Abstract
With recent advances in microarrays and sequencing it is now relatively straightforward to compare pre-mRNA splicing patterns in different cellular conditions on a genome-wide scale. Such studies have revealed extensive changes in cellular splicing programs in response to stimuli such as neuronal depolarization, DNA damage, immune signaling and cellular metabolic changes. However, for many years our understanding of the signaling pathways responsible for such splicing changes was greatly lacking. Excitingly, over the past few years this gap has begun to close. Recent studies now suggest notable trends in the mechanisms that link cellular stimuli to downstream alternative splicing events. These include regulated synthesis or degradation of splicing factors, differential protein–protein interactions, altered nuclear translocation and changes in transcription elongation.
Alternative splicing and the signals that regulate it
In the last decade our knowledge in the field of alternative splicing has increased exponentially due to technical advancements such as splicing-sensitive microarrays and deep sequencing. These studies have revealed that nearly all human genes undergo some form of alternative splicing 1, 2. Most typically, alternative splicing involves the differential inclusion or exclusion of a specific exon in different cell types or growth conditions, although all other imaginable patterns have been observed including retention of introns, exclusion of a portion of an exon, and mutually exclusive inclusion of exons 3. In each case, the pattern of splicing is generally determined by the binding of regulatory proteins to cis-acting auxiliary sequences that in turn control the location of binding and/or activity of the enzymatic complex at neighboring splice sites (Box 1). Importantly, any of these differential patterns have the capacity to alter the open reading frame of the resultant mRNA or alter the presence of cis-regulatory elements that control mRNA stability or translation. Therefore, the precise control of alternative splicing plays an essential role in shaping the proteome of any given cell, and changes in splicing patterns can significantly alter cellular function in response to changing environmental conditions3–5.
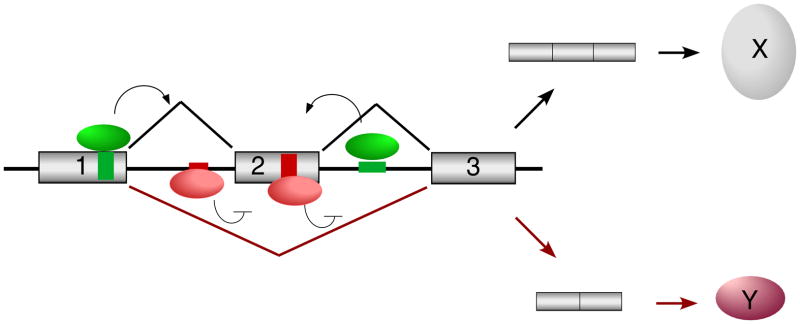
Box 1. Combinatorial Regulation of Alternative Splicing.
The basic joining of exons and removal of introns is catalyzed by a macromolecular complex called the spliceosome47. The precise sites of cleavage and ligation are determined by binding of various subunits of the spliceosome to sequence elements at the exon and intron boundaries in a pre-mRNA; namely the 5 splice site, the branch point sequence, a pyrimidine-rich track, and the 3 splice site. However, mammalian splice sites are poorly conserved and are typically not sufficient to bind the spliceosome with high affinity. Proteins bound to non-splice site sequences within the exon or intron can influence the efficiency of spliceosomal binding via mechanisms that include, but not limited to, spliceosomal recruitment or steric hindrance48,49. Sequences that promote spliceosomal recognition of an exon are called exonic or intronic splicing enhancers (ESE or ISE; Figure I, green boxes), whereas sequences that are required to inhibit recognition of the exon are called splicing silencers (ESS or ISS; Figure I, red boxes). Members of the ubiquitously expressed SRSF protein family typically bind enhancers to promote exon inclusion (green ovals), whereas members of the hnRNP family of proteins typically repress exon usage via silencer elements (red ovals). Other splicing regulators such as the FOX, CELF, neuro-oncological ventral antigen (NOVA) and muscleblind-like (MBNL) proteins are more tissue restricted and function equally as repressors and enhancers of splicing through mechanisms that are still largely undefined. Most transcripts that have been studied contain multiple ISE/ESEs and ISS/ESSs and are bound by multiple regulatory proteins which can antagonize the function of each other directly or indirectly (Figure I). Therefore, subtle changes in the balance of expression or binding of individual regulatory proteins can frequently alter the ratio of mRNA isoform expression.
Fundamental differences in splicing patterns have been observed in epithelial versus mesenchymal cells, neurons before and after depolarization, heart tissue during development and in disease conditions, resting and activated T cells, cells during circadian rhythms and cells before and after initiation of apoptosis, to name but a few examples6–10. In all of these cases, at least some of the differential alternative splicing events have been shown to contribute to the functional outcome of the developmental or signaling processes. Over the past decade we have also learned much about various signaling proteins that catalyze post-translational modifications on splicing proteins, often in response to changes in cellular growth or environmental conditions (Box 2).
Box 2. Post-Translational Modification of Splicing Proteins.
