Abstract
Background and Aims
Genomic instability, as reflected in specific chromosomal aneuploidies and variation in the nuclear DNA content, is a defining feature of human carcinomas. It is solidly established that the degree of genomic instability influences clinical outcome. We have recently identified a 12-gene expression signature that discerned genomically stable from unstable breast carcinomas. This gene expression signature was also useful to predict, with high accuracy, the clinical course in independent multiple published breast cancer cohorts. From a biological point of view, this result confirmed the central role of genomic instability for a tumor's ability to adapt to external challenges and selective pressure and hence, for continued survival fitness. This in turn prompted us to investigate whether this genomic instability signature could also predict clinical outcome in other cancer types of epithelial origins, including colorectal tumors, non-small cell lung carcinomas, and ovarian cancer.
Results
The results show that the gene expression signature that defines genomic instability and poor outcome in breast cancer contributes significantly more accurate (P < 0.05; compared with random prediction) prognostic information in multiple cancer types independent of established clinical parameters. The 12-gene genomic instability signature stratified patients into high- and low-risk groups with distinct post-operative survival in three non-small cell lung cancer cohorts (n = 637) in Kaplan-Meir analyses (log-rank P < 0.05). It predicted recurrence in colon cancer patients (n = 92) with an overall accuracy greater than 69% (P = 0.04) in cross-cohort validation. It quantified relapse-free survival in ovarian cancer (n = 124; log-rank P < 0.05). Functional pathway analysis revealed interactions between the 12 signature genes and well known cancer hallmarks.
Conclusion
The degree of genomic instability has diagnostic and prognostic implications. It is tempting to speculate that pursuing genomic instability therapeutically could provide entry points for a target that is unique to cancer cells.
Keywords: Genomic instability, gene expression signatures, breast cancer, ovarian cancer, colon cancer, non-small cell lung cancer
Introduction
Aneuploidy, chromosomal instability, and resulting genomic imbalances are one of the hallmarks of human cancers of epithelial origin (1). While specific chromosomal imbalances are usually acquired before the transition to invasive disease, global destabilization of the genome occurs at later stages (2). The degree of destabilization, measured for instance by the number of chromosomal aberrations or the amount of variability in the nuclear DNA content from one cell to another is an important predictor of clinical outcome, independent of conventional morphological or clinical parameters. For instance, women with genomically stable breast carcinomas have considerable prolonged disease free survival times compared to women whose tumors are genomically unstable (3). A similar picture emerges in prostate carcinomas (4). More recently, the search for predictors of clinical outcome was extended to embrace global gene expression profiling, and numerous signatures of poor prognosis or treatment failure were described (5-7). In an attempt to understand the biological basis of these signatures we analyzed, using gene expression profiling, breast carcinomas that had been characterized as genomically stable or unstable, hence, of good and poor prognosis, respectively (8). These analyses let us to identify a set of 12 genes, which were differentially expressed between stable and unstable tumors (8). The biological and clinical relevance of these genes, i.e., the gene expression signature of genomic instability, was validated with large independent data sets and resulted in an excellent prediction of the clinical course. Inversely, when using established signatures of poor prognosis to predict the degree of genomic instability in our dataset, the results were equally convincing. That means that the basis for clinically used gene expression signatures of poor prognosis is biologically linked to chromosomal instability, which ultimately determines outcome. Since most solid tumors are defined by aneuploidy, we were interested to explore whether the gene expression signature of genomic instability that determines outcome in breast cancer is also useful for the prediction of the clinical course in other entities of carcinomas, namely colorectal, lung, and ovarian cancer.
Material and Methods
Patients and Samples
Colon Cancer
The first cohort contained 50 patients with stage II colon adenocarcinoma (9). None of the patients had emergency surgery or received any adjuvant chemotherapy. Twenty-five patients developed a distant metastasis (liver in 20 patients, lung in five patients) within 52 months. The other 25 patients remained disease free for at least 60 months, with mean follow-up of 79 months. The second cohort contained 24 patients with stage II colon adenocarcinoma (10). None of these patients received adjuvant chemotherapy. Ten patients developed a liver metastasis within 55 months. The other 14 patients remained disease free for at least 60 months, with mean follow-up of 72.2 months. The third cohort contained 18 patients with colon adenocarcinoma (11). A total of 10 patients had no lymph node metastasis (stage II) and did not receive any chemotherapy. The other eight patients had lymph node metastasis (stage III) and received 6-month adjuvant chemotherapy, with 5-FU and levamisole. Patients were evaluated at 3-month intervals for the first postoperative year and at 6-month intervals thereafter. Nine of the 18 patients (five stage II patients and four stage III patients) developed a distant metastasis within 53 months. The other nine patients remained disease-free for at least 60 months, with mean follow-up of 75 months. A summary of patient clinical characteristics is provided in Table 1.
Table 1.
