Abstract
The stress-activated kinase p38α was used to evaluate a fragment-based drug discovery approach using the BioFocus fragment library. Compounds were screened by surface plasmon resonance (SPR) on a Biacore™ T100 against p38α and two selectivity targets. A sub-set of our library was the focus of detailed follow-up analyses that included hit confirmation, affinity determination on 24 confirmed, selective hits and competition assays of these hits with respect to a known ATP binding site inhibitor. In addition, functional activity against p38α was assessed in a biochemical assay using a mobility shift platform (LC3000, Caliper LifeSciences). A selection of fragments was also evaluated using fluorescence lifetime (FLEXYTE™) and microscale thermophoresis (Nanotemper) technologies. A good correlation between the data for the different assays was found. Crystal structures were solved for four of the small molecules complexed to p38α. Interestingly, as determined both by X-ray analysis and SPR competition experiments, three of the complexes involved the fragment at the ATP binding site, while the fourth compound bound in a distal site that may offer potential as a novel drug target site. A first round of optimization around the remotely bound fragment has led to the identification of a series of triazole-containing compounds. This approach could form the basis for developing novel and active p38α inhibitors. More broadly, it illustrates the power of combining a range of biophysical and biochemical techniques to the discovery of fragments that facilitate the development of novel modulators of kinase and other drug targets.
Electronic supplementary material
The online version of this article (doi:10.1007/s10822-011-9454-9) contains supplementary material, which is available to authorized users.
Keywords: p38α, Surface-plasmon resonance, Fragment screening, X-ray crystallography
Introduction
In the context of preclinical drug discovery, fragments can be defined as partial structures of lead- or drug-like molecules with molecular weights typically ranging from 150 to 300 Da. The quality of a fragment can further be defined by its lipophilicity, conformational flexibility, hydrogen bonding capacity and other parameters. Screening of fragment libraries with subsequent hit optimization has become a validated approach for hit identification and an attractive complementary method to high-throughput screening [1, 2]. It offers the potential for identifying chemical starting points with high ligand efficiency [3] whereby most atoms of the fragment are engaged in target interaction. Due to their size, fragments are inherently more prone to fit in many receptor pockets and therefore, fragment libraries typically yield higher hit rates than collections of more elaborated lead-like molecules. Many screening techniques have been used and extensively described, each displaying potential advantages and disadvantages.
As part of a case study, we herein compare several screening technologies: a high-concentration biochemical method using the Caliper LC3000 mobility shift, a high-concentration biochemical fluorescence lifetime platform and a biophysical binding assay applying surface plasmon resonance. Microscale thermophoresis was used to study a few selected hit compounds.
For this study we selected p38α, a well-characterized protein target, to investigate whether a fragment screen could potentially provide new insights and result in chemical starting points for further optimization. We were particularly interested in identifying ligands for allosteric binding sites. p38α is a serine/threonine protein kinase and a member of the mitogen-activated protein (MAP) kinase family. Four isoforms of p38 are known (α, β, γ, δ), which display different expression and activation patterns. They are all part of signalling pathways that transduce extracellular signals to trigger intracellular responses [4]. The α-isoform is the most intensely investigated subtype and is well known for its role in inflammatory processes; upon stress-induced activation, p38α upregulates several downstream kinases and transcription factors and upregulates the production of pro-inflammatory cytokines such as TNFα, IL1-β and IL-6 [5].
The structures of different activation states of p38α have been extensively investigated (for a landmark publication, see Wang et al. [6]). To date, well over 150 crystal structures of p38α have been deposited in the Protein Data Bank (PDB). By far the majority of p38α inhibitors studied thus far bind either at or proximal to the ATP binding site of the kinase; however, other biologically relevant binding pockets have been reported. Diskin et al. [7] found that the detergent n-octyl-β-glucopyranoside as well as arachidonic acid and derivatives thereof bound to a lipid-binding site within the MAP kinase insert region at the C-terminus of p38α. Following this observation, Jefferson et al. [8] and Hajduk et al. [9] identified the first fragments which bound to this site. Their results will be compared with our findings.
Results and discussion
Description of the fragment library
The current BioFocus fragment library consists of a mixture of commercially available and proprietary compounds whereby roughly 80 percent are commercial. It has been generated to be of high quality in terms of purity, solubility, and diversity. To ensure the best possible starting points for medicinal chemistry projects, the following criteria were applied for fragment selection: the library was generated for broad applicability with no bias applied towards any particular protein class or route of administration. The Rule-of-Three [10] was used as guidance and fragment candidates were further filtered in silico to remove molecules with unwanted reactive functionalities.
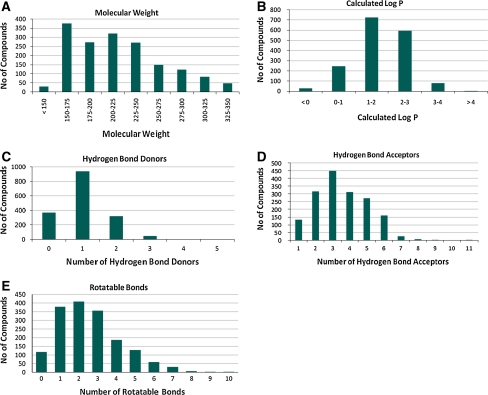
All fragments were assessed for chemical purity with a minimal requirement of 90% as determined by standard analytical methods (HPLC). The aqueous solubility of these fragments was also tested, with a minimum solubility of either 750 μM (LC/MS kinetic solubility determination) or 1 mM (turbidity analysis). This filter process ensured that high quality, soluble fragments would form the basis for binding experiments and minimize false positive outcomes or interference in SPR screens and allow a high success rate in crystallisation experiments. An overview of the StarDrop™ [11] calculated physicochemical properties of this library is illustrated in Fig. 1.
