Abstract
Two common variants (rs1387153, rs10830963) in MTNR1B have been reported to have independent effects on fasting blood glucose (FBG) levels with increased risk to type 2 diabetes (T2D) in recent genome-wide association studies (GWAS). In this investigation, we report the association of these two variants, and an additional variant (rs1374645) within the GWAS locus of MTNR1B with FBG, 2h glucose, insulin resistance (HOMA IR), β-cell function (HOMA B), and T2D in our sample of Asian Sikhs from India. Our cohort comprised 2,222 subjects [1,201 T2D, 1,021 controls]. None of these SNPs was associated with T2D in this cohort. Our data also could not confirm association of rs1387153 and rs10830963 with FBG phenotype. However, upon stratifying data according to body mass index (BMI) (low ≤ 25 kg/m2 and high > 25 kg/m2) in normo-glycemic subjects (n= 1,021), the rs1374645 revealed a strong association with low FBG levels in low BMI group (β= −0.073, p=0.002, Bonferoni p= 0.01) compared to the high BMI group (β= 0.015, p=0.50). We also detected a strong evidence of interaction between rs1374645 and BMI with respect to FBG levels (p= 0.002). Our data provide new information about the significant impact of another MTNR1B variant on FBG levels that appears to be modulated by BMI. Future confirmation on independent datasets and functional studies will be required to define the role of this variant in fasting glucose variation.
Keywords: MTNR1B, Fasting blood glucose, SNP-obesity interaction, Asian Indians
Introduction
Elevated levels of fasting blood glucose (FBG) are hallmarks of type 2 diabetes (T2D). Individual variability in FBG is heritable and around 50% of the variance in FBG levels could be attributed to genetic factors. 1, 2 Recent genome-wide association studies (GWAS) and meta-analysis have identified genes contributing to the variation of FBG levels in populations of European origin. 3–6 Two common variants in the melatonin receptor1-B (MTNR1B) were shown to have independent effects on FBG levels in normoglycemic (NG) individuals with an increased risk for T2D. 3, 7 These results were also later replicated in other independent studies from same or different ethnicities. 8–10 However, these variants explain only a small portion of the heritability of T2D, implying more variants in or around these GWAS signals need to be identified. 11 In this investigation, we report the association of these two previously known variants rs10830963, rs1387153, and another less common variant rs1374645 in MTNR1B with FBG, 2h glucose, homeostasis model assessment (HOMA) for insulin resistance (HOMA IR) and β-cell function (HOMA B), and T2D in this unique sample of Asian Sikhs from India.
Methods
Study Subjects
The study participants are part of our ongoing Sikh diabetes Study. 12 The DNA and serum samples of a total of 2,222 subjects including 1,201 T2D cases [628 male, 573 female, mean age (mean ± SD) 53.9 ± 10.7 yrs.], 1,021 NG controls [535 male, 486 female, mean age 51.9 ± 13.9 yrs.] were used in this investigation. The diagnostic criteria used for recruiting T2D patient and NG control have been described in detail elsewhere. 13 Briefly, the diagnosis of T2D was confirmed by medical records, use of medication, and measuring fasting glucose levels following the guidelines of American Diabetes Association. 14 Impaired glucose tolerance (IGT) was defined as a FBG level > 100.8 mg/dL but < 126.1 mg/dL or a 2 hour oral glucose tolerance test (2h OGTT) > 141.0 mg/dL but ≤ 200 mg/dL. The 2h OGTTs were performed following the criteria of the World Health Organization (WHO) (75 g oral load of glucose). BMI was calculated as weight (kg)/height (meter2). The NG subjects were from the same Punjabi community and from the same geographic location where the T2D patients were recruited. 15 Majority of the subjects were recruited from the state of Punjab from Northern India. Individuals of South, East, and Central Indian origins, or with type 1 diabetes (T1D), or a family member with T1D, rare forms of T2D called maturity-onset diabetes of young (MODYs), and secondary diabetes (e.g., hemochromatosis, pancreatitis) were excluded from the study. The selection of controls was based on a fasting glycemia <100.8 mg/dL or a 2h glucose <141.0 mg/dL. In general, Punjabi Sikhs do not smoke for religious and cultural reasons and about 50% of participants were life-long vegetarians. Subjects with IGT were excluded from this study. All blood samples were obtained at the baseline visit. Insulin was measured by radio-immuno assay (Diagnostic Products, Cypress, USA). HOMA IR was calculated as (fasting glucose mg/dL × fasting insulin μIU/mL)/405 and HOMA B (fasting insulin μIU/mL × 360/fasting glucose mg/dL - 63), as described. 16 All participants provided a written informed consent for investigations. The study was reviewed and approved by the University of Oklahoma Health Sciences Center’s Institutional Review Board, as well as the Human Subject Protection Committees at the participating hospitals and institutes in India.
