Abstract
The innate immune system is a first layer of defense against infection by pathogens. It responds to pathogens by activating host defense mechanisms via interferon and inflammatory cytokine expression. Pathogen associated molecular patterns (PAMPs) are sensed by specific pattern recognition receptors. Among those, the ATP dependent helicase related RIG-I like receptors RIG-I, MDA5 and LGP2 sense the presence of viral RNA in the cytoplasm of host cells. While the precise PAMPs and functions of MDA5 or LGP2 are still unclear, RIG-I senses predominantly viral RNA containing a 5’-triphosphate along with dsRNA regions. Here we review out current knowledge how these PAMPs are sensed and integrated by RIG-I, and how RIG-I’s innate immune function can be used in translational medical approaches.
Introduction
Organisms constantly need to battle against a large variety of environmental stress factors, including physical influences and chemical agents but also infection by a large variety of pathogens. The response to pathogens in humans is governed by the innate and adaptive immune systems. Both types of response systems have a difficult molecular task: to distinguish a large variety of “self” components from an even greater variety of “non-self” components in a highly sensitive yet faithful manner. The innate immune system is hereby a first line of defense against pathogen infection. Its receptors specifically sense pathogen-associated molecular patterns (PAMPs). PAMPs are chemical or structural features present in pathogens but not host cells that serve as alert signals to the innate immune system of the host. Their recognition triggers cellular responses that attempt to counteract the pathogen and initiate other defence responses such as inflammation and adaptive immune reactions.
The response to pathogens by the innate immune system of mammals is initiated by the detection of PAMPs by a variety of host pattern recognition receptors (PRRs). PAMPs include viral RNA, cytoplasmic DNA, prokaryotic nucleotide second messenger, bacterial cell wall components like LPS or flagellar proteins (Akira et al., 2006; Hiscott et al., 2006; Meylan and Tschopp, 2006). PRRs belong to different protein classes such as the Toll like receptor family (TLRs), nucleotide oligomerisation domain (NOD) like receptors (NLRs), C-type (calcium dependent) lectin like receptors (CLRs), retinoic acid inducible gene I (RIG-I) like receptors (RLRs) and members of the HIN200 family of proteins (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009).
While the molecular mechanisms of sensing of some of these PAMPs and signalling by many of these receptors is still unclear, considerable progress has been made towards understanding the recognition of PAMPs by RIG-I (also known as DDX58). RIG-I was initially found as a PRR for viral RNA by screening of cDNA libraries for factors that induce the interferon beta (IFN-β) promoter expression in response to the viral dsRNA mimic polyriboinosinic-polyribocytidylic acid (poly(IC)) (Yoneyama et al., 2004). Type I IFNs (IFN-α and –β families) are key molecules in antiviral innate immunity and collectively orchestrate the antiviral response of cells, such as inhibition of viral replication and signalling to neighboring cells.
A hallmark of RLRs, which belong to superfamily 2 (SF2) helicases/ATPases, is an ATPase domain (Gorbalenya et al., 1988; Gorbalenya et al., 1989). SF2 enzymes are a large class of functionally and mechanistically diverse DNA or RNA dependent ATPases, found in virtually all nucleic acid associated processes. Not all of these enzymes are bona fide “helicases”, as the common name implies, but some simply grip nucleic acids in an ATP dependent fashion or act as molecular motors e.g. in the remodeling of DNA:protein complexes (Hopfner and Michaelis, 2007). Besides the SF2 domain, RIG-I possesses two N-terminally located CARDs (caspase activation and recruitment domain) and a C-terminal RD (regulatory/repressor domain) also called C-terminal domain (CTD) (Fig. 1A). The CARDs are implicated in downstream signalling. The RD turns out to be a major pattern recognition site and senses preferentially dsRNA blunt ends with 5’ triphosphates. The RLR family has two additional members - MDA5 (melanoma differentiation-associated protein 5)/helicard/RH116 and LGP2 (laboratory of genetics and physiology 2) (Andrejeva et al., 2004; Rothenfusser et al., 2005). MDA5 shares the domain architecture with RIG-I (Fig. 1A). RIG-I and MDA5 sense different types of viruses (Kato et al., 2006). RIG-I is a sensor for e.g. hepatitis C virus, Sendai virus, influenza virus, vesicular stomatitis virus, rabies virus and Japanese encephalitis virus, while MDA5 appears to detect picornaviruses (Gitlin et al., 2006; Kato et al., 2006) and noroviruses (McCartney et al., 2008). Overlapping roles of RIG-I and MDA5 were demonstrated for reovirus and dengue virus (Loo et al., 2008). LGP2 has not been shown yet to directly sense a particular viral RNA and was initially characterized as regulatory molecule (Rothenfusser et al., 2005; Yoneyama et al., 2005). Upon activation by PAMPs, RIG-I or MDA5 associate with the adaptor protein IFN-β promoter stimulator 1 (IPS1), also known as virus-induced signalling adapter (VISA), mitochondrial antiviral signalling (MAVS) or CARD adapter inducing IFN-β (CARDIF), located at the outer mitochondrial as well as peroxisomal membranes (Dixit et al., 2010; Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005) (Fig. 1B). IPS1 also possesses an N-terminal CARD, followed by a predicted unstructured proline rich region and a single transmembrane domain. The CARD of IPS1 has been studied by X-ray crystallography and possesses the classic CARD fold, suggesting homotypic CARD-CARD mediated interactions, potentially with the CARDs of RIG-I and MDA5 (Potter et al., 2008). However, a direct interaction of the CARDs between IPS1 and RIG-I or MDA5 has not yet been demonstrated with purified components in vitro and may require additional components or modifications. For instance, activation of RIG-I requires ubiquitylation of CARD2 (Gack et al., 2007) but can also proceed via noncovalent interaction with K63-linked polyubiquitin chains (Zeng et al., 2010). The interaction of RIG-I or MDA5 with IPS1 activates the serine/threonine kinases IKKα/β and IKK-i/TBK-1, involving dimer formation of IPS1 (Baril et al., 2009; Tang and Wang, 2009). IKKα/β then phosphorylates NF-κB, which translocates to the nucleus to activate its target genes. In contrast, IKK-i/TBK-1 phosphorylates IRF3 and IRF7, which then trigger the expression of IFN-α and IFN-β genes (Johnson and Gale, 2006; Maniatis et al., 1998).