Fig. 1.
Calculated physiochemical properties of the BioFocus fragment library: (a) molecular weight, (b) calculated logP, (c) number hydrogen bond donors, (d) number of hydrogen bond acceptors, (e) number of rotatable bonds
The average molecular weight of the library is 198 Da and compares favourably with other libraries that display average molecular weights in the range of 180–250 Da (analysis of commercially available libraries carried out internally). Overall, the profile of calculated properties was similar to that of many fragment libraries [12], although there was a high proportion of compounds with H-bond acceptors greater than three (the average number of H-bond acceptors of other commercial libraries was two). In our view, a higher number of H-bond acceptors is not necessarily unfavourable as such compounds offer several directed points of contacts.
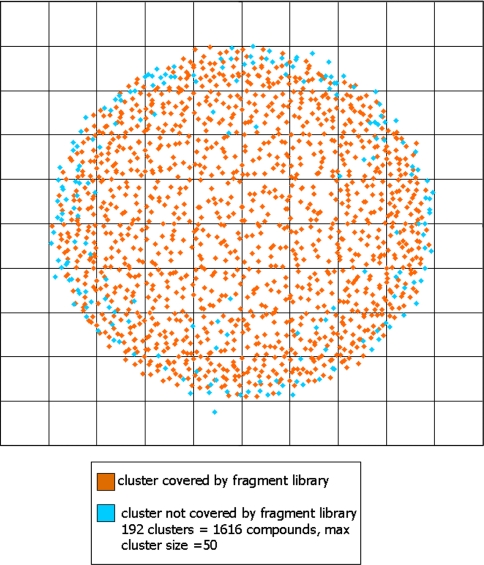
Assessing the coverage of chemical space and, in essence, the applicability of the library is not simple. Quantifying the diversity of any library using computational algorithms can be performed. Such predictions involve a calculation of molecular fingerprints of the library members followed by generation of a matrix of similarities which can be compressed and represented as a plot of diversity. These approaches give an indication of the internal diversity of the library and reveal chemical space that is over or under represented. Generating a true assessment of diversity is challenging: to perform such an analysis, a framework or canvas of chemical space is required upon which the library must be superimposed. We used the ChEMBL database [13], from which we extracted all compounds with a binding affinity of less than 1 μM to their respective biological targets. These compounds were considered as ‘drug-like’ and formed the test set on which the canvas of chemical space was based. In total this set comprised ~190,000 compounds from which we evaluated the coverage of our fragment library as substructures. The internal diversity was assessed using protocols built in Pipeline Pilot [14] and in-house tools. Initially, the FCFP-6 molecular fingerprints were calculated before the set was condensed into 1,500 clusters. A clustering step was required to reduce the intense computational resource that would be necessary to perform a diversity calculation on the entire test set. Using the compound most central as the clusters’ representative in chemical space, a cluster based plot of molecular diversity was then generated. Each fragment was searched for as an exact substructure present within the fully elaborated members of the test set. As a result, 52% of the BioFocus fragment library existed as a substructure of the test set and these results were transferred to the plot giving a visual representation of the diversity (Fig. 2). This exercise was not used to generate a library consisting solely of fragments that exist in known active compounds, but rather to give an overview. In our view, a fragment library should have at most around 50% representation as this ensures sufficient novel starting points for further development.
Fig. 2.
Plot of BioFocus fragment library diversity coverage showing 20,000 clusters compared with clusters representing the ChEMBL database
These results indicated that our library covered the clusters well, with 1,308 of the 1,500 clusters represented, translating into only 1,616 compounds from the 190,000 test set that lacked representation. Using ChEMBL as the test set also gave us a top level indication of the likely areas of success for our library. The hierarchical structure of ChEMBL allowed us to extract the associated protein classes and protein names for the test set. Such information meant that every substructure hit could be traced back to a protein and protein class. Whilst we appreciate that in reality this may not translate to true fragment binding, we found the results of this analysis extremely useful in assessing the evolution of our fragment library.
Description of conducted screens
Subsets of the BioFocus fragment library were selected for screening in various biophysical and biochemical assay formats. This selection process was driven by structural diversity with no particular bias toward partial structures of known kinase inhibitors.
