |
| 29' | 146.3, C | 141.7, C | ||
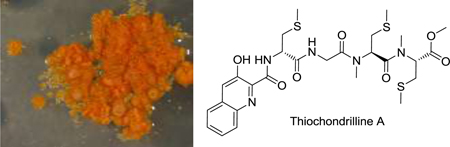
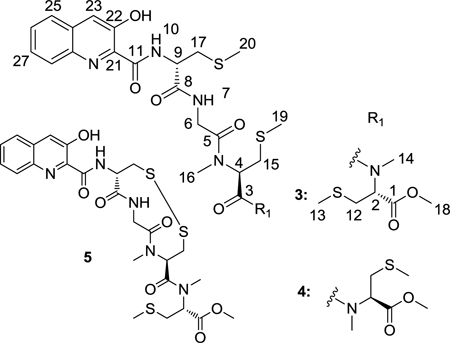
Thiochondrilline A (3) and B (4), isolated in a 3:1 ratio, were inseparable despite extensive HPLC work. HRMS of the mixture helped support the molecular formula of C27H37O7N5S3 for both compounds. 1H NMR (Table 2) showed that the two compounds had nearly identical chemical shifts with the exception of H-2 and H-14. Extensive 1D and 2D NMR in comparison to thiocoraline (1) led to the initial structural assignments of thiochondrilline A (3) and B (4), though ambiguity remained about the conformation around the amide bond at C-3. The 2D ROESY spectrum showed a correlation between H-2 and H-4 for thiochondrilline B (4) and led to the proposal that thiochondrilline A (3) was trans and thiochondrilline B (4) was cis around the amide bond at C-3. Therefore, an ab initio study was performed in order to determine the conformation around the amide bond at C-3.
TABLE 2.
1H and 13C NMR data for 3–5 (600 MHz, CDCl3)
| 3 | 4 | 5 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Position | δC, mult. | δH (J in Hz) | HMBC | δC, mult. | δH (J in Hz) | HMBC | δC, mult. | δH (J in Hz) | HMBC |
| 1 | 169.8, C | 169.8, C | 170.1, C | ||||||
| 2 | 56.4, CH | 5.11, dd (4.4, 11.5) | 58.8, CH | 4.76, dd (4.0, 10.5) | 58.0, CH | 5.04, dd (4.4, 11.2) | |||
| 3 | 169.7, C | 169.6, C | 169.0, C | ||||||
| 4 | 52.1, CH | 5.62, m | 3, 16 | 51.7, CH | 5.63, m | 3, 15, 16 | 58.1, CH | 5.13, dd (3.5, 10.0) | 3 |
| 5 | 167.9, C | 167.9, C | 168.8, C | ||||||
| 6a | 41.5, CH2 | 4.16, m (4.3) | 5 | 41.3, CH2 | 4.06, m (3.7) | 5 | 44.6, CH2 | 5.20, dd (10.3, 15.6) | 5, 8 |
| 6b | 4.20, m (4.3) | 4.12, m (3.7) | 3.79, dd (2.6, 15.6) | ||||||
| 7-NH | 7.27, t (2.7) | 7.20, m | 7.01, br d (9.7) | ||||||
| 8 | 169.4, C | 169.4, C | 169.2, C | ||||||
| 9 | 52.1, CH | 4.84, m (7.7) | 8, 11 | 52.2, CH | 4.81, m (7.3) | 8, 11 | 54.4, CH | 4.80, br t (9.0) | |
| 10-NH | 9.09, d (8.1) | 9.07, d (8.1) | 8.86, d (8.5) | ||||||
| 11 | 169.2, C | 169.2, C | 168.1, C | ||||||
| 12a | 32.6, CH2 | 3.08, m | 2, 13 | 33.3, CH2 | 2.99, m | 33.9, CH2 | 2.92, dd (5.0, 9.4) | 2, 13 | |
| 12b | 2.88, m | 2.84, m | 3.12, m | ||||||
| 13 | 15.2, CH3 | 2.10, s | 12 | 15.4, CH3 | 2.05, s | 12 | 15.9, CH3 | 2.12, s | 12 |
| 14 | 32.5, CH3 | 2.93, s | 2, 3 | 29.3, CH3 | 2.78, s | 2, 3 | 33.2, CH3 | 3.20, s | 2, 3 |
| 15a | 33.2, CH2 | 2.91, m | 19 | 33.2, CH2 | 2.91, m | 19 | 43.2, CH2 | 4.00, br s | |
| 15b | 2.83, m | 2.83, m | 2.70, br s | ||||||
| 16 | 29.1, CH3 | 2.87, s | 4, 5 | 29.0, CH3 | 2.80 ,s | 4, 5 | 30.7, CH3 | 2.86, s | 5, 4 |
| 17a | 36.2, CH2 | 3.08, m | 8, 20 | 36.2, CH2 | 3.08, m | 8, 20 | 43.6. CH2 | 3.66, dd (9.7, 14.4) | 8, 9 |
| 17b | 3.04, m | 3.03, m | 3.13, m | ||||||
| 18 | 52.7, CH3 | 3.72, s | 1 | 52.7, CH3 | 3.71, s | 1 | 52.9, CH3 | 3.75, s | 1 |
| 19 | 15.6, CH3 | 2.08, s | 15 | 15.8, CH3 | 2.14, s | 15 | |||
| 20 | 15.9, CH3 | 2.23, s | 17 | 15.9, CH3 | 2.22, s | 17 | |||