Figure 1. Domain structure of and signalling by the RLR family members.
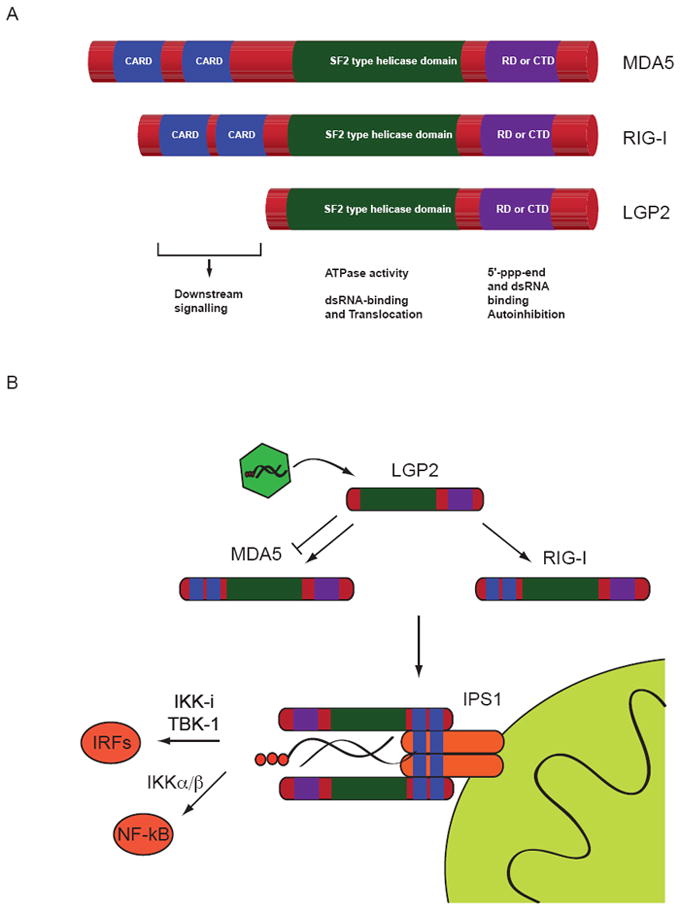
(A) RIG-I, MDA5 and LGP2 share a C-terminal RD domain and an SF2-type ATPase domain. In addition RIG-I and MDA5 have two N-termial CARD domains required for signalling via IPS1 that are absent in LGP2.
(B) Signalling in the RLR pathway is initiated by binding of RNA to the respective receptor (RIG-I or MDA5). LGP2 may assist ligand recognition or inhibit signalling under certain circumstances. Binding of one of the receptor proteins to the mitochondrial adapter IPS1 leads to the activation of transcription factors of the IRF and NF-kB families. This ultimately leads to the production of interferons and inflammatory cytokines.
Here we review our current knowledge regarding the nature of patterns sensed by RIG-I and the other RLRs, and the mechanism how pattern recognition translates molecular changes on the receptor level into an anti-viral signal. Knowledge of the nature of these patterns also offers exciting therapeutic potential to treat e.g. malignant disease.
PAMPs recognised by RIG-I-like helicases
The RLRs have evolved to recognize the presence of viruses in infected cells. RIG-I and MDA5 sense the presence of largely different sets of viruses, but do not always act mutually exclusive (Fredericksen et al., 2008; Kato et al., 2006). The concept of virus recognition by RLRs, depends on PAMPs that are part of the viral nucleic acid repertoire (Fig. 2). Understanding the nature of the PAMPs that are recognised will form the basis of understanding differential virus recognition by RLRs as well as their activation mechanisms.
Figure 2. PAMPs recognised by RIG-I are found in viral genomic RNAs.
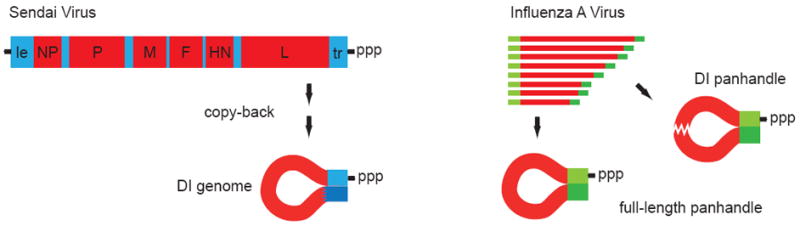
Recent studies suggest that DI-genomic RNA may serve as a principal ligand for RIG-I in the situation of viral infection of a cell. In the case of Sendai Virus DI-genomes arise by copy back mechanisms forming a self-complementary structure (left). Influenza virus genome segments are inherently self-complementary at the ends (panhandle) but may also give rise to internally deleted DI-genomic segments (right).
