Abstract
Efficient electron transfer from reductase domain to oxygenase domain in nitric oxide synthase (NOS) is dependent on the binding of calmodulin (CaM). Rate constants for the binding of CaM to NOS target peptides was only determined previously by surface plasmon resonance (SPR) (Biochemistry 35, 8742–8747, 1996) suggesting that the binding of CaM to NOSs is slow and does not support the fast electron transfer in NOSs measured in previous and this studies. To resolve this contradiction, the binding rates of holo Alexa 350 labeled T34C/T110W CaM (Alexa-CaM) to target peptides from three NOS isozymes were determined using fluorescence stopped-flow. All three target peptides exhibited fast kon constants at 4.5 °C: 6.6 × 108 M−1s−1 for nNOS726–749, 2.9 × 108 M−1s−1 for eNOS492–511 and 6.1 × 108 M−1s−1 for iNOS507–531, 3 – 4 orders of magnitude faster than those determined previously by SPR. Dissociation rates of NOS target peptides from Alexa-CaM/peptide complexes were measured by Ca2+ chelation with ETDA: 3.7 s−1 for nNOS726–749, 4.5 s−1 for eNOS492–511, and 0.063 s−1 for iNOS507–531. Our data suggest that the binding of CaM to NOS is fast and kinetically competent for efficient electron transfer and is unlikely rate-limiting in NOS catalysis. Only iNOS507–531 was able to bind apo Alexa-CaM, but in a very different conformation from its binding to holo Alexa-CaM.
Keywords: calmodulin, nitric oxide synthase, Alexa Fluor 350, fluorescence resonance energy transfer, binding rate constant, fluorescence stopped-flow
1. Introduction
Nitric oxide synthase (NOS) catalyzes the bio-synthesis of nitric oxide (NO), which is simultaneously a cytoprotective molecule and a vital signal for the major downstream target, soluble guanylyl cyclase (sGC), in many physiological processes [1, 2]. Two of the three isoforms of NOS, neuronal NOS (nNOS) and endothelial NOS (eNOS) are constitutively expressed while inducible NOS (iNOS) is expressed in response to various stimulants. All the three isozymes form head-to-tail/antiparallel homodimers [3]. Each monomer contains two major domains, a N-terminal oxygenase domain which contains one complement each of heme and (6R)-5,6,7,8-tetrahydrobiopterin (BH4) and binds substrate L-arginine, and a C-terminal reductase domain which contains one each of FMN, FAD prosthetic group and NADPH binding site. These two domains are connected by a calmodulin (CaM) binding domain. The oxygenase domain catalyzes conversion from L-arginine to NO in two steps: (1) generation of intermediate Nω-hydroxy-L-arginine (NOHA) and (2) further oxidation of NOHA to NO and L-citrulline. Both steps require sequential transfer of reducing equivalents from NADPH to FAD, then to FMN and finally from FMN of one monomer to the oxygenase domain of the other monomer. Binding of CaM is pivotal in the inter-subunit electron transfer from the reductase domain to the oxygenase domain [4–6].
CaM is a small acidic Ca2+ binding protein involved in many physiological processes including neurotransmission, fertilization, memory, motility, cell defense, proliferation and programmed cell death [7]. CaM has 148 amino acid residues and consists of two globular domains (N- and C-lobes) tethered by a central linker (Fig. 1). Each domain contains a pair of Ca2+ binding EF-hands. The C-lobe EF-hands exhibit higher affinities for Ca2+ than those in the N-lobe [8]. In the absence of Ca2+ i.e., apo CaM, the central linker is relaxed in solution (Fig. 1A) [9]. Binding of Ca2+ induces a significant conformational change, where the two helices in each EF-hand switch from near parallel conformation to a more perpendicular conformation for Ca2+ binding and the methionine-rich hydrophobic surfaces are exposed, providing critical van der Waals interactions with the hydrophobic surfaces of many CaM binding targets (Fig. 1B) [10]. In the classical binding conformation, the N- and C-lobes of Ca2+ loaded CaM (holo CaM) clamp in an antiparallel orientation on target peptides which typically adopt a helical conformation [11]. On the other hand, some CaM targets are capable of binding to either apo or holo CaM in a variety of other conformations than the classical mode [12, 13]. In some cases, CaM is shown to bind in a very different conformation to the target peptide fragment from binding to the whole target protein [14, 15].
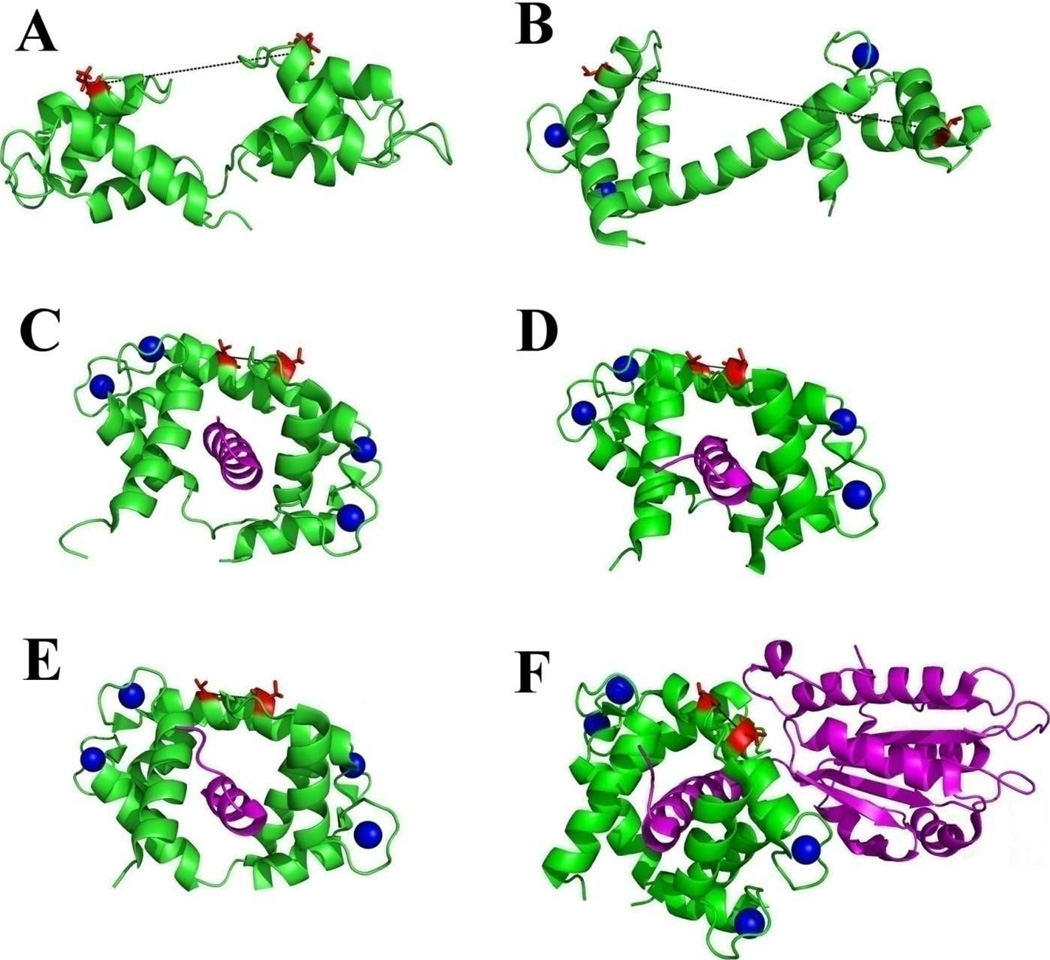
Figure 1. Structures of CaM and its complexes with NOS targets.
A: solution structure of apo CaM, 1CFC [9]; B: crystal structure of holo CaM, 1UP5 [10]; C – E, crystal structures of holo CaM complexed with NOS target peptides, C: nNOS, 2O60; D: eNOS, 1NIW [29] and E: iNOS, 3GOF. F: crystal structure of complex CaM/iNOSCaM-FMN, 3HR4 [30]. CaM molecules are shown with green ribbons and its NOS target molecules are shown with magenta ribbons. Calcium ions are represented with blue spheres. Residues T34 (left in each structure) and T110 (right in each structure) are represented with red sticks. The distances between Cα atoms of T34 and T110 are measured from the structures and indicated in black dash lines: 27.0 Å in apo CaM, 51.5 Å in holo CaM. This distance is 12.4, 13.0 and 13.3 Å in structures 2O60, 1NIW and 3GOF, respectively. In structure 3HR4, this distance is 13.6 Å.
CaM binds NOS isozymes with high affinities, as indicated by the low Kd values (nM) of holo CaM binding to target peptides derived from the CaM-binding domains of constitutive NOSs and ≤ 0.1 nM for the peptide derived from the CaM-binding domain of iNOS [16–23]. In addition to the main CaM binding target peptide, additional CaM-sensitive elements exist in the reductase and oxygenase domains contributing to NOS activation by CaM [2, 24]. The activation of constitutive NOSs (eNOS and nNOS) by the binding of CaM is Ca2+-dependent [25]. On the other hand, iNOS is capable of binding to both holo and apo CaM and its activity is shown to be Ca2+-independent [25–28]. Crystal structures reveal that holo CaM binds in the classical conformation to target peptides of all three NOS isozymes (PDB code: 2O60 (nNOS), 1NIW (eNOS) and 3GOF (iNOS), Fig. 1C–E). Recently, the crystal structure of the CaM-bound iNOS segment containing both FMN and CaM binding domains (3HR4) was published and indicates that holo CaM is bound in the classical conformation [29, 30]. Crystal structures (3GOF and 3HR4) clearly show that holo CaM binds to both iNOS target peptide and a larger segment containing both FMN subdomain and CaM-binding domain of iNOS protein in an anti-parallel orientation and the targets are in a helical structure. Therefore, it is likely that holo CaM binds all three NOS proteins in the classical mode. On the other hand, the binding mode of apo CaM to iNOS is very different from that of holo CaM to iNOS, as indicated by the unique β-turn conformation discovered in the apo CaM/iNOS peptide complex [12]. Previous steady-state fluorescence and circular dichroism studies suggest that both CaM lobes are capable of binding to iNOS target peptide in a Ca2+-independent manner and in the presence of iNOS peptide, the C-lobe exhibits large conformational changes during its association with Ca2+ [27].
The critical dynamic interplay by the binding of CaM to NOS isozymes to facilitate the inter-subunit electron transfer from the reductase domain to the oxygenase heme is not well understood [2, 25, 30, 31]. The rate of the first step of this interplay, the binding of CaM to NOS targets, was measured as the binding rate of CaM to NOS target peptides using SPR method [21]. The rates thus determined likely represent the upper limit for the binding of CaM to NOS proteins since CaM likely binds full length NOS proteins slower than target peptides alone due to the participation of other structural elements of NOS proteins in their binding to CaM [2, 16, 25, 32]. The only measurements on binding rates by SPR method of holo CaM to immobilized nNOS725–750 and iNOS503–528 target peptides are 1.58 × 105 M−1s−1 and 3 × 104 M−1s−1, respectively [21]. The binding rate thus determined for holo CaM to nNOS peptide implies that the binding of CaM to constitutive NOS proteins is slow and may be a rate-limiting step in NOS catalysis. However, such possible overall electron transfer rates limited by the slow CaM binding are inconsistent with the rates measured for heme and flavin reduction in NOS catalysis [25, 33]. Since the immobilization of NOS target peptides on SPR matrix may slow down their binding to CaM [34], it is necessary to reexamine the binding kinetics of CaM to NOS target peptides in solution. It is also necessary to examine the kinetic details of iNOS’s differential binding to holo and apo CaM.