Surface plasmon resonance screen
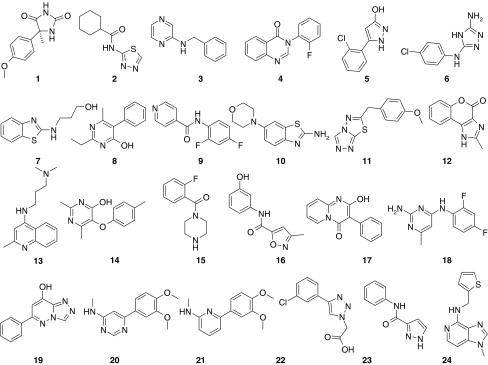
A selection of 266 fragments from the BioFocus fragment library was screened at 200 μM concentration for binding to active p38α using SPR technology (Biacore™ T100) with MKK6 and CA as specificity controls. Hits against p38α were selected on the basis of the following criteria: a binding response greater than 50% of TRmax (102 of the 266 compounds or 38%), specificity for p38α compared to the reference surfaces (binding responses equivalent to less than 50% of TRmax against CA or MKK6) and sensogram shapes consistent with fragment binding (rapid on and off rates). Based on these criteria, 24 confirmed hit fragments were selected. The binding affinities of these fragments were determined by SPR to range from 0.2 to >10 mM (data in Table 1, structures in Fig. 3). The estimated binding stoichiometry of these compounds varied from approximately 1:1 binding to superstoichiometric binding (up to 6:1). Importantly, these ratios are only approximations due to variations in effects on refractive index amongst compounds.
Table 1.
SPR results for the 24 confirmed selective fragment hits (structures in Fig. 3)
| Compound | Mw (Da) | K d (mM) | % TRmax |
|---|---|---|---|
| 1 | 220.2 | 1.80 | 122 |
| 2 | 211.3 | 3.87 | 137 |
| 3 | 185.2 | 1.27 | 221 |
| 4 | 240.0 | 0.78 | 216 |
| 5 | 194.6 | 1.59 | 365 |
| 6 | 210.0 | 1.38 | 434 |
| 7 | 208.3 | >10 | N/A |
| 8 | 214.3 | 2.21 | 333 |
| 9 | 234.0 | 1.97 | 232 |
| 10 | 235.3 | 1.84 | 147 |
| 11 | 246.3 | >10 | 0 |
| 12 | 200.0 | 4.89 | 493 |
| 13 | 243.4 | 2.45 | 312 |
| 14 | 230.0 | >10 | N/A |
| 15 | 208.2 | 0.22 | 198 |
| 16 | 218.2 | 0.79 | 126 |
| 17 | 238.0 | 7.13 | 624 |
| 18 | 236.2 | 1.54 | 255 |
| 19 | 212.2 | 2.34 | 447 |
| 20 | 245.3 | 3.19 | 288 |
| 21 | 244.3 | 0.48 | 125 |
| 22 | 237.6 | >10 | N/A |
| 23 | 187.2 | >10 | N/A |
| 24 | 244.3 | 1.9 | 170 |
Fig. 3.
Structures fragment hits described in Table 1
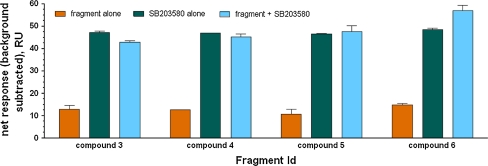
In order to gain insights about the binding mode, we used SPR to conduct a competitive study of the 24 confirmed hits and SB203580, a known ATP site binding inhibitor. Compounds were tested in the presence and absence of SB203580 at a saturating concentration in order to determine whether there is an additive binding response, which would indicate non-competitive binding; i.e. binding at an alternative site remote from the ATP binding site. Under these conditions, compounds that bind at the ATP binding site are expected to compete with SB203580 giving rise to non-additive responses. Eight of the 24 compounds examined were determined to compete with SB203580 for the ATP binding site, four compounds were non-competitive and the mode of binding of the remainder could not be determined conclusively due to poor affinities or discrepancies between independent experiments. Figure 4 shows the data for four compounds which were subsequently analyzed by X-ray crystallography (see below), indicating a non-ATP competitive binding mode for compound 6 and competitive binding by the other three compounds.
Fig. 4.
SPR analysis of binding competition between SB203580 and the four fragments analyzed by X-ray crystallography
Mobility shift assay
To assess the biochemical activity of the fragments, a larger portion (890 compounds) of the BioFocus fragment library was screened in a mobility shift assay format using the LC3000 (Caliper) [15] at 1 mM and 250 μM concentrations. The hit rate was expected to be lower compared to the SPR analysis as only compounds blocking functionally relevant sites can be detected under these conditions. In this screen, the assay quality was high, with Z’ values for all test plates above 0.5.
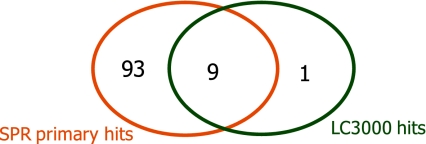
Out of 890 compounds, 39 demonstrated >50% inhibition at 1 mM and >30% inhibition at 250 μM concentration. All of the 39 fragments that exhibited activity in the biochemical assay have also been detected in the initial SPR screen (1 mM), while SPR yielded additional hits that were not identified in the biochemical screen. In order to compare the results of the two technologies, we focused on the 266 fragments which were selected for the earlier screen by SPR. This set included 10 out of the 39 hits identified in the LC3000 MSA screen and 102 fragments that fulfilled the criterion for a primary hit in SPR. The overlap of hits obtained by the two orthogonal technologies is shown in Fig. 5. Nine out of 10 LC3000 fragment hits were also identified in the binding assay.
Fig. 5.