| 21 | 134.0, C | 134.0, C | 134.0, C | ||||||
| 22 | 153.5, C | 153.5, C | 153.7, C | ||||||
| 22-OH | 11.55, s | 21, 23 | 11.53, s | 21, 23 | 11.31, s | 21, 23 | |||
| 23 | 120.5, CH | 7.64, s | 21, 22, 28, 29 | 120.5, CH | 7.64, s | 21, 22, 28, 29 | 120.9, CH | 7.64, s | 21, 22, 28, 29 |
| 24 | 132.1, C | 132.1, C | 132.4, C | ||||||
| 25 | 126.1, CH | 7.70, m | 27 | 126.1, CH | 7.70, m | 27 | 126.6, CH | 7.70, d (7.9) | 24, 27 |
| 26 | 128.7, CH | 7.52, m | 28 | 128.7, CH | 7.52, m | 28 | 129.1, CH | 7.54, m | 28, 29 |
| 27 | 127.3, CH | 7.54, m | 25 | 127.3, CH | 7.54, m | 25 | 129.1, CH | 7.56, m | 24, 25 |
| 28 | 129.7, CH | 8.01, d (8.3) | 26 | 129.7, CH | 8.01, d (8.3) | 26 | 129.8, CH | 7.97, d (8.2) | 26, 29 |
| 29 | 141.4, C | 141.4, C | 141.7, C | ||||||
Thiochondrilline A (3) and B (4) were modeled with molecular modeling software, and ab initio calculations were analyzed with the DP4 probability method32 to determine the conformation around the amide bond at C-3 and the absolute configuration at C-2. Spartan 1033 was used to find the lowest energy conformer through a Monte Carlo conformer search (MMFF), and Gaussian 0934 was used for geometry optimization and NMR calculations (B3LYP/6–31G(d,p)).35 NMR shifts were referenced to TMS and benzene using the multi-standard (MSTD) approach,36 and the DP4 probability method32 was used to compare the calculated NMR shifts for the two proposed structures with the observed chemical shifts. The recently published DP4 method was developed by testing 117 molecules, including 21 natural products that were originally published with misassigned stereochemistry.32 Calculated and observed chemical shifts for thiochondrilline A (3) and B (4) were uploaded to the DP4 method online applet (http://www-jmg.ch.cam.ac.uk/tools/nmr/DP4), which uses a mathematical algorithm to quantify the probability of the correct assignment of each structure. The DP4 method calculated a 72.2% probability that thiochondrilline A (3) was trans around the amide bond at C-5 and a 94.3% probability that thiochondrilline B (4) was the cis isomer (See Supporting Information for all calculated NMR shifts). Absolute configurations at C-2, C-4, and C-9 were assumed to be the same as thiocoraline (1), and the configuration at C-2 was confirmed by ab initio calculations. Each configuration, R and S, at C-2 was modeled for thiochondrilline A (3) and B (4), and the DP4 probability method32 calculated a 100% probability of the R configuration for both thiochondrilline A (3) and B (4) using calculated 1H and 13C NMR shifts. After determining the conformational relationship between thiochondrilline A (3) and B (4), variable temperature 1H NMR experiments (500 MHz, CDCl3) at 5 °C and 45 °C resulted in no change, indicating that the compounds were stable. The insolubility of thiochondrilline A (3) and B (4) in most solvents prevented variable temperature experiments at higher temperatures.