DNA Sequencing
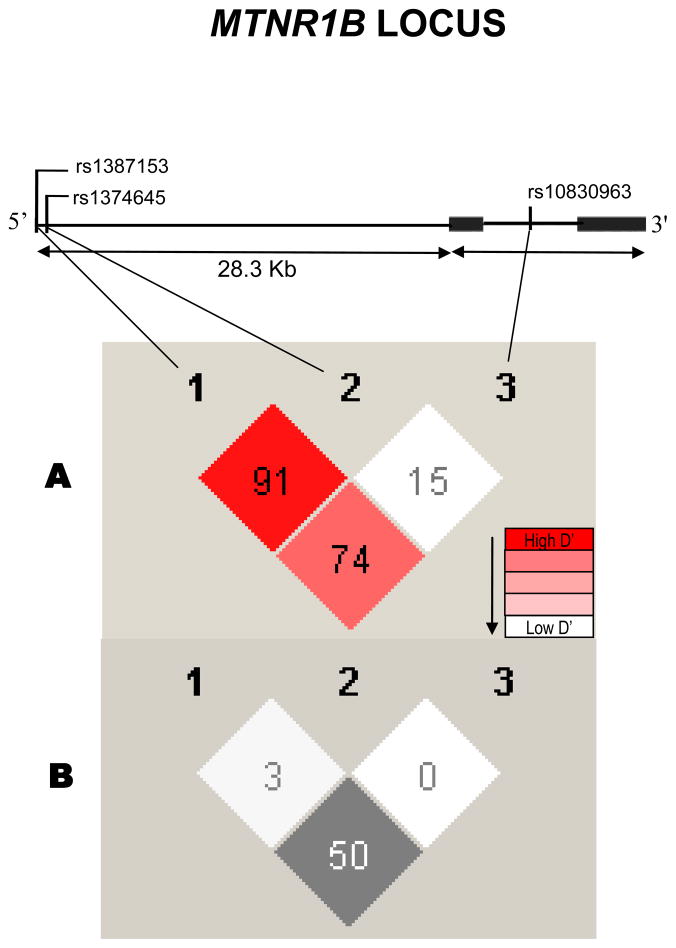
DNA was extracted from buffy coats using QiaAmp blood kits (Qiagen, Chatworth, USA) or by the salting out procedure. 17 In search of putative functional variants in the MTNR1B gene, we explored 5′ UTR region around rs1387153 using PCR and DNA sequencing techniques. A variant detected through sequencing using Applied Biosystems’ automatic sequencer 3730 (ABI, Foster City, USA) was a common point mutation (C→ T) detected at position 92313529, was only 53 bases downstream from rs1387153 and was later found to have a dbSNP number rs1374645 by1000 genomes with a rare minor allele frequency (MAF) in Caucasians as 0.028. Figure 1 shows the location of three investigated SNPs in the MTNR1B gene. We further explored 5′ UTR region to discover more variants using DNA samples from 30 T2D and 30 NG controls from our SDS cohort. However, our initial sequencing results did not reveal the presence of any further common variant (allele frequency >0.05) in this region. Another mutation (G→A) discovered in exon 2 was also not polymorphic and was not further pursued.
Figure 1.
Figure 1 describes human MTNR1B locus showing genomic structure of MTNR1B and 28.3 kb 5′ un-translated region harboring rs1387153, rs1374645, and rs10830963 variants. These variants reside within 62.1 kb linkage disequilibrium (LD) block (CEU, HapMap Phase II) on chromosome 11 near 5′UTR of MTNR1B. Top portion of the Figure 1 shows the position of three MTNR1B SNPs genotyped in this study. Figure 1(A) shows pair-wise LD between SNPs and Figure 1(B) shows pair-wise correlation (r2) between SNPs.
SNP Genotyping
Genotyping for all three SNPs was performed using TaqMan pre-designed or TaqMan made-to order SNP Genotyping Assays from Applied Biosystems Inc. (ABI, Foster City, USA). Genotyping reactions were performed on an ABI 7900 genetic analyzer using 2 ul of genomic DNA (10 ng/ul), following manufacturers’ instructions. For quality control, 8–10% replicative controls and 4–8 negative controls were included in each 384 well plate to match the concordance, and the discrepancy rate of duplicate genotyping was <0.2%. Genotyping call rate was 96% or more in all the SNPs genotyped.
Statistical Analysis
Data quality for SNP genotyping was checked by establishing reproducibility of control DNA samples. Departure from Hardy-Weinberg equilibrium (HWE) in controls was tested using the Pearson chi-square test. The genotype and allele frequencies in T2D cases were compared to those in control subjects using the chi-square test. Statistical evaluation of genetic effects on T2D risk used multivariate logistic regression analysis with adjustments for age, gender, and other covariates. Continuous traits with skewed sampling distributions (e.g. glucose, insulin, HOMA IR, and HOMA B) were log-transformed before statistical analysis. However, for illustrative purposes, values were re-transformed into the original measurement scale. General linear models (GLM) were used to test the impact of genetic variants on transformed continuous traits (FBG, 2h glucose, HOMA IR, and HOMA B) in only NG subjects after adjusting for the effects of covariates. Significant covariates for each dependent trait were identified by using Spearman’s correlation and step-wise multiple linear regression with an overall 5% level of significance using SPSS for Windows statistical package (version 18.0) (SPSS Inc., Chicago, USA). Age, gender, and BMI were used as significant covariates in un-stratified analysis while age and gender were significant covariates in data stratified by BMI. Homozygous minor allele carriers of rs1374645 were analyzed combined with heterozygotes because of low frequency of minor alleles (0.06). Mean values between cases and controls were compared by using an unpaired t-test. The portion of variance on each adjusted quantitative trait (e.g. FBG) attributable to each MTNR1B polymorphism was estimated as R2 for the SNP by comparing the complete model to a model in which that specific SNP was removed. SNP-BMI interaction effects on FBG levels were tested using all three SNPs by applying GLM procedures and using FBG as a dependent variable. Age and gender were included as covariates in the model. Interaction analysis was only performed in un-stratified sample of NG controls.