Clinical characteristics of colon cancer patient cohorts.
| Barrier et al. (9) (n=50) | Barrier et al. (10) (n=24) | Barrier et al. (14) (n=18) | |
|---|---|---|---|
| Mean follow-up (months) | |||
| Patients with recurrence | 52 | 55 | 53 |
| Patients without recurrence | 79 | 72.2 | 75 |
| Tumor Stage | |||
| Stage II | 100% | 100% | 55.6% |
| Stage III | 0% | 0% | 44.4% |
| Recurrence within 5-year after surgery | |||
| Yes | 50% | 58.3% | 50% |
| No | 50% | 41.7% | 50% |
Non-small Cell Lung Cancer
The cohort from Bild et al. (12) contained 111 patients of which 67 were of stage I, 18 of stage II, 24 of stage III, and two of stage IV. There were two cell types in this cohort: lung adenocarcinoma (n = 58) and squamous cell lung cancer (n = 53). The cohort from Bhattacharjee et al. (13) contained 84 patients with lung adenocarcinoma. Sixty-two patients were of stage I, 14 of stage II, and eight of stage III. Twenty-six tumors were well differentiated, 43 moderately differentiated, and 15 poorly differentiated. The cohort from Shedden et al. (14) contained 442 lung adenocarcinomas collected from multiple cancer centers and institutes. Two hundred and seventy-six patients were in stage I, 94 in stage II, and 68 in stage III and four patients with undefined stage. A summary of clinical characteristics including age, gender, and median follow-up time for each cohort is given in Table 2.
Table 2.
Clinical characteristics of lung cancer patient cohorts.
| Bild et al. (12) (n=111) | Bhattacharjee et al. (13) (n=84) | Shedden et al. (14) (n=442) | |
|---|---|---|---|
| Histology | |||
| Adenocarcinoma | 52% | 100% | 100% |
| Squamous cell | 48% | ||
| Median follow-up (months) | 31 | 38 | 47 |
| Age (mean, S.D.*) | 65 (10) | 63 (11) | 64 (10) |
| Sex | |||
| Male | 57% | 43% | 50% |
| Female | 43% | 57% | 50% |
| Tumor Stage | |||
| Stage I | 60% | 74% | 62% |
| Stage II | 16% | 17% | 22% |
| Stage III | 22% | 9 % | 15% |
| Stage IV | 2% | - | - |
| Unknown | - | - | 1% |
S.D. denotes standard deviation
Ovarian Cancer
The ovarian cancer cohort (n = 124) was retrieved from Bild et al. (12) 94.4% (117/124) of these ovarian cancer patients had advanced stages (III and IV). The cohort was randomly partitioned into training and test sets of equal size. Table 3 presents the median follow-up time and the distribution of tumor stage for the training and test cohorts.
Table 3.
Clinical characteristics of ovarian cancer patient cohorts.
DNA Microarray Analysis
The RNA extraction and cDNA preparation in these studies was described in their original publications (9-14). Three colon cancer datasets were all generated with Affymetrix U133A arrays. The lung adenocarcinoma dataset from Bhattacharjee et al. (13) were measured on Affymetrix U95A arrays. The non-small cell lung cancer dataset from Bild et al. (12) was quantified with Affymetrix U133 Pus 2.0 arrays. The lung adenocarcinoma datasets from Shedden et al. (14) were generated with Affymetrix U133A. The ovarian cancer dataset from Bild et al. (12) were assayed with Affymetrix U133A (retrieved with record GSE3149 from Gene Expression Omnibus).
Computational methods for prognostic classifications
RIPPER
RIPPER is a propositional rule learning algorithm proposed by Cohen (15) with improvements over original incremental reduced error pruning (IREP). In RIPPER algorithm, after an initial rule set is learned from IREP, the rule set is further pruned repeatedly based on a different metric and stopping condition on randomized data. The repeated pruning stops when the rule set learned from IREP is refined into a rule set with optimized size and performance. JRip learner with WEKA 3.4 (16) was employed in the analysis.
Cox proportional hazard model
In survival analysis, the hazard function assesses the instantaneous risk of death at time t, condition on survival to that time point:
where T is the variable represents the survival time with cumulative distribution function P(t) = Pr(T ≤ t).
A Cox proportional hazard model defines the relationship between the survival of patients and a set of variables, such as gene expressions. The Cox model gives the hazard at time t for an individual with a given set of predictors denoted by X:
A hazard ratio is defined as the hazard for one individual divided by the hazard for a difference individual:
where X* denotes the set of predictors for one individual and X denotes the set of predictors for the other individual (17).
In our analysis, the hazard ratio represents the ratio of hazard (i.e., death from cancer) between the average risk scores of two prognostic groups.
Prognostic Prediction of Recurrence in Colon Cancer
The matching genes in the 12-gene genomic instability signature were identified with Affymetrix IDs. Nine common genes were found in each of the three colon cancer cohorts with six genes having matches to multiple probes. The mean expression of the duplicate probes for each gene was used in this study. The patient cohort from Barrier et al. (9) was used as training set (n = 50), while the cohorts from another two papers by Barrier et al. (10;11) were combined into an independent validation set (n = 42). A training model was built with the 9 signature genes to classify recurrence in colon cancer patients using a rule-based learner RIPPER. A 10-fold cross validation was used to evaluate the performance of the training model. This training model was used to predict recurrence in each patient in the validation set.
Prognostic Categorization of Non-Small Cell Lung Cancer
The DNA microarray data were generated on three lung cancer cohorts using different Affymetrix platforms. So, gene symbols were used to find the matching genes in the signature. In each patient cohort, a Cox proportional hazard model was constructed by using the matching genes as covariates to predict lung cancer survival after the initial treatment. The non-small cell lung cancer cohort (n = 111) from Bild et al. (12) was used as training set. A risk score was generated for each patient in this cohort. A high risk score represents a high probability of postoperative treatment failure, and similarly for a low risk score. Based on the distribution of the risk scores in this cohort, a cutoff point was identified to stratify patients into high- or low-risk groups. This cutoff risk score was applied in prognostic categorization in the two lung adenocarcinoma patient cohorts from Bhattacharjee et al. (13) (n = 84) and Shedden et al (n = 442) (14). In each cohort, the survival probability of each prognostic group was assessed with Kaplan-Meier analysis. The difference between the survival probabilities in the two groups was estimated with log-rank tests. These analyses were performed with software packages in R (17).