HRMS supported the molecular formula of C25H31O7N5S3 for thiochondrilline C (5). 1H NMR shifts (Table 2) closely resembled thiochondrilline A (3) and B (4) but with the absence of two S-methyl groups. The S-methyl at C-13 was present as evidenced by an HMBC correlation from H-13 to C-12. The absence of two S-methyl groups led to the initial proposal of a disulfide bond linking the two side chains. Without direct evidence of HMBC correlations across the disulfide bond, several measures were taken to confirm the proposed structure. With the possibility that (+)ESI MS created the disulfide bond by the oxidation of two thiol groups, (−)ESI was conducted (608.1274 m/z) and supported the same molecular formula as calculated from (+)ESI MS. Thiochondrilline C (5) and the possible thiol analog were optimized with Spartan 10 and Gaussian 09, and ab initio NMR calculations were compared to observed chemical shifts. Using the DP4 probability method,32 a comparison of all 13C chemical shifts for thiochondrilline C (5) and the thiol analog produced a 97.5% probability of the correct structure being thiochondrilline C (5) rather than the thiol analog. An examination of the calculated chemical shifts at C-15 and C-17 especially supported the assignment of thiochondrilline C (5) (See Supporting Information). The absolute configurations of thiochondrilline C (5) at C-2, C-4, and C-9 were assumed to be the same as thiocoraline (1) based on biosynthetic precedent.18
HRMS and isotopic distribution37 supported the molecular formula of C48H56O13N10S6 for 12'-sulfoxythiocoraline (6), one more oxygen atom than thiocoraline (1). H-13' (2.64 ppm), H-12' (3.13, 3.33 ppm), and H-2' (6.06 ppm) (Table 1) indicated the existence of an S-methyl-containing sulfoxide. The COSY and HMBC spectra confirmed the structural difference between thiocoraline (1) and 12'-sulfoxythiocoraline (6). The absolute configuration of 12'-sulfoxythiocoraline (6) was assumed to be the same as thiocoraline (1) due to the near identical 1H and 13C shifts. Currently, we cannot conclusively state whether 12'-sulfoxythiocoraline (6) originated as a natural product or an oxidized product of thiocoraline (1) as a result of our isolation process.
Each of the five analogs was screened against the A549 human cancer cell line in comparison to thiocoraline (1) (Table 3), and the cytoxicity of 22'-deoxythiocoraline (2) helped provide a better understanding of the SAR of thiocoraline (1). Related analogs synthesized by the Boger group14–15 demonstrated that the absence of both phenol groups reduced the compound’s cytotoxicity. [N-(2-quinoline carboxyl)-d-Cys-Gly-NMe-l-Cys-NMe-l-Cys-(Me)]2 (cysteine thiol) dilactone), identical to thiocoraline (1) except the absence of both phenol groups, was one hundred times less potent than thiocoraline (1). 22'-deoxythiocoraline (2) was fourteen times less potent than thiocoraline (1) against the A549 cell line. Hence, the potency of 22'-deoxythiocoraline (2) lies between the potency of thiocoraline (1) and Boger’s synthetic analog, emphasizing the importance of each phenol group to thiocoraline’s activity. The 3-OH-quinaldic system provides a tricyclic hydrogen-bonded conformation that is proposed to stabilize the complex with DNA.12 One phenol group, in the case of 22'-deoxythiocoraline (2), helps stabilize the complex with DNA, though to a lesser extent than the two phenol groups in thiocoraline (1). Consequently, 22'-deoxythiocoraline (2) reinforces the role of the phenol group in interactions with DNA and effects on the potency.