Haplotype analysis of three MTNR1B SNPs was performed using HAPLOVIEW (version 4.0) (http://www.broadinstitute.org/haploview/haploview) that uses an accelerated expectation maximization algorithm to calculate haplotype frequencies. Effect of three-site haplotypes on quantitative variables were determined using PLINK (version 1.0.6) (http://pngu.mgh.harvard.edu/~purcell/plink/). To adjust for multiple testing, we used Bonferroni’s correction (0.05/ number of observations). Statistical power was assessed using the Genetic Power Calculator. 18 Assuming an additive genetic model, 1,021 unrelated control subjects, and α = 0.05, our study has 80% power to detect a variant (with allele frequency as small as 0.02) that is associated with a difference in FBG of as little as 1 mg/dL. This accounts for an effect size of 0.1, a small effect size for a quantitative trait like FBG corresponding to detecting as significant βs outside of the range of ± 0.05. These effect sizes are determined by differences between the group means relative to within group standard deviation.
Results
Demographics and clinical characteristics of study subjects are summarized in Table 1. The genotype distribution for all three investigated SNPs was in HWE in controls with respective p values as, rs1387153 (p= 0.170), rs1374645 (p= 0.332), and rs10830963 (p= 0.053). The variant rs1374645, despite in tight linkage disequilibrium (LD) with rs1387153 (D′=0.91), was poorly correlated (r2=0.03) to rs1387153 and it had no correlation with rs10830963 (D′= 0.15; r2= 0.00) in this sample (Figure 1). The other two variants (rs1387153 and rs10830963) exhibited relatively stronger correlation with each other (D′= 0.74, r2= 0.50).
Table 1.
Clinical characteristics of study population stratified by gender and disease (Mean ± SD)
| Gender* | NG† Controls (n=1,021) (535 M/486 F) | T2D¥ Cases (n=1,201) (628 M/573 F) | |
|---|---|---|---|
| Age (yr) | M | 51.3 ± 15.2 | 54.1 ± 10.2 |
| F | 50.1 ± 13.3 | 53.7 ± 9.8 | |
| Age at Diagnosis (yr) | M | - | 45.9 ± 10.2 |
| F | - | 47.4 ± 9.4 | |
| Duration of Diabetes (yr) | M | - | 8.0 ± 6.9 |
| F | - | 6.5 ± 5.7 | |
| BMI (kg/m2) | M | 25.7 ± 4.8 | 26.6 ± 4.4 |
| F | 27.1 ± 6.7 | 28.4 ± 5.4 | |
| FBG (mg/dL)€ | M | 95.4 ± 11.2 | 163.8 ± 60.7 |
| F | 93.7 ± 10.9 | 163.3 ± 64.9 | |
| 2h Glucose (mg/dL) | M | 104.0 ± 22.0 | 206.8 ± 67.3 |
| F | 106.6 ± 17.9 | 193.9 ± 67.2 | |
| Fasting Insulin(μIU/mL) | M | 7.5 ± 7.1 | 5.9 ± 5.8 |
| F | 7.7 ± 7.4 | 6.3 ± 5.6 | |
| HOMA IR* | M | 1.7 ± 1.6 | 2.3 ± 2.2 |
| F | 1.7 ± 1.6 | 2.5 ± 2.3 |
M- Males, F- Females;
Normoglycemic;
type 2 diabetes;
fasting blood glucose;
homeostasis model assessment for insulin resistance
None of these SNPs revealed any association with T2D in this Asian Indian cohort (Table 2). In multiple linear regression analysis, a marginal increase in HOMA IR was observed among ‘G’ allele carriers in rs10830963. As shown in Table 3, mean levels of HOMA IR were increased from 1.6 (1.4–1.8) in CC to 1.7 (1.5–1.9) in CG to 2.0 (1.6–2.5) in GG carriers, showing effect size of β= 0.13, p= 0.026, Bonferroni p= 0.005. However, our data failed to confirm significant association of rs10830963 and rs1387153 with FBG concentrations; [β= 0.00 95%CI (−0.01 – 0.02), p= 0.586] and [β= 0.01 95%CI (−0.01 – 0.02), p= 0.338], respectively, unlike seen in Caucasian studies. On the other hand, in rs1374645, mean levels of 2h glucose were significantly reduced [β= −0.06 95%CI (−0.12 – −0.01), p=0.026] in minor ‘T’ allele carriers (CT+TT) 99.7 (94.9–104.8) compared to non-‘T’ allele carriers (CC) 106.0 (103.9 –108.1) in combined (un-stratified) NG cohort after adjusting for age, BMI, and gender (Table 3). However, this association did not remain significant after applying Bonferroni correction.
Table 2.
Genotype distribution and association of candidate SNPs with type 2 diabetes
| SNP | Position | Genotype** | NG (%) | T2D (%) | Odds Ratios (OR)* (95%CI) | P value |
|---|---|---|---|---|---|---|
| rs1387153 | 92313476 | CC | 363 (37) | 459 (40) | 1.0 (0.9–1.1) | 0.668 |
| CT | 479 (50) | 558 (48) | ||||
| TT | 131 (13) | 147 (12) | ||||
| C/T | 0.62/0.38 | 0.63/0.37 | ||||
| rs1374645 | 92313529 | CC | 869 (87.9) | 1054 (88.2) | 1.0 (0.8–1.2) | 0.801 |
| CT | 118 (11.9) | 137 (11.5) | ||||
| TT | 2 (0.2) | 4 (0.3) | ||||
| C/T | 0.94/0.06 | 0.94/0.06 | ||||
| rs10830963 | 92348358 | CC | 393 (40) | 435 (37) | ||
| CG | 445 (44) | 560 (48) | 1.0 (0.9–1.2) | 0.458 | ||
| GG | 163 (16) | 174 (15) | ||||
| C/G | 0.61/0.39 | 0.61/0.39 |
ORs were adjusted for age, sex, and BMI; Position of SNPs on chromosome has been taken from NCBI (Genome Build 36.3);
risk allele is bold faced
Table 3.
