Abstract
The ascomycete Mycosphaerella graminicola is the causal agent of septoria tritici blotch (STB), one of the most destructive foliar diseases of bread and durum wheat globally, particularly in temperate humid areas. A screening of the French bread wheat cultivars Apache and Balance with 30 M. graminicola isolates revealed a pattern of resistant responses that suggested the presence of new genes for STB resistance. Quantitative trait loci (QTL) analysis of a doubled haploid (DH) population with five M. graminicola isolates in the seedling stage identified four QTLs on chromosomes 3AS, 1BS, 6DS and 7DS, and occasionally on 7DL. The QTL on chromosome 6DS flanked by SSR markers Xgpw5176 and Xgpw3087 is a novel QTL that now can be designated as Stb18. The QTLs on chromosomes 3AS and 1BS most likely represent Stb6 and Stb11, respectively, and the QTLs on chromosome 7DS are most probably identical with Stb4 and Stb5. However, the QTL identified on chromosome 7DL is expected to be a new Stb gene that still needs further characterization. Multiple isolates were used and show that not all isolates identify all QTLs, which clearly demonstrates the specificity in the M. graminicola–wheat pathosystem. QTL analyses were performed with various disease parameters. The development of asexual fructifications (pycnidia) in the characteristic necrotic blotches of STB, designated as parameter P, identified the maximum number of QTLs. All other parameters identified fewer but not different QTLs. The segregation of multiple QTLs in the Apache/Balance DH population enabled the identification of DH lines with single QTLs and multiple QTL combinations. Analyses of the marker data of these DH lines clearly demonstrated the positive effect of pyramiding QTLs to broaden resistance spectra as well as epistatic and additive interactions between these QTLs. Phenotyping of the Apache/Balance DH population in the field confirmed the presence of the QTLs that were identified in the seedling stage, but Stb18 was inconsistently expressed and might be particularly effective in young plants. In contrast, an additional QTL for STB resistance was identified on chromosome 2DS that is exclusively and consistently expressed in mature plants over locations and time, but it was also strongly related with earliness, tallness as well as resistance to Fusarium head blight. Although to date no Stb gene has been reported on chromosome 2D, the data provide evidence that this QTL is only indirectly related to STB resistance. This study shows that detailed genetic analysis of contemporary commercial bread wheat cultivars can unveil novel Stb genes that can be readily applied in marker-assisted breeding programs.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-011-1623-7) contains supplementary material, which is available to authorized users.
Introduction
Septoria tritici blotch (STB) is a fungal wheat disease that is caused by the ascomycete Mycosphaerella graminicola (Fuckel) J.Schröt. The disease was first described in France (Desmazieres 1842; Sprague 1938), but was neglected for a long time due to overwhelming stripe rust and powdery mildew epidemics. Thus, STB was long considered as a secondary disease that mostly appeared in years with low levels of other cereal diseases. Nevertheless, it has been present in Europe for over a century, along with Stagonospora nodorum (Bearchell et al. 2005) and is currently considered to be one of the most important wheat diseases. Infections result in severe necrosis of the foliage that is filled with the asexual and sexual fructifications (Eyal 1999; Hunter et al. 1999; Kema et al. 1996c; McDonald et al. 1996; Shaw and Royle 1989). In Europe, STB usually establishes through airborne ascospores that are discharged from wheat debris and deposited in young wheat crops in the fall (Suffert et al. 2010). This is followed by rain splash driven spore dispersal during the growing season (Eriksen and Munk 2003; Halama 1996; Pastircak 2005; Scott et al. 1988; Shaw and Royle 1989, 1993). However, M. graminicola can reproduce sexually throughout the year, which provides the fungus with a mechanism to overcome adverse biotic or abiotic conditions (Kema et al. 1996c; Zhan et al. 2007; Ware et al. unpublished data).
STB management is largely effectuated by the application of fungicides and breeding for resistance. Due to its increased importance in Europe, STB is a main target as well as a serious concern of the agrochemical and breeding industry because of recent outbreaks of resistance to strobilurins (Cools and Fraaije 2008; Fraaije et al. 2005; Fraaije et al. 2007; McCartney et al. 2007; Stammler et al. 2008; Torriani et al. 2009) and steadily increasing levels of resistance to azole fungicides (Cools and Fraaije 2008; Fraaije et al. 2005; Fraaije et al. 2007; Mavroeidi and Shaw 2005; Stergiopoulos et al. 2003). These problems raised questions about the sustainability as well as the environmental impact of crop protection agents (Verweij et al. 2009). Hence, in several European countries, including France, Spain, Germany, Italy, the Netherlands and the UK, pesticide reduction programs have been developed and adopted by policy makers (Anonymous 2009). Therefore, a new focus on host resistance to increase the commercial lifetime of cultivars is required as part of a strategy to control STB.
In the UK, STB was unheard of as a major wheat disease before the late 1970s, but emerged as a major foliar blight in the early 1980s on susceptible cvs. such as cvs. Norman and Longbow. These cultivars were then replaced by others of similar susceptibility, and significant progress in breeding for resistance was not made until the mid-1990s (Paveley 2006). Demands for cultivars with better resistance levels resulted in the release of cv. Claire in 1999 that was replaced by cv. Alchemy (Angus and Fenwick 2008). Currently, other high-yield potential cultivars with moderate to high resistance to STB have been recommended, such as cv. Stigg (Anonymous 2010a; Angus et al. 2010). In France, 15 cultivars covered almost 77% of the total wheat acreage in 2003, in which cv. Apache ranked first with 23.7% and cvs. Isengrain, Tremy, Shango, Orvantis, Soissons, Caphorn and Charger together covered 37.3% (Anonymous 2005). Recent resistance screens indicated that the majority of these cultivars were highly susceptible to a substantial number of isolates in the seedling stage and hence, their resistances have a narrow efficacy (Tabib Ghaffary et al., unpublished data). Consequently, there is an urgent need for new resistance genes (Tabib Ghaffary et al., 2011 submitted to TAG).
The identification, characterization and processing of resistance to STB in practical wheat breeding programs, however, are not routine issues and several parameters can be used for disease scoring, such as the percentage of induced necrosis (N) or the percentage of pycnidia (P), the asexual fructifications of M. graminicola, in the foliage. Both parameters are strongly interwoven, as necrosis is conditional for pycnidia development, but is suggested to be under different genetic control (Kema et al. 1996d; Shetty et al. 2003, 2007, 2009). Currently, most screens involve well-characterized fungal isolates in repeated young plantlet assays and detached leaf assays (Arraiano et al. 2001a; Arraiano and Brown 2006; Kema et al. 1996a; Kema et al. 1996b; Kema and van Silfhout 1997), which have contributed to the data reliability and eventually to the mapping of resistance genes. Seedling screens offer opportunities to identify the efficacy of resistance to a wide panel of isolates, but—due to quarantine limitations—these can only be used to a limited extent under field conditions. Nevertheless, practical resistance breeding is a difficult multi-location, multi-pathogen and multi-pathotype effort responding to actual epidemiological situations—for instance for cereal rust diseases—and even legislation such as for Fusarium head blight (FHB) (Vanloqueren and Baret 2008). This resulted in 73, 89, 61 and 95 identified resistance genes for stripe rust, leaf rust, stem rust and powdery mildew, respectively, but only 17 Stb genes have been reported (Tabib Ghaffary et al. 2011, submitted to TAG). The majority of these Stb genes have a limited efficacy and hence are only sparsely deployed in breeding programs (Arraiano et al. 2007; Chartrain et al. 2009; Goodwin 2007), whereas the resistance genes to other wheat diseases are widely applied in new commercial wheat cultivars.
The apparent need for additional resistance genes prompted us to screen a wide variety of germplasm that resulted in the identification of new Stb genes (Tabib Ghaffary et al. 2011, submitted to TAG; Tabib Ghaffary, unpublished data). Here, we report the characterization of STB resistance in the French winter wheat cvs. Apache and Balance with 30 M. graminicola isolates and the identification of new Stb genes and associated molecular markers that can be readily applied in marker-assisted breeding programs.