In this study, we first measured the heme reduction rate in nNOS by NADPH with and without pre-binding of CaM to test whether the binding of CaM imposes rate-limiting on the electron transfer in nNOS. We then determined and compared the binding kinetics of CaM to peptides derived from the CaM-binding domains of nNOS, eNOS and iNOS by monitoring the changes of the relative distance between CaM’s N- and C-lobes using fluorescence resonance energy transfer (FRET) and stopped-flow spectroscopy.
The distance between residues T34 and T110 of CaM minimizes in its classical binding to its targets and the change of this distance has been used to study the association of CaM to a various targets [14, 35]. Similarly to these studies, we first prepared a CaM double mutant, T34C/T110W CaM, and then site-specifically labeled C34 with Alexa 350 to form Alexa-CaM. The major absorption of Alexa 350, centered at 346 nm, overlaps with the fluorescence emission of W110 (FW110). Change of the distance between the two lobes can be easily followed by the FW110 quenching dependent on the FRET between W110 and Alexa 350. We determined the association and dissociation rate constants, kon and koff, of Alexa-CaM to NOS target peptides. Our measured kon values were much faster than those previously determined by SPR, suggesting that the binding of CaM is unlikely a rate-limiting step in NOS catalysis. Our kinetic data also disclosed great insights into the very different sequential dynamic events between the binding of CaM to the constitutive and inducible NOS targets.
2. Experimental Procedures
2.1. Materials
All the reagents for buffer preparations were from Sigma (St. Louis, MO) and were all of analytical grade. IPTG was from RPI Research Products International Corps (Mt. Prospect, IL). Alexa Fluor 350 and Fura Red were from Invitrogen (Eugene, OR). Dpn I was from BioLabs (Ipswich, MA). 4, 4’-dithiodipyridine was from Aldrich (Milwaukee, WI).
Target peptides corresponding to the NOS CaM-binding domains [27, 29, 36] were custom synthesized and HPLC-purified to purity of 98% or higher at EZBiolab (Westfield, IN). The peptide stock solutions were prepared in water and the concentrations were determined with amino acid analysis method at Molecular Structure Facility, University of California at Davis (Davis, CA). The peptide sequences are:
rat nNOS726–749 RRAIGFKKLAEAVKFSAKLMGGAM;
bovine eNOS492–511 RKKTFKEVANAVKISASLMG;
human iNOS507–531 RPKRREIPLKVLVKAVLFACMLMRK.
2.2. Expression and purification of wt nNOS
Rat brain nNOS cDNA in bluescript (SK(−))TA plasmid (kindly provided by Dr. S. Snyder, The Johns Hopkins University, Baltimore) was used as template for PCR amplification of the DNA fragment encoding amino acids 1–1429 (wt nNOS). The wt nNOS forward primer 5’-CGCCATATGCATCACCATCACGAAGAGAACACGTTTGGG-3’ (translation start codon underlined) also included the NdeI (in italics) site and four-His tag (in bold). The reverse primer 5’-GTCTCTAGATTAGGAGCTGAAAACCTCATC-3’ had an inserted XbaI restriction site (in italics) after the stop codon (underlined). The PCR product was first ligated into the TA vector (Invitrogen) and then introduced into XL10 Gold E. coli cells for propagation according to the manufacturer's recommendation. Grown positive colonies of TA-4His-wt-nNOS plasmid were purified using the Wizard Plus SV Minipreps DNA Purification System (Promega, Madison, WI). The sequence of 4His-wt nNOSDNA was confirmed by primer extension sequencing (University of Texas Health Science Center Sequencing Core). The 4His-wt-nNOS DNA was excised from the plasmid by double digestion with NdeI and XbaI restriction enzymes and subsequently ligated into the corresponding sites of the pCW vector. Purified pCW-4His-wt-nNOS plasmid was transformed into E. coli BL21 Star (DE3) cells containing the pT-groE plasmid with a chloramphenicol resistance gene for protein expression. Wild type nNOS was expressed and purified following the method previously published [37].
2.3. Generation of T34C/T110W CaM
The T34C /T110W CaM mutant was prepared in two steps. First T34C CaM cDNA was generated by PCR using wt human CaM cDNA in pET-23d (+) vector (Novagen) as the template and the primers were:
forward 5’-CCAAGGAGCTGGGGTGCGTGATGAG-3’,
reverse 5’-CTCATCACGCACCCCAGCTCCTTGG-3’.
After Dpn I digestion of the PCR reaction mixture to remove the template, TOP10 cells was transformed with the PCR products to amplify the vector with T34C CaM mutation. The purified vector was then used as the template to prepare T34C/T110W CaM by PCR using the following primers:
forward 5’-GCTACGTCACGTGATGTGGAATCTCGGCGAG-3’,
reverse 5’-CTCGCCGAGATTCCACATCACGTGACGTAGC-3’.
The PCR reaction mixtures were then digested with Dpn I and the vector containing T34C/T110W CaM mutant was propagated in TOP10 cells. The DNA sequences of all the CaM mutants were verified by DNA sequencing (University of Texas Health Science Center Sequencing Core).
2.4. Expression and purification of wt CaM and T34C/T110W CaM
Competent BL21 (DE3) plys S cells were transformed with wt CaM pET-23d (+) or T34C/T110W CaM pET-23d (+) vectors and grown in LB media at 37 °C. Protein expression was induced with 1 mM IPTG when OD595nm reached 0.6 and cells were harvested 6 hours after induction. The proteins were purified following the published protocol [38]. For the purification of T34C/T110W CaM, 10 mM DDT was included in all buffers. Purified wt CaM or T34C/T110W CaM was first dialyzed against 10 mM EDTA, which was later removed by extensive dialysis into 50 mM HEPES, pH 7.8 buffer without EDTA (0.1mM DTT was included in all the dialysis buffers for T34C/T110W CaM). Nucleic acid co-purified with T34C/T110W CaM was removed using Novagen® Benzonase nuclease (EMD Chemicals USA).
2.5. Alexa 350 labeling of T34C/T110W CaM
Concentrated T34C/T110W CaM solution was passed through a 10DG desalting column (Bio-Rad) to remove DTT. The concentration of free thiol group (C34) was verified with 4, 4’-dithiodipyridine titration [39]. The T34C/T110W CaM solution was then mixed with Alexa 350 solution in DMSO, with a final ratio [Alexa 350] : [T34C/T110W CaM] = 9 – 10 : 1. The reaction mixture was incubated for 2 hours on a shaker at room temperature. Un-reacted Alexa 350 was removed using 10DG column chromatography. The disappearance of the free thiol group (C34) was verified using 4, 4’-dithiodipyridine titration.
2.6. NOS activity assay
The stimulatory effects of wt CaM, T34C/T110W CaM and Alexa-CaM on activation of wt nNOS were evaluated by measuring the NO releasing rate of wt nNOS [40, 41]. The released NO was captured by oxyhemoglobin (HbO2) to form methemoglobin and the initial slope of the A400nm change reflected how fast NO was released. The extinction coefficient used was 19.1 mM−1cm−1. The assay was conducted at room temperature in 50 mM HEPES, pH 7.7 buffer containing 100 mM NaCl, 10% glycerol and 100 µM CaCl2 supplemented with 1 mM L-arginine, 100 µM NADPH and 4.4 µM HbO2. The reactions were started by adding wt nNOS together with CaM (wt CaM, T34C/T110W CaM or Alexa-CaM) to the substrate mixture above. The final concentrations were 50 nM for nNOS and 100 nM for CaM species.
2.7. Stopped-flow kinetics of heme and flvain reduction in wt nNOS by NADPH
Heme and flavin reduction in wt nNOS by NADPH was conducted with optical stopped-flow at room temperature. 2µM wt nNOS was mixed with 100 µM L-arginine and 5 µM BH4 in 100 mM HEPES, pH 7.7 buffer containing 100 mM NaCl and 10% glycerol. In one experiment, wt nNOS was reacted with a mixture of 50 µM NADPH and 2 µM wt CaM in the same buffer containing 200 µM CaCl2. In the other experiment, 2 µM wt CaM and 200 µM CaCl2 were added to the wt nNOS solution and then reacted with 50 µM NADPH. Heme reduction was observed at 398 nm and flavin reduction was followed at 480 nm. Reaction rates were obtained by fitting the kinetic data with single-exponential function for heme reduction,
| (1) |
and double-exponential function for flavin reduction,
| (2) |
where ΔA is the change of absorbance.
2.8. Fluorescence spectroscopy
Steady-state fluorescence experiments were conducted at room temperature using a Hitachi (Tokyo, Japan) F4500 fluorescence spectrophotometer. The excitation wavelength was 280nm. The spectral bandwidths for both excitation and emission were kept the same and varied from 2.5 nm to 10 nm. Scan speed was 240 nm/min. Each spectrum was an average of two scans.
The distance, R, between W110 and Alexa 350, was estimated as follows,
| (3) |
where ET is the efficiency of FRET and Ro is the “critical” distance for 50% energy transfer efficiency. ET was defined as 1 − Φ/ Φ0 and the ratio of quantum yields before and after NOS peptide binding, Φ/ Φ0, was estimated using the ratio of fluorescence intensity at 350 nm. Ro of the tryptophan/Alexa350 donor/acceptor pair was 21 Å [42].
For the stoichiometric titrations, the change of FW110 at 350 nm (350nmΔF) for each addition of NOS target peptides to holo Alexa-CaM or apo Alexa-CaM (iNOS507–531 only) was followed. The affinity of iNOS507–531 to apo Alexa-CaM, Kd, was obtained by fitting to the following function [43]:
| (4) |
where
a = [apo Alexa-CaM] + [iNOS507–531] + Kd
and [apo Alexa-CaM/iNOS507–531] was calculated as: (ΔF/350nmΔFmax) × [apo Alexa-CaM].
2.9. Kinetic measurements of the binding of Alexa-CaM to NOS target peptides
The association and dissociation of Alexa-CaM with NOS target peptides were followed with the change of FW110 using an Applied Photophysics (Leatherhead, UK) SX.18MV-R fluorescence stopped-flow system. All the measurements were conducted at 4.5 °C, to slow down the reaction and minimize loss of kinetic data in the dead time, 1.5 ms, of our stopped-flow instrument. The temperature was controlled with a Julabo F25 refrigerated and heating circulator (Vista, CA). The excitation wavelength was 280 nm with 9.3 nm spectral band width (Applied Photophysics SpectraKinetic monochromator). The fluorescence emission was filtered using an Oriel 300 – 400 nm bandpass filter with 90 nm FWHH (Stratford, CT). For the kinetics of association, holo or apo Alexa-CaM was mixed with various concentrations of NOS target peptides at least an order of magnitude higher to meet the pseudo first-order conditions.
The observed rates of the fluorescence change during the binding of target peptides, kobs, were obtained by fitting the experimental data to the single exponential function,
| (5) |
where ΔF and ΔFmax are the changes of fluorescence and F0 is the final fluorescence intensity in arbitrary units, and t is time in second. To obtain kon for NOS target peptides to Alexa-CaM, kobs values obtained above were plotted versus the peptide concentrations, and the slope of the linear relationship represented the second-order kon (in M−1s−1),
| (6) |
where koff is the intercept of y-axis [44].