Splicing regulatory proteins are subject to modification by phosphorylation, acetylation, methylation, sumolylation and hydroxylation, although in many cases the modifying enzymes and/or functional consequence are not fully established26, 50–52. The best characterized modification is the phosphorylation of the extensive Arg-Ser dipeptides found within SR proteins26. For example, AKT-mediated phosphorylation of sites on SFRS1 (also called ASF/SF2 or SRp30a) alters the activity of this protein in fibronectin53 and caspase-9 alternative splicing54. SR proteins are also direct substrates for members of the SRPK family and CLK family of dual-specificity kinases. Interestingly, each of these three kinase families phosphorylate distinct serine residues on SR proteins, with differing functional consequences26. The activity of these kinases is also presumably countered by phosphatases, with at least protein phosphatase 1 (PP1) being known to act on SR proteins55. Given that both cycles of both phosphorylation and dephosphorylation are required for SR protein function, the balance of kinase and phosphatase activity is crucial in the promotion of promote splicing.
HnRNP proteins, as well as other non-SR splicing factors, are also subject to extensive post-translational modifications51. Protein arginine methyltransferases (PRMTs) modify many of the RGG box-containing hnRNPs56, and PKA, casein kinase II and MNK1/2 phosphorylate hnRNP I (PTB), hnRNP C and hnRNP A1, respectively31, 57–59. Additional kinases including phosphoinositide-3-kinase (PI3K), CaMKIV, ATM and ATR also modulate splicing; however, whether these proteins directly modify splicing factors, or function indirectly, has yet to be determined.
For many years, however, our knowledge of the functional consequences of the modification of splicing proteins, and the mechanisms by which signaling pathways lead to inducible changes in splicing, has lagged far behind. Excitingly, this is beginning to change, and although much remains to be learned, recent years have brought notable progress in our understanding of the mechanistic connections between signaling pathways and alternative splicing. In this review we will highlight some emerging themes in the recent literature regarding how signaling pathways alter the activity of splicing regulatory proteins and/or the splicing process. Moreover, our focus will be on acute (fast) signaling events rather than on differentiation programs or disease conditions, which have been reviewed elsewhere11–13.
Making and Breaking of RNA-binding proteins: A prime way to regulate splicing
Arguably the simplest mechanism by which a signaling pathway can affect alternative splicing is by altering the expression level of a critical regulatory protein. Given the complexity of influences on a given transcript (Box 1), even relatively modest changes in the expression of one splicing factor can cause a shift in the balance of forces determining exon inclusion or exclusion. Signaling pathways are well documented to alter transcription programs through regulation of transcriptional activators such as nuclear factor-kappa B (NF-κB), signal transducer and activator of transcription (STATs), nuclear factor of activated T-cells (NFAT), and many others. It is therefore to be expected that one consequence of signaling is the induced transcription of genes encoding SR proteins, heterogeneous nuclear ribonucleoproteins (hnRNPs) or other splicing regulators, which would alter the splicing of genes responsive to such factors. Such a mechanism has been proposed, for instance, to contribute to the signal responsive splicing of the gene encoding the receptor tyrosine phosphatase CD45 in response to the antigen stimulation of T cells. In this case, activation-induced skipping of CD45 exons 4 and 6 is driven by two proteins, PTB-associated splicing factor (PSF) and hnRNP L-like (hnRNPLL)14–16. HnRNPLL protein expression is increased severalfold upon T cell activation, with a corresponding increase in hnRNPLL mRNA. Thus, although formal proof is still lacking, induced expression of hnRNPLL is assumed to result from transcriptional regulation. Increased mRNA expression of additional splicing factors, such as the SR proteins SRSF1, 6 and 11, has also been demonstrated in response to T cell activation17. Furthermore, widespread differences in the mRNA expression of SR and hnRNP proteins has been observed in several differentiation processes18, 19, suggesting that this type of regulation is particularly efficient for establishing a long-term, and less-reversible, splicing pattern in terminally differentiated cells.
Importantly, however, not all inducible changes in protein expression are a result of transcription. This has, for example, been demonstrated for the splicing regulatory protein CELF1 (CUGBP, Elav-like family member 1; also called CUGBP1 or Brunol2). CELF1 protein levels are increased in animal models of Myotonic Dystrophy (DM)20 owing to an increase in the phosphorylation and stability of CELF1, thereby leading to an overall increase in its steady-state levels 21. This protein-stabilizing phosphorylation is attributable to increased protein kinase C (PKC) activity in the DM cells21. Strikingly, increased protein stability is not the only mechanism for regulating CELF1 expression; CELF1 protein expression is also regulated by miRNAs during heart development22. Such combinatorial control of protein expression underscores the idea that precise modulation of regulatory protein expression is critical to maintain appropriate splicing patterns required for cellular function.