Materials and methods
Plant materials and pathogen isolates
A double-haploid (DH) population of 91 lines derived was developed from a cross between cvs. Apache and Balance. Seedling assays were performed in a greenhouse compartment. The parental cvs. Apache and Balance were planted in VQB 7 × 7 × 8 cm TEKU® plastic pots, 10 linearly sown seeds per pot, while the DH lines were planted in 5.5 × 5 cm round Jiffy® pots, five seeds per pot using a steam-sterilized peat/sand mixture. All plants were grown in a controlled greenhouse compartment with 16-h/day light supplemented with Son-T Agro 400 W lamps (Hortilux, Boca Raton, Florida, USA). Pre-inoculation temperature and relative humidity (RH) were 18/16°C (day/night rhythm) and 70% RH, whereas post-inoculation temperature and RH were 22/21°C and ≥85% RH, respectively. Adult plant experiments were carried out in 2007 and 2008 in Cappelle-en-Pévèle and Prémesques in northern France at the breeding stations of Florimond Desprez and Serasem, respectively. Each field plot contained two 1.5-m length rows with 0.3 m spacing.
Seedling evaluations involved deep screening of the parental cultivars with 30 monopycnidial M. graminicola isolates in 2007 and 2008 followed by a progeny evaluation in three replications, in which eight isolates were tested in the first replication (pre-screening) and five in subsequent replications (Table 1). In all seedling experiments, an alpha lattice experimental design was adopted that considered each pot as an experimental unit with random arrangement for each isolate–replication combination on separate parallel tables in the above-mentioned greenhouse compartment. Field evaluations were performed with isolate IPO323 in a single replicated randomized block experiment in 2007 and a double replicated randomized block design at both locations in 2008.
Table 1.
Mycosphaerella graminicola isolate panels and their origin that were used for parental cultivars and Apache/Balance doubled-haploid progeny screening
| Isolate code | Origin | |
|---|---|---|
| Country | Location | |
| IPO94218a | Canada | Saskatoon |
| IPO00003a | USA | Colusa |
| IPO00005a | USA | Colusa |
| IPO90006a | Mexico | Toluca |
| IPO90015a | Peru | Unknown |
| IPO87016a,d | Uruguay | Dolores |
| IPO86068a | Argentina | Balcarce |
| IPO99015a | Argentina | Unknown |
| IPO89011a,d | Netherlands | Barendrecht |
| IPO92004a | Portugal | Casa Valhas |
| IPO95054a | Algeria | Berrahal |
| IPO92034a,c | Algeria | Guelma |
| IPO88018a | Ethiopia | Holetta |
| IPO88004a | Ethiopia | Kulumsa |
| IPO95036a,c | Syria | Minbeg |
| IPO86013a | Turkey | Adana |
| IPO02166a | Iran | Dezful,Safi Abad |
| IPO02159a | Iran | Gorgan, AqQaleh |
| IPO95052a,e | Algeria | Berrahal |
| IPO86022a,e | Turkey | Altinova |
| IPO323b,d | Netherlands | W.Brabant |
| IPO94269b | Netherlands | Kraggenburg |
| IPO98022b,d | France | Villaines la Gonais |
| IPO98046b,d | France | St. Pol de Leon |
| IPO98047b | France | Aire D’Havrincourt |
| IPO98094b,c | France | Aire D’Havrincourt |
| IPO052461b,f | France | Unknown (Biogemma) |
| IPO052462b,f | France | Unknown (Biogemma) |
| IPO052463b,f | France | Unknown (Biogemma) |
| IPO052464b,f | France | Unknown (Biogemma) |
aUsed for parental screen in 2008
bUsed for parental screen in 2007
cUsed in pre-screening
dTriplicated on DH lines
eDurum wheat-adapted strains
fProvided by Biogemma, Clermond-Ferrand, France
Inoculation procedures and scoring
Pre-cultures of each isolate (Table 1) were prepared in an autoclaved 100-ml Erlenmeyer flask containing 50-ml yeast–glucose (YG) liquid medium (30 g glucose, 10 g yeast per liter of dematerialized water). The flasks were inoculated using a small piece of mycelium maintained at −80°C and incubated in a shaker (Innova 4430, New Brunswick Scientific, USA) adjusted at 125 rpm and 18°C for 5–6 days. These pre-cultures were then used to inoculate three 250-ml Erlenmeyer flasks containing 100 ml of YG media per isolate that were incubated under the aforementioned conditions to provide enough inoculum for the seedling inoculation assays at growth stage (GS) 11 (Zadoks et al. 1974). The inoculum concentration was adjusted to 107 spores/ml in a total volume of 40 ml for a set of 18 plastic pots or 24 Jiffy® pots and was supplemented with two drops of Tween 20 (MERCK®, Nottingham, UK).
Field inoculations were performed with a backpack air-pumped sprayer, which was calibrated at a rate of 10 L/100 m2 at flag leaf appearance stage (GS 47–49), using a concentration of 106 spores/ml supplemented with 36 ml of four times diluted Tween 20 (MERCK®, Nottingham, UK) surfactant. Inoculations started when the foliage of the earliest DH lines developed and were subsequently repeated twice at 3- to 5-day intervals to compensate for earliness differences.
Disease severity was evaluated 21 days after inoculation in the seedling and the adult plant stage (with some variation ±2 days depending on weather conditions). In the seedling stage, the percentages of necrosis (N) and pycnidia (P; asexual fructifications) were scored separately on the first leaves, as well as NLP and PLP (days between inoculation and first N and P appearance, respectively). In the adult plant stage, the total percentage of STB symptoms on the flag leaf was recorded in 2007 and 2008, as well as earliness and tallness in 2008. Data loggers were installed at the flag leaf level to monitor the actual field conditions (RH and temperature at 10-min intervals) throughout the experiments.
FHB was established by distributing maize debris among the plants during tillering in the adult plant experiment in 2008. Disease was rated as percentage of infected spikelets per ear during STB assessments.
Mapping and QTL analysis
DNA was extracted from first leaf samples of cvs. Apache, Balance and the DH lines using the Promega Wizard® Magnetic DNA Purification System for Food (blc) according to the manufacturer’s instructions with slight modifications. Genetic polymorphism analyses were performed with Diversity Arrays Technology (DArT) version 2.3 and 3 (Triticarte Pty Ltd, Canberra, Australia) that were supplemented with additional SSR data. Mapping analyses were performed using JoinMap® 4 software with settings LOD ≥ 3 (Log of Odds) for grouping as well as the maximum likelihood mapping option for linkage group generation (Van Ooijen 2006). The DArT markers with low quality parameters (ANOVA based P value < 80) were removed form the data set (Akbari et al. 2006) and marker positions were compared and verified using the publicly available databases at INRA (Anonymous 2010b), Triticarte (Anonymous 2010c, d) and Grain Genes (Anonymous 2010e, f).
QTL analysis was performed using MapQTL® 5.0 (Van Ooijen 2004) using the interval mapping (IM) option for QTL position detection, followed by MQM (Multiple QTL Model) after cofactor selection either by automatic cofactor selection (ACS) or manual investigation of the marker alignment on the linkage groups where the peaks of IM QTLs were detected. Minimum significant LOD values were calculated by 1,000 permutation tests to determine 5% probability thresholds for seedling and adult plant stage experiments. The Excel formula option was used for Bartlett’s χ 2 tests to determine the homogeneity of replication error variances enabling QTL analyses with average or individual replicate disease scores (Chu et al. 2010; Friesen et al. 2009). The QTL profiles were drawn with MapChart 2.2 software (Voorrips 2002).
The explained variance (%) of a detected QTL strongly depends on the size of a tested population. For instance, the probability of detecting a QTL that explains 10% of the total variance in a population of 200 individuals is 0,8 (Van Ooijen 2004), but it decreases almost linearly with smaller populations (Charmet 2000; Cornforth and Long 2003; Dupuis and Siegmund 1999; Knapp et al. 1990; Van Ooijen 1992). Here, the size of the Apache/Balance population was limited (N = 91). To increase the probability of QTL detection: (a) a wide range of isolates was used to screen the parents, and a subset of eight highly distinctive isolates was selected for a pre-screening that was followed by tests with five of these isolates in subsequent replications; (b) the most recent release of DArT markers was used, (DArT marker V.3) that increased the genome coverage from 1,497 to 3431 cM, which strongly contributed to QTL detection; and (c) three replicated data sets were used for final QTL analysis that was preceded by Bartlett’s test for homogeneity of these replicates.