2.10. Stopped-flow dissociation kinetics of Alexa-CaM/NOS target peptide complexes
The complexes of holo Alexa-CaM with NOS target peptides were rapidly mixed with 2 mM EDTA at 4.5 °C and the change of the same fluorescence emission as in binding study was followed. For the dissociation of nNOS and eNOS peptides, the observed rate constants, kobs1 and kobs2, were obtained by fitting the biphasic kinetic data to the double exponential function,
| (7) |
where ΔF, ΔFmax1 and ΔFmax2 are the changes of fluorescence and F0 is the final fluorescence intensity in arbitrary units, and t is time in second. For the dissociation of iNOS peptide, the observed rate, kobs, was obtained by fitting the kinetic data to a single exponential function using time points longer than 2s,
| (8) |
where ΔF and ΔFmax are the changes of fluorescence and F0 is the final fluorescence intensity in arbitrary units, and t is time in second.
3. Results
3.1. Functional integrity of Alexa-CaM and T34C/T110W CaM
The functional integrity of T34C/T110W CaM and Alexa-CaM to stimulate the NO generation by wt nNOS was checked and compared to that of wt CaM. Typical activity of nNOS with wt CaM was around 400 nmol/min/mg, corresponding to a turnover number of ~1.1 s−1. The NO formation activity of wt nNOS complexed with either T34C/T110W CaM or Alexa-CaM were 96.8 ± 5.8% (n = 4) and 92.2 ± 3.4% (n = 4) of that with wt CaM respectively, indicating the mutations and fluorescence probe labeling did not significantly affect CaM’s ability to bind and activate wt nNOS. These results led us to the conclusion that the conformation of CaM was not significantly perturbed by the two mutations and the subsequent fluorescence labeling, suggesting Alexa-CaM bound to NOS targets similarly to wt CaM.
3.2. Stopped-flow kinetics of heme and flavin reduction in wt nNOS by NADPH
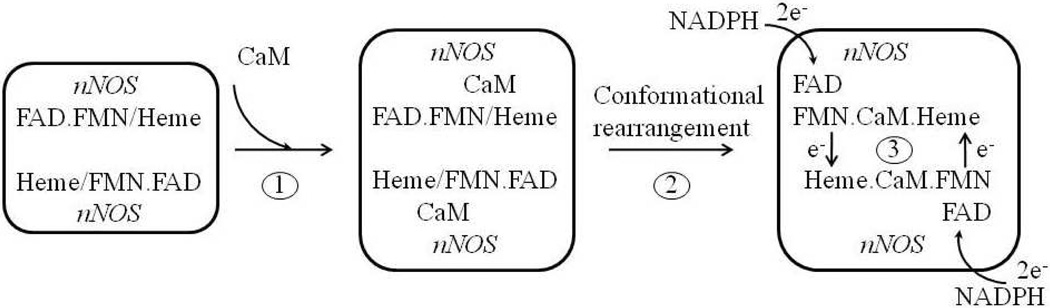
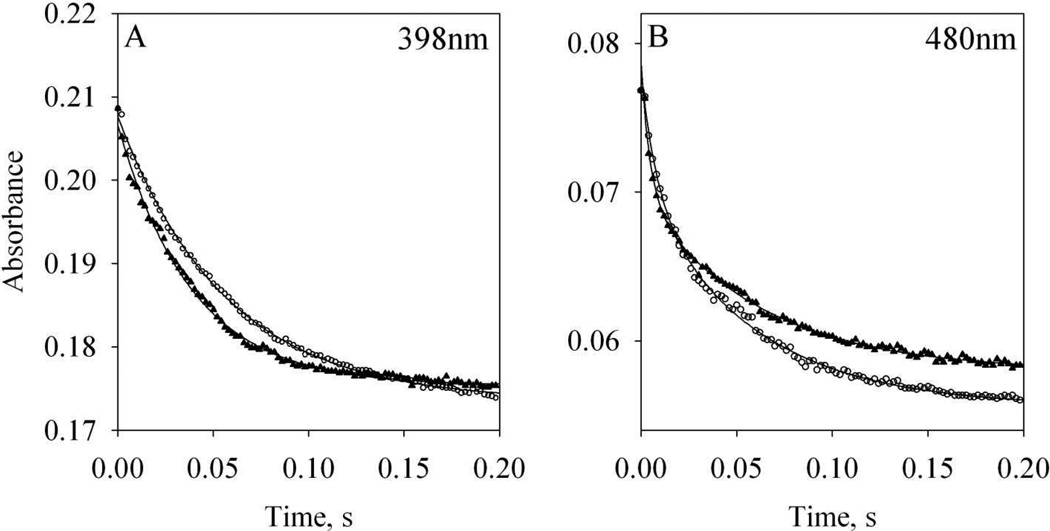
To examine whether the previously published kon value for CaM binding to the target peptide of nNOS [21] matches the rate of electron transfer in nNOS, the rate constants of heme and flavin reduction in wt nNOS by NADPH were measured. When wt NOS (supplemented with L-arginine and BH4) reacted with a mixture of NADPH and holo CaM, the expected sequence of kinetic events was (1) CaM association with nNOS, (2) conformational rearrangement in the CaM bound NOS homodimer and (3) facilitated electron transfer from FMN to heme (Scheme 1). The reduction of heme by FMN exhibited a single exponential decay with rate of 18.2 ± 0.8 s−1 (n = 2, Fig. 2A, circles). On the other hand, the NADPH reduction of flavins was biphasic; a fast phase with a rate of 132.4 ± 1.6 s−1 (n = 2) and a slow phase with a rate of 16.7 ± 1.0 s−1 (n = 2, Fig. 2B, circles). The fast phase was most likely due to electron transfer from NADPH to FAD followed by much faster electron transfer to FMN [33]. The slower phase of flavin reduction was ascribed to the comproportionation of electrons, including the forward and reverse reaction between FAD and FMN as well as electron transfer to the heme [33, 45, 46]. In another study, these two phases were ascribed to FMN reduction gated by NADP+ release after the rapid formation of an equilibrium between an NADPH-NOS charge-transfer species and NADP+-bound two-electron-reduced NOS reductase domain [47].
Scheme 1. CaM binding facilitates electron transfer in nNOS.
Each monomer of nNOS homodimer binds one CaM in the presence of Ca2+ in Step 1. Conformational rearrangement presumably happens subsequently (Step 2) to position FMN properly for fast electron transfer from FMN to heme (Step 3).
Figure 2. Stopped-flow kinetics of heme and flavin reduction in wt nNOS by NADPH.
Open circles: kinetics of 2µM wt nNOS replenished with 100 µM L-arginine and 5 µM BH4 reacted with a mixture of 50 µM NADPH and 2 µM holo wt CaM at room temperature. Solid triangles: 2µM wt nNOS replenished with 100 µM L-arginine and 5 µM BH4 was incubated with 2 µM holo wt CaM and then reacted with 50 µM NADPH. Each kinetic profile was an average of three stopped-flow traces. A: reduction of heme followed at 398 nm; B: reduction of flavin followed at 480 nm. Lines represent the fittings to Eqs. (1) and (2).
Pre-incubating wt nNOS with holo CaM presumably removed the steps 1 and 2 above (Scheme 1), positioning FMN properly for efficient electron transfer to heme. When CaM-bound nNOS was reacted with NADPH, a slightly faster reduction of heme, 26.3 ± 1.1 s−1, was detected (n = 2, Fig. 2A, triangles). The reduction of flavin was still a biphasic process, exhibiting a faster rate for the first phase, 312.7 ± 6.7 s−1 (n = 2) and an almost unchanged rate for the second phase, 18.4 ± 1.3 s−1 (n = 2, Fig. 2B, triangles). Under both experimental conditions, whether 2 µM CaM was or was not pre-bound to nNOS, reduction rates for both heme and flavin were much faster than 0.32 s−1, calculated based on a kon of 1.58 × 105 M−1s−1 for the binding of CaM to nNOS target peptide determined previously by SPR [21], raising a concern about the accuracy of on-rate determination by this method.
3.3. FRET in Alexa-CaM bound to NOS target peptides
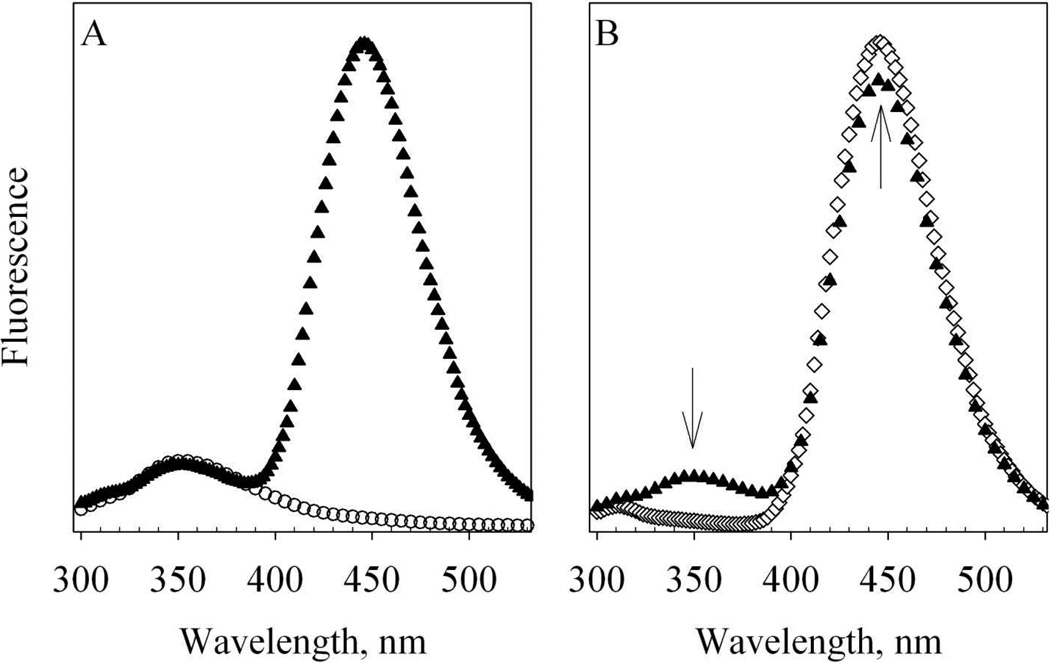
When excited at 280 nm, where Alexa 350 has 20% absorbance as its excitation maximum at 346 nm, Alexa-CaM exhibited two major emission bands. One band centered at 354 – 356 nm corresponding to the emission maximum of W110 and the other band centered at 446 – 448 nm corresponding to the emission maximum of Alexa 350 (Fig. 3A). With 280 nm excitation, the 446 nm emission of Alexa-CaM was about twice as high as that of free Alexa 350 (data not shown). This enhancement of Alexa 350 fluorescence in Alexa-CaM compared to that of free Alexa 350 may be due to the change of its micro-environment by linking to CaM (Fig. 3A). Whereas FW110 of unlabeled T34C/T110W CaM, is essentially the same as that for Alexa-CaM (Fig. 3A), indicating the micro-environment of W110 was not affected by the Alexa 350 labeling.