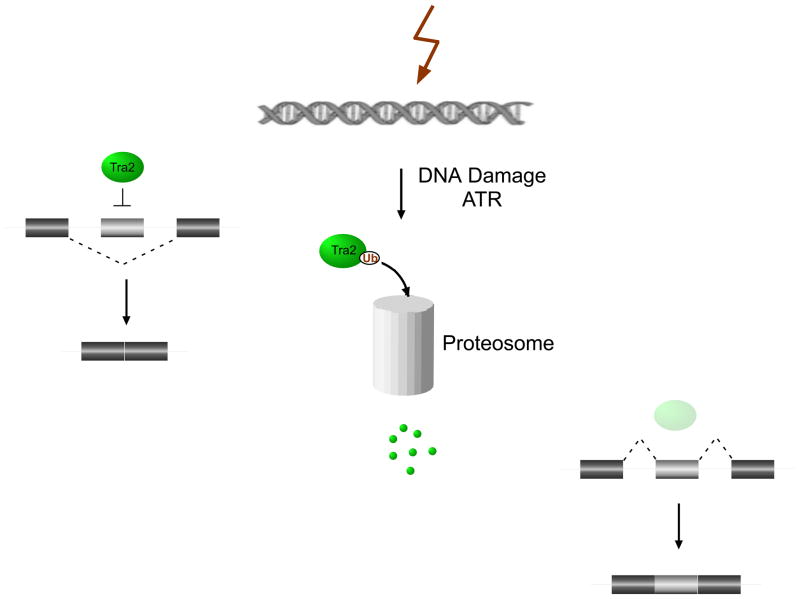
Interestingly, signaling pathways not only can promote protein stability, but also induce protein turn-over. Indeed, several recent papers suggest a common theme of signal-induced protein degradation as a mechanism to decrease expression of a splicing regulator in order to shift splicing patterns. One example comes from DNA damage induced changes in splicing of Drosophila Taf1 (TBP-associated factor 1) (Fig 1). A screen for factors that modulate alternative splicing in response to the chemotherapeutic agent camptothecin (CPT) identified the SR-related protein Transformer-2 (Tra2) as a key regulator in this pathway23, 24. Tra2 is downregulated upon CPT treatment in a manner that is independent of new protein synthesis. Critically, loss of Tra2 protein, as well as the DNA damage-induced change in splicing, could be prevented by treatment with the proteasome inhibitor MG132. These results suggest a role for the proteasome in regulating Tra2 protein levels and, in turn, alternative splicing of Taf1. Further support for this model came from an experiment in which the alteration of a potential ubiquitylation site within Tra2 conferred partial resistance to CPT-induced Tra2 degradation.
Fig.1. Regulated protein turnover as a means to control alternative splicing.
An example of how the regulated degradation of RNA binding proteins can modulate alternative splicing. Tra2 levels are reduced by proteasomal degradation in response to ataxia telangiectasia and Rad3 related (ATR) signaling, most likely through ubiquitin (Ub)-mediated degradation. The reduction in Tra2 protein alleviates normal Tra2 repression of Taf1 alternative exons (light grey boxes). Reproduced, with permission from [23].
Recently, two additional examples have been published in which regulated protein degradation is a determining factor of DNA damage-induced apoptosis in human cells. One case involves the regulated acetylation and stability of the SR protein SRSF2 (also called SC35 or SRp30b)25. Acetylation of SRSF2 on Lys52 in its RNA-binding domain was found to inhibit RNA binding and promote its proteasomal degradation. Interestingly, acetylation of SRSF2 was attributed to the competing activities of the acetyl transferase TIP60 and the deacetylase HDAC6. Strikingly, treatment of cells with DNA damaging agents such as cisplatin inhibits TIP60 expression, thereby decreasing acetylation of SRSF2 and resulting in increased stability and expression of the protein25. Furthermore, TIP60 regulates the activity of SRSF2 by controlling nuclear translocation of the SR kinases SRPK1 and SRPK2, which induce phosphorylations on SR proteins that control their localization and activity26. Thus, cisplatin-induced loss of TIP60 leads to the accumulation of non-acetylated, phosphorylated SRSF2, which in turn promotes splicing of caspase-8 to the proapoptotic isoform25. Together, this work provides an exciting example of how multiple post-translational modifications and regulated proteasomal degradation of a splicing factor together promote apoptosis in response to DNA damage.
Cisplatin and oxaliplatin also control the splicing of the BCLX gene to favor the generation of the proapoptotic Xs over the antiapoptotic XL form27. Formation of the proapoptotic BCL-Xs isoform upon DNA damage requires a pathway involving ataxia telangiectasia mutated (ATM), CHK2, p53, and tyrosine phosphatases, and a cis-regulatory element in the affected BCLX exon termed SB1. Interestingly, previous work showed that SB1 mediates PKC-induced repression of BCL-Xs. Therefore, the PKC and ATM pathways appear to have opposite effects on the same regulatory event. The finding that pharmacological inhibition of the proteasome abrogates the formation of the smaller BCL-X isoform, induced by either DNA damage or PKC inhibition, led to the suggestion that these pathways converge on a regulatory protein stabilized in its phosphorylated form and prone to proteasome-mediated degradation in its non-phosphorylated state27. Therefore, this work provides another example of how regulation of the proteasome can act as a prominent step in signal-induced alternative splicing; however, whether the splicing factor regulated by ATM and/or PKC is SRSF2, CELF1 or some additional target of regulated degradation remains to be determined. Interestingly, the splicing of BCL-X is also switched toward the smaller isoform in response to ceramide signaling through effects on the core splicing factor spliceosome associated protein 155 (SAP155)28. Thus, BCL-X provides a clear example of how a single gene can be regulated downstream of multiple distinct signaling pathways via convergent and/or unique mechanisms.
