Abstract
More than a century ago, it was proposed that mechanical forces could drive tissue formation. However, only recently with the advent of enabling biophysical and molecular technologies are we beginning to understand how individual cells transduce mechanical force into biochemical signals. In turn, this knowledge of mechanotransduction at the cellular level is beginning to clarify the role of mechanics in patterning processes during embryonic development. In this perspective, we will discuss current mechanotransduction paradigms, along with the technologies that have shaped the field of mechanobiology.
Introduction
Why should we study mechanical forces in cell biology? This is a reasonable question given the daunting task of adopting a plethora of theoretical approaches and experimental tools that would be necessary to address the problem. The answer, however, is rather fundamental: a vast body of research has accumulated to illustrate that forces are ubiquitous in vivo and that these forces directly impact cell function. Although the biological effects of forces are perhaps most evident in the context of physical activity - breathing, heart pumping, blood flow, and physical exercise - such forces also regulate morphogenesis, cell migration, and even cell adhesion to extracellular matrix. Importantly, it is now apparent that such forces can regulate a wide variety of biological processes, from cell proliferation and differentiation to tissue mass homeostasis and complex inflammatory cascades.
The idea that forces can regulate tissue remodeling and development was articulated more than a century ago. In 1892, the surgeon and anatomist Julius Wolff postulated that bone tissue adapts its structure to the mechanical environment based on the observation that trabeculae matched the principal stress lines in bones caused by daily physical loading (Wolff, 1892). Although the alignment of trabeculae could have arisen strictly during prenatal development, he reported this remodeling occurred even after healing of misaligned fractures. In the same era, mechanical forces were proposed to shape tissues and organs during embryonic development (Roux, 1895; Thompson, 1917), but the tools were not available to directly test such ideas experimentally. Nearly a century passed before these concepts began to captivate the scientific community once again.
Interestingly, the development of mechanobiology as a field appears to be closely connected to the advent of enabling technologies. For example, the earliest observations suggesting that mechanical forces drives embryogenesis and bone structure were a natural result of newfound microscopy methods. Mechanobiology received relatively little attention for much of the 20th century as scientists focused on developing molecular biology tools to catalogue the genetic basis for life. The recent renaissance in studying mechanics primarily in cell culture has largely been enabled by a suite of tools to measure and manipulate such forces in vitro. For example, in vitro application of strains that would be experienced by bone during physical activity increases cell proliferation (Raab-Cullen et al., 1994), osteogenic differentiation (el Haj et al., 1990; David et al., 2007) and bone matrix deposition (Bancroft et al., 2002), which are all characteristic for mechanically-induced anabolic bone growth in vivo. Mechanical stretch that mimics the effect of pulsating blood flow has been shown to trigger many alterations in endothelial and smooth muscle cell signaling (Chien et al., 1998; Tzima et al., 2005), vascular cell proliferation (Davies et al., 1984; Haudenschild et al., 1985; Sumpio et al., 1987), and expression of inflammatory markers (Yamawaki et al., 2005). Methods to apply shear flow on cell cultures have shown that shifts between steady and turbulent shear flow can prevent or promote inflammatory activation, respectively, and may explain the localization of atherosclerotic plaques to specific regions along the vascular tree (Davies et al., 1986; Shyy et al., 1994). Flow rates during development also can drive arterial versus venous phenotype (le Noble et al., 2004). Bioreactor-type devices have been developed also to model the compression experienced by chondrocytes in the articular joint, tension in muscles, ligaments and tendons, as well as impact forces associated with trauma (Freed et al., 1993; Molnar et al., 1997; Garvin et al., 2003; Frieboes and Gupta, 2009).
In addition to such externally applied forces, non-muscle cells generate contractile forces on their own. This was first illustrated by Harris and Stopak, showing that cells cultured on soft polymer substrates would wrinkle the substrate surface (Harris et al., 1980; Harris et al., 1981). The implication that forces could be ever present, even in settings without an explicit mechanical stimulus, seemed heretical at the time. However, over the past three decades, it has become clear that 1) most eukaryotic cells can generate intracellular forces that act on the surrounding extracellular matrix (ECM) or neighboring cells, and that 2) this contractile activity is critical for a number of biological processes such as cell migration, mitosis, as well as stem cell differentiation and self renewal. For physical functions such as mitosis and migration, force is clearly ‘essential’ in the same way that oxygen is essential for life. That is, without force, mitosis and migration cannot proceed (Civelekoglu-Scholey and Scholey, 2010; Renkawitz and Sixt, 2010). The role of force in genetic responses such as proliferation and differentiation, however, appears to be ‘regulatory’ in the same manner as cytokines might be. Although we have some clues to how forces exert these regulatory functions, clarifying these mechanisms remains a central question for mechanobiology.