Early functional studies on RIG-I, MDA5 and LGP2 used poly(IC), a commercially available double-stranded RNA (dsRNA) analog, as a synthetic ligand to stimulate their activity. Later experiments in mice have shown that MDA5 is responsible for the recognition of poly(IC) in mice (Gitlin et al., 2006). A study that has compared poly(IC) preparations of diffenrent lenghts came to the conclusion that MDA5 preferentially recognises long poly(IC) chains whereas shorter poly(IC) is recognised by RIG-I (Kato et al., 2008). Results obtained with poly(IC) as a surrogate for RNA have been taken as valid, however data on genuine RNA ligands with respect to their recognition by MDA5 are scarce. A study that aimed to identify immunostimulatory RNAs bound to MDA5 under physiological conditions came to the conclusion that it is a complex RNA-structure present in large RNA assemblies that activates MDA5 (Pichlmair et al., 2009). In summary, the exact PAMP that is recognized by MDA5 remains largely uncharacterised so far.
Information on the nature of the ligands for the third member of the RIG-I like helicase family LGP2 is similarly scarce. The regulatory domain of LGP2 was shown to bind the ends of double-stranded RNA independently of a 5’-triphosphate group and with higher affinity than single-stranded RNA (Li et al., 2009b; Pippig et al., 2009). Natural ligands for LGP2 during viral infection have not been identified yet. LGP2 was first described in cell culture experiments to be an (most likely competitive) inhibitor of RIG-I mediated recognition of viruses (Rothenfusser et al., 2005; Yoneyama et al., 2005). This was first confirmed by experiments using knock-out mice (Venkataraman et al., 2007). However the analysis of a different strain of LGP2 knock-out mice unexpectedly revealed, that the presence (and ATPase activity) of LGP2 can in fact be required to elicit a strong signal via MDA5 as well as RIG-I during viral infection (Satoh et al., 2010). The mechanism for this phenomenon is still unclear but as the response to viruses detected via MDA5/RIG-I (but not the response to poly(IC)) required LGP2, it was proposed that LGP2 works upstream of RIG-I and MDA5 perhaps by unwinding or stripping nucleoproteins of viral RNA, thereby making the nucleic acid PAMP accessible for binding.
More detailed information is available on the RNA structures and ligands recognised by RIG-I. The first studies on RIG-I ligand properties have revealed that it is a free 5’-triphosphate on RNAs in the cytoplasm of infected cells that serves as a recognition signal for RIG-I (Hornung et al., 2006; Pichlmair et al., 2006). This fits perfectly with the PAMP concept, because besides very few exceptions free 5’-triphosphate RNA ends are absent from eukaryotic cytoplasm due to RNA metabolism in the nucleus, for example mRNA capping. Two recent studies that have revisited the issue of RIG-I ligand properties using chemically synthesised RNAs however found that 5’-triphosphate is not the whole story (Schlee et al., 2009; Schmidt et al., 2009). A base-paired region in the range of 10 to 20 nucleotides in proximity of the free 5’-triphosphate end of the RNA ligand is essential for immunostimulatory activity via RIG-I .The base-paired stretch may result from annealing of two separate strands or from local hairpin (or panhandle) formation in a single RNA molecule. Schlee et al. stress the importance of a blunt-end conformation proposing that for optimal activation the triphosphate-modified base at the 5’-end itself has to be base-paired and 3’- or 5’-overhangs lead to reduced immunostimulatory activity (Fig. 2).
In addition to 5’- triphosphate dsRNAs other RNA-configurations have been described to be able to activate RIG-I. These include 5’-monophosphate dsRNAs and 3’-monophosphate RNase L cleavage products of cellular RNA (Malathi et al., 2007; Takahasi et al., 2008). However, their ability to directly activate RIG-I was not confirmed in subsequent studies. There are also reports suggesting that base composition e.g. in the case of hepatitis C virus plays a pivotal role in RIG-I-mediated sensing requiring AU-rich sequences to be active (Saito et al., 2008). Apparently, the first base that carries the 5’-triphosphate (C being less active than A, G or U) (Schlee et al., 2009) has some influence. Other studies however found no evidence for a sequence motive detected by RIG-I and it is therefore thought that it is not the primary sequence but the 5’-triphosphate modification and base-paired secondary structure that defines the molecular pattern detected by RIG-I. Thus, much has been learned about patterns that serve as recognition motifs for RIG-I, but these results have not yet lead to a general prediction as to what type of RNA is really recognised in a virus infected cell. The propositions and speculations varied in different studies from viral genomic RNA, replication intermediates, non-coding transcription products (leader RNA), defective interfering (DI)-genomes to endogenous RNA ligands.