Patient Stratification in Ovarian Cancer
The ovarian cancer patient cohort (n =124) from Bild et al. (12) was randomly partitioned into a training set (n = 62) and a testing set (n = 62). Using the training set, a Cox model was built based on the 12-gene genomic instability signature as covariates. A risk score was generated for each patient. Based on the distribution of the risk scores in the training model, a cutoff value was identified for patient stratification. This training model and cutoff value were applied to the testing set.
Biological Pathway Analysis
Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA) is a proprietary web-based curated database which provides contents of gene and protein interactions reported in the literature. In this study, we used IPA to delineate molecular networks of genes interacting with the 12-gene signature. Core analysis was used to identify the most significant biological processes and functions from the merged network related to the 12-gene signature in human tissues and cell lines.
Results
Genomic instability is a defining feature of human cancers of epithelial origin (1;2). We have previously established a biological relationship between the degree of genomic instability, poor prognosis gene expression signatures and clinical outcomes in patients with breast cancer (8). A set of 12 genes that was differentially expressed between genomically stable and unstable tumors predicted recurrence-free survival (including metastasis and relapse) and overall survival in multiple independent breast cancer cohorts. In the present study, we sought to investigate whether this genomic instability gene signature also predicts clinical outcomes in other cancer types of epithelial origins, including colon cancer (n = 92), non-small cell lung cancer (n = 637), and ovarian cancer (n = 124). The signature genes are listed in Table 4.
Table 4.
The 12-gene genomic instability signature.
| Gene symbol | Gene description | Clone ID | Chromosome location |
|---|---|---|---|
| NXF1 | Nuclear RNA report factor 1 | 1722870 | 11q12-q13 |
| cDNA DKFZp762M127 | Homo sapiens mRNA | 1822809 | 11 |
| P28 | Dynein, axonemal, light intermediate | 1998792 | 1p35.1 |
| KIAA0882 | KIAA0882 protein | 2190664 | 4q31.1 |
| v-myb | Avian myeloblastosis viral oncogene | 2555590 | 6q22-q23 |
| CDKN2A | Cyclin-dependent kinase inhibitor | 2740235 | 9p21 |
| unknown | Human clone 23948 mRNA sequence | 3123244 | 15q22.32 |
| RERG | RAS-like, estrogen-regulated; growth inhibitor | 644989 | 12p13.1 |
| SCYA18 | Small inducible cytokine subfamily | 690231 | 17q11.2 |
| STK15 | Serine/threonine kinase 15 | 2007691 | 20q13.2-q13.3 |
| HNF3A | Hepatocyte nuclear factor 3 | 171194 | 12p13.1 |
| unknown | Incyte EST | 88935 | N/A |
Genomic instability signature is an independent predictor of colon cancer recurrence
To construct a molecular classifier to predict colon cancer recurrence, 50 patients with stage II colon adenocarcinoma (9) were used as training cohort. Nine genes within the 12-gene genomic instability signature were identified from the DNA microarray data. These signature genes were used to classify recurrence in each patient with the JRip algorithm. The performance of the classifier was evaluated in a 10-fold cross validation on the training set (Table 5). The genomic instability signature correctly predicted recurrence in 72% (36/50) of patients, with a sensitivity of 68% (17/25) and a specificity of 76% (19/25). The model identified in the training cohort was applied to predict recurrence in each patient in the validation cohort (n = 42), combining patients retrieved from Barrier et al (10;11). In the validation, the genomic instability signature correctly predicted recurrence in 69.1% (29/42) of patients, with a sensitivity of 73.9% (14/19) and a specificity of 65.2% (15/23). The cohorts from Barrier et al. (9;10) contained only stage II lymph node negative colon adenocarcinomas. These results indicate that the 12-gene genomic instability signature provided independent prognostic information in addition to tumor stage. Once validated in larger, independent cohorts this signature could be potentially used to select lymph node-negative patients for receiving adjuvant chemotherapy.
Table 5.
Prediction accuracy of colon cancer recurrence using the 12-gene genomic instability signature.
| Patients | Sensitivity (Recurrence within 5-y) | Specificity (no recurrence within 5-y) | Overall Accuracy | P-Value* |
|---|---|---|---|---|
| Training set (n=50) (9) All stage II | 68% (17/25) | 76% (19/25) | 72% (36/50) | 0.01 |
| Validation set (n=42) (10;11) | 73.9% (14/19) | 65.2% (15/23) | 69.1% (29/42) | 0.04 |
P<0.05 represents the overall accuracy is significantly higher than that of random prediction (two-sided Z-tests).