Association of SNP genotypes with quantitative traits affecting glucose homeostasis in NG Controls (un-stratified for obesity)‡
| rs1387153 | CC | CT | TT | Effect β (95% CI) | P value |
|---|---|---|---|---|---|
| Total (N) | 363 | 479 | 131 | -- | -- |
| FBG (mg/dL)€ | 95.0 (93.5 – 96.5) | 94.9 (93.7 – 96.2) | 96.2 (93.4 – 99.2) | 0.01 (−0.01 – 0.02) | 0.338 |
| 2h Glucose (mg/dL) | 105.8 (102.8 – 108.8) | 104.6 (101.5 – 107.7) | 105.8 (101.5 – 110.3) | 0.00 (−0.03 – 0.02) | 0.841 |
| Fasting Insulin(μIU/mL) | 7.5 (6.6 – 8.5) | 7.6 (6.8 – 8.4) | 7.2 (5.7 – 9.2) | −0.01 (−0.13 – 0.10) | 0.844 |
| HOMA IR* | 1.6 (1.4 – 1.8) | 1.7 (1.5 – 1.9) | 1.8 (1.3 – 2.3) | 0.04 (−0.08 – 0.16) | 0.538 |
| HOMA B£ | 84.5 (72.5 – 98.5) | 86.6 (76.5 – 98.2) | 82.2 (60.8 – 111.2) | −0.01 (−0.15 – 0.12) | 0.813 |
| rs1374645† | CC | CT + TT | Effect β (95% CI) | P value | |
| Total (N) | 869 | 118 + 2 | -- | -- | -- |
| FBG (mg/dL) | 95.4 (94.5 – 96.4) | 93.4 (90.9 – 96.1) | -- | −0.02 (−0.05 – 0.01) | 0.169 |
| 2h Glucose (mg/dL) | 106.0 (103.9 – 108.1) | 99.7 (94.9 – 104.8) | -- | −0.06 (−0.12 – −0.01) | 0.026 |
| Fasting Insulin(μIU/mL) | 7.5 (6.9 – 8.2) | 7.5 (6.1 – 9.2) | -- | −0.03 (−0.26 – 0.21) | 0.826 |
| Homa-IR | 1.7 (1.5 – 1.8) | 1.6 (1.3 – 2.0) | -- | −0.01 (−0.25 – 0.23) | 0.921 |
| Homa B | 84.5 (76.7 – 93.2) | 88.6 (68.4 – 114.8) | -- | 0.00 (−0.25 – 0.27) | 0.935 |
| rs10830963 | CC | CG | GG | Effect β (95% CI) | P value |
| Total (N) | 393 | 445 | 163 | -- | -- |
| FBG (mg/dL) | 95.2 (93.8 – 96.7) | 94.6 (93.3 – 96.0) | 95.8 (93.7 – 98.0) | 0.00 (−0.01 – 0.02) | 0.586 |
| 2h Glucose (mg/dL) | 106.4 (103.6 – 109.3) | 105.3 (102.2 – 108.5) | 102.3 (97.9 – 107.0) | −0.01 (−0.04 – 0.01) | 0.367 |
| Fasting Insulin(μIU/mL) | 7.3 (6.4 – 8.2) | 7.6 (6.8 – 8.5) | 8.4 (6.9 – 10.1) | 0.08 (−0.03 – 0.19) | 0.151 |
| Homa-IR | 1.6 (1.4 – 1.8) | 1.7 (1.5 – 1.9) | 2.0 (1.6 – 2.5) | 0.13 (0.02 – 0.24) | 0.026 |
| Homa B | 80.3 (69.3 – 92.9) | 88.2 (77.1 – 101.0) | 95.0 (75.4 – 119.5) | 0.05 (−0.05 – 0.20) | 0.232 |
fasting blood glucose;
homeostasis model assessment for insulin resistance;
homeostasis model assessment for β cell function;
homozygous minor allele carriers were analyzed combined with heterozygotes because of small number;
Analysis was performed using age, BMI, and gender as covariates in multiple linear regression.