An interesting recent approach involves isolating and analysing RIG-I-stimulatory RNAs from virus infected cells. In an elegant study Rehwinkel et al. did this using an influenza virus expression system and Sendai virus as infection models (Rehwinkel et al., 2010). Here, immunostimulatory RNA associated with RIG-I (and the influenza protein NS1 as a competitive inhibitor) was isolated by pull-down and found by size-fractionation-analysis to correspond to the influenza virus genomic segments. RIG-I-associated preparations isolated from Sendai virus infected cells showed that the active RNA component is contained in genomic fractions with a size > 300 bases as analysed by PCR methods and gel electrophoresis. From their data they could conclude that the natural ligands for RIG-I in their model systems are not transcripts or leader RNAs but 5’-triphosphorylated genomic sequences produced during the replicative cycle. Through complementary sequences at the 5’- and 3’-ends, the genomic-segments of influenza are predicted to form panhandle conformations leading to the base-paired stretch close to the triphosphorylated 5’-end, postulated by the studies defining the RIG-I PAMP with synthetic model-RNAs (Fig. 2 right).
An even more comprehensive approach was taken by Adolfo Garcia-Sastre’s lab. In a very recent set of experiments Baum et al. isolated immunostimulatory RNA associated with RIG-I from cells also infected with Sendai virus and influenza virus and analysed the bound RNAs with second generation-sequencing. Their data indicate that in the case of Sendai virus infection it is the relatively short genomes of DI particles, that are found associated with RIG-I (Baum et al., 2010). This is consistent with earlier observations that copy-back DI particle content determines the immunostimulatory capacity of viral preparations (Strahle et al., 2006). As the copy-back DI-genomes bound to RIG-I in the study from the Garcia-Sastre lab have a length of 546-nts and form a long perfect dsRNA portion (92 nt) at the triphosphorylated 5’-end, their results are consistent with the data from Rehwinkel et al. and the predicted RIG-I PAMP described above (Fig. 2 left). In cells infected with influenza virus Baum et al. found both whole genomic-segments and internal deletion DI-genomes associated with RIG-I thereby at least partly confirming the Rehwinkel study. Further studies however are needed to analyse whether the RIG-I ligands identified in Sendai virus infection and influenza virus infection (DI-genomes and whole genome-segments) are also paradigmatic for infections with other viruses.
Yet another new perspective on RIG-I ligands comes from the recent discovery that in certain cases immune recognition of intracellular non-CpG DNA is RIG-I-dependent. Two studies show that DNA-dependent RNA polymerase III (RNAP III) transcribes DNA into 5’-triphosphate RNA that is then recognised by RIG-I (Ablasser et al., 2009; Chiu et al., 2009). The mechanism seems to be restricted to certain types of DNA and the synthetic model DNA poly-deoxyadenylic-deoxythymidylic acid (poly(dAdT)) is used as a paradigm to study it. Where the recognition by RNAP III takes place in unclear because Chiu et al. show that RNAP III produces the RIG-I ligands in the cytoplasm but Ablasser et al. use EBV-derived nuclear transcripts as a physiological model. So far this RNAP-III dependent production of RIG-I ligands was shown in infection models for EBV, HSV and Legionella pneumophila. More detailed analysis of the RNAP III – RIG-I axis in infection models might provide us with new information on the nature of RNAs recognised by RIG-I in infected cells.
Structural mechanism of pattern sensing by RIG-I and RLRs
RIG-I is a complex multidomain molecule and much work has been done over the past years to address the roles of the individual domains in sensing and signalling. All three domain families of RIG-I i.e. CARDs, SF2 and RD have been found to be involved in determining the specificity of RIG-I for 5’-triphosphate containing dsRNA, suggesting a global structural change in RIG-I from a signal off into signal on states. For instance, although CARDs are the signalling module of RIG-I, they are also important for the RNA selectivity of RIG-I. ΔCARD-RIG-I is much more efficiently stimulated in vitro by the non-phosphorylated dsRNA than wtRIG-I, and looses specificity for 5’-triphosphate-RNA (Cui et al., 2008). The two main RNA binding sites are located on the SF2 domain and RD. The latter senses the 5’-triphosphate moiety on RNAs. The cysteine rich RDs are a unique hallmark and defining feature of RLRs, although only the RD of RIG-I has been found to bind specifically 5′-triphosphate-RNA (Fig. 3). The RD, first described by Michael Gale and coworkers, is important for the activity of the enzyme in vivo and in vitro (Saito et al., 2007). Structural studies indicated that RDs from RIG-I, MDA5 and LGP2 are fold-related to each other (Fig. 3A), but bear little sequence homology to other known protein domains (Cui et al., 2008; Li et al., 2009a; Li et al., 2009b; Lu et al., 2010a; Lu et al., 2010b; Pippig et al., 2009; Takahasi et al., 2009; Takahasi et al., 2008; Wang et al., 2010). However, some structural relatives of RD could be identified in the Protein Data Bank. RD is fold-related to the C-terminal methionine sulfoxide reductase domain of PilB (PDB entry 1L1D) and MSS4, a GDP/GTP exchange factor for small Rab-like GTPases (PDB entry 2FU5). Interestingly, MSS4 stimulates nucleotide exchange by structurally modulating the nucleotide-binding site of the Rab8 GTPase (Itzen et al., 2006) and it will be interesting to analyse whether MSS4 and RD have a related regulatory effect on their functionally associated NTP binding domain (Cui et al., 2008).