Figure 3. Fluorescence spectra of T34C/T110W CaM and Alexa-CaM.
A. 1.3µM apo T34C/T110W CaM (open circles) and 1.3µM apo Alexa-CaM (solid triangles) at room temperature. Buffer: 50 mM HEPES, pH 8.0 with 100µM EDTA. B. 1.3µM holo Alexa-CaM in 50 mM HEPES, pH 8.0 with 100µM EDTA (solid triangles) and mixed with 5.8 µM eNOS492–511 and 300 µM CaCl2 (open diamonds) at room temperature. Each spectrum is the average of two scans and the fluorescence intensities are in arbitrary units. The arrows stand for the changes of fluorescence.
Titration of holo unlabeled T34C/T110W CaM with NOS target peptides resulted in a 7 – 8 nm blue shift and decreased FW110 (Fig. 4, pluses vs. solid triangles), indicating the environment of W110 changed upon binding to NOS target peptides. Addition of EDTA reversed these changes for nNOS726–749 and eNOS492–511, but not for iNOS507–531 (Fig. 4, open star vs. pluses), indicating Ca2+-dependence of the binding of nNOS and eNOS peptides and Ca2+-independence of the binding of iNOS507–531. Compared to FW110 of T34C/T110W CaM, FW110 of Alexa-CaM exhibited a much larger change titrated with NOS target peptides, as illustrated by eNOS492–511 in Fig. 3B, due to efficient FRET from W110 to Alexa 350. This efficient FRET almost completely quenched FW110 and enhanced the fluorescence of Alexa 350. Based on the efficiency of FRET, the distance between W110 and Alexa 350 was estimated to be 17.5 Å, agreeing reasonably well with the distance between Cα atoms of T34 and T110 in crystal structure 1NIW (Fig.1D). The FRET in Alexa-CaM due to the binding of NOS target peptides provided us a good tool to follow the binding kinetics. In determination of kon constants for the binding of NOS target peptides to CaM, concentration of CaM species had to be kept low so that the pseudo first order kobs constants were not too large to be measured reliably by our rapid-mixing device (Fig. 7). We focused on the FW110 change as it was more dramatic in percentage change than that of Alexa 350 emission, which had a large background emission.
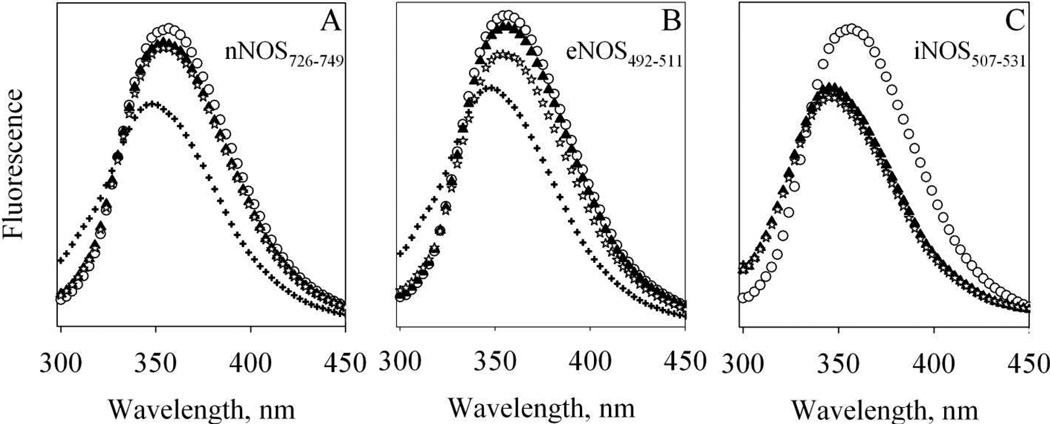
Figure 4. Fluorescence of T34C/T110W CaM and its complexes with NOS target peptides.
Each spectrum is the average of two scans obtained at room temperature and the fluorescence intensities are in arbitrary units. Buffer: 50 mM HEPES, pH 7.8 with 100 mM NaCl and 50 µM EDTA. 8.7µM Apo T34C/T110W CaM (open circles) was first mixed with (A) 39.3 µM nNOS726–749 (B) 35.3 µM eNOS492–511 or (C) 31 µM iNOS507–531 (solid triangles); then mixed with 450 µM CaCl2 (plus) and finally mixed with 4 mM EDTA (stars).
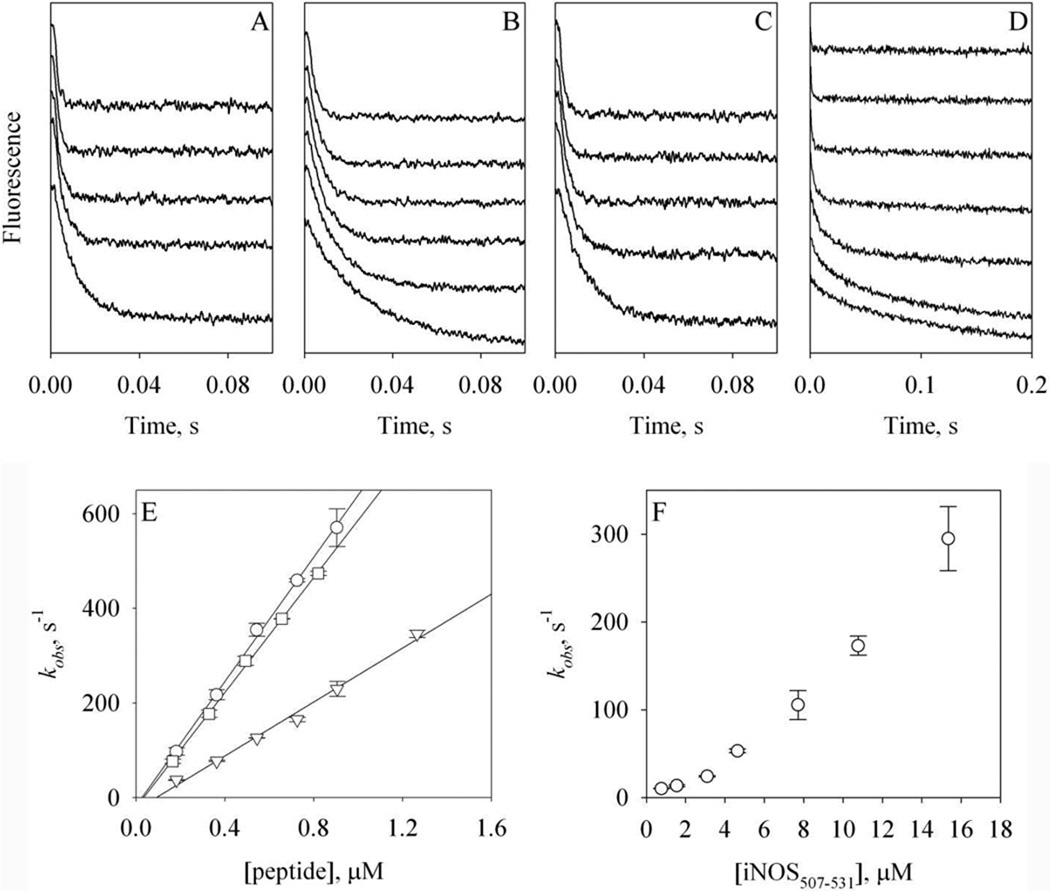
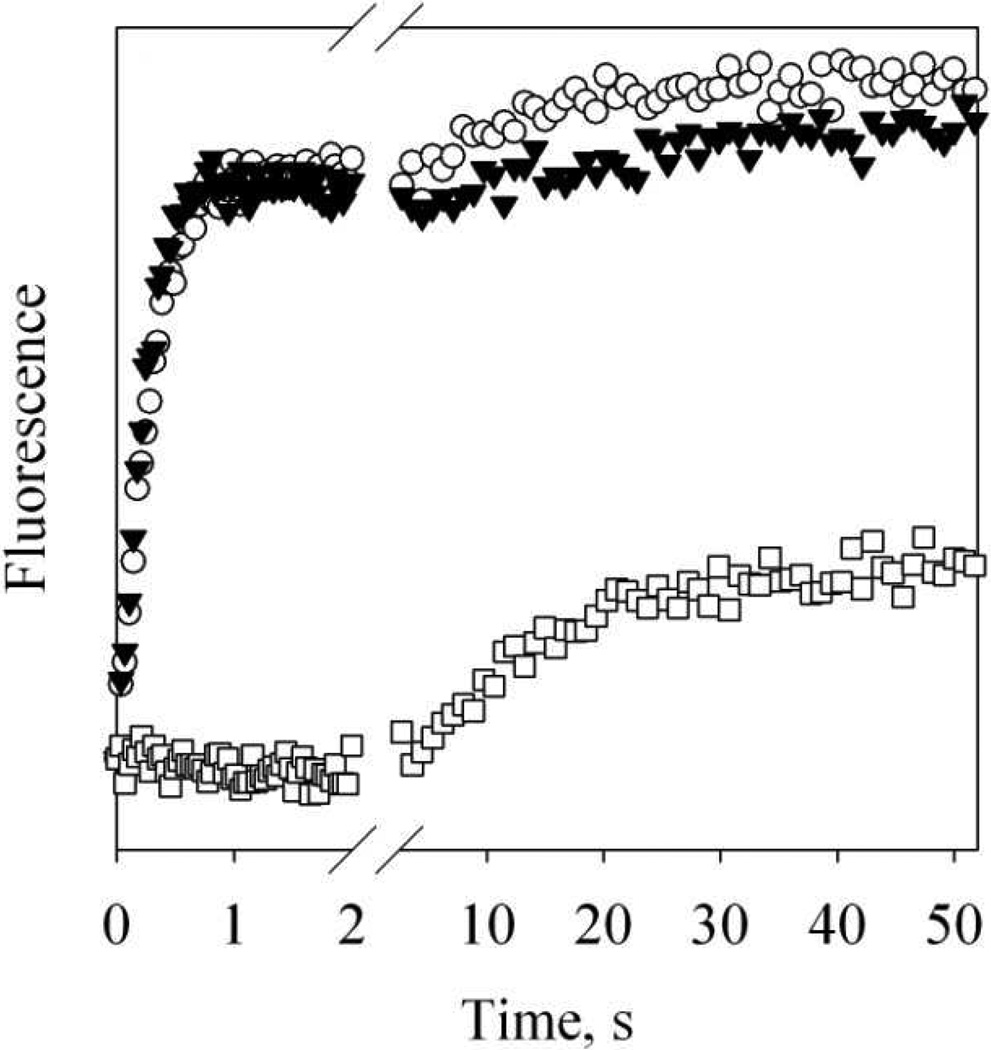
Figure 7. Kinetics of the binding of Alexa-CaM to NOS target peptides.
A – D: stopped-flow time-dependent FW110 change at 4.5 °C during the association of holo Alexa-CaM with nNOS726–749 (A), eNOS492–511 (B) or iNOS507–531 (C), or the association of apo Alexa-CaM with iNOS507–531 (D). A – C: [holo Alexa-CaM] = 44 nM and [peptide] = 0.18 to 1.3 µM (bottom to top). D: [apo Alexa-CaM] = 44 nM and [peptide] = 0.8 to 15.3 µM (bottom to top). Traces are vertically offset for clarity. Buffer: 50 mM HEPES, pH 8.0 with 100 mM NaCl and 200 µM CaCl2 (A –C) or 50 mM HEPES, pH 8.0 with 100 mM NaCl and 500 µM EDTA (D). Each trace was an average of 8 – 14 stopped-flow shots. E: dependence of the rate constants kobs of holo Alexa-CaM on the concentrations of the peptides: nNOS726–749 (circles), eNOS492–511 (triangles) or iNOS507–531 (squares). F: dependence of the rate constants kobs of apo Alexa-CaM on the concentrations of iNOS507–531. The error bars are from two individual measurements.