Given the dependence of this field on enabling technologies, this perspective will present (1) our current understanding of the role of mechanical forces in cell biology, (2) the techniques that are being developed to enable such studies, and (3) recent efforts to consider mechanical forces in development. Due to space constraints, this perspective is not an exhaustive review of mechanobiology, but rather examines the current thinking and directions in the field. We begin by describing the cellular context in terms of the physical structures through which forces are transmitted.
The mechanotransduction machinery in a cell-ECM unit
Mechanical forces are calculated in engineering devices by measuring deformation of materials, be they membranes under pressure or changes in electrical current in a narrowing metallic strip under stretch (strain gauge). By analogy, knowing which cellular structures bear force is a starting point for understanding where forces are transmitted and potentially transduced into biochemical signals. In discussing how mechanical forces regulate cells, it is essential that we do not consider the cell in isolation, but rather in direct physical contact with the ECM. This is because adhesion of cells to matrix results in the structural organization of the cell itself. Integrin binding and clustering against ECM ligands leads to changes in cell shape and cytoskeletal architecture, anchoring the actin cytoskeleton to sites of adhesion. The cytoskeleton is linked to the nuclear envelope, thus forces experienced or generated by the cell-ECM module (Figure 1) are therefore transmitted and sensed throughout this module as a coordinated system. In this section, we will examine where and how those forces might be transduced to regulate cell function. Along the way, we will describe the tools used to study these mechanotransduction processes.
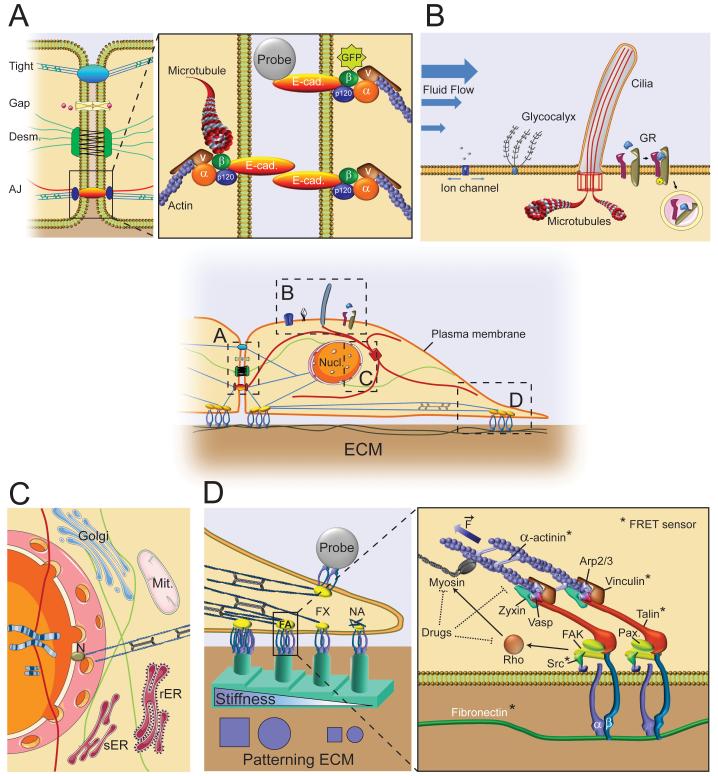
Figure 1. Mechanotransduction in a Cell-ECM unit.
Center: An overview of a cell connected to ECM and a neighboring cell. The boxes (A - D) zoom in on areas of interest where mechanotransduction in the cell-ECM unit occurs. The blue lines represent actomyosin filaments, green lines embody intermediate filaments and the red lines correspond to microtubules. This color code is maintained in all panels. The blue structures linking the cell with the ECM represent integrins (detailed in D). nucl.: nucleus; pm: plasma membrane.