Moving RNA-binding proteins
As an alternative to altering protein stability, some splicing events are controlled by signal-induced changes in the localization or accessibility of critical regulatory proteins. Because many RNA-binding proteins (e.g., members of the SR and hnRNP families) shuttle between the nucleus and the cytoplasm, it is not surprising that some signaling pathways accomplish splicing regulation by controlling the nuclear-cytoplasmic distribution of splicing regulatory proteins4. For example, as mentioned above SRPK1 intracellular distribution is highly regulated. Indeed, two studies have shown that different SRPK1 regulatory proteins, TIP6025 and the heat-shock protein (HSP) co-chaperones HSP40 and activator of heat shock 90kDa protein ATPase homolog 1 (AHA1)29, can regulate the nuclear translocation of SRPK1 and, therefore, the phosphorylation of SRSF2. In the latter study29, SRPK1 is sequestered in the cytoplasm by interaction with the heat shock protein complex under normal conditions, but is released upon osmotic shock and translocates into the nucleus. Once in the nucleus, SRPK1 induces hyperphosphorylation of several SR proteins, which in turn alters their activity on downstream target genes26 and causes changes in splicing patterns.
HnRNP proteins also exhibit signal-responsive changes in localization. The best documented example is hnRNP A1, which is sequestered in the cytoplasm in response to osmotic shock through phosphorylation by the MAP kinase interacting serine/threonine kinases, MNK1/2. This phosphorylation of hnRNP A1 blocks its association with the nuclear transport factor KAPB2, thus preventing its nuclear import30, 31. Given the antagonistic relationship between hnRNP A1 and SR proteins, it is interesting to note that osmotic stress has an opposite effect on the nuclear localization of SRPK1 and hnRNP A1, suggesting that the reciprocal movement of these two proteins might amplify their differential effects on splicing. Regulation of hnRNP A1 nuclear transport has also been proposed to explain the increase in hnRNP A1 expression and RNA-binding that can drive a subset of alternative splicing events upon neuronal depolarization10. Finally, it is worth emphasizing that in the aforementioned phosphorylation-dependent regulation of CELF1, some conditions stabilize only nuclear, and not cytoplasmic, CELF121, thus further highlighting the importance of compartment-specific modifications and localization patterns of modifying enzymes.
Changing partners: modulating splicing through altered protein interactions
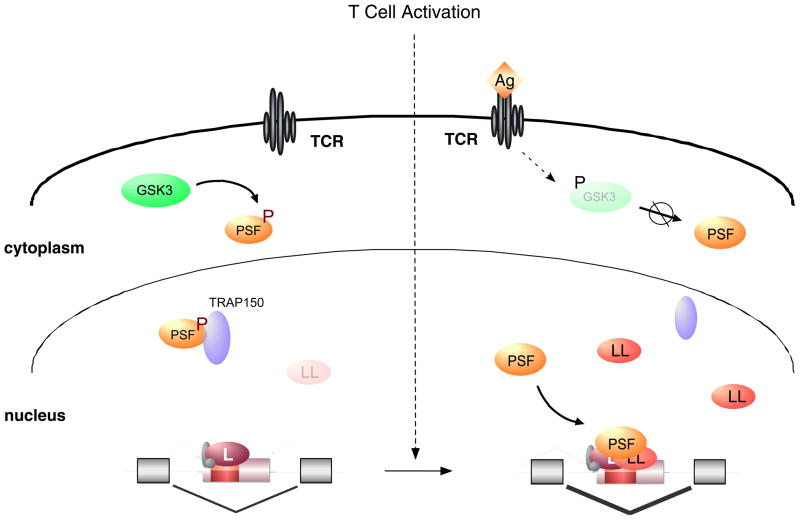
Recently, a new mechanism of signal-induced change in protein function was demonstrated in the regulation of CD45 alternative splicing in response to T cell activation (Fig. 2). Some of the antigen-induced skipping of CD45 exons is attributable to the increased expression of hnRNPLL, as described above; however, the multifunctional RNA-binding protein PSF also is necessary16. PSF only binds CD45 RNA in activated cells, but curiously, neither the nuclear-cytoplasmic distribution nor the abundance of PSF changes between cells grown under resting or stimulated conditions16. Further investigation revealed that Thr 687 of PSF is differentially phosphorylated in resting and activated T cells through the signal-regulated activity of the glycogen synthase kinase-3 (GSK3)32. Strikingly, the phospho-T687 form of PSF associates tightly with thyroid-hormone receptor associated protein 150 (TRAP150), which in turns prevents PSF–RNA binding. Upon T cell activation, GSK3 activity is reduced; de novo protein synthesis leads to gradual replacement of T687-phosphorylated PSF with the non-phosphorylated form, which is not bound by TRAP150 and thus is able to participate in CD45 exon exclusion32.
Fig. 2. Altered protein–protein interactions and induced protein expression regulate CD45 alternative splicing during T cell activation.