TABLE 3.
A549 Cancer Cell Line Cytotoxicity Data
| Compound | EC50, µM |
|---|---|
| 1 | 0.0095 |
| 2 | 0.13 |
| 3&4 | >10 |
| 5 | 2.86 |
| 6 | 1.26 |
22'-Deoxythiocoraline (2), thiochondrilline A (3), and thiochondrilline B (4) also provided insight into the biosynthetic pathway of thiocoraline. 22'-Deoxythiocoraline (2) suggested that the putative loading module, consisting of the TioJ and TioO proteins, can be promiscuous with respect to the starter unit. In the biosynthesis of thiocoraline (1), TioJ and TioO are proposed to load 3-OH-quinaldic acid as the starter unit.18 However, the absence of one phenol group in 22'-deoxythiocoraline (2) suggested that TioJ and TioO also have the ability to load quinaldic acid as the starter unit.
As open chain monomers of thiocoraline (1), thiochondrilline A (3) and B (4) provided insight into the mechanisms by which thiocoraline (1) is cyclized. Thiochondrilline A (3) and B (4) revealed that cyclization of thiocoraline most likely occurs before methylation of the thiol groups. In thiocoraline, only the C-12 thiol groups are methylated. However, all three thiol groups are methylated in thiochondrilline A (3) and B (4), suggesting that the putative methylation protein, TioN,38 has the ability to methylate multiple thiol groups and potentially carboxylic acids. Alternatively, if methylation were to occur prior, the expected product would contain C-15 S-Me Cys and not a disulfide bridge. Hence, cyclization most likely occurs before methylation in the biosynthesis of thiocoraline (1).
CONCLUSION
22'-deoxythiocoraline (2) reinforced the importance of both phenol groups in contributing to the bioactivity of thiocoraline (1) and, along with thiochondrilline A (3) and B (4), provided additional insight into the biosynthesis of thiocoraline (1). This study also demonstrated the utility of the DP4 probability method for analyzing ab initio NMR calculations to solve stereochemical problems in an efficient manner.
EXPERIMENTAL SECTION
General Experimental Procedures
NMR spectra were obtained in CDCl3 with a 600 MHz spectrometer with 1H{13C/15N} cold probe and 500 MHz spectrometer with 13C/15N{1H} cryoprobe.
Biological Material
Sponge specimens were collected on February 10, 2010, in the Florida Keys (24° 39’ 17.90”, 81° 17’ 51.09”). A voucher specimen for Chondrilla caribensis f. caribensis (FLK-10-4-24), identified by Mary Kay Harper-Ireland (University of Utah), is housed at the University of Wisconsin-Madison. For cultivation, a sample of sponge (1 cm3) was rinsed with sterile seawater, macerated using a sterile pestle in a micro-centrifuge tube, and dilutions were made in sterile seawater, with vortexing between steps to separate bacteria from heavier tissues. Dilutions were separately plated on two distinct media that have yielded diverse actinomycetes from both marine sponges and sediment: M1 and M4.39 M1 was made using artificial seawater. Both were supplemented with 50 µg/mL cycloheximide and 25 µg/mL nalidixic acid. Plates were incubated at 31 °C for 28 days.