A: Mechanotransduction at Adherens Junctions. (Left panel) Different cell-cell junctions connect neighboring cells. Tight junctions (Tight) constitute of sealing strands of protein complexes (blue ellipse) in the intercellular space that are anchored to the actomyosin skeleton in the cytoplasm. Gap junctions (GAP) are clusters of connexon channels that allow ion exchange (pink circles) and electrochemical communication between two cells. Desmosomes (Desm.) link intermediate filaments through adhesion plaques (green). Adherens junctions (AJ) tether actomyosin skeleton and microtubules via cadherin complexes (orange). (Right panel) Molecular structure of AJ: E-cad: E-Cadherin, α: Alpha Catenin, β: Beta Catenin, p120: p120 Catenin, v: Vinculin, Probe: Atomic Force Microscope, Optical tweezers, and Magnetic tweezers can be used to manipulate and measure force at the AJ. In addition, labeling molecules such as beta catenin with GFP allows measuring of the size of the AJs, which can be used to estimate the force acting on the AJs.
B: Mechanoreceptors at the cell membrane. Fluid flow or stretch deforms the plasma membrane which can lead to activation of ion channels that results in an influx of ions. In addition, fluid flow directly impacts glycocalyx and cilia movement which triggers diverse downstream signaling cascades. Moreover, mechanical forces mediate growth factor receptor (GR) clustering and endocytosis, and thus affect GR signal transduction as well.
C: Mechanotransduction at the nucleus. The nucleus and surrounding organelles (Golgi apparatus, Mit: Mitochondria, rER/sER: rough and smooth endoplasmic reticulum) are interconnected by intermediate filaments and microtubules. Nesprins (N) tether the nucleus with the actomyosin cytoskeleton. Changing cell shape/contractility can alter spatial localization of organelles and induce conformational changes of nuclear pores.
D: Mechanotransduction at the Focal Adhesion (FA). (Left): Integrin clustering develops Nascent Adhesions (NA) that mature to Focal Complexes (FXs) and Focal Adhesions (FA), a process controlled by actomyosin contractility which can be modified by stiffness (step rigidity posts or crosslinked polymer substrates, in green), cell shape (patterning, in blue) or external application of force with atomic force microscopy, optical tweezers, or magnetic tweezers (Probe). (Right): simplified molecular structure of FA. α/β: alpha and beta unit of integrins, Pax: paxillin, F: Force delivered by actomyosin contraction. Clustering of integrins can induce RhoA signaling, thereby increasing myosin contractility which leads to unfolding of proteins as observed by FRET. The molecules for which a FRET sensor has been developed are marked with an asterisk (*). Pharmacological drugs can be used to alter myosin contractility and actin polymerization.
Structural basis of force transmission in cells
In the cytoplasm, the cytoskeleton is a fundamental structure for mediating force transmission (Wang et al., 1993). The cytoskeleton is a highly dynamic cellular scaffolding structure composed of filamentous actin (6nm in diameter), intermediate filaments (10nm) and microtubules (23nm). These three cytoskeletal elements are not single proteins, but consist of many monomers able to span large distances within the cell. Tubulins polymerize to form hollow cylinders known as microtubules and provide a structure for motor proteins such as kinesins and dyneins to travel between different cell compartments (Sheetz, 1996). Vimentin, keratin, and lamin monomers form intermediate filaments that connect the nucleus with the endoplasmic reticulum, mitochondria, and Golgi apparatus, providing structural integrity to the cell. Actin monomers assemble into filamentous actin (F-actin) and together with myosin filaments, form the cytoskeletal contractile apparatus. The actomyosin cytoskeleton connects multiple parts of the cell membrane as well as the cell membrane to the nucleus (Sims et al., 1992). At the cell membrane, these filaments anchor into clusters of proteins that include focal adhesions (FAs) which link the cytoskeleton through transmembrane integrin receptors with the ECM. In the extracellular space, the ECM materializes as a mesh of cross-linked proteins and carbohydrates, and depending on the tissue, can include different constituents including collagen, laminin, elastin and fibronectin fibers interlocked with hyaluronic acid and proteoglycans.