(i) In resting T cells, the RNA binding protein PSF is phosphorylated (P) by GSK3 and forms a tight complex with TRAP150. In this complex, PSF is unable to bind CD45 RNA and therefore does not participate in splicing regulation. (ii) Upon T cell activation by antigen (Ag) binding to the T cell receptor (TCR), GSK3 activity decreases due to an inhibiting phosphorylation. This leads to accumulation of newly translated, hypophosphorylated PSF which is not bound to TRAP150 and thus is recruited to the CD45 exon silencer to increase exon exclusion in activated T cells. HnRNP LL also contributes to increased CD45 exon exclusion in activated T cells. hnRNP LL mRNA and protein levels increase upon T cell activation, thus providing an example of a splicing regulator that is controlled by signal-induced changes in transcription.
Exactly how phosphorylation of PSF regulates its association with TRAP150, and why this interaction inhibits RNA-binding remains to be determined, but this system presents a mechanism distinct from altered expression or localization as a means to control the accessibility and functionality of a splicing regulatory protein. Of note, GSK3 has been implicated in TAU exon 10 splicing and in the phosphorylation of SRSF233, thus suggesting a broader role of GSK3-mediated signaling events in controlling alternative splicing through pathways in addition to its control of PSF. Moreover, a recent study of neuronal depolarization also hints at a model of regulated accessibility. HnRNP L binds calcium/calmodulin-dependent protein kinase-IV (CaMKIV)-responsive elements (CaRREs) to control depolarization-induced splicing events34. CaMKIV signaling increases hnRNP L–CaRRE binding without any apparent change in total nuclear expression34. Because CaMKIV also changes hnRNP L phosphorylation concomitant with binding, altered protein interactions might control hnRNP L accessibility, similar to that observed with PSF control. However, other mechanisms have not been ruled out, including the possibility that phosphorylation directly alters the affinity of hnRNP L for the CaRRE RNA.
A further example for signal-modified protein–protein interactions has been shown for the alternative splicing of the E3 ubiquitin ligase murine double minute-2 (MDM2) in response to DNA damage35. MDM2 controls p53 levels by targeting it for proteasomal degradation. Upon genotoxic stress, several Mdm2 exons are skipped thereby reducing functional MDM2 protein levels and allowing the accumulation of p53. Exon inclusion in Mdm2 is regulated cotranscriptionally, and this has been linked to an interaction between the DNA Polymerase II (Pol II) complex and the splicing machinery via Ewings sarcoma breakpoint region 1 (EWS1) and Y-box binding protein 1 (YB1), as depletion of either protein increases Mdm2 exon skipping in a way that mimics DNA damage. Specifically, EWS is part of the Pol II complex and is thought to recruit the spliceosome via an interaction with the splicing factor YB1, so as to facilitate the inclusion of exons with suboptimal splice sites. Consistent with the reduction of Mdm2 exon inclusion upon genotoxic stress, treatment with CPT to induce DNA damage reduces the EWS–YB1 interaction, thus rendering spliceosome recruitment and splicing less efficient. Notably, loss of the EWS–YB1 interaction by depletion of EWS and/or YB1 alters not only Mdm2 splicing, but also the splicing of at least five other genes that are regulated in response to DNA damage35. Therefore, the model of regulated protein interaction mediating signal-induced changes in splicing is probably a widespread phenomena of which we have only seen the tip of the iceberg.
Pol II modifications and chromatin structure: Regulating splicing cotranscriptionally
An increasing number of studies have demonstrated functional cross-talk between transcription and splicing. For example, numerous RNA-binding proteins, such as SR proteins and Sam68 can interact with the Pol II complex or transcription factors to facilitate their recruitment to nascent RNA36–38. Alternatively, altering the speed of Pol II, e.g. by changed chromatin structure or differential Pol II phosphorylation, can influence kinetically driven choices in splicing by allowing time for a weak exon to be bound by the spliceosome39.
Given the extent to which signaling pathways are known to influence transcription, it is not surprising that co-transcriptional regulation of splicing is frequently modulated in response to signaling events. The DNA damage-induced regulation of MDM2 provides an example of how transcription-coupled splicing can be influenced by altering the efficiency of Pol II-mediated recruitment of splicing factors35. Similarly, at least two other mechanisms of signal-induced changes in Pol II elongation rates leading to alternative splicing patterns have been uncovered recently.