Results
Mapping
A total of 962 polymorphic markers between cvs. Balance and Apache, including 169 SSR and 793 DArT markers (231 and 562 DArT markers of polymorphic chip versions 2.3 and 3, respectively), were used for mapping. A genetic map with 36 linkages group was constructed, containing 786 DArT and SSR markers (428 and 205 DArT markers of V3 and V2.3, respectively; as well as 153 SSR markers) covering 3,431 cM of the total wheat genome. A total of 176 markers (134 and 26 DArT markers of V3 and V2.3, respectively; plus 16 SSR markers) were excluded from mapping due to marker similarity (109 loci), significant segregation distortions or unreliable DArT scores (67 loci).
Isolate selection and QTL analyses for seedling resistance to septoria tritici blotch
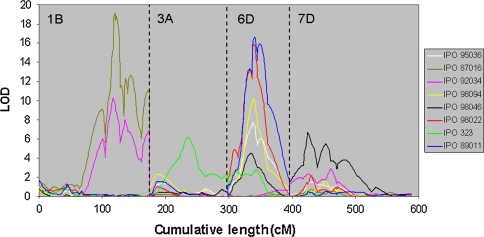
Disease development in all seedling assays was excellent with maxima of 100% N and 83% P on the susceptible checks. The field evaluations were prone to strong environmental fluctuations, but resulted in adequate STB levels in 2007 and 2008 at both locations. The initial screening of parental cvs. Apache and Balance with 30 M. graminicola isolates showed a clear contrast (P = 0.05) with 15 isolates (Tables 1, 2). Nine isolates differentiated the parents for N, and 12 showed significant differences for P. Finally, isolates, IPO87016, IPO92034, IPO323, IPO98022, IPO89011and IPO98094, as well as IPO95036 and IPO98046, were selected for a single replicated pre-screening of the DH lines. QTL analysis with P phenotypic data resulted in five significant QTLs on chromosomes 3AS, 1BS, 6DS and 7D (7DS/7DL switch) with higher LOD values than the threshold (LOD = 3.5) that was determined by permutation test at P = 0.05 (Fig. 1; Table 3). The highest LOD values per QTL were obtained with isolates IPO323, IPO98022, IPO98046 and IPO87016 (Table 4); hence, these isolates were selected, along with IPO89011 that also detected a major QTL on chromosome 6DS, to complete the data set with two additional replications.
Table 2.
Screening with 30 Mycosphaerella graminicola isolates from diverse origin resulted in significant differences (Δ) between the parental cvs. Apache and Balance (* P = 0.05, labeled)
| Isolate | Necrosis % | Pycnidia % | ||||
|---|---|---|---|---|---|---|
| Apache | Balance | Δ | Apache | Balance | Δ | |
| IPO00003 | 90 | 65 | ns | 7 | 5 | ns |
| IPO00005 | 3 | 8 | ns | 0 | 1 | ns |
| IPO02159 | 84 | 89 | ns | 0 | 20 | * |
| IPO02166 | 62 | 33 | ns | 2 | 11 | ns |
| IPO86013 | 91 | 77 | ns | 23 | 25 | ns |
| IPO86022 | 6 | 27 | ns | 0 | 0 | ns |
| IPO86068 | 4 | 57 | * | 1 | 2 | ns |
| IPO87016 | 10 | 90 | * | 0 | 51 | * |
| IPO88004 | 98 | 71 | * | 18 | 14 | ns |
| IPO88018 | 13 | 97 | * | 1 | 14 | * |
| IPO89011 | 54 | 15 | ns | 23 | 1 | * |
| IPO90006 | 16 | 15 | ns | 0 | 2 | ns |
| IPO90015 | 26 | 58 | ns | 4 | 7 | ns |
| IPO92004 | 18 | 85 | * | 0 | 16 | * |
| IPO92034 | 30 | 84 | ns | 0 | 27 | * |
| IPO94218 | 5 | 22 | ns | 0 | 4 | * |
| IPO95036 | 52 | 79 | ns | 8 | 31 | ns |
| IPO95052 | 16 | 2 | ns | 0 | 0 | ns |
| IPO95054 | 16 | 80 | * | 0 | 4 | * |
| IPO99015 | 3 | 98 | * | 0 | 23 | * |
| IPO323 | 100 | 96 | ns | 25 | 0 | * |
| IPO94269 | 100 | 100 | ns | 13 | 19 | ns |
| IPO98022 | 100 | 86 | * | 32 | 8 | * |
| IPO98046 | 100 | 100 | ns | 24 | 44 | ns |
| IPO98047 | 100 | 100 | ns | 16 | 10 | ns |
| IPO98094 | 96 | 100 | ns | 32 | 10 | * |
| IPO052461 | 100 | 98 | ns | 0 | 0 | ns |
| IPO052462 | 100 | 100 | ns | 0 | 0 | ns |
| IPO052463 | 100 | 58 | * | 0 | 0 | ns |
| IPO052464 | 100 | 96 | ns | 0 | 0 | ns |
(Bold) Resistance source cv. Apache, (Italic) Resistance source cv. Balance
Fig. 1.
Interval mapping LOD profile of the Apache/Balance DH mapping population using eight Mycosphaerella graminicola isolates in a pre-screening test (P)
Table 3.
Summary of detected quantitative trait loci for necrosis (N) and pycnidia (P) in the Apache/Balance mapping population with five Mycosphaerella graminicola isolates
| Chromosomal position | IPO 323 | IPO 98022 | IPO89011 | IPO98046 | IPO 87016 |
|---|---|---|---|---|---|
| 3AS | N P | ||||
| 6DS | P | N P | P | P | |
| 7Da | P | P | P | ||
| 1BS | N P |
aThe QTLs detected on 7D vary over isolates. Isolate IPO98022 detected a QTL on 7DS, while IPO89011 detected a QTL on 7DL and isolate IPO98046 detected QTLs on both 7DS and 7DL
Table 4.
Quantitative trait loci (QTLs) associated with necrosis (N) and pycnidia development (P) in the Apache/Balance DH population after inoculation with five Mycosphaerella graminicola isolates in the seedling stage
| Isolate | Closest marker | Chromosome position | Phenotypic data seta | Resistance source | N | P | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| PDb (cM) | LOD | Exp. (%) | PD (cM) | LOD | Exp. (%) | |||||
| IPO323 | wPt-0836 | 3AS | R1 | Balance | 0 | 12.2 | 46.1 | 1 | 7.3 | 27.7 |
| R2 | 1 | 25.5 | 73.7 | 1 | 11.1 | 39.7 | ||||
| R3 | 1 | 25.6 | 73.1 | 1 | 10.8 | 38.7 | ||||
| Xgpw5176-Xgpw3087 c | 6DS | R1 | Balance | 3.2–5 | 3.6 | 12.7 | ||||
| R2 | 0.3–8 | 3.1 | 8.9 | |||||||
| R3 | 4.3–4 | 3.5 | 11 | |||||||
| IPO98022 | Xgpw5176-Xgpw3087 c | 6DS | R1 | Balance | 6.3–2 | 6.4 | 30.4 | 5.3–3 | 16.3 | 47 |
| R2 | 8.3–0 | 5.4 | 21.6 | 5.3–3 | 13.1 | 47.4 | ||||
| R3 | 0.3–8 | 4.4 | 18.8 | 5.3–3 | 12.3 | 48 | ||||
| Xgwm111 | 7DS | R1 | Apache | 1.1 | 6.2 | 11.8 | ||||
| R2 | 0 | 5.2 | 11.2 | |||||||
| R3 | 0 | 2.2d | 5.9 | |||||||
| IPO89011 | Xgpw5176-Xgpw3087 c | 6DS | Ave. | Balance | 5.3–3 | 23.16 | 67.5 | |||
| wPt-1859 | 7DL | Ave. | Apache | 0 | 4.5 | 8 | ||||
| IPO98046 | Xgwm111 | 7DS | R1 | Apache | 0 | 9.5 | 27.5 | |||
| R2 | −e | – | – | |||||||
| R3 | 0 | 6.2 | 20.8 | |||||||
| Xgpw313 | 7DL | R1 | Apache | – | – | – | ||||
| R2 | 5 | 6.8 | 20.5 | |||||||
| R3 | – | – | – | |||||||
| Ave. | ||||||||||
| Xgpw5176-Xgpw3087 c | 6DS | R1 | Balance | 8.3–0 | 7 | 19 | ||||
| R2 | 8.3–0 | 7.9 | 24.2 | |||||||
| R3 | 6.3–2 | 7.4 | 27.1 | |||||||
| IPO87016 | wPt-2019 | 1BS | R1 | Apache | 2 | 19.1 | 63.3 | |||
| R2 | 1 | 21.1 | 68.3 | |||||||
| R3 | 0 | 17.8 | 59.3 | |||||||
| Ave. | 1 | 21.11 | 67.3 | |||||||
aR1, R2 and R3 represent first, second and third replicate data. QTL analysis was performed on averaged data (Ave) when Bartlett’s χ 2 test indicated nonsignificant phenotypic variation over replicates, otherwise replicates were processed individually
bPD, QTL peak distance in cM
cFlanking markers
dNot significant, but consistent QTL position
eNo QTL detected
Not all isolates detected all QTLs, which underscores the specificity in the M. graminicola–wheat pathosystem. The results clearly show that P is the most efficient parameter for QTL detection as nine QTLs were detected using this parameter compared to three for N (Table 3). Isolates IPO323 and IPO87016 specifically detected the 3AS and 1BS QTLs, respectively. With the exception of IPO87016, all isolates detected the 6DS QTL. The 7D QTLs were detected by isolates IPO98022, IPO89011 and IPO98046, but the genomic position of the associated marker as not consistent. Moreover, despite the fact that some isolates did not show a significant difference between both parents, DH analyses detected QTLs for N and/or P. For instance, IPO98046 induced a non-significantly different P level in both parents (Table 2), but in the DH analysis it detected the QTLs on 6DS and 7D. The 7D QTL, however, was not consistent in all replications (7DS or 7DL). Isolate IPO323 did not differentiate the parents for N, but still detected the 3AS QTL in the DH analysis. NLP data enabled the detection of more QTLs than N, but PLP reduced their number compared to P (not shown). NLP and PLP also detected two additional minor QTLs with LODs of 4.8 and 3.9 on chromosomes 5A and 2B, respectively (not shown).