From a mechanical standpoint, applying force to this cell-ECM unit leads to structural deformations and rearrangements of the ECM, force transmission through the FA, and (given the highly interconnected nature of the cytoskeleton) deformation of nearly every aspect of intracellular structure, including the position of mitochondria, endoplasmic reticulum, and the nucleus (Figure 1C). However, “outside-in” force transmission is only half the tale, as cells also generate force. Polymerization and de-polymerization of microtubules drive pushing and pulling forces, respectively, to control the position of mitotic spindles, chromosomes and nuclei (Dogterom et al., 2005). The head domain of myosin II pulls on actin filaments to generate traction forces, which are transmitted to focal adhesions and deforms the ECM via an ‘inside-out’ transmission path (Vicente-Manzanares et al., 2009). Hence, mechanical forces are experienced throughout the cell via the integrated cytoskeleton-focal adhesion-ECM architecture.
Structural basis for mechanotransduction
The integrated nature of this cell-ECM structure raises the possibility that many anatomical sites could be involved in transducing such forces into biochemical signals. Beginning with the ECM itself, it has been shown that forces can induce opening of quaternary domain-domain structures, as well as direct unfolding of unstable domains, such as in fibronectin (Krammer et al., 1999). These events in turn can reveal cryptic sites for engagement and signaling of cellular receptors. Similarly, it has been shown that ECM-bound growth factors such as TGFβ can be activated and released via mechanical forces to trigger cellular signaling (Wipff et al., 2007). Given the complexity of characterizing changes in molecular structure and multi-molecular organization, especially in disordered materials like the ECM, it remains a major challenge to systematically examine the role of forces in modulating the ‘bioactivity’ of ECM.
Intracellular mechanisms of mechanotransduction have been even more difficult to demonstrate. For example, it has been postulated that force-induced nuclear deformations can directly alter genomic structure and accessibility of transcription factors to specific genetic targets, but no direct demonstration of such a mechanism has yet been described. Nonetheless, circumstantial evidence for a nuclear mechanosensor is provide by recent studies showing that lamins and nesprins - scaffolding proteins critical to the integrity of nuclear structure - can impact force transduction (Chancellor et al., 2010; Lammerding et al., 2004). New molecular-resolution imaging approaches (e.g., super-resolution microscopy) that can track chromatin changes following force application may soon be ready to begin to address this impasse. One recent approach to provide an unbiased examination of mechanotransduction involves ‘shotgun’ labeling of cryptic cysteines that are revealed upon mechanical loading. Inspired perhaps by the findings of cryptic site unmasking in fibronectin, Discher and colleagues used this approach to identify intracellular proteins that undergo conformational change when in cells that are mechanically stretched (Johnson et al., 2007). The Sheetz group also screened for stretch-dependent binding of cytoplasmic proteins on Triton X-100-insoluble cytoskeletons (Sawada and Sheetz, 2002) before using more targeted approaches to demonstrate force-induced unfolding of talin and the Src family kinase substrate p130cas (del Rio et al., 2009; Sawada et al., 2006). It remains to be seen to what extent such unfolding events contribute to the many force-induced changes in cellular signaling.
Although cytoskeletal and ECM-based mechanotransduction has been difficult to demonstrate, receptor mediated transduction of forces has been convincingly shown for stretch-activated ion channels (Lansman et al., 1987; Sadoshima et al., 1992) and for integrins (Wang et al., 1993). For stretch-activated receptors, mechanical forces appear to alter the conformation of the Transient Receptor Potential (TRP) family of channels, allowing for rapid signaling responses such as calcium influx (< 4 ms) following mechanical perturbations (Figure 1B) (Matthews et al., 2010). In contrast to this relatively straightforward mechanism, integrins appear to be involved in a complex response to force, perhaps resulting from their central position in dynamically regulating the interactions between the cytoskeleton and the ECM.