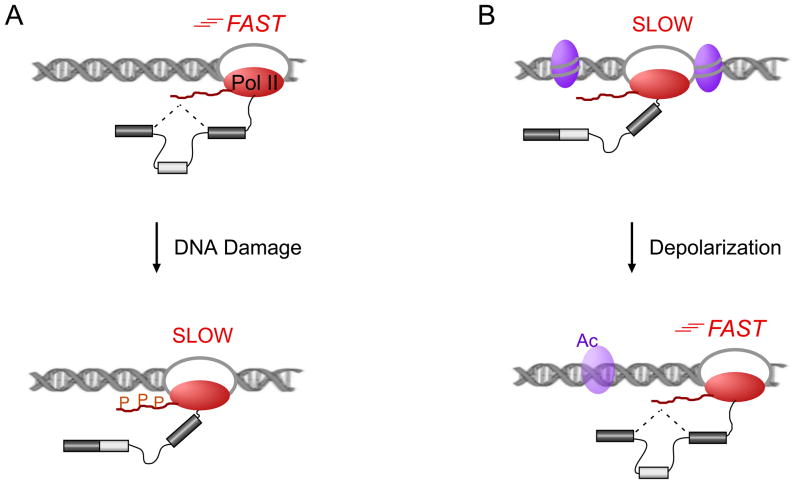
In a case of signaling pathways directly altering the speed of Pol II elongation, a recent study demonstrated that p53-independent hyperphosphorylation of the Pol II C-terminal domain (CTD) in response to UV-induced DNA damage led to a slower elongation rate and affected the splicing of key apoptotic regulators such as caspase-9 and BCLX (Fig. 3)40. This effect on splicing was strictly cotranscriptional and amino acid substitutions in the CTD that either mimic or prevent phosphorylation also mimicked or prevented the effect of UV treatment, respectively, thus providing compelling evidence for this model. Even though Pol II is also subjected to proteasome-mediated degradation after DNA damage, the cotranscriptional splicing regulation observed here seemed to be solely dependent on the phosphorylation, and not the ubiquitylation, of Pol II, thus setting the mechanism apart from the DNA damage-induced protein stability pathways discussed above. In addition, large-scale analysis of UV-treated and control cells showed an overlay between genes with reduced transcription and genes with altered splicing patterns40, pointing to a concerted regulation of many pre-mRNAs in the DNA damage response.
Fig. 3. Signal-induced changes in Pol II processivity regulate cotranscriptional splicing.
In the kinetic coupling model, alternative splicing can be regulated by the speed of RNA polymerase II (Pol II; red oval) such that a slower polymerase leaves the spliceosome more time to recognize exons with suboptimal splice sites (light grey box), thus increasing inclusion. A) A model for DNA damage-induced alternative splicing of several apoptotic regulators, showed that phosphorylation (P) within the C-terminal domain (CTD, red wavy line) of Pol II leads to reduced Pol II speed and changed patterns of alternative splicing40. B) Another mechanism to change Pol II processivity has been seen in neurons. Depolarization leads to loosening of the chromatin structure by changing the acetylation (Ac) of specific histones (purple ovals). This allows for increased Pol II elongation rates and leads to a consistent change in NCAM1 alternative splicing41.
Studies of neuronal depolarization revealed another mechanism for regulation of co-transcriptional splicing (Fig. 3). A study investigating neural cell adhesion molecule 1 (NCAM1) exon 18 skipping upon depolarization, found that histone modifications changed around the alternatively spliced exon in a way that increases chromatin accessibility41. This was accompanied by enhanced PolII processivity, which in the model of kinetic coupling favors exon exclusion; indeed, exon exclusion was observed in this NCAM1 system. Further supporting this model, Trichostatin A, a drug widely used to alter histone acetylation, induced consistent splicing changes, and the expression of a slow Pol II mutant strongly increased exon inclusion, thus linking Pol II processivity with splicing41.
Feedback loops in signal-induced alternative splicing
Thus far we have described multiple mechanisms by which DNA damage, T cell activation and neuronal depolarization alter splicing, emphasizing that cells invoke complex and tight control of alternative splicing in response to changing cellular conditions to precisely regulate protein expression and cellular function. Consistently, in order to maintain homeostasis cells need to turn off induced splicing programs once conditions return to baseline (e.g. repolarization of neurons, clearance of antigen, DNA repair, etc). One mechanism to reset gene expression is to interrupt the signaling cascade by removal or inactivation of the initiating receptors or signaling molecules themselves. Indeed, many receptors, kinases and phosphatases undergo signal-induced alternative-splicing in an autoinhibitory manner. For example, alternative splicing of CD45 in response to T cell activation attenuates the sensitivity of the cell to further antigen stimulation42. Similarly, extracellular signal-regulated kinase-1 (ERK1), the FYN proto-oncogene and protein tyrosine kinase 2 beta (PYK2), which all encode kinases involved in propagating T cell signaling, undergo alternative splicing in response to T cell activation to reduce expression, modulate substrate specificity or alter localization patterns17.