The details of the mapped QTLs in seedling experiments are shown in Table 4. The LOD values and explained variances vary substantially with the applied isolates and also with the presence of additional QTLs. For instance the 6DS QTL explains only approximately 10% of the observed variation in the presence of the 3AS QTL in tests with IPO323, but close to 68% in the presence of the 7DL QTL in tests with IPO89011. In tests with isolate IPO98046, both the 6DS and 7D (7DS/7DL switch) QTLs explain about 20% of the observed variation. Remarkably, in the case of the resistance to isolate IPO98046, the 6DS QTL is expressed in all replications, whereas the QTL on 7DS is identified in two of the replications (Table 4).
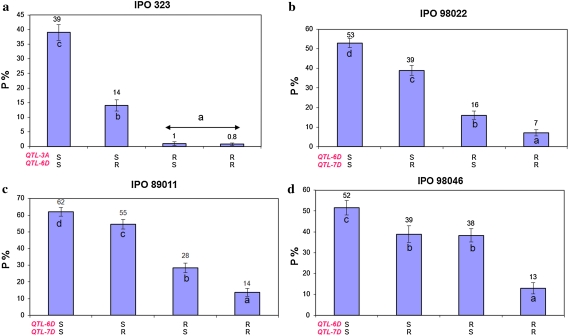
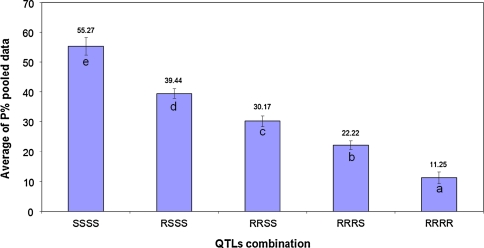
Since multiple QTLs in the Apache/Balance DH population (Tables 3, 4) were detected, additional analyses of the interaction between these QTLs were performed (Figs. 2, 3). Isolate IPO323 detected the 3AS and 6DS QTLs. Four groups that significantly differed in P were generated by averaging the P levels for all DH lines with and without the resistant and susceptible alleles of DArT marker wPt-0836 and flanking SSR markers, Xgpw5176-Xgpw3087. Marker wPt-0836 is present in cv. Apache and diagnostic for its susceptibility. The absence of the resistance alleles from both parents resulted in 39% of P. The presence of the resistance alleles of the flanking markers linked to the 6DS QTL reduced it to 14%, but without the DArT marker wPt-0836 (the QTL on 3AS), the average of P dropped to just 1%. Accumulation of both resistance alleles associated with the 6DS and 3AS QTLs did not significantly lower P. Hence, the 3AS QTL is epistatic over the 6DS QTL in the analysis with isolate IPO323. In tests with isolates IPO98022 and IPO89011, the 6DS QTL had a larger effect than the 7D QTL (7DS/7DL switch), but the presence of both QTLs lowered P to 7%. This shows that 6DS and 7D had an additive effect, but the additive effect of the former QTL is much stronger as it has a higher LOD value. This was also shown for tests with isolate IPO98046 where the individual QTLs contributed equally to disease reduction, but the combination of both QTLs minimized the disease level. Eventually, the accumulation of four QTLs in the Apache/Balance DH population for average STB levels over all used isolates was tested, which clearly demonstrated that the pyramiding of the associated markers gradually and significantly reduced disease levels (Fig. 3).
Fig. 2.
Various interactions between QTLs in the Apache/Balance DH mapping population detected by single isolates controlling P. SS, individual DH lines merely carrying susceptibility alleles of two markers associated with QTLs; RS and SR, individual DH lines carrying a resistance allele of a marker linked to one QTL and a susceptibility allele of a marker of another QTL; RR, individual DH lines with both resistance alleles. Same letters in the columns indicate not significantly different P values (P = 0.05). a Epistatic effect of the major QTL-3AS over the minor QTL-6DS detected by IPO323; b and c QTLs detected by IPO98022 and IPO89011, respectively, show a mutual additive effect; d Additive interaction between two QTLs with almost equal LOD scores
Fig. 3.
Stacking effect of individual QTLs that were detected in the Apache/Balance DH mapping population. Overall resistance to the five employed M. graminicola isolates (P) significantly (P = 0.01) increased with the number of QTLs in DH lines. SSSS and RRRR, representative of individual DH lines merely carrying markers of susceptibility or resistance alleles associated with QTLs on chromosomes 3AS, 6DS, 7DS (only the most common marker Xgwm111) and 1BS, respectively. RSSS and RRRS, individual DH lines with combinations of one resistance allele linked to a QTL and three susceptibility alleles or vice versa. RRSS, individual DH lines carrying two QTLs associated with resistance and two QTLs associated with susceptibility
Detection of QTLs associated with resistance to STB in the adult plant stage
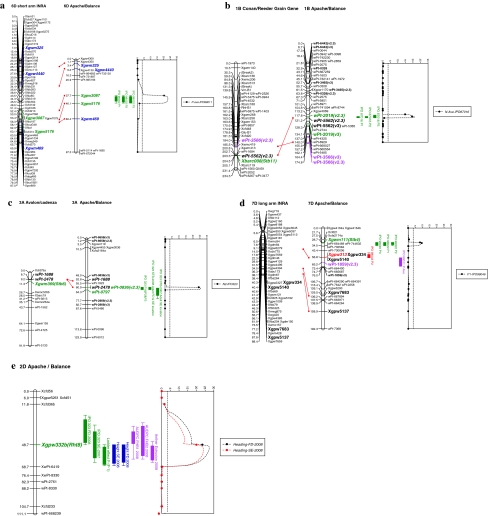
Due to field size limitations, the Apache/Balance DH population was only tested with isolate IPO323 in both years. The weather conditions for STB development were conducive in both years, resulting in differentiating STB levels (quantified by the average severity of DH lines with/without 3AS associated DArT markers and with/without 6DS flanking SSR markers that were identified in the seedling stage with isolate IPO323). QTL analyses revealed three QTLs on chromosomes 3AS, 2DS and 6DS that were associated with STB resistance (Table 5; Fig. 4). The 3AS and 6DS QTLs were also detected at the seedling stage. The former QTL was consistently expressed at both locations in both years, but the latter QTL was only detected in 2008 at the Serasem location and, similar to the seedling analyses, explained a lower percentage of the observed variation. Interestingly, the 2DS QTL was exclusively and consistently detected throughout all adult plant tests, but was also significantly correlated with earliness (−0.48 and −0.25, P = 0.05 at Florimond Desprez and Serasem, respectively), tallness (−0.36, P = 0.05 at Serasem) and resistance to FHB (Fig. 4e). Subsequent regression analyses that fitted means of logit-transformed STB values on earliness and tallness left no residual STB resistance effect for the 2D locus (P = 0.359).