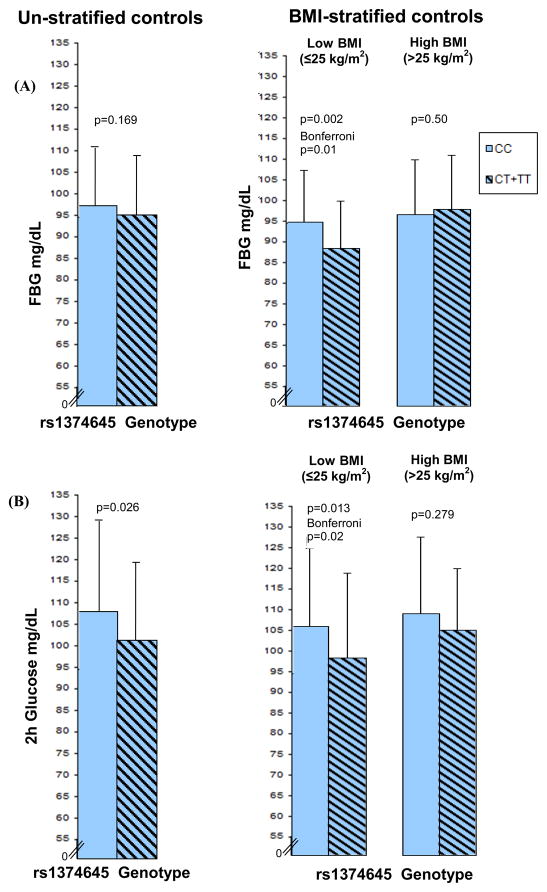
On the other hand, upon stratifying data by low (BMI≤25 kg/m2) and high (BMI>25 kg/m2), the minor ‘T’ allele carriers of rs1374645 revealed a strong association with lower FBG levels only in low BMI group [β= −0.073 95%CI (−0.119 – −0.028), p=0.002, Bonferroni p=0.01], as the mean levels of FBG were significantly higher among CC genotypes (non-‘T’ allele carriers) (94.5 ± 12.5 mg/dL) compared to CT+TT genotypes (‘T’ allele carriers) (88.0 ± 11.4 mg/dL) (Table 4, Figure 2A). However, this association was not observed among high BMI individuals [β= 0.015 95%CI (−0.028 – 0.057), p=0.50], as the mean FBG did not differ among CC (96.2 ± 13.3 mg/dL) versus CT+TT individuals (97.5 ± 13.1 mg/dL). As shown in the Table 4, the observed association did not disappear even after including covariates (age and gender) and other two SNPs in the model. Similarly, upon analyzing the effect of this SNP rs1374645 on 2h glucose in BMI-stratified sample, the significance was again confined to the low BMI group. The mean levels of 2h glucose were also moderately higher among CC (105.3 ± 18.8 mg/dL) compared to CT+TT (97.7 ± 20.4 mg/dL) individuals in non-obese group (≤25 kg/m2, p=0.013, Bonferroni p=0.02), while there was no significant difference in 2h glucose among CC (108.3 ± 18.5 mg/dL) versus CT+TT (104.4 ± 14.9 mg/dL) individuals in high BMI group (>25 kg/m2, p=0.279) (Table 4, Figure 2B).
Table 4.
Multiple linear regression analysis showing the impact of MTNR1B genotypes on FBG and 2h glucose levels in NG cohort in data stratified by obesity
| Non-Obese (BMI ≤ 25) | Obese (BMI >25) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| FBG in NG Controls | |||||||||
| β | 95% CI | P value | β | 95% CI | P value | ||||
| Lower | Upper | Lower | Upper | ||||||
| Age | 0.001 | 0.001 | 0.002 | 0.001 | Age | 0.002 | 0.000 | 0.003 | 0.004 |
| Gender | −0.011 | −0.040 | 0.018 | 0.444 | Gender | −0.003 | −0.029 | 0.024 | 0.839 |
| rs1387153 | 0.020 | −0.009 | 0.048 | 0.173 | rs1387153 | 0.013 | −0.015 | 0.040 | 0.367 |
| rs1374645 | −0.073 | −0.119 | −0.028 | 0.002* | rs1374645 | 0.015 | −0.028 | 0.057 | 0.500 |
| rs10830963 | −0.024 | −0.063 | 0.015 | 0.224 | rs10830963 | 0.003 | −0.034 | 0.039 | 0.884 |
| Age | 0.161 | 0.000 | 0.002 | 0.002 | Age | 0.119 | 0.000 | 0.002 | 0.011 |
| Gender | −0.052 | −0.042 | 0.013 | 0.315 | Gender | 0.007 | −0.023 | 0.027 | 0.880 |
| rs1374645 | −0.152 | −0.102 | −0.021 | 0.003* | rs1374645 | 0.038 | −0.022 | 0.052 | 0.413 |
|
2h glucose in NG controls | |||||||||
| Age | 0.079 | −0.001 | 0.002 | 0.226 | Age | 0.081 | 0.000 | 0.003 | 0.171 |
| Gender | 0.052 | −0.028 | 0.066 | 0.424 | Gender | 0.095 | −0.007 | 0.075 | 0.108 |
| rs1387153 | 0.119 | −0.022 | 0.087 | 0.239 | rs1387153 | −0.058 | −0.065 | 0.034 | 0.538 |
| rs1374645 | −0.129 | −0.146 | 0.006 | 0.070 | rs1374645 | −0.080 | −0.112 | 0.023 | 0.198 |
| rs10830963 | −0.060 | −0.065 | 0.035 | 0.550 | rs10830963 | 0.044 | −0.036 | 0.059 | 0.636 |
| Age | 0.071 | −0.001 | 0.002 | 0.269 | Age | 0.080 | 0.000 | 0.003 | 0.162 |
| Gender | 0.059 | −0.025 | 0.068 | 0.361 | Gender | 0.110 | −0.001 | 0.079 | 0.057 |
| rs1374645 | −0.160 | −0.154 | −0.018 | 0.013* | rs1374645 | −0.062 | −0.097 | 0.028 | 0.279 |
FBG- fasting blood glucose; NG- normoglycemic;
p-value are significant after Bonferroni correction
Figure 2.
Figure 2(A) shows distribution of FBG in combined controls (un-stratified) and stratified by obesity (low BMI ≤ 25 kg/m2 and high BMI>25 kg/m2). Analysis was performed using BMI, age and gender as covariates in un-stratified sample and age and gender as covariates in BMI-stratified sample. Figure 2(B) shows the distribution of 2h glucose in combined controls (unstratified) and stratified by obesity status. P values are from linear regression models.