Comparison of hits identified in the SPR and LC3000 MSA screens. Two hundred and sixty-six fragments were tested in SPR at 200 μM and in the LC3000 MSA screen at 250 μM and 1 mM. Of those fragments, 102 were primary hits from SPR with TRmax > 50%, 10 were (confirmed) hits in LC3000 (<50% Ar at 1 mM and <70% Ar at 250 μM)
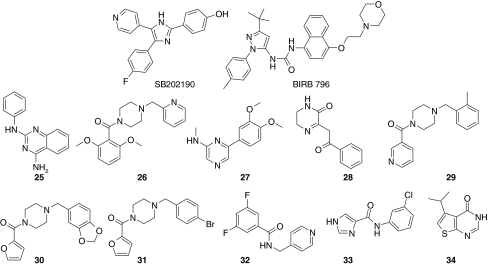
From the top 24 compounds which were selected based on affinity, binding stoichiometry and competition determinations in the SPR screen (Table 2), 11 did not show significant activity at 1 mM concentration in the biochemical assay, whereas 13 showed at least 50% inhibition.
Table 2.
Activities for 14 fragments and two known control compounds obtained in the LC3000 MSA screen: percent inhibition (PIN) at 1 and 0.25 mM concentrations (primary screen), IC50 and pIC50 values (dose response experiment), and ligand efficiency (LE) [3]
| Compound | PIN (1 mM) [%] | PIN (0.25 mM) [%] | IC50 (μM) | pIC50 | LE |
|---|---|---|---|---|---|
| SB 202190 | 0.014 | 7.86 | 0.31 | ||
| BIRB 796 | 0.028 | 7.55 | 0.19 | ||
| 3 | 55.4 | 28.5 | 778 | 3.11 | 0.22 |
| 4 | 67.6 | 50.0 | 411 | 3.39 | 0.19 |
| 5 | 19.4 | 10.9 | 2,600 | 2.58 | 0.20 |
| 6 | 7.9 | −0.8 | 3,250 | 2.49 | 0.18 |
| 25 | 90.0 | 81.3 | 65 | 4.18 | 0.23 |
| 26 | 89.1 | 71.9 | 150 | 3.82 | 0.15 |
| 27 | 62.3 | 41.8 | 224 | 3.65 | 0.20 |
| 28 | 92.3 | 79.2 | 263 | 3.58 | 0.22 |
| 29 | 67.0 | 40.5 | 555 | 3.26 | 0.15 |
| 30 | 69.4 | 42.1 | 655 | 3.18 | 0.14 |
| 31 | 76.1 | 69.6 | 686 | 3.16 | 0.15 |
| 32 | 57.3 | 33.2 | 766 | 3.12 | 0.17 |
| 33 | 57.6 | 33.4 | 955 | 3.02 | 0.20 |
| 34 | 58.9 | 36.0 | 1,536 | 2.81 | 0.22 |
Fluorescence lifetime assay
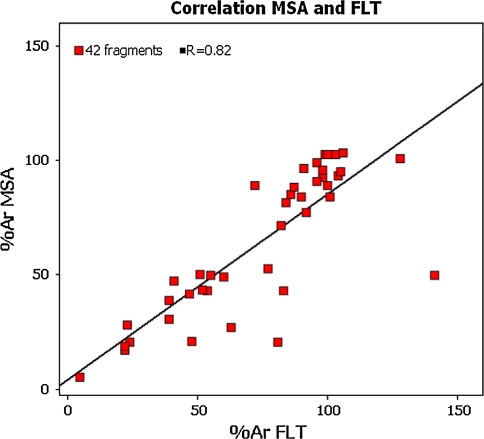
Fluorescence Lifetime Technology was employed as an additional method to assess biochemical activity of compound fragments. A p38α FLT assay was developed using the FLEXYTE™ technology from Almac utilising a 9-aminoacridine labelled substrate tailored for p38α [16, 17]. A subset of 42 fragments was assayed in this format at 250 μM concentration. The test set included the 24 confirmed hit fragments identified in the SPR screen (Table 1); in addition, we tested fragments that showed activity in the LC3000 MSA assay. As illustrated in Fig. 6, the FLT activity data was compared to the screening data obtained from the mobility shift assay (LC3000).
Fig. 6.
Comparison of activities obtained for 42 fragments tested in the LC3000 MSA and FLT assays. Fragment concentration was 250 μM in both assays. ATP concentration was 10 μM in the MSA and 150 μM in FLT. % Ar are relative activities with regards to no enzyme (0%) and full reaction (100%) control raw data
For FLT, a weak cut-off criterion of 30% inhibition (70% residual activity, % Ar) was chosen for hit definition due to the higher ATP concentration applied in the assay.
A reasonable overall correlation between Caliper LC3000 MSA and FLT screening data was observed for the 42 compound fragments that were tested in both assay formats. Of the 42 fragments, 20 were confirmed hits in the LC3000 MSA testing (i.e. part of the hit list of 39 fragments). Sixteen of those were confirmed by FLT. Only one fragment of the 24 SPR confirmed hit fragments was confirmed in FLT and MSA biochemical assays with around 49% inhibition in both biochemical assays.
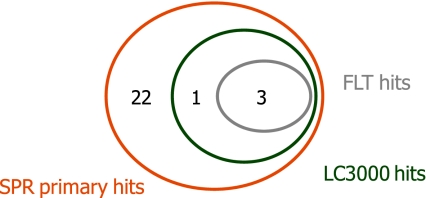
Figure 7 summarizes the overlap of hit fragments for all three technologies. For this comparison, we considered the 26 fragments that were assessed in all three assays. Due to the selection process, all of those were primary hits in the SPR assay and included the 24 confirmed hits selected based on selectivity, binding affinity and stoichiometry analysis. Four fragments were included in the 39 hit fragments identified in the LC3000 MSA screen, three of which were also confirmed in the FLT assay.