ECM-bound integrins cluster to initiate the formation of highly organized protein assemblies composed of multiple protein-specific strata. At the cell membrane, the integrin layer is oriented with the head domains connecting to the ECM and the cytoplasmic tails binding to focal adhesion kinase (FAK) and paxillin. Recently revealed with super-resolution methods, this cytoplasmic layer assembles with a stratum containing talin and vinculin, and an uppermost actin-regulatory sheet consisting of zyxin, VASP (vasodilator stimulating phosphoprotein) and α-actinin which tethers the FA to the actomyosin cytoskeleton (Kanchanawong et al., 2010) (Figure 1D). Nascent adhesions consisting of integrin clusters either turn over rapidly (~60 seconds) or mature into larger Focal Complexes (FX) of which some assemble into larger FAs (Zimerman et al., 2004; Vicente-Manzanares et al., 2008; Choi et al., 2008).
Importantly, the morphogenesis of these adhesions requires force (Parsons et al., 2010). Force appears to unfold several protein domains of talin, paxillin and p130cas exposing cryptic binding and/or phosphorylation sites (del Rio et al., 2009; Sawada et al., 2006; Zaidel-Bar et al., 2007) which leads to stabilization of nascent adhesions into FXs. Further application of force drives the maturation of FXs into FAs, now tethered to thick actin stress fibers. Recent evidence suggests that the integrin receptor itself switches to a high-affinity state in response to force, at least for the ubiquitous α5β1 isoform (Friedland et al., 2009). Conversely, inhibition of myosin II and reducing cytoskeletal tension also leads to disassembly of FAs (Kirfel et al., 2004; Rid et al., 2005). Although it remains largely unclear whether nascent adhesions, FXs, and FAs are actually distinct states or lie on a single continuum, it is apparent that small and large adhesions have distinct molecular signatures. Recent proteomic studies of blebbistatin-treated (myosin inhibited) small adhesions compared to native adhesions display a large number of compositional differences (Byron et al., 2011; Kuo et al., 2011). These early studies will likely provide important handles to begin to explore the nature of these complex force-responsive structures.
Perhaps the most interesting feature of these adhesions is that they not only perform a physical-anchoring function, but also are biochemical signaling centers. FAs contain many signaling proteins such as FAK, ERK, JNK, Src, MEK, Ras, and Raf (Miyamoto et al., 1995), all of them involved in a myriad of pathways regulating diverse cell functions including migration, proliferation, and differentiation. As such, changes to composition of such adhesions mediated by contractility can impact localization of members within a signaling cascade, thereby orchestrating cellular responses to force.
Because adhesion dynamics and related signaling are regulated by mechanical stress at the cell-ECM interface, it appears that one can modulate adhesion signaling through multiple distinct means. That is, manipulating the density of ECM ligand deposited on a substrate impacts adhesion signaling by controlling the amount of integrin ligation and clustering from the outside-in. Adhesion signaling can also be modulated by mechanical stresses at adhesions, for example, by manipulating cellular contractility or the mechanical stiffness of the ECM. Interestingly, functional studies thus far would suggest that these different approaches to impact adhesions lead to indistinguishable consequences. Increasing ECM density, RhoA-mediated myosin activity, or ECM stiffness all suppress cellular apoptosis and enhance proliferation (Chen et al., 1997; Wozniak et al., 2003; Pirone et al., 2006). Although such links would suggest the possibility that focal adhesions use a common mechanism to integrate adhesive and mechanical cues, a clearer molecular mechanism describing the transduction of ligand density, traction force, and substrate mechanics is required to convincingly explain the basis for similarity or uniqueness of each of these cues.
Even with these mechanisms described, it is equally important to not simply describe what cells could transduce, but what cues they do transduce in different physiologic or pathologic settings. Indeed, recent studies have begun to link these fundamental mechanotransduction processes to clinically important settings. For example, the stiffening of arteries associated with atherosclerosis appears to be involved in promoting neointimal smooth muscle hyperproliferation and vascular lumen narrowing (Klein et al., 2009). Similarly, the enhanced tissue stiffening due to the fibrotic response of tumors plays a role in tumor proliferation, progression, and metastasis (Paszek and Weaver, 2004; Levental et al., 2009). In contrast, tissue angiogenesis appears to be enhanced with decreased tissue stiffness and cellular contractility (Ingber et al., 1995). Together, these studies are beginning to illustrate the ubiquitous and critical role of mechanical forces in biological systems.