Table 5.
Quantitative trait loci (QTL) associated with resistance to STB evoked by inoculations with Mycosphaerella graminicola IPO323, earliness and tallness in the adult plant stage under field conditions
| Flag leaf lesion | Earliness | Plant tallness | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Seasem 2007 |
Serasem 2008 |
Florimond Desprez 2008 |
Serasem 2008 |
Florimond Desprez 2008 |
Serasem 2008 |
Florimond Desprez 2008 |
|||||||||||||||
| Closest marker | Chromosome | PDa | LOD | Exp % | PD | LOD | Exp % | PD | LOD | Exp % | PD | LOD | Exp % | PD | LOD | Exp % | PD | LOD | Exp % | PD | LOD | Exp % |
| Xgpw332b | 2DS | 2 | 8.9 | 30.9 | 6 | 3.5 | 7.8 | 11.9 | 6.7 | 30.9 | 5 | 24 | 77.5 | 3 | 27.3 | 79.3 | 6 | 5.6 | 24.8 | 7 | 6.56 | 23.3 |
| wPt-0797 | 3AS | 0 | 6.2 | 19.1 | ||||||||||||||||||
| wPt-0836 | 3AS | 1 | 14.2 | 35.6 | 1 | 8.6 | 28.6 | |||||||||||||||
| Xgpw5176-Xgpw3087 b | 6DS | 8.3–0 | 5.9 | 12.5 | ||||||||||||||||||
aPD, QTL peak distance in cM
bFlanking markers
Fig. 4.
LOD profiles of QTL sections involved in STB resistance in the seedling (a, b, c, d) and adult plant stage (e) after individual inoculations with five Mycosphaerella graminicola isolates as well as earliness, tallness and Fusarium head blight (FHB) severity in the adult plant stage (e) in the Apache/Balance DH population. P and N are disease parameters obtained from replicates 1, 2 and 3 or from the average (Ave) based on Bartlett’s test (see “Materials and methods”)
Discussion
The present data show that both cvs. Apache and Balance contributed specific resistance to the DH population. The resistance in both parents could be easily differentiated using the 30 M. graminicola isolates panel and enabled the selection of multiple isolates with significant differences that increased the detection of QTLs and helped to understand both the specificity of and interactions between these QTLs. So far, only 17 resistance genes and QTLs have been reported to STB (Arraiano et al. 2007; Chartrain et al. 2009; Goodwin 2007; Tabib Ghaffary et al. 2011, submitted to TAG) and there is a clear need for an extended arsenal of resistance genes to support resistance breeding. The QTL on chromosome 6DS is a new resistance gene, as no other Stb gene has been mapped to this chromosome, except for the erroneous location of Stb3 (Adhikari et al. 2004) that was later correctly assigned to chromosome 7AS (Goodwin 2007). Hence, the 6DS QTL that was detected in the French winter wheat cv. Balance with four M. graminicola isolates and flanked by the SSR markers Xgpw3087 and Xgpw5176 is associated with a new resistance gene to STB that is designated as Stb18.
This is an isolate-specific resistance gene that was detected with the French M. graminicola isolates, IPO98022 and IPO98046, and with the Dutch isolates, IPO89011 and IPO323. Isolate IPO89011 detected Stb18 at the seedling stage, whereas IPO323 identified it in both the seedling and adult plant stage. IPO89011 is also avirulent on Stb9 (Chartrain et al. 2009) and Stb5 (Arraiano et al. 2001b), confirming the presence of multiple avirulence factors in M.graminicola isolates. Isolate IPO87016 from Uruguay is specifically virulent to Stb18 as no QTL other than the 1BS QTL was detected with this isolate, which was also confirmed by additional phenotyping assays. In the adult plant stage, Stb18 was detected only in 2007, but this is most likely due to the epistatic effect of the QTL on chromosome 3AS. All other QTLs also demonstrated gene-for-gene interactions that were operational in the M. graminicola–wheat pathosystem (Brading et al. 2002). Earlier findings that P rather than N is a reliable disease parameter (Kema et al. 1996a) are supported by the current data, as Stb18 was only detected once for N but multiple times for P. Previously, Kema et al. (1996a) concluded that N and P are under different genetic control, which is in accordance with the current MapQTL analyses.
The publicly available map databases show that the flanking markers of Stb18 on 6DS, Xgpw3087 and Xgpw5176, have also been mapped on chromosomes 6A and 2D, respectively. However, in the Apache/Balance population, these markers were linked with Xgpw4440, Xgwm325, Xgpw4350, Xgpw43 and Xgm469, which are positioned on chromosome 6DS in the aforementioned linkage map databases. In the mapping process, the marker alignment of chromosome 6DS was sorted by an LOD score of 4, indicating a 10,000-fold higher likelihood of linkage. It is therefore concluded that Stb18 and its closest flanking markers, Xgpw3087 and Xgpw5176, are mapped on chromosomes 6DS.
Another major QTL was detected and mapped on chromosome 1BS using isolate IPO87016. Previously, (Chartrain et al. 2005c) mapped Stb11 on chromosome 1BS in the wheat line TE9111 and determined the linked SSR marker Xbarc008 using the Mexican isolate IPO90012. In the Apache/Balance map, the identified 1BS QTL is associated with DArT marker wPt-2019 (v2.3) that is mapped next to DArT marker wPt-5562 (v2.3) (3.4 cM), which is tightly linked to Xbarc008 (1.2 cM) in the Conan/Reeder reference map at the Grain Genes database (Anonymous 2010e, f). Phenotypic interaction between IPO87016 and the Stb differential set of cultivars, also confirmed that IPO87016 is avirulent on TE9111 (Tabib Ghaffary et al. unpublished data) that is reported to carry Stb11, Stb6 and Stb7, which are mapped on chromosomes 1BS, 3AS and 4AL, respectively (Chartrain et al. 2005c). The isolate IPO87016 is virulent on Stb6, but avirulent on Stb7 (Tabib Ghaffary et al. unpublished data). Therefore, the observed resistance in cv. Apache can be due to Stb7 or Stb11. As the only detected QTL was positioned on chromosome 1BS and not on chromosome 4AL, we conclude that the QTL in cv. Apache represents Stb11, which was also confirmed by map comparison and additional phenotypic data. The QTL associated with Stb11 in the Apache/Balance population is linked to DArT marker wPt-2019 that can be used in addition to Xbarc008 as an alternative for marker-assisted selection.
The QTL on chromosome 3AS is associated with DArT marker wPt-0836 (v2.3). This marker is clustered with wPt-2478 that is also mapped in the Avalon/Cadenza reference map (Anonymous 2010d) close (2.1 cM) to marker Xgwm369 that was determined as a closely linked marker of Stb6 (Brading et al. 2002). This gene confers resistance to isolate IPO323 and is prevalent among a worldwide set of cultivars and breeding lines (Arraiano and Brown 2006; Chartrain et al. 2005b; Eriksen et al. 2003). As no other gene has been mapped on chromosome 3AS in the Apache/Balance population, the 3AS QTL must represent Stb6 in cv. Balance. Additional evidence is provided by screening with the other isolates that did not detect the 3AS QTL and are all virulent on cv. Shafir that carries Stb6 (Tabib Ghaffary et al. unpublished data). Unfortunately, the DArT marker wPt-0836 cannot be used for detecting Stb6, as it is associated with susceptibility to isolate IPO323 in cv. Apache.
Adhikari et al. (2004) and Arraiano et al. (2001b) have reported Stb4 and Stb5 on chromosome 7DS, respectively, and linkage with SSR marker Xgwm111, which is recognized as a specific marker for Stb4 (0.7 cM). This marker is also present on the Apache/Balance 7D linkage group and is associated with QTLs that were detected with isolates, IPO98046 and IPO98022. IPO98046 is avirulent on cv. Tadinia (Tabib Ghaffary et al. unpublished data) that carries Stb4 and Stb6 (Arraiano and Brown 2006; Chartrain et al. 2005b), but virulent on cv. Shafir that carries Stb6 (Brading et al. 2002, Tabib Ghaffary et al. unpublished data). The present data confirm this observation, as IPO98046 did not, but IPO323 did detect Stb6 on chromosome 3AS. Hence, the 7DS QTL that was detected with IPO98046 in cv. Apache seems identical to Stb4. Stb5, present in CS/synthetic 6x, is also reported on chromosome 7DS (Arraiano et al. 2001b). Despite that isolate IPO98022 is avirulent on CS/synthetic 6× and detected a QTL on chromosome 7DS that is linked to SSR marker Xgwm111, it is dissimilar with Stb5 as this isolate is virulent on cv. Tadinia that carries both Stb4 and Stb6 (Tabib Ghaffary et al. unpublished data). The QTL detected with IPO89011 is associated with DArT marker wPt-1859 that is positioned amidst SSR markers on chromosome 7DL (Fig. 4d). No Stb genes have been mapped to this chromosome arm and, hence, cv. Apache carries one or more unknown Stb genes on chromosome 7DL that require further characterization.