Figure 3. Structures and mechanisms of RLR activation.
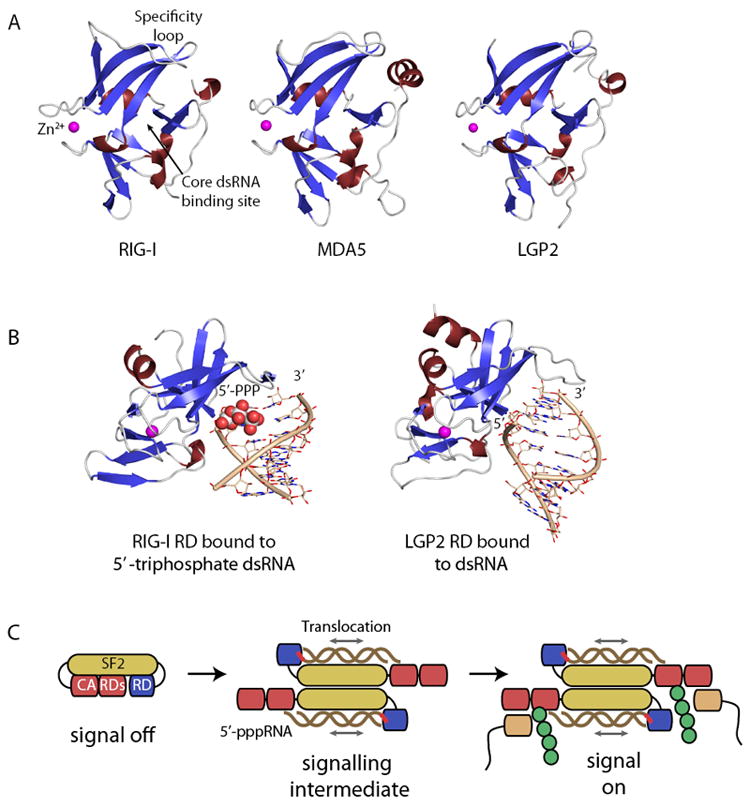
(A) Regulatory/Repressor domains (ribbon models with highlighted secondary structure) of RIG-I, MDA5 and LGP2 possess the same fold, including a zinc (magenta sphere) binding site. Nucleic acids bind to a central core RNA binding site, while specificity for different types of RNA ends is provided by a specificity loop.
(B) RDs emerge as dsRNA end binding domains. RIG-I RD preferentially binds to dsRNA ends containing a triphosphate moiety (color coded spheres) while LGP2 appears to recognise unphosphorylated ends.
(C) Possible model for PAMP integration and signalling by RIG-I. Binding of pppRNA to RD and the SF2 domain could switch the enzyme into an active dimer. Translocation of the SF2 domain on dsRNA may induce a conformational change in RIG-I with exposed CARDs, leading to ubiquitylation and interaction with the IPS1 adaptor.
After it was established that RIG-I RD senses the 5’-triphosphate moiety, further work established that RDs of RIG-I, LGP2 and MDA5 are in fact RNA binding domains with specificity for dsRNA ends (Li et al., 2009b; Lu et al., 2010a; Lu et al., 2010b; Wang et al., 2010). In the case of RIG-I these are dsRNA ends bearing a 5’-triphosphate moiety, while the RDs of LGP2 and MDA5 bind dsRNA ends with or without 5’-triphosphates. The binding activity of MDA5’s RD has not been analysed in more detail because it shows comparatively weak affinity for RNA, but there is circumstantial evidence that MDA5 and LGP2 might cooperate and also MDA5 binds dsRNA ends (Takahasi et al., 2009). Structural studies indicate that RDs from all three RLRs possess a three-leafed β-sheet structure with short connecting helices. The three leaves are held together and stabilised by a metal binding site, formed by four invariant cysteines that coordinate a zinc ion. The arrangement of the three leaves creates a shallow groove on the concave side of RDs that carries the main positive electrostatic potential and forms the binding site for RNA ends. Structures of RNA complexes are now available for RIG-I’s RD in complex with 5’-tri/diphosphate dsRNA as well as blunt end dsRNA without phosphate moieties and show how RD’s sense nucleic acids (Fig. 3B).
These studies indicated that RDs bind dsRNA with a fairly conserved mode of recognition, although some differences can be observed. The core interaction is formed by recognising predominantly the backbone of the last two (RIG-I) or three (LGP2) bases from the 5’-end, while only the very end of the 3’ contributes to binding. Here, a tighter interaction of LGP2 with the 3’ terminus is observed, while RIG-I appears to bind the 5’-terminus more tightly. The 5’-terminal strand is mainly bound at the core binding site, while the recognition of the RNA end structures (5′-triphosphates, pi-system of the bases, sugar of the 3’-end) is mediated to a substantial extent by a specificity loop that is markedly distinct between RIG-I and LGP2. RIG-I is specifically adapted by having several positively charged residues in this region that are critical for counteracting the negative charge of the triphosphate moiety (Li et al., 2009b; Lu et al., 2010a; Lu et al., 2010b; Wang et al., 2010). Binding of RNAs containing 5’-tri/diphosphates is somewhat different to recognition of unphosphorylated RNA ends (Lu et al., 2010a; Lu et al., 2010b; Wang et al., 2010). This different recognition could, in addition to a diminished binding energy, add to the enzymes specificity. This is consistent with observations that different ligands differentially affect the overall conformation of RIG-I (Ranjith-Kumar et al., 2009).