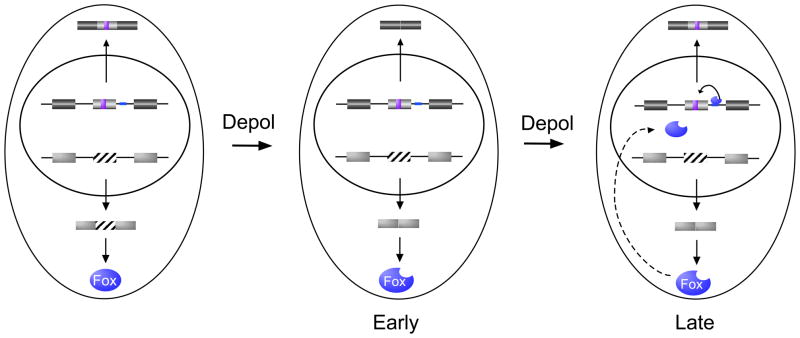
An alternative approach to resetting the splicing profile is to induce expression of an antagonistic regulatory factor. Exactly such a mechanism is seen upon neuronal chronic depolarization (Fig. 4)43. As described above, neuronal depolarization results in widespread changes in splicing, in many cases involving increased skipping of exons that are regulated by CaRREs9. Interestingly, some of these CaRRE-repressed exons are once again included upon prolonged depolarization. This reversion of splicing pattern is attributable to CaMK-induced alternative splicing of FOX1 which encodes an RNA-binding protein43. FOX1 controls the splicing patterns of numerous genes implicated in synaptic activity. Importantly, many genes regulated by CaRREs also contain a FOX1 binding site which typically exerts an opposite effect on exon inclusion as does the CaRRE sequence. Strikingly, nuclear localization of FOX1 is dependent on skipping of exon 19, which itself is repressed upon CaMK signaling. Therefore, neuronal depolarization not only represses CaRRE-regulated exons, but also generates a FOX1 isoform that migrates efficiently to the nucleus. Once at sufficient concentration in the nucleus, FOX1 antagonizes the activity of the CaRRE to restore exon inclusion43. This work thus provides an elegant example of the complexity of splicing networks that can exist downstream of individual stimuli and the importance of fully understanding the program of induced splicing events and cross-talk between such events in order to delineate programmed changes in alternative splicing and predict how specific signaling pathways will impact protein expression within a cell.
Fig. 4. A feedback loop in signal-induced alternative splicing.
CaRREs (purple box) and binding sites for FOX1 (blue line) represent two splicing regulatory elements known to regulate neural-specific genes. Upon neural depolarization (Depol), the CaRRE-responsive exons and exon 19 of FOX1 (striped box) are initially repressed. The skipping of FOX1 exon 19 results in expression of a form of FOX1 that is localized to the nucleus. For exons that are controlled by both CaRRE and FOX1 elements, this increase in nuclear FOX1 counters the activity of the CaRRE, thereby restoring exon inclusion.
Timing Issues
One final point raised by the FOX1 feedback loop is the issue of timing in alternative splicing regulation. For many, if not most, of the examples of signal-induced alternative described herein, the splicing changes are most readily observed hours or days following stimulation. This is in striking contrast to the minute-to-hour time scale in which signal-induced protein modification or transcription is typically studied. In some cases this delay in alternative splicing stems from the inherent time required for protein degradation and/or de novo synthesis. Such delays can be biologically important “timers” as in the case of FOX1 feedback to CaRRE-regulated exons or changes in CD45 splicing to attenuate T cell responsiveness43,44. However, even in cases in which modifications to the relevant regulatory protein are manifest quickly, alterations in splicing are observed more slowly than instances of induced transcription due to half-life of the pre-existing RNA. For example, even a complete switch in isoform expression in an RNA with a typical half-life of 4 hours will be observed as at most a 50% change if bulk message is assayed 4 hours after stimulation. For RNAs with longer half-lives or smaller splicing changes, this issue of residual message has an even larger impact. Therefore, a 12 hour delay between stimuli and splicing change might not indicate a complex, indirect signaling pathway (as is sometimes implied), but rather be a result of a highly stable message.
Concluding remarks
In summary, we present here an overview of recent progress in uncovering the mechanisms by which signaling pathways impinge on alternative splicing regulation. Far from a simple one-mechanism-fits-all, signaling pathways can alter the stability, expression, interaction partners and/or location of a splicing regulatory protein so as to alter the splicing pattern of their target genes. This review is not intended to be fully comprehensive, rather our goal is to provide an indication of where the field is heading in the hopes of encouraging further study in this area as clearly there remains much to be learned. Most of the mechanisms described herein have only been observed thus far to regulate one or two genes. More global studies are therefore needed to catalogue the full scope of alternative splicing that occurs downstream of a given stimulus or pathway. Moreover, we need to understand how many divergent mechanistic pathways are triggered by a single stimulus. For example, T cell signaling not only induces the PSF-hnRNPLL regulatory program described above15,17, but also activates at least two other splicing regulatory mechanisms which regulate a non-overlapping set of exons45,46. Similarly, DNA damage triggers multiple splicing-relevant pathways as delineated above. Therefore, even once we know the profile of splicing changes triggered by a particular stimuli, dissecting specific regulatory pathways and ultimately being able to predict which genes are regulated by a given pathway will require significantly more investigation, and will keep researchers busy for much time to come.
Figure I.