Fig. 7.
Comparison of the three screen technologies. Twenty-six compounds tested in all technologies were considered. Due to the selection process followed, all were primary hits in the SPR screening. Hit criterion SPR: TRmax > 50% (includes the 24 confirmed hits), Hit criteria LC3000 MSA: > 50% inhibition at 1 mM and >30% inhibition at 250 μM, hit criterion FLT: > 30% inhibition at 250 μM
Potency determination in both biochemical assay formats
In a next step, ten fragments inhibiting p38α activity in the MSA and the four fragments selected for X-ray crystallography were further tested in concentration response analysis experiment in the MSA. A good correlation of activities in the primary screen and potency testing was observed as shown in Table 2 (Fig. 8).
Fig. 8.
Structures fragments described in Table 2
For all 14 compounds tested in this experiment, IC50 values could be determined. The weakest activity was observed for the non ATP-site binding compound 6, for which an IC50 of about 3 mM was observed. The sensitivity of the assay was confirmed by testing serial dilutions of reference compounds SB202190 and BIRB796 in parallel.
Microscale thermophoresis, nanotemper
Three fragments that were identified in the SPR based fragment screen and one inactive negative control compound within the same chemical series were selected for analysis by microscale thermophoresis. The activities of the fragments determined by different biophysical and biochemical assay technologies correlated well and are summarized in Table 3.
Table 3.
Comparison of assay results for compounds 4–6
| Compound | Binding site | SPR K d (mM) | MSA IC50 (mM) | FLT IC50 (mM) | MST K d (mM) |
|---|---|---|---|---|---|
| 4 | ATP | 0.78 | 0.41 | 0.52 | 0.89 |
| 5 | ATP | 1.59 | 2.6 | n.a.b | 1.15 |
| 6 | Distal site | 1.38 | 3.24 | n.a. | >0.3a |
| Neg. control | – | TRmax 25.4% (0.2 mM) | 2.2% inh. (1 mM) | Not tested | >2 |
aConcentration dependent binding for compound 6 has been detected by MST, but the K d could not be exactly determined due to a high Hill slope; an estimated K d for 6 is within the range of those determined for compounds 4 and 5
bDose response curves were started at 1.5 mM maximal concentration and IC50 values are not applicable
Using microscale thermophoresis, binding to p38α was detected for compounds 4, 5 and 6. While the binding curves for compounds 4 and 6 had positive amplitudes, the curve for compound 5 surprisingly displayed a negative amplitude. The relevance of the inverse amplitude of the binding curve observed for 5 in the MST assay is not yet understood, but may reveal new information regarding the molecular interaction between the compound and protein that could not be obtained by the use of alternative technologies.
Due to solubility limits, the fragments could not be tested at concentrations above 2 mM and thus saturation binding could not be determined. Nevertheless, for compounds 4 and 5 K d values of 893 and 1,152 μM were estimated based on fitted sigmoidal binding curves. No significant interaction with p38α was observed for the inactive control compound. Table 3 summarizes the IC50 values obtained in the LC3000 MSA and FLT assays as well as the K d values obtained in the SPR and MST assays for four fragments.
X-ray analysis of selected hits
Selection of hits for crystallization
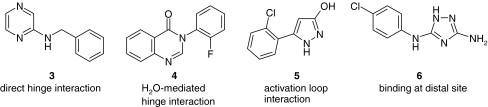
Of the hits identified by SPR, 12 molecules were taken forward into crystallization/soaking trials and crystal structures were obtained for the four compounds shown in Fig. 9 (for experimental details, see Supplementary materials and references [18–25]).
Fig. 9.
Structures of compounds (3–6) analyzed by X ray crystallography with binding mode
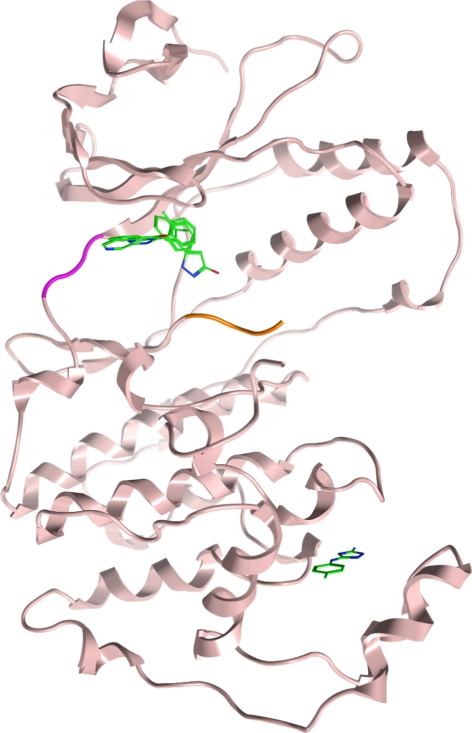
As illustrated in Fig. 10, three of these compounds were shown to bind within the ATP-binding site (compounds 3–5), whereas one of the compounds (6) unexpectedly occupied a distal, potentially allosteric site within the C-terminal MAP kinase insert region. This binding site has recently been described as a lipid binding pocket [7–9].