Genomic instability signature predicts lung cancer survival
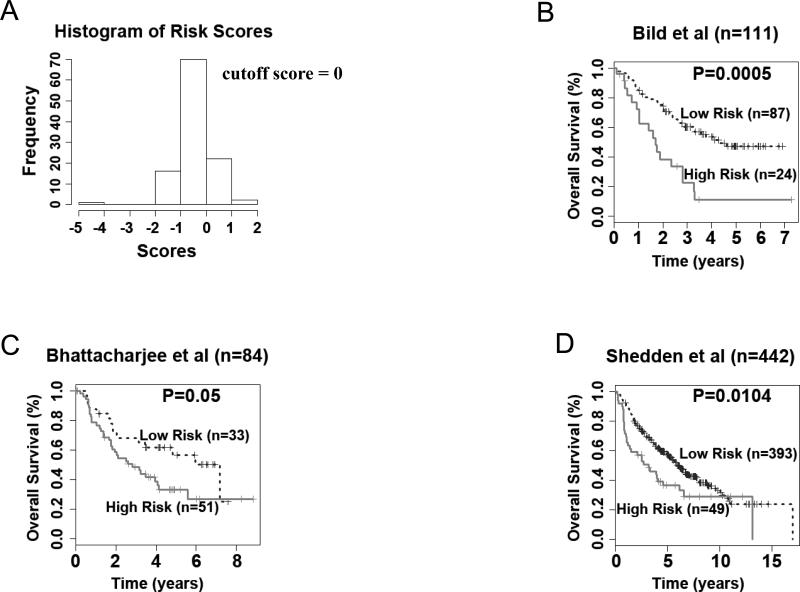
To explore the clinical relevance of the 12-gene genomic instability signature for the prognostication of patients with non-small cell lung cancer, the lung cancer cohort retrieved from Bild et al. (12) was used as a training cohort. A Cox model of overall survival was constructed based on the 12-gene signature, with each gene variable as a covariate. A survival risk score was generated for every patient, with a higher risk score representing a greater probability of treatment failure (i.e., death). Based on the histogram representing distribution of gene expression-defined risk scores in this cohort (Fig. 1A), a cutoff value of zero, the peak value in the histogram, was used to stratify patients into high- and low-risk groups. This cutoff value represents the linear additive expression levels of all the signature genes in lung cancer patients. This stratification separated patients into two groups with distinct overall survival (log-rank P = 0.0005) in Kaplan-Meier analysis (Fig. 1B). This cutoff risk score was applied to two additional cohorts from Bhattacharjee et al. (13) (n = 84) and Shedden et al. (14) (n = 442). The 12-gene signature generated significant prognostic categorization in both lung adenocarcinoma cohorts from Bhattacharjee et al. (13) (log-rank P = 0.05; Fig 1C) and Shedden et al. (14) (log-rank P = 0.01; Fig. 1D) in Kaplan-Meier analyses. In all three studied non-small cell lung cancer cohorts, the low-risk groups had above 80% of 2-year postoperative survival rate, representing a significantly better prognosis compared with the corresponding high-risk groups for which the 2-year survival was ranging from 38% to 58%. Furthermore, the 12-gene genomic instability signature had a significant hazard ratio (HR ≥ 1.55, p <0.05) in predicting poor-prognosis in all three studied lung cancer cohorts (Table 6). The results show that the expression patterns of the 12-gene genomic instability signature could be used to predict postoperative survival in non-small cell lung cancer patients.
Figure 1.
The 12-gene genomic instability signature predicts overall survival in non-small cell lung cancer. (A) Histogram of gene expression-defined risk scores in the training cohort from Bild et al. (12). The peak value with risk score zero in the histogram was defined as the cutoff in prognostic categorization. Gene expression defined-high and low risk groups had significant post-operative survival in patient cohorts from (B) Bild et al. (12), (C) Bhattacharjee et al. (13), and (D) Shedden et al. (14) in Kaplan-Meier analyses.
Table 6.
Hazard ratio of prognostic model for lung cancer and ovarian cancer presented in Figure 1 and Figure 2.
| Patient Cohort | Hazard Ratio [95% CI] |
|---|---|
| Lung Cancer | |
| Bild et al. (12) (n=111) | 2.87 [1.69, 4.87] |
| Bhattacharjee et al. (13) (n=84) | 2.63 [1.61, 4.29] |
| Shedden et al. (14) (n=442) | 1.55 [1.23, 1.96] |
| Ovarian Cancer | |
| Bild et al. (12) (Training, n=62) | 16.82 [5.05, 56.06] |
| Bild et al. (12) (Testing, n=62) | 2.08 [1.00, 4.29] |
Genomic instability signature predicts ovarian cancer outcome
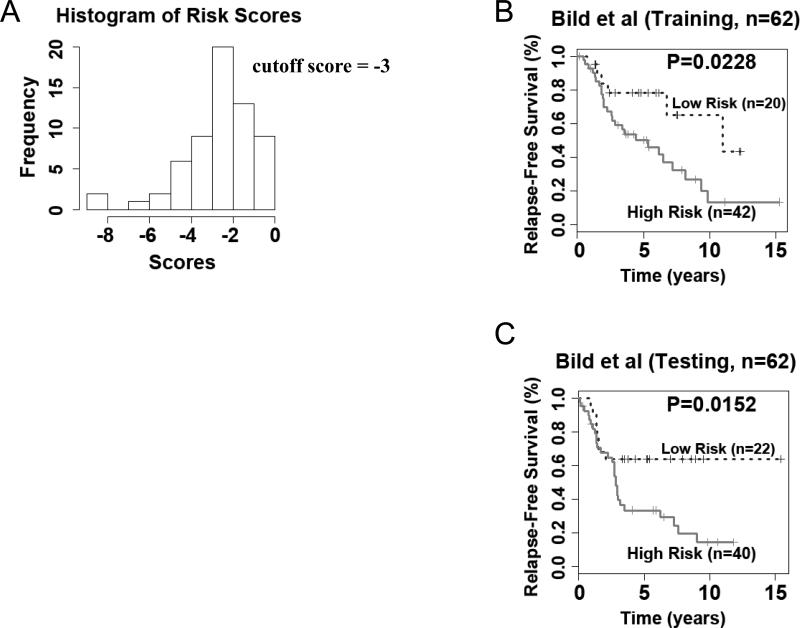
Ovarian cancer is a common malignancy in women, whose prognosis is bleak due to a usually advanced disease stage at the time of diagnosis. In order to extent the potential usefulness of our genomic instability signature we explored its value for predicting clinical outcome in patients with ovarian cancer. The ovarian cancer cohort (n = 124) from Bild et al. (12) was randomly split into a training set (n = 62) and a testing set (n = 62). A Cox model was built on the training set using the signature genes as covariates. A survival risk score was generated for each patient. A cutoff value was identified based on the histogram of the risk scores in the training set (Fig. 2A). Patients with a risk score greater than the cutoff were stratified into the high-risk group, and otherwise, into the low-risk group. The high- and low-risk groups had significantly (log-rank P = 0.02) different relapse-free survival in the training cohort in Kaplan-Meier analysis (Fig. 2B). This training model and stratification scheme were applied to the testing set, and generated significant prognostic stratification (log-rank P = 0.015) in Kaplan-Meier analysis (Fig. 2C). The survival risk score estimated by the model also showed strong association with the ovarian cancer relapse-free survival (hazard ratio = 2.08, 95% CI: [1.00-4.29]) in the test set (Table 6). These results demonstrate that the 12-gene genomic instability signature could indentify more aggressive ovarian cancers that were more likely to develop recurrence after surgical resections and initial treatment. The high risk patients defined with this genomic instability gene signature might benefit from second line chemotherapy.