The binding of Alexa-CaM to the three NOS target peptides in the presence and absence of Ca2+ was compared in more details (Fig. 5). Mixing either nNOS726–749 or eNOS492–511 peptide with apo Alexa-CaM did not cause any significant change of FW110 (Fig. 5A & B, open circles vs. solid triangles). Further addition of 200 µM Ca2+ almost completely quenched FW110 leaving small residual emission (Fig. 5A & B, pluses). The observed FRET clearly indicated that the distance between Alexa 350 and W110 decreased greatly upon holo Alexa-CaM binding to either nNOS726–749 or eNOS492–511. This quenching of FW110 was reversed by chelation of Ca2+ from Alexa-CaM with EDTA (Fig. 5A & B, stars). These fluorescence changes verified again that the binding of either nNOS726–749 or eNOS492–511 to CaM was Ca2+-dependent, consistent with previous observations and Ca2+-dependence of the activation of nNOS and eNOS by CaM [25, 27, 28].
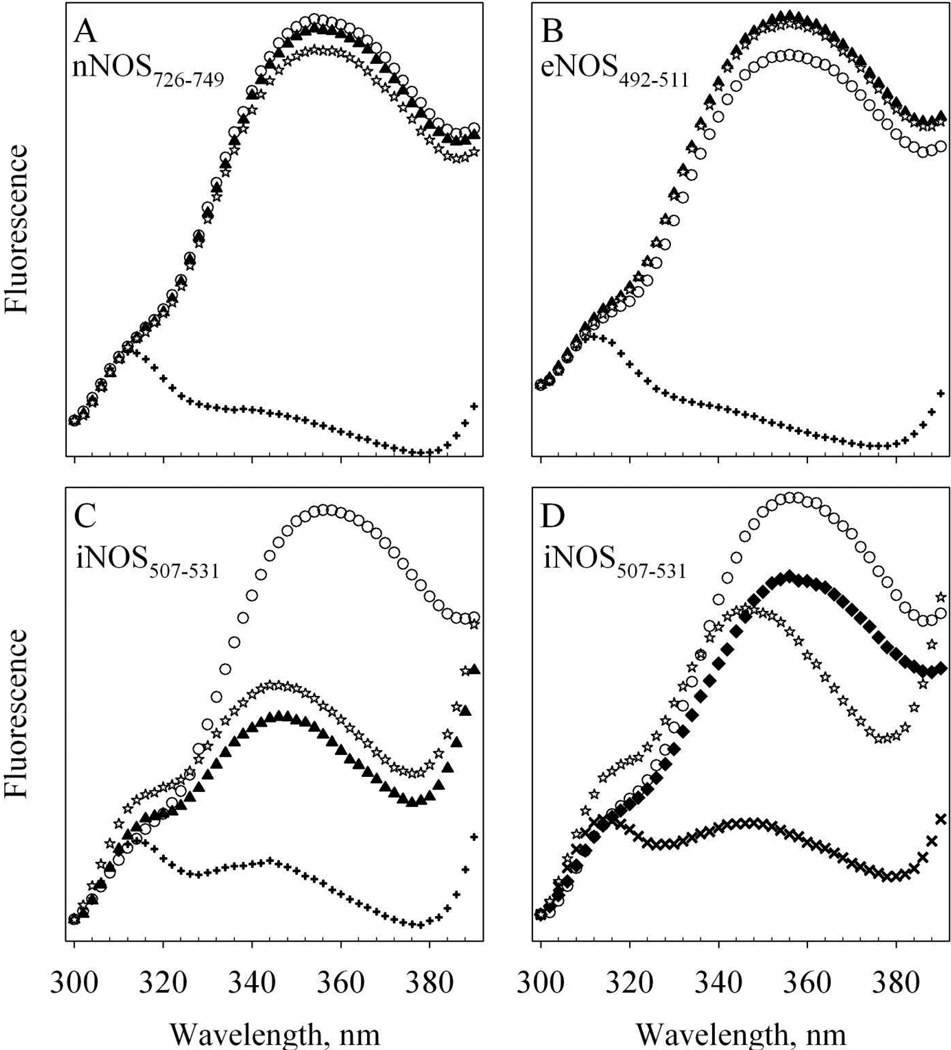
Figure 5. Fluorescence of Alexa-CaM and its complexes with NOS target peptides.
The change of FW110 between 300 – 390 nm at room temperature is shown. Each spectrum is the average of two scans and the fluorescence intensities are in arbitrary units. Buffer: 50 mM HEPES, pH 7.8 with 100 mM NaCl and 100 µM EDTA. A – C: apo 53 nM Alexa-CaM (open circles) was first mixed with (A) 3.4 µM nNOS726–749 (B) 3.4 µM eNOS492–511 or (C) 3.1 µM iNOS507–531 (solid triangles); then mixed with 300 µM CaCl2 (plus) and finally mixed with 2 mM EDTA (stars). D: apo 53 nM Alexa-CaM (open circles) was first mixed with 300 µM CaCl2 (solid diamonds); then mixed with 3.1 µM iNOS507–531 (cross) and finally with 2 mM EDTA (stars).
In contrast to nNOS726–749 and eNOS492–511, the iNOS507–531 peptide was able to bind both apo Alexa-CaM (Fig. 5C) and holo Alexa-CaM (Fig. 5D). In the absence of Ca2+, mixing of iNOS507–531 peptide with Alexa-CaM led to significant yet incomplete quenching of FW110 and a blue shift of FW110 emission max to 346 nm (Fig. 5C, solid triangles vs. open circles). This indicated that the iNOS507–531 was able to bind to apo Alexa-CaM, as demonstrated previously by other studies [21, 27]. The formation of apo Alexa-CaM/iNOS507–531 complex may introduce a conformational rearrangement in which the distance between Alexa 350 and W110 is shortened leading to partial quenching of FW110 by Alexa 350. The blue shift of FW110 maximum indicated that the environment around W110 became somewhat more hydrophobic. Upon addition of 200 µM Ca2+, FW110 was close to be completely quenched (Fig. 5C, plus). This latter quench of FW110 was most likely due to the conformational rearrangement during the formation of holo Alexa-CaM/iNOS507–531 complex, where Alexa-CaM bound to iNOS507–531 in classical conformation and FRET from W110 to Alexa 350 was more efficient in this conformation. Ca2+ induced conformational rearrangement in the complex of apo CaM/iNOS507–531 was also demonstrated previously [27]. The extent of quenching induced by Ca2+ could be reversed by EDTA while the emission peak remained at 346 nm, blue-shifted from FW110 in free Alexa-CaM (Fig. 5C, stars), indicating that iNOS507–531 stayed bound to apo Alexa-CaM (and unlabeled T34C/T110W CaM, Fig. 4C) when Ca2+ was removed as observed before [27].
The environment of residue W110 in free apo Alexa-CaM exhibited some conformational rearrangement during Ca2+ loading to Alexa-CaM, as indicated by the slight decrease of FW110 in holo Alexa-CaM (Fig. 5D, open circles vs. solid diamonds). When iNOS507–531 was mixed with holo Alexa-CaM, FW110 was almost completely quenched (Fig. 5D, cross). FW110 only partially recovered from its basal state by chelation of Ca2+ with EDTA. The emission maximum remained at 346 nm (Fig. 5D, stars), demonstrating again that in the presence of iNOS507–531, Ca2+-depletion did not dissociate Alexa-CaM from iNOS507–531. Overall, the association of all three target peptides with holo Alexa-CaM appeared to induce similar conformational change in Alexa-CaM. These observations are in line with the crystal structures which show that CaM binds its NOS targets in classical mode (Fig. 1, C–F)
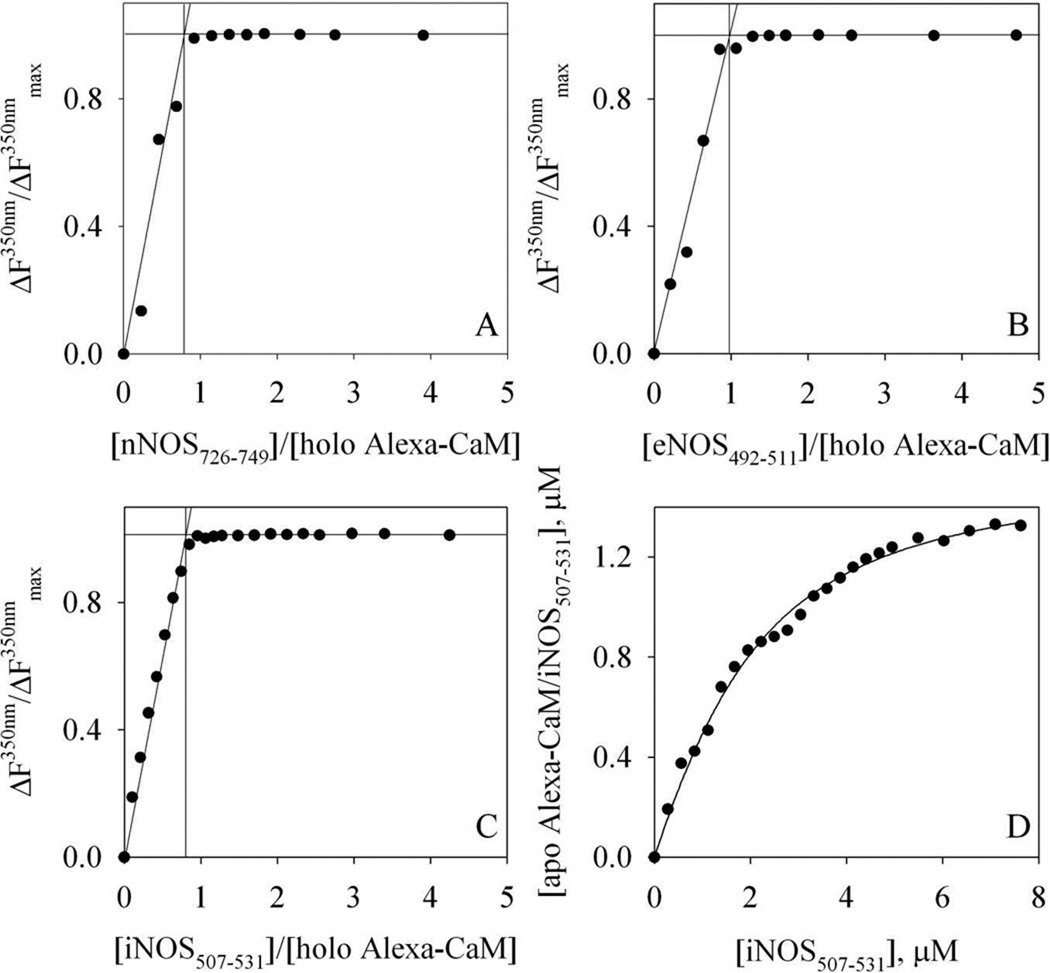
We next determined the binding stoichiometry of NOS target peptides to holo Alexa-CaM by fluorescence titration. Quenching of FW110 during serial addition of NOS target peptides to holo Alexa-CaM was linear and exhibited a clear break point at the concentration ratio close to 1 (Fig. 6A–C), indicating a stoichiometric binding of all three NOS target peptides with holo Alexa-CaM. On the other hand, the affinity of iNOS507–531 for apo Alexa-CaM appeared to be much weaker (Fig. 6D) compared to its binding to holo Alexa-CaM (Fig. 6C). The observed non-linear binding isotherm (Fig. 6D) could be fitted reasonably well by Equation (4) to yield a Kd of ~1.1 µM for iNOS507–531 to apo Alexa-CaM.