Fig. 10.
Overlay of fragments binding to p38α; hinge (magenta) and activation loop (orange). Compounds 3, 4 and 5 bound to the active site and compound 6 bound to a distal site
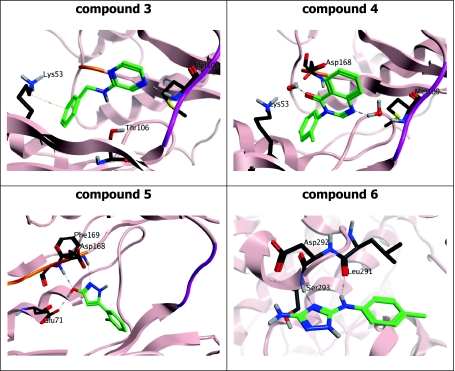
The structure and exact binding mode of these compounds is shown in Fig. 11. The benzylaminopyrazine (3) undergoes a direct interaction with the hinge element of p38α: a hydrogen bond is observed from N(1) of the pyrazine ring of the fragment to the backbone of Met109 on the hinge (residues 107–109). A lipophilic contribution between the side chain of Lys53 and the phenyl ring stabilizes the rear of the fragment. The O···N distance between the side chain of gatekeeper Thr106 and the aniline linker is measured at 3.6 Å, providing a key hydrogen bond interaction.
Fig. 11.
Binding mode of compounds 3–6 in complex with p38α
In contrast, the bicyclic pyrimidinone (4) displays an indirect, water-mediated interaction to Met109 and Lys53/Asp168 at the rear of the pocket. Val38 provides a lipophilic interaction to the plane of the bicyclic core. The hydroxypyrazole (5) does not interact with the hinge region of the kinase. Here, the electron density maps show clear features of ligand binding near the DFG motif of the activation loop (residues 168–170). The hydroxyl group of compound 5 serves as an H-bond donor to Glu71 and acceptor from Phe169, whereas the pyrazole nitrogen interacts with the backbone amide group of Asp168. There is a π-cation interaction between the chloro-phenyl ring and the side chain of Lys53 (the invariant lysine). Notably, additional peaks in the electron density between this binding site and the hinge region of the kinase indicate additional fragment binding in two distinct parts of the pocket, possibly due to disorder and low ligand occupancy as the electron density is not well defined and the additional sites could not be included in the model. This observation is consistent with the SPR analysis that indicates super-stoichiometric binding for this fragment.
Interestingly, the aminotriazole 6 binds in a pocket within the C-lobe formed by an opening of the helix-loop-helix motif comprising residues Ala244 to Ser261 and induced a translation of the Trp197 side chain. Crystal structures of other small molecules binding to this pocket have recently been published [7–9]. In contrast to the previously reported structures, the aminotriazole 6 induces a unique conformation of Trp197 which modifies the protein surface within this region. The fragment undergoes key interactions with the protein, including a hydrogen bond with the backbone of Ser293 and another with Leu291. Aromatic ring binding is driven by a lipophilic interaction with the Cβ atom on the side chain of Glu192.
Initial ligand optimization
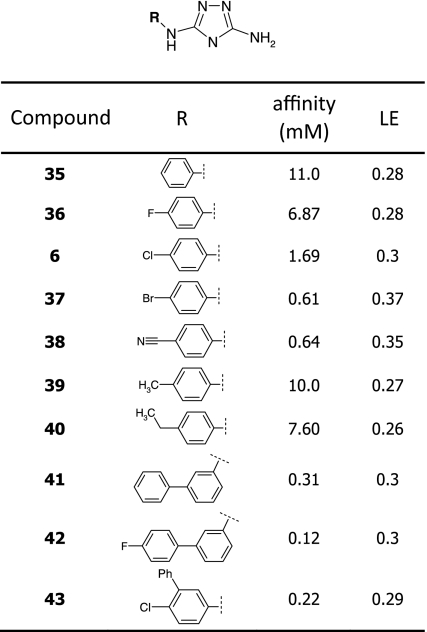
Whereas many p38α inhibitors that bind proximal to the ATP-binding site have been described, only a few small molecules are known that interact within the C-lobe of the MAP kinase insert region. Importantly, the 4-chlorophenyl triazole (6) displayed a slightly different binding mode compared to the known ligands as it induced a conformational change of Trp197 in a different manner than has been observed previously. A ligand with improved affinity to this lipophilic pocket may interfere more specifically with the biological function of p38α. For these reasons, we commenced an optimization process aiming at improved affinity of analogs of compound 6.
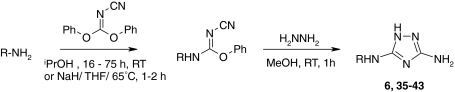
The triazole hit 6 and close analogs were prepared according to a method described by Webb and Labaw [26] (Scheme 1). Conversion of the appropriate aniline with diphenyl cyanocarbonimidate gave the cyano intermediates, which upon treatment with hydrazine cyclised to the corresponding diamino-triazoles (6, 35–43). All final compounds were purified either by crystallization, column chromatography or reverse-phase HPLC and showed purity levels in excess of 95% as determined by analytical HPLC and H-NMR spectroscopy.