Fermentation, Extraction, and Isolation
Strain WMMA107 was fermented in 25 × 150 mm culture tubes (4 × 10 mL) in medium ASW-A (20g soluble starch, 10g glucose, 10g peptone, 5g yeast extract, 5g CaCO3 per liter of artificial seawater) for one week at 28 °C. 250 mL baffled flasks (16 × 50 mL) were inoculated with 2 mL from the culture tube and shaken at 200 RPM at 28 °C for seven days. 2 L flasks (32 × 500 mL) containing medium ASW-A with Diaion HP20 (4% by weight) were inoculated with 25 mL and shaken at 200 RPM at 28 °C for seven days. Filtered HP20 was washed with water and extracted with acetone. The acetone extract (35 g) was subjected to a liquid-liquid partitioning using 30% aqueous methanol and chloroform (1:1). The chloroform soluble partition (10 g) was subjected to silica gel (SiO2) column chromatography (350 g, 40–60 µm particle size) with hexanes and ethyl acetate (0–100%). Fractions containing 1–6 were combined and subjected to RP HPLC (10–100% CH3CN-H2O, 30 min) using a Phenomenex Luna C18 column (250 × 10 mm, 5 µm), yielding 5 (2.0 mg, tR 23.2 min), 6 (1.6 mg, tR 23.6 min), 3 and 4 (1.5mg, tR 24.2 min), 2 (0.5 mg, tR 25.6 min), and 1 (80 mg, tR 27.5 min), respectively. Fraction 8, containing thiochondrilline A and B, was subjected to 11 isocratic and gradient methods of HPLC separation testing different solvents and solvent compositions. Insufficient separation was achieved. The inability to separate amide rotamers chromatographically is consistent with past literature,40,41 though amide rotamers have been chromatographically separated in some cases.42
Sequencing
Genomic DNA was extracted using the UltraClean Microbial DNA Isolation kit (Mo Bio Laboratories, Inc.). 16S rDNA genes were amplified using 100–200 ng genomic DNA template with the primers 8–27F (5’ to 3’ GAGTTTGATCCTGGCTCAG) and 1492R (5’ to 3’ GGTTACCTTGTTACGACTT). The following PCR conditions were used: 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 55 °C for 1 min, 72 °C for 1.5 min, with a final step of 72 °C for 5 min. The PCR bands were excised from the gel and purified using the QIAquick Gel Extraction kit (QIAGEN). One µL purified product was sequenced. Sequencing reactions were performed by the UW Biotechnology Center and reactions were sequenced with an ABI 3730xl DNA Analyzer. WMMA107, WMMA102, WMMA105, WMMA110, and WMMA111 were identified as a Verrucosispora sp. by 16S sequencing, and WMMA 107 demonstrated 99% sequence similarity to Verrucosispora sp. CNP-852 SD01 (accession number EU 214938.1). The 16S sequence was deposited in GenBank (accession number JF520832-JF520836).
Cytotoxicity
Human lung adenocarcinoma A549 cells were obtained from the American Tissue Culture Collection (CCL-185) and were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin (Cellgro) at 37 °C in a humidified atmosphere containing 5% CO2. Cytoxicity of the compounds was determined using the resazurin assay.43 The EC50 was calculated using GraphPad Prism Version 5.0.
Molecular Modeling Calculations
Molecular modeling calculations were performed on a Dell Precision T5500 Linux workstation with a Xeon processor (3.3 GHz, 6-core). Low energy conformers were obtained using Spartan 10 software (MMFF). The low energy conformer for each compound was analyzed by Gaussian 09 for geometry optimization and NMR calculations (B3LYP/6–31G(d,p)). Molecules were modeled in gas phase.
22'-Deoxythiocoraline (2): white solid; [α]25D -98 (c 0.0005, CHCl3); UV (MeOH) λmax (log ε) 210 (4.96), 230 (4.57), 299 (3.72), 360 (3.62) nm; IR (ATR) υmax 3355, 1657, 1519, 1225, 772 cm−1; 1H and 13C NMR (See Table 1); HRMS [M+H]+ m/z 1141.2533 (calcd for C48H57O11N10S6, 1141.2527).
Thiochondrilline A & B (3,4): white solid; [α]25D -128 (c 0.0013, CHCl3); UV (MeOH) λmax (log ε) 210 (4.87), 230 (4.46), 298 (3.57), 360 (3.24) nm; IR (ATR) υmax 3344, 1651, 1519, 757 cm−1; 1H and 13C NMR (See Table 2); HRMS [M+Na]+ m/z 662.1745 (calcd for C27H37O7N5S3Na, 662.1747).