Mechanotransduction in a multicellular context
In vivo, cells generally are not solitary but are tightly connected to each other via cell-cell junctions (Figure 1A). Adherens junctions (AJ) link the cytoskeletons of adjacent cells via clusters of cadherins, of which the cytoplasmic tails are tethered to cortical actin filaments by protein complexes harboring vinculin, p120, alpha and beta catenin (Yamada et al., 2005). From a mechanical point of view, AJs are well placed to act as mechanosensing complexes for multicellular architectures. Indeed, similar to FAs, AJs grow when an external force is applied (outside-in) or when actomyosin contraction is enhanced (inside-out), leading to strengthening of the AJ (le Duc et al., 2010; Liu et al., 2010; Smutny et al., 2010). Such a positive feedback mechanism may be in place in order to prevent cell-cell ruptures. Part of this feedback mechanism may involve force-induced unfolding of alpha catenin, exposing cryptic binding sites to vinculin, which in turn supports recruitment of more actomyosin fibers (Yonemura et al., 2010). In addition, Rac1 activated by cadherin-catenin clustering (Nakagawa et al., 2001) can recruit p190RhoGAP to p120 catenin, resulting in inhibition of Rho activity (Wildenberg et al., 2006). Hence, the ultimate size of AJs is partially determined by the balance between Rho and Rac signaling at the AJ (Liu et al., 2010).
The presence of a cell-cell pathway for transmitting force has numerous implications. In the context of multicellular sheets, uniform contractile activity of cells can lead to non-uniform distributions of cell-ECM stress that are determined by the geometry of the cell sheet (Nelson et al., 2005). Furthermore, these underlying stress distributions triggered spatially localized patterns of cell proliferation. These patterns of proliferation were disrupted when cells expressed a dominant negative cadherin, suggesting that changes in AJs could guide multicellular patterning. Similarly, geometrically-imposed patterns of intercellular stress direct lineage commitment of mesenchymal stem cells which preferentially differentiate to adipocytes in low tensile zones while regions of higher tension are characterized by alkaline phosphatase positive pre-osteoblasts (Ruiz and Chen, 2008). Recent work suggests that these localized patterns of force are also present in models of 3D morphogenesis (Nelson et al., 2006; Gjorevski and Nelson, 2010).
Thus, these early studies suggest that our insights into multicellular mechanics and mechanotransduction will require a deeper understanding of the mechanical and signaling interdependencies between cell-cell and cell-ECM adhesions. Recent studies have begun to illustrate examples of direct mechanical connections between these two types of adhesions, as well as indirect connections including cadherin engagement-induced activation of RhoGTPases to impact focal adhesions (Maruthamuthu et al., 2011; de Rooij et al., 2005; Nelson et al., 2004; Weber et al., 2011). Taken together, it appears that long range force transmission through AJs can potentially play a distinct role in guiding patterning processes during embryonic development (Martin et al., 2010), a topic we will examine momentarily after discussing some of the techniques that have been developed to measure and to manipulate force in cells.
Approaches to measure and to manipulate cellular forces
Cells are constantly probing, pushing, and pulling on their microenvironment. A growing body of work is investigating what regulates the cellular generation of contractile forces. To identify when such forces are invoked, several approaches have been developed that allow quantitative measurement of cellular traction forces. In addition, numerous methods are being used to manipulate cell-ECM forces, all of which have been critical in enabling the growing field of mechanobiology. In this section we will examine the tools used to measure and manipulate cellular forces, and how they have contributed to our understanding of the role of mechanical events in both physiological and pathological settings.