Figure 2.
The 12-gene genomic instability signature predicts overall survival in ovarian cancer. (A) Histogram of gene expression-defined risk scores in the training cohort from Bild et al. (12). The peak value with risk score of -3 in the histogram was defined as the cutoff in prognostic categorization. Gene expression defined-high and low risk groups had significant post-operative ovarian cancer survival in both (B) training and C) testing cohorts.
Functional pathway analysis
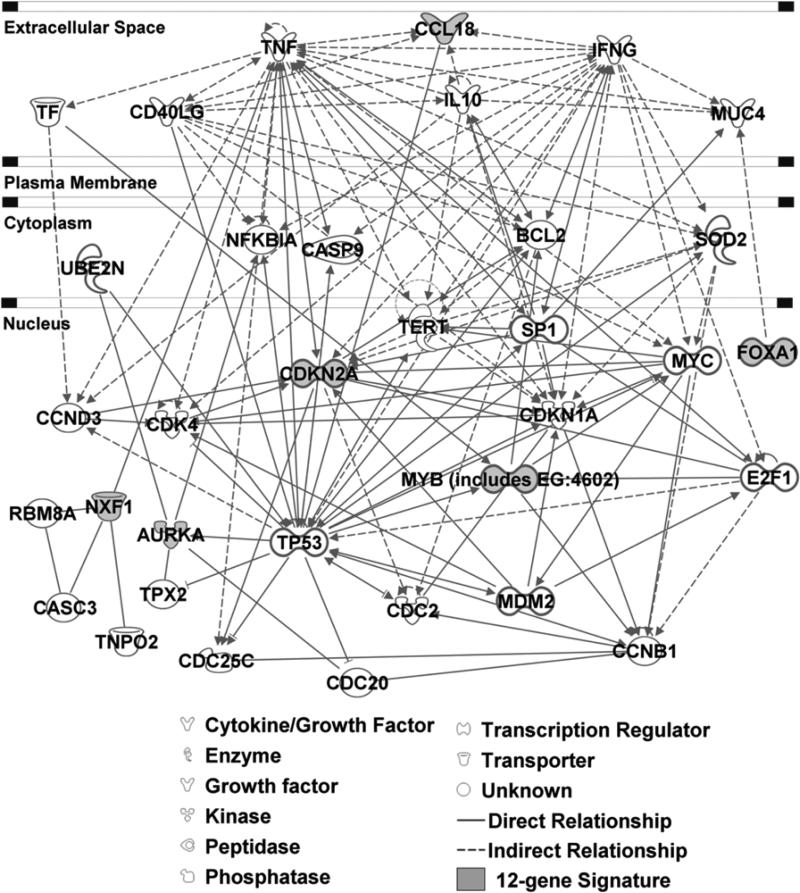
The 12-gene genomic instability signature was able to distinguish more aggressive tumors in multiple cancer types, indicating that this signature might be involved in important mechanisms of tumor genesis and progression. Functional pathway analysis was performed based on curated database of molecular interactions reported in the literature using Ingenuity Pathway Analysis. The results show that the signature genes interact with multiple prominent cancer signaling pathways, including the oncogenes NFKBIA, MYC, BCL2, CDK1 (CDC2), E2F1, and SOD2 as well as the tumor suppressor genes TP53 and CASP9 (Fig. 3). Specifically, Aurora-A (AURKA) is an essential regulator of mitosis and is frequently amplified in human cancer. Aberrant expression of Aurora-A (AURKA) in mammalian cells induces centrosome amplification, genomic instability, and transformation. The E2 ubiquitin ligase UBE2N binds specifically to the Aurora-A (AURKA) Phe-31 variant (18) and stimulates ubiquitination of Aurora-A (AURKA) both in vitro and in vivo (19). In MCF7 cells, human Aurora-A (AURKA) protein increases polyubiquitination of human IKBA (NFKBIA) protein (20). The p14ARF (CDKN2A)-induced G2 arrest and subsequent apoptosis inhibits the growth of human tumoral cells lacking functional TP53, which correlated with inhibition of CDC2 (CDK1) activity (21). Studies showed that human p14ARF (CDKN2A) protein activates Caspase9 (CASP9) and mitochondrial apoptosis pathway entirely independent of TP53, and these caspase-9-like activities were greatly enhanced in cells lacking functional p21 (CDKN1A) (22). Furthermore, overexpression of SOD2 decreases accumulation of CDKN2A during confluent growth of fibroblasts (23). BCL-2 is a direct target of c-Myb, and overexpression of c-Myb was accompanied by up-regulation of BCL-2 expression (24). Methylation of E2F elements derived from the dihydrofolate reductase, E2F1, and CDC2 promoters prevents the binding of all E2F family members tested (E2F1 through E2F5) (25). In contrast, methylation of the E2F elements derived from the c-Myc and c-Myb promoters minimally affects the binding of E2F2, E2F3, E2F4, and E2F5 but significantly inhibits the binding of E2F1 (26). CCL18 is one of the most abundant chemokines produced by immature dendritic cells, and may be part of an inhibitory pathway to limit specific immune responses at peripheral sites. In maturing human dendritic cells (27). TNF-α (TNF) protein decreases expression of CCL18 mRNA (28).