Finally, the data show that accumulation of QTL-associated markers incrementally contributes to higher and broader levels of STB resistance. Chartrain et al. (2004, 2005a, c) thoroughly analyzed STB resistance in cvs. KK4500 and TE9111. They described several Stb genes in these cultivars and suggested that gene pyramiding might be an effective method of resistance breeding, but neither interactions between these genes nor phenotype/genotype associations were addressed. Still, KK4500 and TE9111 have relatively broad efficacy (Kema et al. 1996a, b). This was in accordance with our findings that Stb resistance gene accumulation is a valid strategy to breed for wide efficacy resistance in wheat to STB, as was also shown in many other breeding programs dealing with other crops and various single or multiple biotic stresses (Barloy et al. 2007; Song et al. 2009). Therefore, a detailed characterization of known and new Stb genes is indispensable and contributes greatly to their deployment in marker-assisted stacking strategies in commercial breeding programs.
QTL analysis in adult plants
The field experiments confirmed the presence of the 3AS and 6DS QTLs that were identified as Stb6 and Stb18. The latter is inconsistently expressed in the presence of Stb6, which also provides mature plant resistance to specific M. graminicola isolates, as reported earlier (Arraiano and Brown 2006; Brading et al. 2002; Chartrain et al. 2005b). The new QTL on chromosome 2D was consistently and exclusively expressed in adult plants in both years at both locations. However, this QTL is strongly associated with earliness and tallness, and regression analyses did not show a significant residual effect on STB resistance. We are therefore reluctant to assign STB resistance to the 2D QTL and rather suggest that it indirectly influences STB resistance by regulating earliness and tallness that are known to affect STB severity (Arama et al. 1999; Arraiano et al. 2009; Simon et al. 2005). The associated SSR marker Xgpw332 is also associated with Rht8 and Pp1 that are involved in the regulation of wheat tallness and earliness (Korzun et al. 1998; Worland et al. 1988; Anounymous 2010a). These physiological parameters also influence FHB resistance (Somers et al. 2003; Steiner et al. 2004). Interestingly, a QTL for FHB resistance was mapped on the same position in the Apache/Balance population. Previously, Handa et al. (2008) identified a possible multidrug resistance associated protein (MRP) at this 2D chromosomal location that is involved in the wheat–Fusarium interaction. We tentatively conclude that the 2D QTL confers earliness/tallness in wheat and therefore indirectly contributes to multiple pathogen resistance.
This project showed that new Stb loci can still be identified in contemporary commercial wheat cultivars by using panels of carefully characterized M. graminicola isolates. Such screens also demonstrate the efficacy of Stb genes in various production environments and therefore contribute to STB resistance management.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This project is financially supported by the Fonds de Soutien à l’ObtentionVégétale (FSOV2008B), France and Bioexploit (EC Integrated Project FOOD-CT2005-513959). S.M. Tabib Ghaffary acknowledges partial financial support from the Agricultural Research and Education Organization (AREO) of Iran. We hereby acknowledge the important contribution of Drs. Maarten van Ginkel (ICARDA), Abbas Keshavarz (Former Head of AREO-SPII) and Mohammad Reza Jalal Kamali (CIMMYT-Iran) in conceiving this project. We also thank Dr. Thierry Marcel for his worthwhile analytical suggestions. We thank Els Verstappen for excellent M. graminicola maintenance and greenhouse management.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Adhikari TB, Wallwork H, Goodwin SB. Microsatellite markers linked to the Stb2 and Stb3 genes for resistance to septoria tritici blotch in wheat. Crop Sci. 2004;44:1403–1411. doi: 10.2135/cropsci2004.1403. [DOI] [Google Scholar]
- Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. TAG Theor Appl Genet. 2006;113:1409–1420. doi: 10.1007/s00122-006-0365-4. [DOI] [PubMed] [Google Scholar]
- Angus W, Fenwick P (2008) Using genetic resistance to combat pest and disease threats. HGCA conference ‘Arable cropping in a changing climate’, pp 23–24
- Angus WJ, Fenwick PM, Berry ST (2010) A breeder’s perspective on disease resistance breeding. The Dundee conference Proceedings Crop Protection in Northern Britain, pp 69–74
- Anonymous (2005) French wheat classes (http://www.muehlenchemie.de/downloads-future-of-flour/FoF_Kap_11.pdf
- Anonymous (2009) Reducing pesticides use: the Ecophyto 2018 plan. The role of usage indicators in evaluating the achievement of targets (http://agriculture.gouv.fr/IMG/pdf/Analyse_n4_Anglais.pdf)
- Anonymous (2010 g) GrainGene-2D (http://wheat.pw.usda.gov/cgi-bin/graingenes/quickquery.cgi?query=nearbyloci&arg1=Xgpw332&arg2=10
- Anonymous (2010b) Inra-genoplant (http://grain.jouy.inra.fr/grain/export/home/infoservices/htdocs/ggpages/SSRclub/GeneticPhysical/textggmsatgpw.html)
- Anonymous (2010c) Triticarte (http://www.triticarte.com.au/content/wheat_diversity_analysis.html)
- Anonymous (2010d) DArTmap (http://www.triticarte.com.au/pdf/WheatDArTmapsVersion1.2.xls)
- Anonymous (2010e) GrainGene (http://wheat.pw.usda.gov/cgi-bin/cmap/viewer?submit=Data+Source%3A&data_source=GrainGenes)
- Anonymous (2010f) GrainGene-1B (http://wheat.pw.usda.gov/cgi-bin/cmap/download_data?format=tab&map_acc=Conan%2FReeder-1B&download=1)
- Anounymous (2010a) HGCA Recommended List® winter wheat 2010/11 (http://www.hgca.com/document.aspx?fn=load&media_id=5665&publicationId=240)
- Arama PF, Parlevliet JE, van Silfhout CH. Heading date and resistance to septoria tritici blotch in wheat not genetically associated. Euphytica. 1999;106:63–68. doi: 10.1023/A:1003593218761. [DOI] [Google Scholar]
- Arraiano LS, Brown JKM. Identification of isolate-specific and partial resistance to septoria tritici blotch in 238 European wheat cultivars and breeding lines. Plant Pathol. 2006;55:726–738. doi: 10.1111/j.1365-3059.2006.01444.x. [DOI] [Google Scholar]
- Arraiano LS, Brading PA, Brown JKM. A detached seedling leaf technique to study resistance to Mycosphaerella graminicola (anamorph Septoria tritici) in wheat. Plant Pathol. 2001;50:339–346. doi: 10.1046/j.1365-3059.2001.00570.x. [DOI] [Google Scholar]
- Arraiano LS, Worland AJ, Ellerbrook C, Brown JKM. Chromosomal location of a gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in the hexaploid wheat ‘Synthetic 6x’. Theor Appl Genet. 2001;103:758–764. doi: 10.1007/s001220100668. [DOI] [Google Scholar]
- Arraiano LS, Chartrain L, Bossolini E, Slatter HN, Keller B, Brown JKM. A gene in European wheat cultivars for resistance to an African isolate of Mycosphaerella graminicola. Plant Pathol. 2007;56:73–78. doi: 10.1111/j.1365-3059.2006.01499.x. [DOI] [Google Scholar]
- Arraiano LS, Balaam N, Fenwick PM, Chapman C, Feuerhelm D, Howell P, Smith SJ, Widdowson JP, Brown JKM. Contributions of disease resistance and escape to the control of septoria tritici blotch of wheat. Plant Pathol. 2009;58:910–922. doi: 10.1111/j.1365-3059.2009.02118.x. [DOI] [Google Scholar]