Thiochondrilline C (5): white solid; [α]25D -77 (c 0.0011, CHCl3); UV (MeOH) λmax (log ε) 209 (4.84), 230 (4.43), 298 (3.47), 360 (3.43) nm; IR (ATR) υmax 3354, 1656, 1519, 1229, 782 cm−1; 1H and 13C NMR (See Table 2); HRMS [M+Na]+ m/z 632.1297 (calcd for C25H31O7N5S3Na, 632.1278).
12'-Sulfoxythiocoraline (6): white solid; [α]25 d -96 (c 0.0013, CHCl3); UV (MeOH) λmax (log ε) 209 (5.07), 230 (4.65), 299 (3.76), 360 (3.69) nm; IR (ATR) υmax 3359, 1656, 1519, 747 cm−1; 1H and 13C NMR (See Table 1); HRMS [M+H]+ m/z 1173.2424 (calcd for C48H57O13N10S6, 1173.2425).
Supplementary Material
ACKNOWLEDGMENT
This work was supported by funding from the University of Wisconsin-Madison School of Pharmacy, the Graduate School at the University of Wisconsin, and the UW College of Agriculture and Life Sciences. This work was also funded in part by the NIH, NIGMS Grant R01 GM092009. We would like to thank the Analytical Instrumentation Center at the University of Wisconsin-Madison for the facilities to acquire spectroscopic data, especially NMR and MS. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grants P41RR02301 (BRTP/NCRR) and P41GM66326 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin, the NIH (RR02781, RR08438), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA. We would like to thank Dr. Kim Ritchie and the Mote Marine Laboratory for assistance with collection and Mary Kay Harper (University of Utah) for sponge taxonomy.
Footnotes
Supporting Information Available 1D and 2D NMR spectra, ab initio calculation summary. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Berdy J. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Zäehner H, Fiedler HP. The Need for New Antiobiotics. In: Hunter PA, Darby GK, Russel NJ, editors. Fifty Years of Antimicrobials. Cambridge, England: Cambridge University Press; 1995. pp. 67–84. [Google Scholar]
- 3.Qui J. Nature News. 2010 Apr 18; http://www.nature.com/news/2010/100418/full/news.2010.190.html.
- 4.Feling RH, Buchanan GO, Mincer TJ, Kaufmann CA, Jensen PR, Fenical W. Angew. Chem. Int. Ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi M, Natsume T, Tamaoki S, Watanabe J, Asano H, Mikami T, Miyasaka K, Miyazaki K, Gondo M, Sakakibara K, Tsukagoshi S. Jpn. J. Cancer Res. 1997;88:316–327. doi: 10.1111/j.1349-7006.1997.tb00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mita AC, Hammond LA, Bonate PL, Weiss G, McCreery H, Syed S, Garrison M, Chu QS, DeBono JS, Jones CB, Weitman S, Rowinski EK. Clin. Cancer Res. 2006;12:5207–5215. doi: 10.1158/1078-0432.CCR-06-0179. [DOI] [PubMed] [Google Scholar]
- 7.Mayer AM, Glaser KB, Cuevas C, Jacobs RS, Kem W, Little RD, McIntosh JM, Newman DJ, Potts BC, Shuster DE. Trends Pharmacol. Sci. 2010;31:255–265. doi: 10.1016/j.tips.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Faircloth G, Jimeno J, D’lncalci M. Eur. J. Cancer. 1997;33:S175. [Google Scholar]