Figure 3.
Functional pathway analysis of the 12-gene genomic instability signature using Ingenuity Pathway Analysis. The biological network showed genes interacting with the signature genes as reported in the literature.
Discussion
Genomic instability is a defining and ubiquitous feature of human cancers (1;2). In many instances the degree of genomic instability is correlated to the clinical course, with highly unstable tumors showing an in general poorer prognosis (3;29;30). Also, the degree of genomic instability increases with cancer progression: pre-invasive dysplastic lesions usually carry only few genomic imbalances, often in the form of gains and losses of entire chromosomes or chromosome arms (2;31). These early changes can occur in an otherwise stable genome. During tumor progression, additional imbalances accumulate, including regional high-level copy number genomic amplification. Specific tumor entities can present with different degrees of genomic instability. For instance, HNPCC-associated colorectal carcinomas are karyotypically stable and present in general with a better prognosis compared to sporadic tumors that are usually highly aneuploid (32-34). A similar situation can be observed in breast carcinomas: here, patients with tumors that are diploid or close to diploid fare considerably better then patients with aneuploidy carcinomas (3;30). In a recent publication we explored whether that difference in the degree of genomic instability and its profound correlation to prognosis is biologically related to gene expression signatures of poor prognosis that were identified over the past few years in large cohorts of breast cancer patients. In order to test this hypothesis, we profiled 48 breast tumors that were defined as being genomically stable and unstable. This resulted in a 12-gene signature of genomic instability that clearly separated the two groups (8). In addition, the 12-gene signature predicted outcome in multiple independent data sets, and, conversely, the clinically used poor prognosis signature correctly predicted the degree of genomic instability in our dataset. This established a biological link between independently derived prognostic signatures (5-7). We were now interested in exploring whether this signature would also assist prognostication of disease outcome in cancer entities other then breast. In order to test this hypothesis, we applied the signature to published datasets from lung (12-14), colorectal (9-11), and ovarian carcinomas (12).
In each studied cancer type, a patient stratification scheme was developed based on the expression of the 12-gene genomic instability signature, and was validated on independent patient cohorts. Based on the clinical outcome provided in three colon cancer cohorts, a machine learning algorithm JRip was used in the model construction on the training set (n = 50) with stage II colon carcinoma to predict patients’ recurrence after surgery. The model accuracy was 72% on the training cohort in a 10-fold cross validation. This prognostic model was applied to the combined testing set (n = 42) and achieved an overall accuracy of 69.1% in the cross cohort validation. These results are more accurate (P < 0.05) compared with random predictions. In the prognostic validation of non-small cell lung cancer, a prognostic model was built with Cox model using the gene expression profiles as covariates. The cutoff point for prognostic categorization was defined based on histogram of gene expression defined-risk scores on the training cohort (n = 111). This stratification scheme was applied to two additional patient cohorts (n = 526). In each patient cohort, the gene signature separated patients into different prognostic groups with remarkably different (log-rank P < 0.05) clinical outcomes in Kaplan-Meier analyses. Similarly, this scheme was used in the prognostic validation on ovarian cancer. In both training and testing cohorts (n = 124), the gene expression defined-model provided significant (log-rank P < 0.02) post-operative prognostic stratification in Kaplan-Meier analyses. The 12-gene instability signature had significantly higher hazard ration in poor prognosis groups over good prognosis groups in all studied lung cancer and ovarian cancer cohorts.
Genome-wide association studies utilizing human tissue samples have enhanced the prognostic capacity of cancer outcomes. Four breast cancer signatures, including intrinsic subtypes (35), poor prognosis signature (MammaPrint®) (7), recurrence score (Oncotype DX®) (36), and wound response (37), represent largely the same prognostic space (38). Our identified 12-gene genomic instability signature predicted disease-free survival and overall survival in multiple breast cancer patient cohorts with heterogeneous disease stage, including both early stage and advanced breast cancers (8). In the evaluation, the 12-gene genomic instability signature is comparable as Oncotype DX® and could potentially be more accurate than the other above mentioned signatures in terms of predicting disease-free survival and overall survival in van de Vijver's cohort (7). More importantly, the 12-gene signature showed prognostic ability beyond early-stage breast cancer that constitutes the patient group for MammaPrint® and Oncotype DX®. The 12-gene genomic instability signature quantified disease-free survival and overall survival in a broad patient population including those with advanced stage (T3/T4), tumor grade III, lymph node metastasis, or negative estrogen receptor status (ER-) (8). The present study confirms that the identified 12-gene genomic instability signature predicted clinical outcomes in multiple tumor types. Together, this 12-gene signature could extend breast cancer prognostic space defined by MammaPrint® and Oncotype DX®, among other breast cancer signatures with potential clinical utility (12;39-41).