In vivo, RIG-I is found in multimeric complexes as it signals downstream but due to the many proteins involved in such structures, it is unclear whether these multimers present a defined protein complex or are the result of co-localising to viral RNA (Saito et al., 2007). In vitro studies found that in the presence of 5’-triphophate-dsRNA as well as unphosphorylated dsRNA, RIG-I forms a homogenous, stable dimer, while in the absence of the ligand, RIG-I is monomeric (Cui et al., 2008; Ranjith-Kumar et al., 2009). ΔCARD-RIG-I also shows pppRNA dependent dimerisation (Cui et al., 2008), suggesting that dimer formation occurs via a CARD-independent mechanism (Ranjith-Kumar et al., 2009). A low-resolution structure of the ΔCARD-RIG-I dimer was obtained using negative stain electron microscopy (Ranjith-Kumar et al., 2009). Thus, a correct dimer with multiple protein RNA interactions might be a prerequisite for appropriate signal transduction. It is worth noting, that downstream signalling as well as RNA binding by other members of the RLR-pathway also involves protein dimerization (Baril et al., 2009; Murali et al., 2008; Tang and Wang, 2009). Thus, ligand induced self-association of pattern receptors, followed by association-stimulated activation of signal transduction / effector enzymes could be a central activation mechanism also employed by RLRs.
If PAMPs dimerise RIG-I much like PAMPs dimerise TLRs, why does RIG-I contain a SF2 domain in addition to RDs? There are several possibilities. For instance, the ATPase function could provide improved specificity, because RIG-I has a more difficult sensing task to perform in the RNA rich environment of the cytoplasm than TLRs in the endosome, which is devoid of self nucleic acids. Another function could be direct competition with viral proteins, such as a protein displacement activity on RNA or RNA unwinding. Indeed, some unwinding activity has been demonstrated by RIG-I, but it appears to be less robust than one would expect it to be for a bona fide helicase (Takahasi et al., 2008). SF2 enzymes share conserved sequence motifs (somehow often misnamed as “helicase motifs”) that mediate ATP and nucleic acid binding. In general, ATP-binding and –hydrolysis induced conformational changes between two subdomains of the SF2-fold repositions two nucleic acid binding sites on the surface of the SF2 domain that leads to dynamic, differential nucleic acid interactions. Depending on the precise biochemical mechanism this conformational power stroke leads to a directional transport of nucleic acids, unwinding of nucleic acid structures, or grip of nucleic acids. For instance, in NS3 of hepatitis C virus, the ATP cycle directionally advances the enzyme by single base(pair) steps on product ssRNA, leading to unwinding of adjacent nucleic acid structures or displacement of proteins bound to substrate RNA (Dumont et al., 2006; Myong et al., 2007).
While multimerisation is probably a key part of the RIG-I activation mechanism, for instance by subsequent multimerisation of IPS1 and downstream factors (Baril et al., 2009), dimer formation in vitro does not depend on ATP (Cui et al., 2008). Based on this, an additional activation step is required for downstream signalling by RIG-I, because RIG-I signalling in general requires intact ATP-binding motifs (Bamming and Horvath, 2009; Yoneyama et al., 2004). What is the functional role of the SF domain and its RNA stimulated ATPase activity? A first hint came from the observation that the ATPase activity of the isolated RIG-I SF2 domain is stimulated very efficiently by dsRNA but not by ssRNA (Cui et al., 2008). Often, “helicases” translocate on the DNA/RNA substrate that activates their ATPase activity, suggesting that RIG-I is a dsRNA translocase. In fact, a robust dsRNA translocation activity of RIG-I could be shown using single-molecule studies (Myong et al., 2009). While double-stranded RNA translocation activity of both wtRIG-I and ΔCARD-RIG-I are observed, these experiments did not reveal substantial RNA unwinding activity. Interestingly, RIG-I is much slower on a generic dsRNA substrate than ΔCARD-RIG-I, which confirms the inhibitory role of the CARDs. In the presence of 5’-triphosphate, this inhibition is lifted and the enzyme translocates efficiently on dsRNA. Thus, RIG-I integrates both 5’-triphosphate and dsRNA patterns by a cooperation of RD and SF2 domain, consistent with its optimal ligands in cellular assays.