Approaches to measure cell-generated traction forces
Owing to the evolution of biomaterials and polymer chemistry, the conception of soft substrates linked with ECM matrices has enabled the measurement of forces generated by even single cells. The basic concept of using an elastic substrate for force measurement was originally conceived by Harris et al. -- when adherent to a thin silicone membrane, non-muscle fibroblasts can cause wrinkles in the substrate (Harris et al., 1981). However, due to the inherent non-linearity of wrinkling and the complexity of the displacement field generated by a single cell, this technique was not applicable to accurately quantify cell forces. The development of traction force microscopy by Dembo and Wang has been a significant improvement to measure cellular forces. This method uses fluorescent microbeads embedded in a polyacrylamide hydrogel as markers for tracking the deformation of the gel caused by the adherent cell. After obtaining the displacement vector for every bead, the inverse problem is solved to calculate the cell generated force field (Dembo and Wang, 1999; Butler et al., 2002; Yang et al., 2006). With this technique, several groups have demonstrated that in addition to tangential forces, cells also exert forces normal to the 2D planar adherent surface (Hur et al., 2009; Delanoe-Ayari et al., 2010; Maskarinec et al., 2009). However, the calculation of forces is computationally intensive, because deformations propagate on these continuous substrates. Furthermore, changing the cross-linking chemistry to manipulate the rigidity of hydrogels may inadvertently affect surface hydration, chemistry, and adhesiveness. To address these limitations in the design of soft substrates, micro-fabricated arrays of elastomeric cantilever posts have been developed to quantify cell forces by measuring the deflection of the posts under cell tension (Tan et al., 2003; du Roure et al., 2005).
These two force measurement techniques have demonstrated that (1) cells pull harder on stiffer surfaces, affecting cell shape, motility, growth rate, and intracellular signaling and that (2) adherent cells continuously apply tensile forces to substrates that are directed towards the centroid of the cell (Tan et al., 2003; Dembo and Wang, 1999; Balaban et al., 2001). Interestingly, similar observations have recently been reported in cells embedded in a 3D hydrogel, suggesting that mechanotransduction mechanisms might be conserved between 2D and 3D (Legant et al., 2010).
Ultimately, these methods were critical in identifying a role for cellular forces in several important settings. For example, it had been observed that cells restricted from spreading against extracellular matrix become growth arrested, regardless of whether that restriction is due to cell crowding upon reaching confluence (Nelson and Chen, 2002), decreased ECM ligand coating density on a surface (Ingber and Folkman, 1989), or micropatterning to define the area of spreading of a cell (Chen et al., 1997). These changes in cell spreading were later found to impact cell contractility. Restricting cell spreading on micropost arrays revealed that decreased spreading prevented cells from generating traction forces (Tan et al., 2003). Conversely, upregulating contractility by activating RhoA rescued proliferation even in unspread cells (Pirone et al., 2006), thus demonstrating that the mechanism by which cell shape regulates contractility is a mechanotransduction event. One could consider these contractile forces as being part of an ‘autocrine’ loop.
Approaches to manipulate cell-ECM forces
Intracellular forces can be modulated by using traditional molecular methods to directly target the force-generating apparatus. Several pharmacologic agents are available for inhibiting modulators of contractility, including the molecular motor myosin II (blebbistatin), the upstream regulators of myosin phosphorylation Myosin Light Chain Kinase (ML-6, ML-9, and the Rho/ROCK signaling pathway (fasudil, Y27639, C3 botulinum exotoxin), as well as the polymerization processes of actin (latrunculin, cytochalasin D). Similarly, molecular-genetic methods have also been used effectively to target these pathways, including the non-muscle myosin II isoforms themselves, and have been a mainstay in identifying force in a regulatory pathway (Figure 1D).
In addition to molecular methods, recent attention has turned toward the use of biophysical approaches to manipulate cell-ECM mechanics. One such method is to vary the mechanical stiffness of the substrate to which cells attach. Most commonly, stiffness is manipulated by changing the degree of cross-linking of polymeric hydrogels, the most widely available of which is polyacrylamide. Depending on the chemistry, cross-linking is controlled by the ratio of polymer to cross-linking agents, the duration of exposure to a light source (photopolymerization) or heat, among other factors. Interestingly, whereas the stiffness of most substrates is determined upfront, substrate stiffness of collagen can be altered during the course of the experiment by supplementing ribose to the medium (Girton et al., 1999).