- 9.Pérez Baz J, Cañedo LM, Fernández-Puentes JL, Silva Elipe MV. J. Antibiot. 1997;50:738–741. doi: 10.7164/antibiotics.50.738. [DOI] [PubMed] [Google Scholar]
- 10.Romero F, Espliego F, Pérez Baz J, García de Quesada T, Grávalos D, De la Calle F, Fernández-Puentes JL. J. Antibiot. 1997;50:734–737. doi: 10.7164/antibiotics.50.734. [DOI] [PubMed] [Google Scholar]
- 11.Erba E, Bergamashi D, Ronzoni S, Faretta M, Taverna S, Bonfanti M, Catapano CV, Faircloth G, Jimeno J, D’Incalci M. Br. J. Cancer. 1999;80:971. doi: 10.1038/sj.bjc.6690451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Negri A, Marco E, García-Hernández V, Domingo A, Llamas-Saiz AL, Porto-Sandá S, Riguera R, Laine W, David-Cordonnier MH, Bailly C, García-Fernández LF, Vaquero JJ, Gago F. J. Med. Chem. 2007;50:3322–3333. doi: 10.1021/jm070381s. [DOI] [PubMed] [Google Scholar]
- 13.Boger DL, Ichikawa S. J. Am. Chem. Soc. 2000;122:2956–2957. [Google Scholar]
- 14.Boger DL, Ichikawa S, Tse WC, Hedrick MP, Jin Q. J. Am. Chem. Soc. 2001;123:561–568. doi: 10.1021/ja003602r. [DOI] [PubMed] [Google Scholar]
- 15.Boger DL. Analogues of Thiocoraline and BE-22179. 20040072738. U.S. Patent Application. 2004 April 15; URL: http://ip.com/patapp/US20040072738.
- 16.Tulla-Puche J, Bayó-Puxan N, Moreno JA, Francesch AM, Cuevas C, Álvarez M, Albericio F. J. Am. Chem. Soc., 2007;129:5322–5323. doi: 10.1021/ja0686312. [DOI] [PubMed] [Google Scholar]
- 17.Tulla-Puche J, Marcucci E, Prats-Alfonso E, Bayó-Puxan N, Albericio F. J. Med. Chem. 2009;52:834–839. doi: 10.1021/jm800784k. [DOI] [PubMed] [Google Scholar]
- 18.Lombó F, Velasco A, Castro A, de la Calle F, Braña AF, Sánchez-Puelles JM, Méndez C, Salas JA. Chembiochem. 2006;7:366–376. doi: 10.1002/cbic.200500325. [DOI] [PubMed] [Google Scholar]
- 19.Sheoran A, King A, Velasco A, Pero JM, Garneau-Tsodikova S. Mol. Biosyst. 2008;4:622–628. doi: 10.1039/b801391h. [DOI] [PubMed] [Google Scholar]
- 20.Robbel L, Hoyer KM, Marahiel MA. FEBS J. 2009;276:1641–1653. doi: 10.1111/j.1742-4658.2009.06897.x. [DOI] [PubMed] [Google Scholar]
- 21.Biswas T, Zolova OE, Lombó F, de la Calle F, Salas JA, Tsodikov OV, Garneau-Tsodikova S. J. Mol. Biol. 2010;397:495–507. doi: 10.1016/j.jmb.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 22.Mady AS, Zolova OE, San Millán MA, Villamizar G, De la Calle F, Lombó F, Garneau-Tsodikova S. Mol. BioSyst. 2011;7:1999–2011. doi: 10.1039/c1mb05044c. [DOI] [PubMed] [Google Scholar]
- 23.Waring MJ. Echinomycin and Related Quinoxaline Antibiotics. In: Neidle S, Waring MJ, editors. Molecular Aspects of Anticancer Drug-DNA Interactions. Vol. 1. London: Macmillan & Co.; 1993. pp. 213–242. [Google Scholar]
- 24.Rheims H, Schumann P, Rohde M, Stackebrandt E. Int. J. Syst. Bacteriol. 1998;48:1119–1127. doi: 10.1099/00207713-48-4-1119. [DOI] [PubMed] [Google Scholar]