To examine, if the effect of genetic variation at the MTNR1B locus on FBG is modulated by obesity status, we tested SNP-BMI interactions of all three SNPs in NG controls using GLM procedure and including FBG as dependent variable. We also included age and gender as covariates in the model. As shown in Table 5, a significant evidence of interaction was seen between BMI*1374645 (p=0.002), whereas no evidence of interaction was observed between BMI and other two SNPs (rs1387153 and rs10830963). In order to study the effect of all three SNPs together on T2D and other related quantitative traits, we performed haplotype analysis in the MTNR1B locus. No haplotype was associated with T2D or FBG, 2h glucose, insulin, HOMA IR, or HOMA B levels (data not shown).
Table 5.
Intraction between MTNR1B SNPs and BMI for affecting FBG levels
| Model | Sum of Squares | Mean Square | F | P value |
|---|---|---|---|---|
| BMI | 0.275 | 0.275 | 15.159 | 0.000 |
| Gender | 0.007 | 0.007 | 0.408 | 0.523 |
| Age | 0.257 | 0.257 | 14.189 | 0.000 |
| rs1387153 | 0.046 | 0.023 | 1.270 | 0.281 |
| rs1374645 | 0.199 | 0.199 | 11.002 | 0.001 |
| rs10830963 | 0.021 | 0.010 | 0.570 | 0.566 |
| rs1387153 * BMI | 0.059 | 0.029 | 1.625 | 0.198 |
| rs1374645 * BMI | 0.170 | 0.170 | 9.408 | 0.002 |
| rs10830963 * BMI | 0.019 | 0.010 | 0.533 | 0.587 |
Bonferroni p=0.0055
Discussion
The earlier reported associations of rs1387153 and rs10830963 with FBG concentrations could not be confirmed in our Asian Indian study. 4, 5 However, the ‘G’ (rs10830963) allele-associated non-significant increase in HOMA IR levels was observed in this sample [β= 0.13, p= 0.026, Bonferroni p= 0.005] in the same direction, as reported previously. 5, 19 Another publication on Asian Indians from UK reported association of these MTNR1B SNPs with FBG levels. 8 Incidentally, the allelic distribution in rs1387153 in our Punjabi cohort was comparable with the UK cohort [0.38 vs. 0.38], respectively, while the frequency information on rs10830963 was not available on the UK cohort. Despite similar allele frequencies, we were unable to replicate the association of these SNPs with FBG levels in our Indian Sikh cohort. Perhaps, comparatively smaller size of our cohort could be the reason of non-replication or due to weak LD between these variants and causative SNP in this population. 20 However, considering the common occurrence of these SNPs (MAF in both being >0.35) in our cohort, the sample of 1,021 NG controls would have captured the quantitative trait locus (QTL) effect with >85% power, had the association been robust. Alternatively, the differences in the ethnic composition of the UK cohort, and phenotype heterogeneity and trait characterization could have lead to these differences as our non-smoking Punjabi Sikh cohort is relatively homogenous.
However, the most interesting observation in this study is the strong and independent association of another variant within the same LD block of a GWAS-guided MTNR1B locus for affecting the same trait (FBG) in the absence of obesity. These results suggest that the same loci with common variation may harbor additional independent variants (e.g. low frequency variants like rs1374645, previously not captured by GWAS) to also influence the same trait. 21 Furthermore, the observed association of this variant with FBG levels was again confirmed in SNP*BMI interaction where a strong evidence of interaction was observed in rs1374645 with BMI (p=0.002, Bonferroni p=0.0055). At the same time, the other two SNPs showed no evidence of interaction with BMI for affecting FBG levels. Even the association of rs1374645 with 2h glucose was only confined to the low BMI group (p=0.013, Bonferroni p= 0.02) and not in high BMI group (p=0.279) when the NG sample was stratified by BMI. None of the remaining two SNPs revealed any association with 2h glucose levels when tested in the data stratified by obesity (data not presented). Nevertheless, the evidence of association of rs1374546 with 2h glucose was not as strong as seen with FBG in our stratified data. Likewise, no strong association with 2h glucose with MTNR1B locus has been reported in other studies published previously. 6
From these results it appears that the rs1374645 SNP truly affects fasting glucose concentrations before the onset of obesity. Even adjustment of age and gender did not alter the observed association. Apparently, relatively small size of our sample may be a limitation in this study. However, our data have 80% power to detect a difference in FBG as small as 1mg/dL (corresponding to detecting as significant βs outside of the range of ± 0.05) between genotypes at allele frequency of 0.06 and α =0.05 even in our stratified NG cohort by low and high BMI. Also, this deeply phenotyped homogenous cohort is collected from one geographic location and possibility of false positive associations due to population structure is less likely. Moreover, measurements on BMI and FBG levels are available in over 96% of our entire cohort, which would further strengthened the power.