Figure 6. Titration of Alexa-CaM with NOS target peptides.
A – C: titration of holo Alexa-CaM with NOS target peptides at room temperature. The percentage of FW110 (in arbitrary units) change at 350 nm is plotted versus the ratio, [NOS peptide] : [holo Alexa-CaM]. A: nNOS726–749; B: eNOS492–511; C: iNOS507–531. The lines are the visual guides for determining the breaking points of the titrations. The concentrations of holo Alexa-CaM were 1.3 µM for A and B, and 0.9 µM for C. Buffer: 50 mM HEPES, pH 8.0 with 100 mM NaCl and 200 µM CaCl2. D: titration of 1.3 µM apo Alexa-CaM with iNOS507–531 at room temperature. Buffer: 50 mM HEPES, pH 8.0 with 100 mM NaCl and 2 mM EDTA. The concentration of complex apo Alexa-CaM/iNOS507–531 is plotted versus [iNOS507–531] and the line represents the fit to Equation (4).
3.4. Kinetics of the binding of Alexa-CaM to NOS target peptides
Steady-state fluorescence data shown above provided basis for the measurements of binding kinetics by fluorescence stopped-flow. The experiments were conducted at 4.5 °C to minimize loss of kinetic data in the dead time, 1.5 ms, of the instrument. FW110 exhibited time-dependent quenching when holo Alexa-CaM was quickly mixed with NOS target peptides (Fig. 7A – C). For the association of holo Alexa-CaM to all three NOS target peptides, the observed rate, kobs, exhibited a linear dependence on peptide concentrations (Fig. 7E). The slopes of the linear regressions gave the second-order binding rate constants (kon): nNOS726–749, 6.6 (± 0.6) × 108 M−1s−1 (n = 2); eNOS492–511, 2.9 (± 0.1) × 108 M−1s−1(n = 2) and iNOS507–531, 6.1 (± 0.1) × 108 M−1s−1(n = 2) (Table 1).
Table 1.
kon and koff rate constants of NOS target peptides to holo Alexa-CaM.
| kon (M−1s−1)a | koff (s−1)b | Kd, apparent (nM)c | Kd (nM)d | |
|---|---|---|---|---|
| nNOS726–749 | 6.6 (± 0.6) × 108 | 3.7 ± 0.1 | 5.6 | 1 – 5.0 [18, 19, 21] |
| eNOS492–511 | 2.9 (± 0.1) × 108 | 4.5 ± 0.3 | 1.6 | 2.9 – 4.0 [16, 23] |
| iNOS507–531 | 6.1 (± 0.1) × 108 | 0.063 ± 0.007 | 0.1 | < 0.1 – 3.3 [12, 16, 21–23] |
kon constants were measured with fluorescence stopped-flow at 4.5 °C.
koff constants were measured with fluorescence stopped-flow at 4.5 °C.
Kd, apparent was calculated as koff /kon.
From literatures.
On the other hand, the observed rate of iNOS507–531 binding to apo Alexa-CaM exhibited a non-linear dependence on the concentration of iNOS507–531. Higher iNOS507–531 concentration led to much faster kobs (Fig. 7F). This non-linear dependence prevented us from obtaining the kon for iNOS507–531 to apo Alexa-CaM. The kinetic data also confirmed that iNOS507–531 bound to apo and holo Alexa-CaM in different conformations.
3.5. Kinetics of the dissociation of Alexa-CaM from NOS target peptides
The y-intercept, or k−1, of the secondary plot for kon determination usually provides an estimate for the dissociation rate constant, koff. However, in our case the intercepts were all close to zero or even negative (Fig. 7E). We therefore determined koff by following the recovery of FW110 upon mixing holo Alexa-CaM/NOS target peptides complexes with EDTA by fluorescence stopped-flow (Fig. 8). Upon mixing EDTA with Alexa-CaM complex with either nNOS726–749 or eNOS492–511, FW110 exhibited a biphasic increase (Fig. 8, open circles and solid triangles). The fast phase represented 81% of the overall increase of FW110 {ΔFmax1 + ΔFmax2 in Eq. (7)} in Alexa-CaM/nNOS726–749 complex and 83% in Alexa-CaM/eNOS492–511 complex. The extent of FW110 increase in this phase {ΔFmax1 in Eq. (7)} was similar to the extent of FW110 quenching {ΔFmax in Eq. (5)} during holo Alexa-CaM association with either nNOS726–749 or eNOS492–511. The first phase may therefore represent the kinetics of the departure of W110 from Alexa 350 together with the release of either nNOS726–749 or eNOS492–511. The rate constants of the fast phase were measured 3.7 ± 0.1s−1 (n = 2) and 4.5 ± 0.3 s−1 (n = 2) for complexes of Alexa-CaM with nNOS726–749 and eNOS492–511 respectively (Fig. 8), and were taken as de facto the koff rate constants for the dissociation of either nNOS726–749 or eNOS492–511 from apo Alexa-CaM (Table 1). The extent of FW110 increase of the second phase {ΔFmax2 in Eq. (7)} was small and may correspond to the recovery of the small FW110 decrease when Ca2+ was added to apo Alexa-CaM (Fig. 5D, solid diamonds vs. open circles). The rate constants of slow phase, 0.071 ± 0.001 s−1 (n = 2) for Alexa-CaM/nNOS726–749 complex and 0.038 ± 0.001 s−1 (n = 2) for Alexa-CaM/eNOS492–511 complex (Fig. 8), may reflect how fast the conformation rearrangement of the C-lobe in free apo Alexa-CaM was.
Figure 8. Dissociation of Alexa-CaM from NOS target peptides.
Time-dependent changes of FW110 upon mixing holo Alexa-CaM/NOS target peptide complexes with 2 mM EDTA at 4.5 °C. Starting complexes: 44 nM holo Alexa-CaM with 2.5 µM NOS peptide: nNOS726–749 (open circles), eNOS492–511 (solid triangles) or iNOS507–531 (open squares). Each trace was an average of 8 – 14 stopped-flow shots. Data between 2 s and 2.5 s were omitted for clarity.
The dissociation kinetics of Alexa-CaM/iNOS507–531 complex was very different from the complexes of Alexa-CaM with either nNOS726–749 or eNOS492–511 (Fig. 8, open squares). When holo Alexa-CaM/iNOS507–531 was mixed with EDTA, FW110 essentially remained unchanged in the initial 2 s, suggesting that the distance between the N- and C-lobes stayed unchanged or a slow Ca2+-depletion by EDTA. Then FW110 slowly increased with a kobs of 0.063 ± 0.007 s−1 (n = 2, Table 1) indicating that Ca2+-depleted N- and C-lobes slowly separated from each other likely due to the conformational rearrangement of apo Alexa-CaM. However, compared to the FW110 change {ΔFmax in Eq. (5)} observed during the binding of holo Alexa-CaM to iNOS507–531, only about 30% of the quenched FW110 was recovered {ΔFmax in Eq. (8)}. This suggested that Ca2+ removal did not free up Alexa-CaM from iNOS507–531 and the conformational rearrangement of apo Alexa-CaM occurred in the presence of the associated iNOS507–511.
4. Discussion
4.1. Binding of CaM to NOS proteins unlikely a rate-limiting step of NOS catalysis
A body of experimental evidence shows that the association of CaM with NOS switches on the electron transfer from FMN to heme [3, 48]. The measured rates of electron transfer to heme from the interacting FMN are 40 – 50 s−1 for nNOS, 2 – 5 s−1 for eNOS and 11 s−1 for iNOS [49]. Interdomain electron transfer from FMN to heme was recently measured more accurately using laser flash photolysis and the rates are the same for rat nNOS in the presence of CaM: 36 s−1 and human iNOS: 34 s−1 or murine iNOS: 35 s−1 [50, 51]. No such redox event was observed in the absence of CaM for nNOS [51]. Among the kinetic steps leading to efficient electron transfer in NOSs, the binding of CaM (Scheme 1, Step 1) and its effects on the conformational equilibrium of FMN subdomain may impose a limit on the rate of electron transfer in NOSs. Relatively slow kon constants determined in a previous SPR study, 1.58 × 105 M−1s−1 and 3 × 104 M−1s−1, for the binding of CaM to the nNOS and iNOS target peptides respectively [21], suggest that the binding of CaM to NOS is a rate-limiting step in electron transfer from NADPH to heme in NOSs. However, the heme and flavin reduction rates in nNOS measured in this study (Fig. 2) and previously [33] are much faster than the binding of CaM to NOSs based on the rate constants measured in the SPR study, ~0.3 and 0.06 s−1 for nNOS and iNOS, respectively, at µM level of CaM used in our study. The 0.3 s−1 value of nNOS is even smaller than the turnover numbers, ~1.1 s−1, estimated from steady-state essay for NO formation where full-length nNOS was used. One possible explanation for this large discrepancy is that the binding constants of CaM to NOS target peptides measured by SPR were significantly slower than the true values.
It has been well known that binding kinetics determined using SPR method may be biased by mass transport limitations within immobilization matrix or other factors unrelated to the intrinsic binding kinetics such as the thickness of the matrix, the diffusion coefficient within the matrix, the partition coefficient of the ligand from bulk into the matrix, the sensitivity distribution of the sensor, and the distribution of the immobilized receptor within the matrix [34, 52]. These possible limitations of SPR measurements are likely more severe in measuring fast kon constants, leading to measured rates much lower than the true values, demonstrated by the variation of kon constants of CaM to nNOS peptide using different peptide immobilization chips [21, 53].
The fluorescence stopped-flow method avoided the immobilization of either NOS target peptides or CaM required in SPR method and allowed us to determine the true kon constants of NOS target peptides to Alexa-CaM in solution. All three target peptides bound to holo Alexa-CaM rapidly with kon > 1×108 M−1s−1 (Table 1). This rate reflected how fast the conformations of NOS target peptides rearranged into helical structures and bound to holo Alexa-CaM. The kon values were essentially the same for nNOS726–749 and iNOS507–531, and slower by factor of about 2 for eNOS492–511 (Table 1). The kon values of nNOS or iNOS target peptide to Alexa-CaM are 4.2 × 103 and 2 ×104 fold faster than those determined previously by SPR. The binding rate of CaM to nNOS based on the kon values measured in this study, 1.3 × 103 s−1 at 2 µM CaM, was much faster than the measured rates of flavin and heme reduction, ~ 20 s−1 (Fig. 2), suggesting that the binding of holo CaM to NOS isozymes is fast to allow efficient electron transfer and probably not imposing a rate-limiting step in NOS catalysis.