- Barloy D, Lemoine J, Abelard P, Tanguy AM, Rivoal R, Jahier J. Marker-assisted pyramiding of two cereal cyst nematode resistance genes from Aegilops variabilis in wheat. Mol Breeding. 2007;20:31–40. doi: 10.1007/s11032-006-9070-x. [DOI] [Google Scholar]
- Bearchell SJ, Fraaije BA, Shaw MW, Fitt BDL. Wheat archive links long-term fungal pathogen population dynamics to air pollution. Proc Nat Acad Sci USA. 2005;102:5438–5442. doi: 10.1073/pnas.0501596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brading PA, Verstappen ECP, Kema GHJ, Brown JKM. A gene-for-gene relationship between wheat and Mycosphaerella graminicola, the Septoria tritici blotch pathogen. Phytopathology. 2002;92:439–445. doi: 10.1094/PHYTO.2002.92.4.439. [DOI] [PubMed] [Google Scholar]
- Charmet G. Power and accuracy of QTL detection: simulation studies of one-QTL models. Agronomie. 2000;20:309–323. doi: 10.1051/agro:2000129. [DOI] [Google Scholar]
- Chartrain L, Brading P, Makepeace J, Brown J. Sources of resistance to septoria tritici blotch and implications for wheat breeding. Plant Pathol. 2004;53:454–460. doi: 10.1111/j.1365-3059.2004.01052.x. [DOI] [Google Scholar]
- Chartrain L, Berry ST, Brown JKM. Resistance of wheat line Kavkaz-K4500 L.6.A.4 to Septoria tritici blotch controlled by isolate-specific resistance genes. Phytopathology. 2005;95:664–671. doi: 10.1094/PHYTO-95-0664. [DOI] [PubMed] [Google Scholar]
- Chartrain L, Brading PA, Brown JKM. Presence of the Stb6 gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in cultivars used in wheat-breeding programmes worldwide. Plant Pathol. 2005;54:134–143. doi: 10.1111/j.1365-3059.2005.01164.x. [DOI] [Google Scholar]
- Chartrain L, Joaquim P, Berry ST, Arraiano LS, Azanza F, Brown JKM. Genetics of resistance to septoria tritici blotch in the Portuguese wheat breeding line TE 9111. Theor Appl Genet. 2005;110:1138–1144. doi: 10.1007/s00122-005-1945-4. [DOI] [PubMed] [Google Scholar]
- Chartrain L, Sourdille P, Bernard M, Brown JKM. Identification and location of Stb9, a gene for resistance to septoria tritici blotch in wheat cultivars Courtot and Tonic. Plant Pathol. 2009;58:547–555. doi: 10.1111/j.1365-3059.2008.02013.x. [DOI] [Google Scholar]
- Chu CG, Faris JD, Xu SS, Friesen TL. Genetic analysis of disease susceptibility contributed by the compatible Tsn1-SnToxA and Snn1-SnTox1 interactions in the wheat–Stagonospora nodorum pathosystem. Theor Appl Genet. 2010;120:1451–1459. doi: 10.1007/s00122-010-1267-z. [DOI] [PubMed] [Google Scholar]
- Cools HJ, Fraaije BA. Are azole fungicides losing ground against Septoria wheat disease? Resistance mechanisms in Mycosphaerella graminicola. Pest Manag Sci. 2008;64:681–684. doi: 10.1002/ps.1568. [DOI] [PubMed] [Google Scholar]
- Cornforth TW, Long AD. Inferences regarding the numbers and locations of QTLs under multiple-QTL models using interval mapping and composite interval mapping. Genet Res. 2003;82:139–149. doi: 10.1017/S0016672303006396. [DOI] [PubMed] [Google Scholar]
- Desmazieres J. Neuvieme notice sur quelques plantes cryptogames. Ann des Sci Nat, Bot Ser. 1842;2:91–118. [Google Scholar]
- Dupuis J, Siegmund D. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics. 1999;151:373. doi: 10.1093/genetics/151.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksen L, Munk L. The occurrence of Mycosphaerella graminicola and its anamorph Septoria tritici in winter wheat during the growing season. Eur J Plant Pathol. 2003;109:253–259. doi: 10.1023/A:1022851502770. [DOI] [Google Scholar]
- Eriksen L, Borum F, Jahoor A. Inheritance and localisation of resistance to Mycosphaerella graminicola causing septoria tritici blotch and plant height in the wheat (Triticum aestivum L.) genome with DNA markers. Theor Appl Genet. 2003;107:515–527. doi: 10.1007/s00122-003-1276-2. [DOI] [PubMed] [Google Scholar]
- Eyal Z. The septoria tritici and stagonospora nodorum blotch diseases of wheat. Eur J Plant Pathol. 1999;105:629–641. doi: 10.1023/A:1008716812259. [DOI] [Google Scholar]
- Fraaije BA, Cools HJ, Fountaine J, Lovell DJ, Motteram J, West JS, Lucas JA. Role of ascospores in further spread of QoI-resistant cytochrome b alleles (G143A) in field populations of Mycosphaerella graminicola. Phytopathology. 2005;95:933–941. doi: 10.1094/PHYTO-95-0933. [DOI] [PubMed] [Google Scholar]
- Fraaije BA, Cools HJ, Kim SH, Motteram J, Clark WS, Lucas JA. A novel substitution I381V in the sterol 14 alpha-demethylase (CYP51) of Mycosphaerella graminicola is differentially selected by azole fungicides. Mol Plant Pathol. 2007;8:245–254. doi: 10.1111/j.1364-3703.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- Friesen TL, Chu CG, Liu ZH, Xu SS, Halley S, Faris JD. Host-selective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theor Appl Genet. 2009;118:1489–1497. doi: 10.1007/s00122-009-0997-2. [DOI] [PubMed] [Google Scholar]
- Goodwin SB. Back to basics and beyond: increasing the level of resistance to Septoria tritici blotch in wheat. Australas Plant Pathol. 2007;36:532–538. doi: 10.1071/AP07068. [DOI] [Google Scholar]
- Halama P. The occurrence of Mycosphaerella graminicola, teleomorph of Septoria tritici in France. Plant Pathol. 1996;45:135–138. doi: 10.1046/j.1365-3059.1996.d01-103.x. [DOI] [Google Scholar]
- Handa H, Namiki N, Xu D, Ban T. Dissecting of the FHB resistance QTL on the short arm of wheat chromosome 2D using a comparative genomic approach: from QTL to candidate gene. Mol Breeding. 2008;22:71–84. doi: 10.1007/s11032-008-9157-7. [DOI] [Google Scholar]
- Hunter T, Coker RR, Royle DJ. The teleomorph stage, Mycosphaerella graminicola, in epidemics of septoria tritici blotch on winter wheat in the UK. Plant Pathol. 1999;48:51–57. doi: 10.1046/j.1365-3059.1999.00310.x. [DOI] [Google Scholar]
- Kema GHJ, van Silfhout CH. Genetic variation for virulence and resistance in the wheat Mycosphaerella graminicola pathosystem. 3. Comparative seedling and adult plant experiments. Phytopathology. 1997;87:266–272. doi: 10.1094/PHYTO.1997.87.3.266. [DOI] [PubMed] [Google Scholar]
- Kema GHJ, Annone JG, Sayoud R, VanSilfhout CH, VanGinkel M, deBree J. Genetic variation for virulence and resistance in the wheat–Mycosphaerella graminicola pathosystem. 1. Interactions between pathogen isolates and host cultivars. Phytopathology. 1996;86:200–212. doi: 10.1094/Phyto-86-200. [DOI] [Google Scholar]
- Kema GHJ, Sayoud R, Annone JG, VanSilfhout CH. Genetic variation for virulence and resistance in the wheat–Mycosphaerella graminicola pathosystem. 2. Analysis of interactions between pathogen isolates and host cultivars. Phytopathology. 1996;86:213–220. doi: 10.1094/Phyto-86-213. [DOI] [Google Scholar]
- Kema GHJ, Verstappen ECP, Todorova M, Waalwijk C. Successful crosses and molecular tetrad and progeny analyses demonstrate heterothallism in Mycosphaerella graminicola. Curr Genet. 1996;30:251–258. doi: 10.1007/s002940050129. [DOI] [PubMed] [Google Scholar]
- Kema GHJ, Yu DZ, Rijkenberg FHJ, Shaw MW, Baayen RP. Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology. 1996;86:777–786. doi: 10.1094/Phyto-86-777. [DOI] [Google Scholar]