Scheme 1.
Synthesis of triazole analogs. R is defined in Table 4
Triazole 6 demonstrated a binding affinity of about 1.4 mM and a ligand efficiency of 0.34 as determined by SPR. As illustrated in Fig. 11, one of the triazole ring nitrogens and the NH group provide an ideal hydrogen bond donor/acceptor motif, which locks the compound into its binding position, whereas the chlorophenyl group is embedded into a small hydrophobic pocket. The unsubstituted amino group appeared to point toward the solvent. In a first approach, we started exploring whether the ligand/protein interaction can be improved within the hydrophobic pocket. Hence, we investigated analogs with differing substitution patterns at the phenyl moiety as given in Table 4. Reducing the size of the C(4) substituent led to diminished affinity whilst replacement of the chloro- by the larger bromo-substituent resulted in an improvement (612 μM). Nitrile 38 displayed similar affinity (635 μM) whereas larger groups at C(4) such as an ethyl group (40) were not well tolerated.
Table 4.
Structures and binding affinities of a first generation of triazole analogs
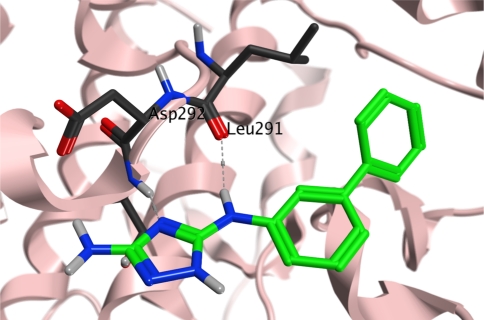
In a second approach we investigated meta-substituted biphenyl triazoles. We hypothesized that this additional phenyl group may directly interact with Trp197. Indeed, biphenyl 41 displayed a threefold improved binding affinity. The 4-fluorobiphenyl 42 and the disubstituted 4-chloro-3-phenyl (43) analogs interacted with similar affinities (0.12 and 0.22 mM, respectively). To confirm the binding mode of these analogs, we solved a crystal structure of p38α in complex with biphenyl 41. While this compound still bound to the same lipophilic pocket at the C-terminus of the kinase, as shown in Fig. 12, an additional weak peak in the electron density map was observed at the hinge region indicating that the compound may bind simultaneously to both sites (not observed with initial hit 6). The overall ligand occupancy at the hinge remains significantly lower than at the allosteric site. Consistent with this finding, super-stoichiometric binding of this fragment has been observed in the SPR experiments.
Fig. 12.
Crystal structure of biphenyl 41 bound to the allosteric pocket
Relative to the position of the initial ligand (6), the triazole ring of the biphenyl compound 41 is shifted towards Asp294: the ring nitrogen and the bridging NH function of 6 still provide an H-bond donor/acceptor motif and interact with the protein backbone, whereas the second phenyl moiety roughly adopts the position of the chlorine atom in 6. Binding of the biaryl ring is driven by lipophilic interaction in the form of two hydrophobic clamps between each ring that feature pairs Ile250/Glu192 and Pro191/Ile259. Trp197 adopts the same conformation as in the p38α/6 complex and alters the protein surface in this area.
Conclusions
A brief description of the BioFocus fragment library is given, which includes key physicochemical properties as well as a novel measure of diversity relative to an arbitrarily defined ‘drug-like molecular space’. A subset of the BioFocus fragment library has been screened for binding affinity and potency for p38α kinase using several assay platforms.
The biochemical assay formats employed in this case study were chosen as detection methods that are relatively insensitive to compound interference [27] while SPR was used as a well-established biophysical method for examining binding interactions. Overall, a good correlation of the data from the different assays was found. As expected, the hit rate observed by SPR was higher than in the biochemical assays; with one exception, all active compounds identified by the biochemical methods were detected by SPR. In addition, SPR identified fragment hits that were not detected in the biochemical screens. This discrepancy is likely due to binding of the compounds at sites that have no influence on the catalytic activity of the enzyme or non-specific binding at the protein surface. The affinity obtained for few compounds that were tested in microscale thermophoresis correlated well with the other technologies, indicating that this may be an attractive alternative biophysical method for fragment testing. However, further validation studies will be critical to understand the positive and negative amplitudes of the individual ligands.
Although the four hit fragments that were followed up further by X-ray crystallographic analysis would not have been selected with the LC3000 MSA assay alone, their inhibitory activity on p38α was confirmed in a concentration–response experiment when higher concentrations of the fragments were used, underlining the value of both technologies for fragment-based drug discovery. Neither of the two biochemical assays appeared to be superior to the other. Similar conclusions were drawn by Böttcher et al. [28] who compared NMR- and SPR-based assay formats with activity assays using fluorescence intensity, fluorescence lifetime or MSA readouts. Purely biophysical techniques such as SPR have the advantage of being able to identify unique low affinity fragment hits, as exemplified in the current study, representing potential starting points for the development of compounds with measurable biochemical inhibition of the target. They could also serve as ‘diagnostic tools’ to identify novel binding sites with yet unknown function.