Our study reveals, for the first time, a significant protective association of a less common variant in the MTNR1B with FBG levels in the absence of obesity. These findings are somewhat in agreement with a prior study where the genetic variants acting on insulin secretion in GCK, HNF1A, and SLC30A8 were associated with T2D only in non-obese individuals and the genetic variants linked with insulin resistance in PPARG2, ADIPOQ were associated with T2D among obese individuals in two European populations 22. In fact, more recently, rare alleles of two variants in PPARGC1A were associated with lower glucose levels in subjects with low BMI and with higher glucose levels in subjects with high BMI. 23 Notably, MTNR1B receptor is abundantly expressed in hypothalamus and It is speculated that the MTNR1B receptor could indirectly regulate glucose levels and insulin secretion through brain control center of circadian clock. 24 A recent study also confirmed the expression of MTNR1B in both islets and sorted pancreatic β cells. 5 These studies support putative direct role of MTNR1B in regulation of insulin secretion. Therefore, it is possible that this variant rs1374645 (being located near 5′ region of the MTNR1B) may be involved in affecting its expression, or the expression of other genes that may influence insulin signaling and insulin response mediated by obesity. 25
As the overall trait variance explained by this SNP is 2.9%, perhaps a causal variant in this gene with a greater risk is yet to be identified. Comprehensive deep sequencing is necessary to identify common and rare putative functional variants in this locus. Our data provide new information about the significant impact of a less common variant in MTNR1B on FBG levels that appears to be modulated by BMI. Future confirmation on independent datasets and functional studies will be required to define the role of this variant in fasting glucose variation, and whether this gene-interaction with obesity status can over time predispose to T2D.
Acknowledgments
This work was supported by National Institute of Health grant numbers KO1 TW006087, funded by Fogarty International Center; R01 DK082766, funded by National Institute of Diabetes and Digestive and Kidney Diseases; and a seed grant from University of Oklahoma Health Sciences Center, Oklahoma City, USA. This study was also supported in part by grant number M01 RR14467 from the National Center for Research Resources, National Institutes of Health, USA. Technical assistance provided by Ms. Bansari Mehta is duly acknowledged. We also warmly thank Ms. Dr. Alix Darden for her excellent editorial support. Authors also thank all the study participants and are grateful for their contribution in this study.
Abbreviations
- T2D
Type 2 diabetes
- MTNR1B
Melatonin receptor 1-B
- GWAS
Genome-wide association study
- HOMA IR
Homeostasis model assessment for insulin resistance
- HOMA B
Homeostasis model assessment for β cell function
- SNP
Single nucleotide polymorphism
- FBG
Fasting blood glucose
- BMI
Body mass index
Footnotes
Declaration of interest: We declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Watanabe RM, Valle T, Hauser ER, Ghosh S, Eriksson J, Kohtamaki K, Ehnholm C, Tuomilehto J, Collins FS, Bergman RN, Boehnke M. Familiality of quantitative metabolic traits in Finnish families with non-insulin-dependent diabetes mellitus. Finland-United States Investigation of NIDDM Genetics (FUSION) Study investigators. Hum Hered. 1999;49:159–168. doi: 10.1159/000022865. [DOI] [PubMed] [Google Scholar]
- 2.Snieder H, Boomsma DI, van Doornen LJ, Neale MC. Bivariate genetic analysis of fasting insulin and glucose levels. Genet Epidemiol. 1999;16:426–446. doi: 10.1002/(SICI)1098-2272(1999)16:4<426::AID-GEPI8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 3.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, Potter SC, Erdos MR, Sanna S, Hottenga JJ, Wheeler E, Kaakinen M, Lyssenko V, Chen WM, Ahmadi K, Beckmann JS, Bergman RN, Bochud M, Bonnycastle LL, Buchanan TA, Cao A, Cervino A, Coin L, Collins FS, Crisponi L, de Geus EJ, Dehghan A, Deloukas P, Doney AS, Elliott P, Freimer N, Gateva V, Herder C, Hofman A, Hughes TE, Hunt S, Illig T, Inouye M, Isomaa B, Johnson T, Kong A, Krestyaninova M, Kuusisto J, Laakso M, Lim N, Lindblad U, Lindgren CM, McCann OT, Mohlke KL, Morris AD, Naitza S, Orru M, Palmer CN, Pouta A, Randall J, Rathmann W, Saramies J, Scheet P, Scott LJ, Scuteri A, Sharp S, Sijbrands E, Smit JH, Song K, Steinthorsdottir V, Stringham HM, Tuomi T, Tuomilehto J, Uitterlinden AG, Voight BF, Waterworth D, Wichmann HE, Willemsen G, Witteman JC, Yuan X, Zhao JH, Zeggini E, Schlessinger D, Sandhu M, Boomsma DI, Uda M, Spector TD, Penninx BW, Altshuler D, Vollenweider P, Jarvelin MR, Lakatta E, Waeber G, Fox CS, Peltonen L, Groop LC, Mooser V, Cupples LA, Thorsteinsdottir U, Boehnke M, Barroso I, Van Duijn C, Dupuis J, Watanabe RM, Stefansson K, McCarthy MI, Wareham NJ, Meigs JB, Abecasis GR. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spegel P, Bugliani M, Saxena R, Fex M, Pulizzi N, Isomaa B, Tuomi T, Nilsson P, Kuusisto J, Tuomilehto J, Boehnke M, Altshuler D, Sundler F, Eriksson JG, Jackson AU, Laakso M, Marchetti P, Watanabe RM, Mulder H, Groop L. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, Marchand M, Delplanque J, Lobbens S, Rocheleau G, Durand E, De Graeve F, Chevre JC, Borch-Johnsen K, Hartikainen AL, Ruokonen A, Tichet J, Marre M, Weill J, Heude B, Tauber M, Lemaire K, Schuit F, Elliott P, Jorgensen T, Charpentier G, Hadjadj S, Cauchi S, Vaxillaire M, Sladek R, Visvikis-Siest S, Balkau B, Levy-Marchal C, Pattou F, Meyre D, Blakemore AI, Jarvelin MR, Walley AJ, Hansen T, Dina C, Pedersen O, Froguel P. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 6.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparso T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proenca C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jorgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martinez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orru M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvanen AC, Tanaka T, Thorand B, Tichet J, Tonjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Rios M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, Cavalcanti-Proenca C, Marchand M, Hartikainen AL, Sovio U, De Graeve F, Rung J, Vaxillaire M, Tichet J, Marre M, Balkau B, Weill J, Elliott P, Jarvelin MR, Meyre D, Polychronakos C, Dina C, Sladek R, Froguel P. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 8.Chambers JC, Zhang W, Zabaneh D, Sehmi J, Jain P, McCarthy MI, Froguel P, Ruokonen A, Balding D, Jarvelin MR, Scott J, Elliott P, Kooner JS. Common genetic variation near melatonin receptor MTNR1B contributes to raised plasma glucose and increased risk of type 2 diabetes among Indian Asians and European Caucasians. Diabetes. 2009;58:2703–2708. doi: 10.2337/db08-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronn T, Wen J, Yang Z, Lu B, Du Y, Groop L, Hu R, Ling C. A common variant in MTNR1B, encoding melatonin receptor 1B, is associated with type 2 diabetes and fasting plasma glucose in Han Chinese individuals. Diabetologia. 2009;52:830–833. doi: 10.1007/s00125-009-1297-8. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi F, Katsuya T, Chakrewarthy S, Yamamoto K, Fujioka A, Serizawa M, Fujisawa T, Nakashima E, Ohnaka K, Ikegami H, Sugiyama T, Nabika T, Kasturiratne A, Yamaguchi S, Kono S, Takayanagi R, Yamori Y, Kobayashi S, Ogihara T, de Silva A, Wickremasinghe R, Kato N. Common variants at the GCK, GCKR, G6PC2-ABCB11 and MTNR1B loci are associated with fasting glucose in two Asian populations. Diabetologia. 53:299–308. doi: 10.1007/s00125-009-1595-1. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy MI, Hirschhorn JN. Genome-wide association studies: past, present and future. Hum Mol Genet. 2008;17:R100–101. doi: 10.1093/hmg/ddn298. [DOI] [PubMed] [Google Scholar]
- 12.Sanghera DK, Demirci FY, Been L, Ortega L, Ralhan S, Wander GS, Mehra NK, Singh J, Aston CE, Mulvihill JJ, Kamboh IM. PPARG and ADIPOQ gene polymorphisms increase type 2 diabetes mellitus risk in Asian Indian Sikhs: Pro12Ala still remains as the strongest predictor. Metabolism: clinical and experimental. 2009 doi: 10.1016/j.metabol.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, Mehra NK, Mulvihill JJ, Ferrell RE, Nath SK, Kamboh MI. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet. 2008;9:59. doi: 10.1186/1471-2350-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diagnosis and classification of diabetes mellitus. Diabetes Care. 2004;27(Suppl 1):S5–S10. doi: 10.2337/diacare.27.2007.s5. [DOI] [PubMed] [Google Scholar]
- 15.Sanghera DK, Bhatti JS, Bhatti GK, Ralhan SK, Wander GS, Singh JR, Bunker CH, Weeks DE, Kamboh MI, Ferrell RE. The Khatri Sikh Diabetes Study (SDS): study design, methodology, sample collection, and initial results. Hum Biol. 2006;78:43–63. doi: 10.1353/hub.2006.0027. [DOI] [PubMed] [Google Scholar]
- 16.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 17.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell SCS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 19.Sparso T, Bonnefond A, Andersson E, Bouatia-Naji N, Holmkvist J, Wegner L, Grarup N, Gjesing AP, Banasik K, Cavalcanti-Proenca C, Marchand M, Vaxillaire M, Charpentier G, Jarvelin MR, Tichet J, Balkau B, Marre M, Levy-Marchal C, Faerch K, Borch-Johnsen K, Jorgensen T, Madsbad S, Poulsen P, Vaag A, Dina C, Hansen T, Pedersen O, Froguel P. G-allele of intronic rs10830963 in MTNR1B confers increased risk of impaired fasting glycemia and type 2 diabetes through an impaired glucose-stimulated insulin release: studies involving 19,605 Europeans. Diabetes. 2009;58:1450–1456. doi: 10.2337/db08-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruglyak L. Genetic isolates: separate but equal? Proc Natl Acad Sci U S A. 1999;96:1170–1172. doi: 10.1073/pnas.96.4.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cauchi S, Nead KT, Choquet H, Horber F, Potoczna N, Balkau B, Marre M, Charpentier G, Froguel P, Meyre D. The genetic susceptibility to type 2 diabetes may be modulated by obesity status: implications for association studies. BMC Med Genet. 2008;9:45. doi: 10.1186/1471-2350-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Povel CM, Feskens EJ, Imholz S, Blaak EE, Boer JM, Dolle ME. Glucose levels and genetic variants across transcriptional pathways: interaction effects with BMI. Int J Obes (Lond) 34:840–845. doi: 10.1038/ijo.2009.302. [DOI] [PubMed] [Google Scholar]
- 24.Van Cauter E. Putative roles of melatonin in glucose regulation. Therapie. 1998;53:467–472. [PubMed] [Google Scholar]
- 25.Ruan H, Lodish HF. Insulin resistance in adipose tissue: direct and indirect effects of tumor necrosis factor-alpha. Cytokine Growth Factor Rev. 2003;14:447–455. doi: 10.1016/s1359-6101(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]