4.2. Difference between the binding of CaM to NOS peptides and full-length NOSs
We are aware of that CaM may bind to full-length NOS proteins slower than to NOS target peptides, as CaM was previously observed to bind to target peptides faster than to the corresponding segments in whole proteins [35]. Other structural elements of NOS proteins participate in the binding of CaM to NOS proteins [2, 16, 24, 32] and can introduce additional rate-limiting step(s). The conformational rearrangement of oxygenase and reductase domains may also occur in concert with the formation of helical structure in CaM-binding domain for binding to CaM and slow down the overall rate of binding. If the binding of CaM to NOS proteins is considerably slower than to NOS target peptides, it can potentially be a rate-limiting step in NOS catalysis. This possibility was hinted by the observation of the slightly faster heme and flavin reduction rates for nNOS pre-bound with CaM than those for nNOS without pre-bound CaM (Fig. 2). It is still possible that the binding of CaM is fast but followed by a slow step of conformational rearrangement (Scheme 1, Step 2) and therefore appeared as a pre-equilibrium step contributing to the overall electron transfer rates in nNOS. These issues will remain unresolved until the determination of the binding kinetics of CaM to NOS proteins. Unfortunately the binding of Alexa-CaM to full-length NOS proteins cannot be monitored by the change of FW110 used in this study due to the large background tryptophan fluorescence from NOS proteins. We are currently trying to determine the binding rates of CaM to full-length NOS proteins by spin-labeling both cysteine residues in T34C/T110C CaM mutant and following the time-dependent change of spin-spin dipolar interactions upon binding of CaM to NOS proteins using electron paramagnetic resonance (EPR).
4.3. Dissociation of Alexa-CaM from NOS target peptides
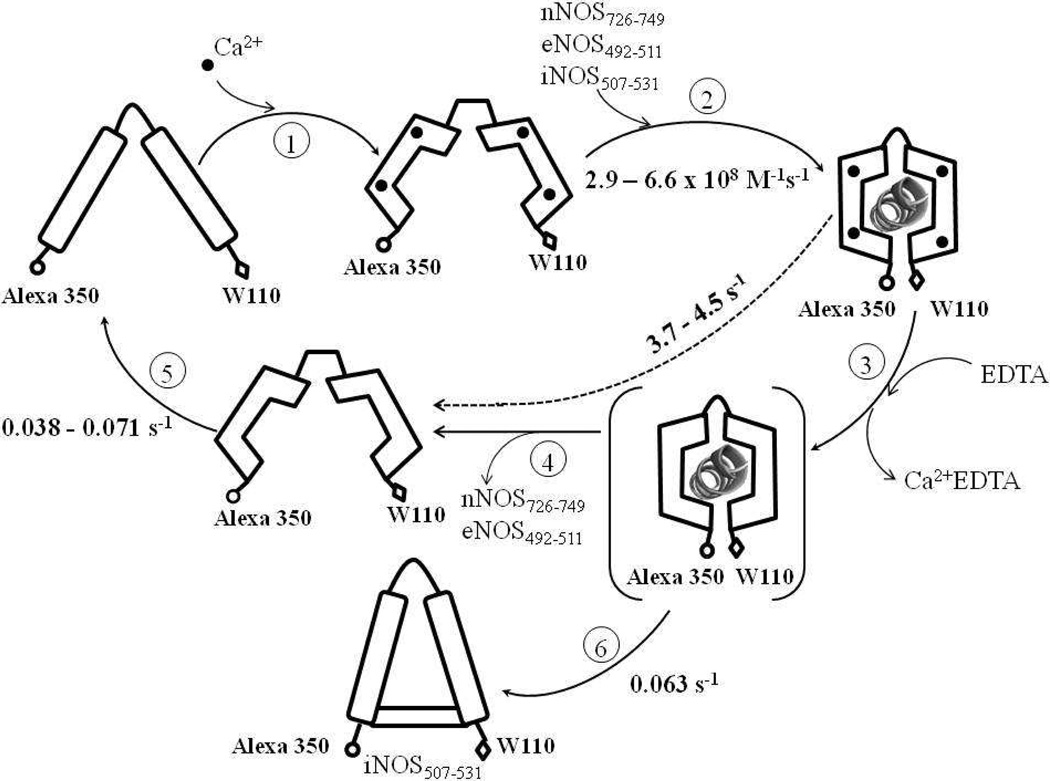
Alexa-CaM dissociated from the target peptides of constitutive NOSs upon removal of Ca2+, giving a biphasic FW110 recovery (Fig. 8). The fast phase with large increase of FW110 likely represented how fast C- and N-lobes departed from each other, the apparent reverse step of the distance shortening during the binding to target peptides. However, the kinetic data did not distinguish between Ca2+ removal from holo Alexa-CaM (Scheme 2, Step 3) and release of either nNOS or eNOS peptides from apo Alexa-CaM (Scheme 2, Step 4) being the rate-limiting step. The dissociation rates of Ca2+ from the complexes of Alexa-CaM with NOS target peptides were directly measured by quickly mixing the complexes with Fura Red and following its time-dependent fluorescence change upon binding to Ca2+ (data not shown). Fura Red is a Ca2+ indicator with strong Ca2+ affinity (140 nM) and its fluorescence (λex 474 nm/λem 660 nm) has no overlap with that of either Alexa 350 or tryptophan [54]. However, due to the low quantum yield of Fura Red (Φ = 0.013), the signal-to-noise ratio of the data was low thus did not lead to accurate rate determinations. Nonetheless, a rough estimate of ~1 – 3 s−1 was made for the dissociation rates of Ca2+ from the complexes of Alexa-CaM with peptides from the three NOS isoforms. The similarity of the Ca2+ dissociation rates from the Alexa-CaM/constitutive NOS peptide complexes to the recovery rates of FW110 (Fig. 8) prevented us from assigning rate constants to either Step 3 or 4 in Scheme 2. The rate constant, measured 3.7 s−1 for nNOS726–749 and 4.5 s−1 for eNOS492–511 respectively, therefore represented the apparent koff of the composite step including Steps 3 and 4 (Scheme 2) and set the lower limit for how fast peptides from constitutive NOSs dissociate from Alexa-CaM.
Scheme 2. Kinetic events during the binding of Alexa-CaM to NOS target peptides.
Values of all rate constants were determined in this study. The species in parenthesis is unresolved intermediate for Steps 3 and 4. The dashed line with rate 3.7 – 4.5 s−1 represents the composite step including Steps 3 and 4 for either nNOS726–749 or eNOS492–511 (see Discussion). The rate of Ca2+ binding to apo Alexa-CaM (Step 1) was not measured in this study but assumed to be as fast as that determined for other CaM species [57]. Holo Alexa-CaM is assumed to bind NOS peptides in classic conformation (Step 2). Distances between Alexa 350 and W110 are purely illustrative and by no means to indicate real scales. The conformation of the apo Alexa-CaM/iNOS507–531 complex is only descriptive to demonstrate that Alexa 350 and W110 are not as separated as in free apo Alexa-CaM.
Dissociation of Alexa-CaM from target peptides of constitutive NOSs by Ca2+-depletion is relatively fast, suggesting that regulation of cellular [Ca2+] is an efficient way to modulate the dynamic interaction between CaM and constitutive NOSs in vivo. The apparent koff constants may provide an estimate for how fast constitutive NOSs are “switched-off” when [Ca2+] drops under physiological conditions. Apparent Kd constants of Alexa-CaM to peptides of constitutive NOSs were calculated as apparent koff/kon, 5.6 nM for nNOS726–749 and 1.6 nM for eNOS492–511, respectively, agreeing reasonably well with the affinities of CaM for NOS target peptides measured previously using various methods (Table 1) [16–23]. The kon and apparent koff constants are the rates of an apparently reversible process: the closing and opening of N- and C-lobes. However, they are not associated with a true reversible chemical step. Apparent Kd constants thus measured likely represent the upper limit of the true Kd constants for Alexa-CaM to peptides of constitutive NOSs and may approximate the apparent affinity of CaM to constitutive NOSs in vivo since the dissociation of CaM from constitutive NOSs is likely controlled by the level of cellular [Ca2+].
Different from peptides of constitutive NOSs, Ca2+-depletion did not release iNOS507–531 from Alexa-CaM (Fig. 8). In the presence of iNOS507–531, Ca2+-depleted N- and C-lobes separated partially with a rate of 0.063 s−1 (Scheme 2, Step 6). This partial separation between N- and C-lobes represented an apparent kinetic reverse step of the distance shortening between the two lobes during the binding of holo Alexa-CaM to iNOS507–531, an apparent Kd as the ratio of apparent koff versus kon, 0.1nM, could be calculated for iNOS507–531 for Alexa-CaM (Table 1). iNOS peptide therefore exhibited significantly lower apparent Kd to Alexa-CaM than peptides of constitutive NOSs.
4.4. Different binding modes of iNOS peptide to apo and holo Alexa-CaM
Other than the higher affinity to Alexa-CaM, the major difference between iNOS507–531 and either nNOS726–749 or eNOS492–511 is its capability to bind both apo and holo Alexa-CaM, demonstrated by both kinetics and steady-state fluorescence data. The binding affinity of iNOS507–531 to apo Alexa-CaM (Fig. 6D) was much weaker than its affinity to holo Alexa-CaM (Table 1). FRET analysis showed that N- and C-lobes are more separated in apo Alexa-CaM/iNOS507–531 complex than in holo Alexa-CaM/iNOS507–531 complex (Fig. 5C and D). The conformation of iNOS peptide has been shown to adopt a type II β turn structure binding to apo CaM [12] and a α-helix to holo CaM [30]. Our data verified that the Ca2+-independent binding of iNOS to CaM was primarily due to the primary sequence of its CaM binding domain [55, 56], although other structural factors may contribute to iNOS activity in the absence of Ca2+, including the lack of the autoinhibitory loop inside the FMN subdomain and a C-terminal tail domain in iNOS [25].
In summary, the binding of Alexa-CaM to NOS target peptides determined in this study by stopped-flow is several order faster than that by SPR [21]. This suggested that the binding of CaM to NOS proteins is competent in supporting efficient electron transfer in NOS catalysis and does not appear to be rate-limiting. Alexa-CaM dissociated promptly from target peptides of constitutive NOSs with the removal of Ca2+, indicating that cellular Ca2+ level is an important controller of the catalysis of constitutive NOSs in vivo. The target peptide of iNOS, on the other hand, is capable of binding to both holo and apo Alexa-CaM but in different conformations, providing mechanism for the Ca2+-independence of iNOS catalysis.
Highlights.
> The binding kinetics of Alexa 350 labeled calmodulin to nitric oxide synthase target peptides in solution was measured using fluorescence stopped-flow. > The binding of calmodulin to nitric oxide synthase is fast and unlikely a rate-limiting step in the catalysis of nitric oxide synthase. > Only inducible nitric oxide synthase is capable of binding to both apo and holo calmodulin, but in different modes.
Acknowledgement
We thank Dr. M. Neal Waxham for critical comments on the manuscript.
Supported by NIH RO1 HL095820 (A.-L.T).