Crystal structures were obtained for four hit fragments, all of which adopted unique binding modes. Three molecules bound at the ATP binding site; compound 5 bound close to the activation loop, while compounds 3 and 4 underwent either a direct or a water-mediated interaction with the hinge element. In contrast, triazole 6 was found to bind at a distal lipid binding pocket and induced a novel conformation of the Trp197 residue. With a first set of analogs, a structure-affinity relationship within this chemical series has been established and the binding affinity was improved by about tenfold. At this stage, it remains unclear whether the inhibitory activity of 6 observed in the biochemical mobility shift assay is indeed due to a truly allosteric effect or if at high concentrations and in solution, this compound simultaneously binds and blocks the ATP binding site. Crystallographic analysis confirmed the interaction of biphenyl analog 41 at the same distal binding site.
Further optimization of the binding affinity will be required to achieve molecular tools that will allow an interrogation of the biological function of this distal binding site. Overall this study illustrates the power of combining a range of biophysical and biochemical techniques to the discovery of fragments that facilitate the development of novel modulators of other kinase and non-kinase drug targets.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to acknowledge the contributions of Marieke Lamers (X-ray crystallography) and Richard Martin (data handling), Almac for providing the FLEXYTE™ peptide as well as support for FLT assay development, Markus Sanner, Céline Klein and Virginie Vögtlin for excellent technical assistance and Angus MacLeod for helpful suggestions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- FLT
Fluorescence lifetime
- SPR
Surface plasmon resonance
- MSA
Mobility shift assay
- MST
Microscale thermophoresis
- PIN
Percent inhibition
- CA
Carbonic anhydrase
References
- 1.Hesterkamp T, Whittaker M. Curr Opin Chem Biol. 2008;12:260–268. doi: 10.1016/j.cbpa.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Congreve M, Chessari G, Tisi D, Woodhead AJ. J Med Chem. 2008;51:3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- 3.Hopkins AL, Groom CR, Alex A. Drug Discov Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Gibson TB, Robinson F, Silvestro L, Pearson G, Xu B, Wright A, Vanderbilt C, Cobb MH. Chem Rev. 2001;101:2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 5.Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW, Strickler JE, McLaughlin MM, Siemens IR, Fisher SM, Livi GP, White JR, Adams JL, Young PR. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Harkins PC, Ulevitch RJ, Han J, Cobb MH, Goldsmith EJ. Proc Natl Acad Sci USA. 1997;84:2327–2332. doi: 10.1073/pnas.94.6.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diskin R, Engelberg D, Livnah O. J Mol Biol. 2008;375:70–79. doi: 10.1016/j.jmb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Jefferson J, Perry P, Harris RM, Moiani D, Olson AJ, Tainer JA. J Mol Biol. 2009;391:1–11. doi: 10.1016/j.jmb.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comess K, Sun C, Abad-Zapatero C, Goedken ER, Gum R, Borhani DW, Argiriadi M, Groebe D, Jia Y, Clampit J, Haasch D, Smith H, Wang S, Song D, Coen M, Cloutier T, Tang H, Cheng X, Quinn C, Liu B, Xin Z, Liu G, Fry E, Stoll V, Ng T, Banach D, Marcotte D, Burns D, Calderwood DJ, Hajduk PJ. ACS Chem Biol. 2011;6:234–244. doi: 10.1021/cb1002619. [DOI] [PubMed] [Google Scholar]
- 10.Congreve M, Carr R, Murray C, Jhoti HH. Drug Discov Today. 2003;8:876–877. doi: 10.1016/S1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 11.StarDrop Version 4.3. Optibrium. http://www.optibrium.com/stardrop
- 12.For a comparison of commcercial libraries, see http://homepage.mac.com/swain/CMC/DDResources/Hit_iden/frag_coll_profiles.html
- 13.ChEMBL_05 release: https://www.ebi.ac.uk/chembldb/index.php. Accessed 7 Apr 2011
- 14.Accelrys pipeline pilot http://accelrys.com/products/pipeline-pilot. Accessed 7 Apr 2011
- 15.Caliper LifeSciences Application note 210 http://www.caliperls.com/assets/009/5902.pdf. Accessed 7 Apr 2011
- 16.Paterson MP, Dusnmore CJ, Hurteaux R, Maltman BA, Cotton GJ, Gray A. Anal Biochem. 2010;402:54–64. doi: 10.1016/j.ab.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Maltman BA, Dunsmore CJ, Couturier SCM, Tirnaveanu AE, Delbederi Z, McMordie RAS, Naredo G, Ramage R, Cotton G. Chem Commun. 2010;46:6929–6931. doi: 10.1039/c0cc01901a. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W (1997) In: Carter CW, Sweet RM (ed) Methods in enzymology, volume 276: macromolecular crystallography, part A. Academic Press, New York, pp. 307–326
- 19.Leslie AGW. Acta Cryst. 2006;D62:48–57. [Google Scholar]
- 20.Evans P. Acta Cryst. 2006;D62:72–82. [Google Scholar]
- 21.Collaborative Computational Project, Number 4 Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Lovell SC, Davis IW, Arendall WB, III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Proteins Struct Funct Genet. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 26.Webb RL, Labaw CS (1982) J Het Chem 19:1205–1206
- 27.Comley J, Drug Discovery World (2003) 2(Summer):91–98
- 28.Böttcher A, Ruedisser S, Erbel P, Vinzenz D, Schiering N, Hassiepen U, Rigollier P, Mayr LM, Woelcke J. J Biomol Screen. 2010;15:1029–1041. doi: 10.1177/1087057110380455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.