- Knapp S, Bridges W, Birkes D. Mapping quantitative trait loci using molecular marker linkage maps. TAG Theor Appl Genet. 1990;79:583–592. doi: 10.1007/BF00226869. [DOI] [PubMed] [Google Scholar]
- Korzun V, Röder M, Ganal M, Worland A, Law C. Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.) TAG Theor Appl Genet. 1998;96:1104–1109. doi: 10.1007/s001220050845. [DOI] [Google Scholar]
- Mavroeidi VI, Shaw MW. Sensitivity distributions and cross-resistance patterns of Mycosphaerella graminicola to fluquinconazole, prochloraz and azoxystrobin over a period of 9 years. Crop Protect. 2005;24:259–266. doi: 10.1016/j.cropro.2004.07.014. [DOI] [Google Scholar]
- McCartney C, Mercer PC, Cooke LR, Fraaije BA. Effects of a strobilurin-based spray programme on disease control, green leaf area, yield and development of fungicide-resistance in Mycosphaerella graminicola in Northern Ireland. Crop Protect. 2007;26:1272–1280. doi: 10.1016/j.cropro.2006.10.027. [DOI] [Google Scholar]
- McDonald BA, Mundt CC, Chen RS. The role of selection on the genetic structure of pathogen populations: Evidence from field experiments with Mycosphaerella graminicola on wheat. Euphytica. 1996;92:73–80. doi: 10.1007/BF00022831. [DOI] [Google Scholar]
- Pastircak M. Occurrence of Mycosphaerella graminicola, teleomorph of Septoria tritici, in Slovakia. Phytoparasitica. 2005;33:377–379. doi: 10.1007/BF02981305. [DOI] [Google Scholar]
- Paveley N (2006) Is disease management sustainable? HGCA conference Arable crop protection in the balance: profit and the environment. pp 5.1–5.12
- Scott PR, Sanderson FR, Benedikz PW. Occurrence of Mycosphaerella-graminicola, teleomorph of Septoria-tritici, on wheat debris in the UK. Plant Pathol. 1988;37:285–290. doi: 10.1111/j.1365-3059.1988.tb02076.x. [DOI] [Google Scholar]
- Shaw MW, Royle DJ. Airborne inoculum as a major source of septoria-tritici (Mycosphaerella-graminicola) infections in winter-wheat crops in the UK. Plant Pathol. 1989;38:35–43. doi: 10.1111/j.1365-3059.1989.tb01425.x. [DOI] [Google Scholar]
- Shaw MW, Royle DJ. Factors determining the severity of epidemics of Mycosphaerella-graminicola (septoria-tritici) on winter-wheat in the UK. Plant Pathol. 1993;42:882–899. doi: 10.1111/j.1365-3059.1993.tb02674.x. [DOI] [Google Scholar]
- Shetty NP, Kristensen BK, Newman MA, Moller K, Gregersen PL, Jorgensen HJL. Association of hydrogen peroxide with restriction of Septoria tritici in resistant wheat. Physiol Mol Plant Pathol. 2003;62:333–346. doi: 10.1016/S0885-5765(03)00079-1. [DOI] [Google Scholar]
- Shetty NP, Mehrabi R, Lutken H, Haldrup A, Kema GHJ, Collinge DB, Jorgensen HJL. Role of hydrogen peroxide during the interaction between the hemibiotrophic fungal pathogen Septoria tritici and wheat. New Phytol. 2007;174:637–647. doi: 10.1111/j.1469-8137.2007.02026.x. [DOI] [PubMed] [Google Scholar]
- Shetty NP, Jensen JD, Knudsen A, Finnie C, Geshi N, Blennow A, Collinge DB, Jorgensen HJL. Effects of beta-1, 3-glucan from Septoria tritici on structural defence responses in wheat. J Exp Bot. 2009;60:4287–4300. doi: 10.1093/jxb/erp269. [DOI] [PubMed] [Google Scholar]
- Simon MR, Perello AE, Cordo CA, Larran S, van der Putten PEL, Struik PC. Association between Septoria tritici blotch, plant height, and heading date in wheat. Agron J. 2005;97:1072–1081. doi: 10.2134/agronj2004.0126. [DOI] [Google Scholar]
- Somers D, Fedak G, Savard M. Molecular mapping of novel genes controlling Fusarium head blight resistance and deoxynivalenol accumulation in spring wheat. Genome. 2003;46:555–564. doi: 10.1139/g03-033. [DOI] [PubMed] [Google Scholar]
- Song W, Xie CJ, Du JK, Xie H, Liu Q, Ni ZF, Yang TM, Sun QX, Liu ZY. A “one-marker-for-two-genes” approach for efficient molecular discrimination of Pm12 and Pm21 conferring resistance to powdery mildew in wheat. Mol Breeding. 2009;23:357–363. doi: 10.1007/s11032-008-9235-x. [DOI] [Google Scholar]
- Sprague R. The status of Septoria graminum. Mycologia. 1938;30:672–678. doi: 10.2307/3754365. [DOI] [Google Scholar]
- Stammler G, Carstensen M, Koch A, Semar M, Strobel D, Schlehuber S. Frequency of different CYP51-haplotypes of Mycosphaerella graminicola and their impact on epoxiconazole-sensitivity and -field efficacy. Crop Protect. 2008;27:1448–1456. doi: 10.1016/j.cropro.2008.07.007. [DOI] [Google Scholar]
- Steiner B, Lemmens M, Griesser M, Scholz U, Schondelmaier J, Buerstmayr H. Molecular mapping of resistance to Fusarium head blight in the spring wheat cultivar Frontana. Theor Appl Genet. 2004;109:215–224. doi: 10.1007/s00122-004-1620-1. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I, van Nistelrooy JGM, Kema GHJ, De Waard MA. Multiple mechanisms account for variation in base-line sensitivity to azole fungicides in field isolates of Mycosphaerella graminicola. Pest Manag Sci. 2003;59:1333–1343. doi: 10.1002/ps.766. [DOI] [PubMed] [Google Scholar]
- Suffert F, Sache I, Lannou C (2010) Early stages of septoria tritici blotch epidemics of winter wheat: build-up, overseasoning, and release of primary inoculum. Plant Pathol 60:166–177. doi:10.1111/j.1365-3059.2010.02369.x
- Tabib Ghaffary SM, Faris JD, Friesen TL, Visser RGF, Van der Lee TAJ, Robert O, Kema GHJ (2011) New broad-spectrum resistance to septoria tritici blotch derived from synthetic hexaploid wheat. Submitted to TAG [DOI] [PMC free article] [PubMed]
- Torriani SFF, Brunner PC, McDonald BA, Sierotzki H. QoI resistance emerged independently at least 4 times in European populations of Mycosphaerella graminicola. Pest Manag Sci. 2009;65:155–162. doi: 10.1002/ps.1662. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW. Accuracy of mapping quantitative trait loci in autogamous species. TAG Theor Appl Genet. 1992;84:803–811. doi: 10.1007/BF00227388. [DOI] [PubMed] [Google Scholar]
- Van Ooijen J. MapQTL®5, Software for the mapping of quantitative trait loci in experimental populations. Wageningen: Kyazma BV; 2004. [Google Scholar]
- Van Ooijen J. JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma BV; 2006. [Google Scholar]
- Vanloqueren G, Baret P. Why are ecological, low-input, multi-resistant wheat cultivars slow to develop commercially? A Belgian agricultural lock-in case study. Ecol Econ. 2008;66:436–446. doi: 10.1016/j.ecolecon.2007.10.007. [DOI] [Google Scholar]
- Verweij P, Snelders E, Kema G, Mellado E, Melchers W. Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infect Dis. 2009;9:789–795. doi: 10.1016/S1473-3099(09)70265-8. [DOI] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Worland A, Petrovic S, Law C. Genetic analysis of chromosome 2D of wheat. Plant Breeding. 1988;100:247–259. doi: 10.1111/j.1439-0523.1988.tb00250.x. [DOI] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF. Decimal code for growth stages of cereals. Weed Res. 1974;14:415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x. [DOI] [Google Scholar]
- Zhan J, Mundt CC, McDonald BA. Sexual reproduction facilitates the adaptation of parasites to antagonistic host environments: Evidence from empirical study in the wheat–Mycosphaerella graminicola system. Int J Parasitol. 2007;37:861–870. doi: 10.1016/j.ijpara.2007.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.