What is the biological relevance of RIG-I translocation activity? RIG-I preferentially binds dsRNA that contain 5’-triphosphate. Both PAMPs are features of many replicating RNA viruses. The dsRNA translocation activity on 5’-triphosphate RNA would serve as a dual layer read out of viral PAMPs. Initial recognition is governed by the 5’-triphosphate. Subsequently, RIG-I forms dimers and starts to translocate on a nearby dsRNA stretch. This translocation might lead to exposed CARDs, thus creating a signalling conformation for downstream interactions (Fig. 3C). The signal strength may be related to the amount of time spent in the translocation mode and therefore to the length of RNA. Such a model might explain how RIG-I and MDA5 may differentially read out very long dsRNA regions. However, RIG-I efficiently detects also rather short RNA, so only a local translocation step seems to be required for activity. We currently favor a model where a short translocation by few base pairs switches CARDs into a conformation that is recognised by factors such as Trim25 and IPS1 (Fig. 3C). Alternatively, the translocation could be a proofreading activity to enable repeated binging to the 5’-triphosphate moiety. This would explain why mutations in some helicase motifs induces constitutive signalling, however, it does not explain why mutations in other ATPase motifs disrupt signalling (Bamming and Horvath, 2009). Finally, ATP driven protein translocation on viral dsRNA might effectively interfere with viral proteins by preventing them from binding, blocking their progression, or displacing them, thus actively interfering with viral replication. Such a function might be important for LGP2, because it appears to act upstream of RIG-I and MDA5 for viruses with a tightly packaged genomic RNA and less for viruses with a more loosely packaged genomic RNA. Further studies that correlate the features of the SF2 and its translocation activity with RIG-I dependent interferon stimulation are required.
Signalling by RIG-I also depends on ubiquitylation of CARDs and RD by TRIM25 and RFN135, respectively, presumably manifesting a signalling conformation or providing an additional recognition platform for downstream factors (Gack et al., 2007; Oshiumi et al., 2009). Using a reconstituted system with partially purified components, it was also found that K63-linked ubiquitin chains can also stimulate RIG-I mediated signalling and that RIG-I interacts with these chains (Zeng et al., 2010). A three-layered read out (5′-triphosphate, dsRNA, ubiquitylation/ubiquitin chain association) is presumably necessary to sense virus replication robustly and to avoid premature innate immune responses due to sporadic dsRNA or sporadic 5’-triphosphates on normal cellular RNA. Alternatively, a replicating virus may only be detectable in a very short time window and a variety of viral countermeasures interfere with RIG-I sensing. Thus, it is possible that this short time window needs manifestation i.e. through an ubiquitylation reaction.
Several key questions, however, need to be addressed to fully understand the mechanism of RIG-I signalling. For instance, what are the two signal-off and signal-on conformations of RIG-I, what is the molecular basis for the RIG-I:IPS1 interaction, how does this interaction activate IPS1 or recruit downstream factors. Furthermore, what is the exact molecular role of ubiquitin chains or ubiquitylation? What is the function of the RIG-I translocase activity and how does the translocation reaction lead to IPS1 activation?
Translational implications of viral pattern recognition
Further structure-function analyses of RLRs will also be important to facilitate the use of RLR ligands for clinical applications like immunotherapy. The basic idea of immunotherapy is harnessing the power of the immune system to fight diseases. In malignant disease the major goal of therapy is to destroy cancerous cells. Here, the link to viral defense mechanisms (including those initiated by RLRs) is obvious, because also in viral infections the immune system’s strategy is to kill the virus-infected cells. This has lead to the concept of using targeted application of PAMPs to mimic a situation of viral infection in the tumor tissue and trigger cell autonomous responses in tumor cells along with cytotoxic T-cell responses that target tumor cells.
With respect to RLR ligands first steps in this direction have been taken. Two reports describe a pathway in which transcriptional activation of the proapoptotic gene NOXA after ligation of MDA5 with poly(IC) directly drives melanoma cells into apoptosis and autophagic cell death, respectively (Besch et al., 2009; Tormo et al., 2009). Besch et al. show that this is also true for RIG-I ligation with 5’-triphosphate dsRNA. It is important to note that they observe that tumor cells are much more sensitive to cytotoxic effects after RLR ligation than are untransformed cells, which allows for tumor-specific effects despite systemic application of the ligands in the mouse models they use. A very recent study by Kübler et al. makes the interesting observation that the cell death that is triggered in ovarian cancer cells by RIG-I ligation is immunogenic in the sense that it facilitates phagocytic uptake of the dying cells (Kubler et al., 2010). If this is a general phenomenon it will certainly add to the immunotherapeutic potential of RLR ligation.
A whole new twist to using RIG-I ligands as immunotherapeutics against cancer came from the interesting idea of combining the immunostimulatory potential of 5’-triphosphate dsRNA with its ability to function as an siRNA. The strategy is to target expression of an oncogenic gene in addition to inducing immune stimulation by 5’-triphosphate dsRNA. In an elegant proof-of-principle study Poeck et al. demonstrated that this works in several mouse models of cancer targeting the anti-apoptotic protein B-cell lymphoma 2 (Bcl-2) (Poeck et al., 2008). Importantly, the study showed that immunostimulation by 5’-triphosphate dsRNA and knock-down of Bcl-2 in the tumor both contributed to the anti-tumoral effect.
In addition to being interesting targets for the immunotherapy of cancer RLRs have been found to play a role in other disease conditions. For example, several genetic studies recently identified IFIH1, the gene coding for MDA5, as a locus involved in type I diabetes and other autoimmune diseases (Psoriasis, selective IgA-deficiency) (Ferreira et al., 2010; Nejentsev et al., 2009; Smyth et al., 2006; Strange et al., 2010). Rare functionally inactive variants of MDA5 are protective, while other (presumably overactive) variants increase the risk for diabetes type I. The mechanistic details are not worked out so far, however these findings make the MDA5 pathway a highly interesting research target also in autoimmunity. In summary, knowledge on RLR ligands and the way they activate the innate immune system begins to be exploited for the immunotherapy of cancer and certainly will in the future be important for other translational research fields like autoimmunity or vaccine development.