This study demonstrates that our identified 12-gene genomic instability signature could predict clinical outcomes in multiple cancer types with epithelial origins. Thus, it provides further evidence linking genomic instability and associated gene expression, and disease courses in human cancers. The functional pathway analysis with curated IPA database delineated a biological network with tight connections between the signature genes and numerous well established cancer hallmarks, indicating important roles of the genomic instability gene signature in tumor genesis and progression. While it is clear that the degree of genomic instability has diagnostic and prognostic implications, it is tempting to speculate that pursuing genomic instability therapeutically could provide entry points for a target that is unique to cancer cells.
Acknowledgements
The authors express their gratitude to Mr. Buddy Chen for editing the manuscript and illustrations. T.R. is supported by National Cancer Institute (Intramural Research Program). N.L.G. is supported by R01LM009500 and NCRR P20 RR16440 and Supplement.
Grant Sponsors: This research is supported by National Cancer Institute ((Intramural Research Program), National Library of Medicine (R01LM009500) and NCRR (P20 RR16440 and Supplement) from the NIH.
References
- 1.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors 7. Nat.Genet. 2003 Aug;34(4):369–76. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 2.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation 13. Genes Chromosomes.Cancer. 1999 Jul;25(3):195–204. doi: 10.1002/(sici)1098-2264(199907)25:3<195::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 3.Auer G, Eriksson E, Azavedo E, Caspersson T, Wallgren A. Prognostic significance of nuclear DNA content in mammary adenocarcinomas in humans 12. Cancer Res. 1984 Jan;44(1):394–6. [PubMed] [Google Scholar]
- 4.Kronenwett U, Huwendiek S, Ostring C, Portwood N, Roblick UJ, Pawitan Y, Alaiya A, Sennerstam R, Zetterberg A, Auer G. Improved grading of breast adenocarcinomas based on genomic instability 2. Cancer Res. 2004 Feb 1;64(3):904–9. doi: 10.1158/0008-5472.can-03-2451. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de RM, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc.Natl.Acad.Sci.U.S.A. 2001 Sep 11;98(19):10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc.Natl.Acad.Sci.U.S.A. 2003 Sep 2;100(18):10393–8. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Vijver MJ, He YD, van 't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N.Engl.J.Med. 2002 Dec 19;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 8.Habermann JK, Doering J, Hautaniemi S, Roblick UJ, Bundgen NK, Nicorici D, Kronenwett U, Rathnagiriswaran S, Mettu RK, Ma Y, et al. The gene expression signature of genomic instability in breast cancer is an independent predictor of clinical outcome. Int.J Cancer. 2009 Apr 1;124(7):1552–64. doi: 10.1002/ijc.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrier A, Boelle PY, Roser F, Gregg J, Tse C, Brault D, Lacaine F, Houry S, Huguier M, Franc B, et al. Stage II Colon Cancer Prognosis Prediction by Tumor Gene Expression Profiling. J Clin Oncol. 2006 Oct 10;24(29):4685–91. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 10.Barrier A, Roser F, Boelle PY, Franc B, Tse C, Brault D, Lacaine F, Houry S, Callard P, Penna C, et al. Prognosis of stage II colon cancer by non-neoplastic mucosa gene expression profiling. Oncogene. 2006 Oct 9;26(18):2642–8. doi: 10.1038/sj.onc.1210060. [DOI] [PubMed] [Google Scholar]
- 11.Barrier A, Lemoine A, Boelle PY, Tse C, Brault D, Chiappini F, Breittschneider J, Lacaine F, Houry S, Huguier M, et al. Colon cancer prognosis prediction by gene expression profiling. Oncogene. 2005 Aug 1;24(40):6155–64. doi: 10.1038/sj.onc.1208984. [DOI] [PubMed] [Google Scholar]
- 12.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006 Jan 19;439(7074):353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc.Natl.Acad.Sci.U.S.A. 2001 Nov 20;98(24):13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat.Med. 2008 Aug;14(8):822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen WW. Fast Effective Rule Induction. Proceedings of the Twelfth International Conference on Machine Learning. 1995:115–23. [Google Scholar]
- 16.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2nd Edition Morgan Kaufmann; 2005. [Google Scholar]
- 17.Everitt B, Hothorn T. A Handbook of Statistical Analyses Using R. Chapman & Hall/CRC; Boca Raton, FL: 2006. [Google Scholar]
- 18.Ewart-Toland A, Briassouli P, de Koning JP, Mao JH, Yuan J, Chan F, Carthy-Morrogh L, Ponder BA, Nagase H, Burn J, et al. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human 1. Nat.Genet. 2003 Aug;34(4):403–12. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- 19.Briassouli P, Chan F, Linardopoulos S. The N-terminal domain of the Aurora-A Phe-31 variant encodes an E3 ubiquitin ligase and mediates ubiquitination of IkappaBalpha. Hum.Mol.Genet. 2006 Nov 15;15(22):3343–50. doi: 10.1093/hmg/ddl410. [DOI] [PubMed] [Google Scholar]