Abbreviations
- CaM
calmodulin
- apo CaM
CaM without bound Ca2+
- holo CaM
CaM with four bound Ca2+
- Alexa 350
Alexa Fluor 350
- Alexa-CaM
Alexa Fluor 350 labeled T34C/T110W CaM at C34
- NOS
nitric oxide synthase
- eNOS
endothelial NOS
- iNOS
inducible NOS
- nNOS
neuronal NOS
- DTT
dithiothreitol
- EGTA
ethyleneglycoltetraacetic acid
- FRET
fluorescence resonance energy transfer
- FW110
intrinsic fluorescence of W110 in T34C/T110 CaM and Alexa-CaM
- SPR
surface plasmon resonance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- 1.Alderton WK, Cooper CE, Knowles RG. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daff S. Nitric Oxide. 2010;23:1–11. doi: 10.1016/j.niox.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Stuehr DJ, Tejero J, Haque MM. FEBS J. 2009;276:3959–3974. doi: 10.1111/j.1742-4658.2009.07120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panda K, Ghosh S, Stuehr DJ. J. Biol. Chem. 2001;276:23349–23356. doi: 10.1074/jbc.M100687200. [DOI] [PubMed] [Google Scholar]
- 5.Su Z, Blazing MA, Fan D, George SE. J. Biol. Chem. 1995;270:29117–29122. doi: 10.1074/jbc.270.49.29117. [DOI] [PubMed] [Google Scholar]
- 6.Tejero J, Haque MM, Durra D, Stuehr DJ. J. Biol. Chem. 2010;285:25941–25949. doi: 10.1074/jbc.M110.126797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klee CB, Crouch TH, Richman PG. Annu. Rev. Biochem. 1980;49:489–515. doi: 10.1146/annurev.bi.49.070180.002421. [DOI] [PubMed] [Google Scholar]
- 8.Weinstein H, Mehler EL. Annu. Rev. Physiol. 1994;56:213–236. doi: 10.1146/annurev.ph.56.030194.001241. [DOI] [PubMed] [Google Scholar]
- 9.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Nat. Struct. Biol. 1995;2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 10.Rupp B, Marshak DR, Parkin S. Acta Crystallogr. D Biol. Crystallogr. 1996;52:411–413. doi: 10.1107/S0907444995011826. [DOI] [PubMed] [Google Scholar]
- 11.Crivici A, Ikura M. Annu. Rev. Biophys. Biomol. Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 12.Yuan T, Vogel HJ, Sutherland C, Walsh MP. FEBS Lett. 1998;431:210–214. doi: 10.1016/s0014-5793(98)00750-9. [DOI] [PubMed] [Google Scholar]
- 13.Vetter SW, Leclerc E. Eur. J. Biochem. 2003;270:404–414. doi: 10.1046/j.1432-1033.2003.03414.x. [DOI] [PubMed] [Google Scholar]
- 14.Drum CL, Yan SZ, Sarac R, Mabuchi Y, Beckingham K, Bohm A, Grabarek Z, Tang WJ. J. Biol. Chem. 2000;275:36334–36340. doi: 10.1074/jbc.M004778200. [DOI] [PubMed] [Google Scholar]
- 15.Drum CL, Yan SZ, Bard J, Shen YQ, Lu D, Soelaiman S, Grabarek Z, Bohm A, Tang WJ. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- 16.Venema RC, Sayegh HS, Kent JD, Harrison DG. J. Biol. Chem. 1996;271:6435–6440. doi: 10.1074/jbc.271.11.6435. [DOI] [PubMed] [Google Scholar]
- 17.Vorherr T, Knopfel L, Hofmann F, Mollner S, Pfeuffer T, Carafoli E. Biochemistry. 1993;32:6081–6088. doi: 10.1021/bi00074a020. [DOI] [PubMed] [Google Scholar]
- 18.Sheta EA, McMillan K, Masters BS. J. Biol. Chem. 1994;269:15147–15153. [PubMed] [Google Scholar]
- 19.Zhang M, Vogel HJ. J. Biol. Chem. 1994;269:981–985. [PubMed] [Google Scholar]
- 20.Censarek P, Beyermann M, Koch KW. Biochemistry. 2002;41:8598–8604. doi: 10.1021/bi025681k. [DOI] [PubMed] [Google Scholar]
- 21.Zoche M, Bienert M, Beyermann M, Koch KW. Biochemistry. 1996;35:8742–8747. doi: 10.1021/bi960445t. [DOI] [PubMed] [Google Scholar]
- 22.Anagli J, Hofmann F, Quadroni M, Vorherr T, Carafoli E. Eur. J. Biochem. 1995;233:701–708. doi: 10.1111/j.1432-1033.1995.701_3.x. [DOI] [PubMed] [Google Scholar]
- 23.Matsubara M, Hayashi N, Titani K, Taniguchi H. J. Biol. Chem. 1997;272:23050–23056. doi: 10.1074/jbc.272.37.23050. [DOI] [PubMed] [Google Scholar]
- 24.Roman LJ, Masters BS. J. Biol. Chem. 2006;281:23111–23118. doi: 10.1074/jbc.M603671200. [DOI] [PubMed] [Google Scholar]
- 25.Roman LJ, Martasek P, Masters BS. Chem. Rev. 2002;102:1179–1190. doi: 10.1021/cr000661e. [DOI] [PubMed] [Google Scholar]
- 26.Gerber NC, Nishida CR, Ortiz de Montellano PR. Arch. Biochem. Biophys. 1997;343:249–253. doi: 10.1006/abbi.1997.0187. [DOI] [PubMed] [Google Scholar]
- 27.Spratt DE, Taiakina V, Palmer M, Guillemette JG. Biochemistry. 2007;46:8288–8300. doi: 10.1021/bi062130b. [DOI] [PubMed] [Google Scholar]
- 28.Stuehr DJ. Biochim. Biophys. Acta. 1999;1411:217–230. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 29.Aoyagi M, Arvai AS, Tainer JA, Getzoff ED. EMBO J. 2003;22:766–775. doi: 10.1093/emboj/cdg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia C, Misra I, Iyanagi T, Kim JJ. J. Biol. Chem. 2009;284:30708–30717. doi: 10.1074/jbc.M109.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daff S. Biochem. Soc. Trans. 2003;31:502–505. doi: 10.1042/bst0310502. [DOI] [PubMed] [Google Scholar]
- 32.Ruan J, Xie Q, Hutchinson N, Cho H, Wolfe GC, Nathan C. J. Biol. Chem. 1996;271:22679–22686. doi: 10.1074/jbc.271.37.22679. [DOI] [PubMed] [Google Scholar]
- 33.Roman LJ, Martasek P, Miller RT, Harris DE, de La Garza MA, Shea TM, Kim JJ, Masters BS. J. Biol. Chem. 2000;275:29225–29232. doi: 10.1074/jbc.M004766200. [DOI] [PubMed] [Google Scholar]
- 34.Schuck P, Minton AP. Anal. Biochem. 1996;240:262–272. doi: 10.1006/abio.1996.0356. [DOI] [PubMed] [Google Scholar]
- 35.Torok K, Tzortzopoulos A, Grabarek Z, Best SL, Thorogate R. Biochemistry. 2001;40:14878–14890. doi: 10.1021/bi010920+. [DOI] [PubMed] [Google Scholar]
- 36.Spratt DE, Newman E, Mosher J, Ghosh DK, Salerno JC, Guillemette JG. FEBS J. 2006;273:1759–1771. doi: 10.1111/j.1742-4658.2006.05193.x. [DOI] [PubMed] [Google Scholar]
- 37.Berka V, Wang LH, Tsai AL. Biochemistry. 2008;47:405–420. doi: 10.1021/bi701677r. [DOI] [PubMed] [Google Scholar]
- 38.Putkey JA, Donnelly PV, Means AR. Methods Enzymol. 1987;139:303–317. doi: 10.1016/0076-6879(87)39094-9. [DOI] [PubMed] [Google Scholar]
- 39.Riener CK, Kada G, Gruber HJ. Anal. Bioanal. Chem. 2002;373:266–276. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- 40.Murphy ME, Noack E. Methods Enzymol. 1994;233:240–250. doi: 10.1016/s0076-6879(94)33027-1. [DOI] [PubMed] [Google Scholar]
- 41.Chen PF, Berka V, Wu KK. Arch. Biochem. Biophys. 2003;411:83–92. doi: 10.1016/s0003-9861(02)00729-4. [DOI] [PubMed] [Google Scholar]
- 42.Villamil Giraldo AM, Lopez Medus M, Gonzalez Lebrero M, Pagano RS, Labriola CA, Landolfo L, Delfino JM, Parodi AJ, Caramelo JJ. J. Biol. Chem. 2009;285:4544–4553. doi: 10.1074/jbc.M109.034512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Copeland RA. Enzymes A Pratical Introduction to Structure, Mechanism and Data Analysis. New York: Wiley-VCH; 2000. [Google Scholar]
- 44.Waxham MN, Tsai AL, Putkey JA. J. Biol. Chem. 1998;273:17579–17584. doi: 10.1074/jbc.273.28.17579. [DOI] [PubMed] [Google Scholar]
- 45.Dunford AJ, Rigby SE, Hay S, Munro AW, Scrutton NS. Biochemistry. 2007;46:5018–5029. doi: 10.1021/bi7001339. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto K, Kimura S, Shiro Y, Iyanagi T. Arch. Biochem. Biophys. 2005;440:65–78. doi: 10.1016/j.abb.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 47.Knight K, Scrutton NS. Biochem. J. 2002;367:19–30. doi: 10.1042/BJ20020667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Welland A, Daff S. FEBS J. 2010;277:3833–3843. doi: 10.1111/j.1742-4658.2010.07787.x. [DOI] [PubMed] [Google Scholar]
- 49.Stuehr D, Pou S, Rosen GM. J. Biol. Chem. 2001;276:14533–14536. doi: 10.1074/jbc.R100011200. [DOI] [PubMed] [Google Scholar]
- 50.Feng C, Tollin G, Holliday MA, Thomas C, Salerno JC, Enemark JH, Ghosh DK. Biochemistry. 2006;45:6354–6362. doi: 10.1021/bi060223n. [DOI] [PubMed] [Google Scholar]
- 51.Feng C, Tollin G, Hazzard JT, Nahm NJ, Guillemette JG, Salerno JC, Ghosh DK. J. Am. Chem. Soc. 2007;129:5621–5629. doi: 10.1021/ja068685b. [DOI] [PubMed] [Google Scholar]
- 52.Schuck P. Biophys. J. 1996;70:1230–1249. doi: 10.1016/S0006-3495(96)79681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fischer T, Beyermann M, Koch KW. Biochem. Biophys. Res. Commun. 2001;285:463–469. doi: 10.1006/bbrc.2001.5206. [DOI] [PubMed] [Google Scholar]
- 54.Poenie M. In: Calcium Signaling, Fluorescent Calcium Indicators Based on BAPTA. Putney JW Jr, editor. Boca Raton: CRC Press; 2000. pp. 1–45. [Google Scholar]
- 55.Censarek P, Beyermann M, Koch KW. FEBS Lett. 2004;577:465–468. doi: 10.1016/j.febslet.2004.10.048. [DOI] [PubMed] [Google Scholar]
- 56.Zoche M, Beyermann M, Koch KW. Biol. Chem. 1997;378:851–857. doi: 10.1515/bchm.1997.378.8.851. [DOI] [PubMed] [Google Scholar]
- 57.Johnson JD, Tikunova SB. Methods Mol. Biol. 2002;173:89–102. doi: 10.1385/1-59259-184-1:089. [DOI] [PubMed] [Google Scholar]