Reciprocal regulation of alternative splicing by activator and repressor proteins
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pan Q, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol. 2007;623:161–174. doi: 10.1007/978-0-387-77374-2_10. [DOI] [PubMed] [Google Scholar]
- 5.Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nature reviews. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- 6.Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Molecular cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 7.Bland CS, et al. Global regulation of alternative splicing during myogenic differentiation. Nucleic acids research. 38:7651–7664. doi: 10.1093/nar/gkq614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warzecha CC, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JA, et al. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS biology. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An P, Grabowski PJ. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS biology. 2007;5:e36. doi: 10.1371/journal.pbio.0050036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes & development. 24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 13.Cooper TA, et al. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Topp JD, et al. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. Rna. 2008;14:2038–2049. doi: 10.1261/rna.1212008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oberdoerffer S, et al. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melton AA, et al. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Molecular and cellular biology. 2007;27:6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ip JY, et al. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warzecha CC, et al. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Molecular cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grosso AR, et al. Tissue-specific splicing factor gene expression signatures. Nucleic acids research. 2008;36:4823–4832. doi: 10.1093/nar/gkn463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang GS, et al. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuyumcu-Martinez NM, et al. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Molecular cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalsotra A, et al. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes & development. 2010;24:653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katzenberger RJ, et al. Control of alternative splicing by signal-dependent degradation of splicing-regulatory proteins. The Journal of biological chemistry. 2009;284:10737–10746. doi: 10.1074/jbc.M809506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katzenberger RJ, et al. ATM and ATR pathways signal alternative splicing of Drosophila TAF1 pre-mRNA in response to DNA damage. Molecular and cellular biology. 2006;26:9256–9267. doi: 10.1128/MCB.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edmond V, et al. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 30:510–523. doi: 10.1038/emboj.2010.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S, Fu XD. SR proteins and related factors in alternative splicing. In: Graveley BJBaBR., editor. Alternative Splicing in the Postgenomic Era. Landes Biosciences; 2007. [Google Scholar]
- 27.Shkreta L, et al. The DNA damage response pathway regulates the alternative splicing of the apoptotic mediator Bcl-x. The Journal of biological chemistry. 286:331–340. doi: 10.1074/jbc.M110.162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massiello A, et al. SAP155 Binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 2006;20:1680–1682. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- 29.Zhong XY, et al. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes & development. 2009;23:482–495. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Houven van Oordt W, et al. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. The Journal of cell biology. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guil S, et al. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Molecular and cellular biology. 2006;26:5744–5758. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heyd F, Lynch KW. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Molecular cell. 40:126–137. doi: 10.1016/j.molcel.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez F, et al. Glycogen synthase kinase-3 plays a crucial role in tau exon 10 splicing and intranuclear distribution of SC35. Implications for Alzheimer’s disease. The Journal of biological chemistry. 2004;279:3801–3806. doi: 10.1074/jbc.M311512200. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, et al. The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. The Journal of biological chemistry. 2009;284:1505–1513. doi: 10.1074/jbc.M805113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dutertre M, et al. Cotranscriptional exon skipping in the genotoxic stress response. Nature structural & molecular biology. 17:1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- 36.Das R, et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Molecular cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 37.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nature structural & molecular biology. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 38.Rajan P, et al. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J Pathol. 2008;215:67–77. doi: 10.1002/path.2324. [DOI] [PubMed] [Google Scholar]
- 39.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 40.Munoz MJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Schor IE, et al. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hermiston ML, et al. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J Clin Invest. 2002;109:9–14. doi: 10.1172/JCI14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JA, et al. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes & development. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lynch KW, Weiss A. A model system for the activation-induced alternative-splicing of CD45 implicates protein kinase C and Ras. Molecular and cellular biology. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matter N, et al. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 46.Mallory MJ, et al. Signal- and developmental-dependent alternative splicing of LEF1 in T cells is controlled by CELF2. Molecular and cellular biology. doi: 10.1128/MCB.05170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahl MC, et al. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature reviews. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.House AE, Lynch KW. Regulation of Alternative Splicing: More than just the ABCs. The Journal of biological chemistry. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- 50.Lynch KW. Regulation of Alternative Splicing by Signal Transduction Pathways. In: Graveley BJBaBR., editor. Alternative Splicing in the Postgenomic Era. Landes Biosciences; 2007. [DOI] [PubMed] [Google Scholar]
- 51.Martinez-Contreras R, et al. hnRNP Proteins and Splicing Control. Landes Biosciences; 2007. [DOI] [PubMed] [Google Scholar]
- 52.Webby CJ, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 53.White ES, et al. Control of fibroblast fibronectin expression and alternative splicing via the PI3K/Akt/mTOR pathway. Exp Cell Res. 316:2644–2653. doi: 10.1016/j.yexcr.2010.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shultz JC, et al. Alternative splicing of caspase 9 is modulated by the phosphoinositide 3-kinase/Akt pathway via phosphorylation of SRp30a. Cancer Res. 70:9185–9196. doi: 10.1158/0008-5472.CAN-10-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Misteli T, Spector DL. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Molecular biology of the cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Molecular cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Magistrelli G, et al. A soluble form of CTLA-4 generated by alternative splicing is expressed by nonstimulated human T cells. European journal of immunology. 1999;29:3596–3602. doi: 10.1002/(SICI)1521-4141(199911)29:11<3596::AID-IMMU3596>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 58.Mayrand SH, et al. Serine/threonine phosphorylation regulates binding of C hnRNP proteins to pre-mRNA. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:7764–7768. doi: 10.1073/pnas.90.16.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie J, et al. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]