- 20.Briassouli P, Chan F, Linardopoulos S. The N-terminal domain of the Aurora-A Phe-31 variant encodes an E3 ubiquitin ligase and mediates ubiquitination of IkappaBalpha. Hum.Mol.Genet. 2006 Nov 15;15(22):3343–50. doi: 10.1093/hmg/ddl410. [DOI] [PubMed] [Google Scholar]
- 21.Eymin B, Leduc C, Coll JL, Brambilla E, Gazzeri S. p14ARF induces G2 arrest and apoptosis independently of p53 leading to regression of tumours established in nude mice. Oncogene. 2003 Mar 27;22(12):1822–35. doi: 10.1038/sj.onc.1206303. [DOI] [PubMed] [Google Scholar]
- 22.Hemmati PG, Normand G, Verdoodt B, von HC, Hasenjager A, Guner D, Wendt J, Dorken B, Daniel PT. Loss of p21 disrupts p14 ARF-induced G1 cell cycle arrest but augments p14 ARF-induced apoptosis in human carcinoma cells. Oncogene. 2005 Jun 9;24(25):4114–28. doi: 10.1038/sj.onc.1208579. [DOI] [PubMed] [Google Scholar]
- 23.Sarsour EH, Agarwal M, Pandita TK, Oberley LW, Goswami PC. Manganese superoxide dismutase protects the proliferative capacity of confluent normal human fibroblasts. J.Biol.Chem. 2005 May 6;280(18):18033–41. doi: 10.1074/jbc.M501939200. [DOI] [PubMed] [Google Scholar]
- 24.Salomoni P, Perrotti D, Martinez R, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells constitutively expressing c-Myb is associated with induction of BCL-2 expression and Myb-dependent regulation of bcl-2 promoter activity 1. Proc.Natl.Acad.Sci.U.S.A. 1997 Apr 1;94(7):3296–301. doi: 10.1073/pnas.94.7.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc.Natl.Acad.Sci.U.S.A. 2000 Jun 6;97(12):6481–6. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc.Natl.Acad.Sci.U.S.A. 2000 Jun 6;97(12):6481–6. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vulcano M, Struyf S, Scapini P, Cassatella M, Bernasconi S, Bonecchi R, Calleri A, Penna G, Adorini L, Luini W, et al. Unique regulation of CCL18 production by maturing dendritic cells. J.Immunol. 2003 Apr 1;170(7):3843–9. doi: 10.4049/jimmunol.170.7.3843. [DOI] [PubMed] [Google Scholar]
- 28.Vulcano M, Struyf S, Scapini P, Cassatella M, Bernasconi S, Bonecchi R, Calleri A, Penna G, Adorini L, Luini W, et al. Unique regulation of CCL18 production by maturing dendritic cells. J.Immunol. 2003 Apr 1;170(7):3843–9. doi: 10.4049/jimmunol.170.7.3843. [DOI] [PubMed] [Google Scholar]
- 29.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat.Genet. 2006 Sep;38(9):1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 30.Kronenwett U, Ploner A, Zetterberg A, Bergh J, Hall P, Auer G, Pawitan Y. Genomic instability and prognosis in breast carcinomas 1. Cancer Epidemiol.Biomarkers Prev. 2006 Sep;15(9):1630–5. doi: 10.1158/1055-9965.EPI-06-0080. [DOI] [PubMed] [Google Scholar]
- 31.Ried T. Homage to Theodor Boveri (1862-1915): Boveri's theory of cancer as a disease of the chromosomes, and the landscape of genomic imbalances in human carcinomas 2. Environ.Mol.Mutagen. 2009 Oct;50(8):593–601. doi: 10.1002/em.20526. [DOI] [PubMed] [Google Scholar]
- 32.Eshleman JR, Casey G, Kochera ME, Sedwick WD, Swinler SE, Veigl ML, Willson JK, Schwartz S, Markowitz SD. Chromosome number and structure both are markedly stable in RER colorectal cancers and are not destabilized by mutation of p53 1. Oncogene. 1998 Aug 13;17(6):719–25. doi: 10.1038/sj.onc.1201986. [DOI] [PubMed] [Google Scholar]
- 33.Ghadimi BM, Sackett DL, Difilippantonio MJ, Schrock E, Neumann T, Jauho A, Auer G, Ried T. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations 2. Genes Chromosomes.Cancer. 2000 Feb;27(2):183–90. [PMC free article] [PubMed] [Google Scholar]
- 34.Schlegel J, Stumm G, Scherthan H, Bocker T, Zirngibl H, Ruschoff J, Hofstadter F. Comparative genomic in situ hybridization of colon carcinomas with replication error 9. Cancer Res. 1995 Dec 15;55(24):6002–5. [PubMed] [Google Scholar]
- 35.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc.Natl.Acad.Sci.U.S.A. 2003 Jul 8;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N.Engl.J.Med. 2004 Dec 30;351(27):2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 37.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, Dai H, He YD, van 't Veer LJ, Bartelink H, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc.Natl.Acad.Sci.U.S.A. 2005 Mar 8;102(10):3738–43. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massague J. Sorting Out Breast-Cancer Gene Signatures. N Engl J Med. 2007 Jan 18;356(3):294–7. doi: 10.1056/NEJMe068292. [DOI] [PubMed] [Google Scholar]
- 39.Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland A, Borresen-Dale AL, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS.Med. 2006 Mar;3(3):e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007 Jan 18;356(3):217–26. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 41.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005 Jul 28;436(7050):518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]