Acknowledgments
We thank Moritz Rapp for his help in the figure preparation. Work in K.-P. H.’s laboratory on RIG-I and helicases is supported by the Deutsche Forschungsgemeinschaft (SFB 455, GK1202 and HO2489/3) and the National Institutes of Health (U19AI83025). Work in S.R.’s laboratory is supported by the Deutsche Forschungsgemeinschaft (DFG GK1202-A3, DFG RO2525/3-1)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamming D, Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral RNA helicase proteins MDA5, RIG-I and LGP2. J Biol Chem. 2009;284:9700–9712. doi: 10.1074/jbc.M807365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril M, Racine ME, Penin F, Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83:1299–1311. doi: 10.1128/JVI.01659-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besch R, Poeck H, Hohenauer T, Senft D, Hacker G, Berking C, Hornung V, Endres S, Ruzicka T, Rothenfusser S, Hartmann G. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J Clin Invest. 2009;119:2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5’-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RC, Pan-Hammarstrom Q, Graham RR, Gateva V, Fontan G, Lee AT, Ortmann W, Urcelay E, Fernandez-Arquero M, Nunez C, et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat Genet. 2010;42:777–780. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 1988;235:16–24. doi: 10.1016/0014-5793(88)81226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J, Lin R, Nakhaei P, Paz S. MasterCARD: a priceless link to innate immunity. Trends Mol Med. 2006;12:53–56. doi: 10.1016/j.molmed.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Michaelis J. Mechanisms of nucleic acid translocases: lessons from structural biology and single-molecule biophysics. Curr Opin Struct Biol. 2007;17:87–95. doi: 10.1016/j.sbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding--structure of Rab8 in complex with MSS4. Embo J. 2006;25:1445–1455. doi: 10.1038/sj.emboj.7601044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Gale M., Jr CARD games between virus and host get a new player. Trends Immunol. 2006;27:1–4. doi: 10.1016/j.it.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kubler K, Gehrke N, Riemann S, Bohnert V, Zillinger T, Hartmann E, Polcher M, Rudlowski C, Kuhn W, Hartmann G, Barchet W. Targeted activation of RNA helicase retinoic acid-inducible gene-I induces proimmunogenic apoptosis of human ovarian cancer cells. Cancer Res. 2010;70:5293–5304. doi: 10.1158/0008-5472.CAN-10-0825. [DOI] [PubMed] [Google Scholar]
- Li X, Lu C, Stewart M, Xu H, Strong RK, Igumenova T, Li P. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys. 2009a;488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Li X, Ranjith-Kumar CT, Brooks MT, Dharmaiah S, Herr AB, Kao C, Li P. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem. 2009b;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ranjith-Kumar CT, Hao L, Kao CC, Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5’ triphosphate. Nucleic Acids Res. 2010a Oct 20; doi: 10.1093/nar/gkq974. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P. The structural basis of 5’ triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure. 2010b;18:1032–1043. doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, Parekh BS, Wathelet MG. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. MDA-5 recognition of a murine norovirus. PLoS Pathog. 2008;4:e1000108. doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol Cell. 2006;22:561–569. doi: 10.1016/j.molcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Murali A, Li X, Ranjith-Kumar CT, Bhardwaj K, Holzenburg A, Li P, Kao CC. Structure and function of LGP2, a DEX(D/H) helicase that regulates the innate immunity response. J Biol Chem. 2008;283:15825–15833. doi: 10.1074/jbc.M800542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T. Cytosolic viral sensor RIG-I is a 5’-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-beta induction during the early phase of viral infection. J Biol Chem. 2009;284:807–817. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippig DA, Hellmuth JC, Cui S, Kirchhofer A, Lammens K, Lammens A, Schmidt A, Rothenfusser S, Hopfner KP. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009;6:2014–2025. doi: 10.1093/nar/gkp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS, Kirschnek S, Gaffal E, Landsberg J, Hellmuth J, et al. 5’-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- Potter JA, Randall RE, Taylor GL. Crystal structure of human IPS-1/MAVS/VISA/Cardif caspase activation recruitment domain. BMC Struct Biol. 2008;8:11. doi: 10.1186/1472-6807-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar CT, Murali A, Dong W, Srisathiyanarayanan D, Vaughan R, Ortiz-Alacantara J, Bhardwaj K, Li X, Li P, Kao CC. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J Biol Chem. 2009;284:1155–1165. doi: 10.1074/jbc.M806219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 5’-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- Strahle L, Garcin D, Kolakofsky D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology. 2006;351:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, Barton A, Band G, Bellenguez C, Bergboer JG, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T, Inagaki F. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem. 2009;284:17465–17474. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Tang ED, Wang CY. Mavs Self-Association Mediates Antiviral Innate Immune Signaling. J Virol. 2009;83:3420–3428. doi: 10.1128/JVI.02623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormo D, Checinska A, Alonso-Curbelo D, Perez-Guijarro E, Canon E, Riveiro-Falkenbach E, Calvo TG, Larribere L, Megias D, Mulero F, et al. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell. 2009;16:103–114. doi: 10.1016/j.ccr.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, et al. Structural and functional insights into 5’-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]