- 25.Bister B, Bischoff D, Ströbele M, Riedlinger J, Reicke A, Wolter F, Bull AT, Zäehner H, Fiedler HP, Süssmuth RD. Angew. Chem. Int. Ed. 2004;43:2574–2576. doi: 10.1002/anie.200353160. [DOI] [PubMed] [Google Scholar]
- 26.Fiedler HP, Bruntner C, Riedlinger J, Bull AT, Knutsen G, Goodfellow M, Jones A, Maldonado L, Pathom-aree W, Beil W, Schneider K, Keller S, Sussmuth RD. J. Antibiot. 2008;61:158–163. doi: 10.1038/ja.2008.125. [DOI] [PubMed] [Google Scholar]
- 27.Shirai M, Okuda M, Motohashi K, Imoto M, Furihata K, Matsuo Y, Katsuta A, Shizuri Y, Seto H. J. Antibiot. 2010;63:245–250. doi: 10.1038/ja.2010.30. [DOI] [PubMed] [Google Scholar]
- 28.Jiang S, Sun W, Chen M, Dai S, Zhang L, Liu Y, Lee KJ, Li X. Antonie Van Leeuwenhoek. 2007;92:405–416. doi: 10.1007/s10482-007-9169-z. [DOI] [PubMed] [Google Scholar]
- 29.Yi-lei N, Ru L, Yong-biao Z, Hui Z, Yuan-rong C, Wei Z, Hong J. Chin. J. Antibiot. 2010;6:426–430. [Google Scholar]
- 30.Schmitz FJ, McDonald FJ. J. Lipid Res. 1974;15:158–164. [PubMed] [Google Scholar]
- 31.Zierer MS, Mourão PA. Carbohydr. Res. 2000;328:209–216. doi: 10.1016/s0008-6215(00)00076-8. [DOI] [PubMed] [Google Scholar]
- 32.Smith SG, Goodman JM. J. Am. Chem. Soc. 2010;132:12946–12959. doi: 10.1021/ja105035r. [DOI] [PubMed] [Google Scholar]
- 33.Spartan 10, v. 1.0.2. Wavefunction Inc.; 2011. [Google Scholar]
- 34.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09, Revision A.1. Wallingford CT: Gaussian, Inc.; 2009. [Google Scholar]
- 35.Stappen I, Buchbauer G, Robien W, Wolschann P. Magn. Reson. Chem. 2009;47:720–726. doi: 10.1002/mrc.2452. [DOI] [PubMed] [Google Scholar]
- 36.Sarotti AM, Pellegrinet SC. J. Org. Chem. 2009;74:7254–7260. doi: 10.1021/jo901234h. [DOI] [PubMed] [Google Scholar]
- 37.Ojanperä S, Pelander A, Pelzing M, Krebs I, Vuori E, Ojanperä I. Rapid Commun. Mass Spectrom. 2006;20:1161–1167. doi: 10.1002/rcm.2429. [DOI] [PubMed] [Google Scholar]
- 38.Zolova OE, Mady AS, Garneau-Tsodikova S. Biopolymers. 2010;93:777–790. doi: 10.1002/bip.21489. [DOI] [PubMed] [Google Scholar]
- 39.Maldonado LA, Fragoso-Yáñez D, Pérez-García A, Rosellón-Druker J, Quintana ET. Antonie Van Leeuwenhoek. 2009;95:111–120. doi: 10.1007/s10482-008-9294-3. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Wu H, Shen L, Qin Y. J. Am. Chem. Soc. 2007;129:13794–13795. doi: 10.1021/ja075705g. [DOI] [PubMed] [Google Scholar]
- 41.Seong CM, Park CM, Choi J, Park NS. Tetrahedron Lett. 2009;50:1029–1031. [Google Scholar]
- 42.McCombie SW, Tagat JR, Vice SF, Lin S, Steensma R, Palani A, Neustadt BR, Baroudy BM, Strizki JM, Endres M, Cox K, Dan N, Chou C. Bioorg. Med. Chem. Lett. 2003;13:567–571. doi: 10.1016/s0960-894x(02)00918-6. [DOI] [PubMed] [Google Scholar]
- 43.O’Brien J, Wilson I, Orton T, Pognan F. Eur. J. Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.