Recent studies have used these methods to reveal a role for ECM stiffness in regulating cell function. ECM stiffness has been shown to affect migration, proliferation and differentiation (Peyton and Putnam, 2005; Lo et al., 2000; Yeung et al., 2005). Whereas increasing stiffness enhances spreading and proliferation of many cell types (Pelham, Jr. and Wang, 1997; Yeung et al., 2005; Thompson et al., 2005) and facilitates tumor growth (Wozniak et al., 2003; Georges and Janmey, 2005; Levental et al., 2009), compliant substrates appear to promote branching of neurons (Flanagan et al., 2002; Willits and Skornia, 2004) or adhesion of and albumin secretion by hepatocytes (Chen et al., 2009; Semler et al., 2005). The mechanism by which cells transduce alterations in substrate stiffness is not fully understood, but it has been shown that cells attached to more compliant substrates exhibit suppressed integrin activation, decreased traction force generation, and immature focal adhesions. As such, it is thought that at least in part, the effects of ECM stiffness on cells are similar to effects of decreased integrin-mediated adhesion and cellular force generation. Several technical challenges, however, remain. Besides stiffness, cross-linking chemistry can also alter the internal structure, surface topology and growth factor adhesion of the hydrogels (Houseman and Mrksich, 2001; Keselowsky et al., 2005; Crouzier et al., 2011). An alternative to overcome these limitations is to seed cells on posts of different height. By varying the height of the microposts, the stiffness is altered even though the material properties are held constant (Fu et al., 2010) (Figure 1D).
A large array of mechanotransduction studies have relied on the local application of force at single sites of adhesion. Magnetic tweezers (MTs), optical tweezers (OTs) and atomic force microscopy (AFM) are the most common tools for investigating the mechanical properties of biological molecules, cells, and tissues. MTs can exert calibrated forces from as little as 0.05 pN up to 150 pN, and as such has mostly been used to manipulate single molecules and to apply forces to transmembrane integrins (Alenghat et al., 2000; Matthews et al., 2004; Wang et al., 1993; del Rio et al., 2009). OTs, exerting forces from 0.1pN to 100pN, have been utilized to characterize molecular motors as kinesin, myosin, and dynein (Asbury et al., 2003; Altman et al., 2004; Gross et al., 2000), and to restrain movement of membrane bound beads on fibroblasts, demonstrating that cells strengthen their cytoskeleton linkages corresponding to the force needed to restrain the beads (Choquet et al., 1997). Because of its wide force range (5-10,000 pN), AFM has been applied to study force induced unfolding of proteins (Rief et al., 1999; Dietz and Rief, 2004; Puchner et al., 2008), as well as to characterize the mechanical and adhesive properties of single cells on substrates or cell-cell adhesion (Puech et al., 2006; Helenius et al., 2008). Recently, Webster et al. have developed an AFM feedback algorithm to rapidly change the stiffness presented to a cell while accurately measuring force and deformation (Webster et al., 2011). This “stiffness clamp” demonstrated that fibroblasts can exhibit a sub-second change in traction rate and contraction velocity in response to step changes in stiffness. This response was independent of the absolute contractile force and cell height, demonstrating that cells can react directly to changes in stiffness alone.
Mechanotransduction in Stem Cells and Development
Multicellular organisms develop distinct shapes that emerge from continuous pulling, buckling, twisting and bending interactions between cells and matrix. Following the genomic revolution, biologists have begun to consider the importance of forces during development as many of the developmental mechanisms involved in sculpting the metazoan body plan remain unexplained by genetics alone. A number of recent studies suggest that mechanical context may be a critical cue in guiding stem cell behavior, perhaps underscoring the link between stem cell function and the mechanical changes occurring during the development of the body plan. Here, we will discuss these stem cell studies as well as traditional embryological studies.
Mechanotransduction in stem cells
Although often generalized in a single term, stem cells represent a wide range of cell types that can be ranked on a continuum of stemness with the zygote and morula derived cells as totipotent stem cells on one end, and terminally differentiated cells, such as red blood cells and osteocytes, on the other. In between, a full range of pluripotent, multipotent and bipotent progenitor cells have been described with different proliferation and differentiation capacities. Importantly, much attention has recently focused on transcriptional and microenvironmental contexts that can drive stem cells into different states, including the reprogramming of a differentiated cell into a more naïve cell (Gurdon et al., 1958; Takahashi and Yamanaka, 2006).
Given the surprisingly plastic nature of stemness and that stem cells live through a constantly changing dynamic mechanical environment (whether in embryogenesis or in the transit of adult stem cells through different tissues), a number of studies have begun to explore whether mechanical forces may be involved in the regulation of stem cell processes. Indeed, recent evidence has shown that compliant but not stiff substrates, create permissive environments promoting self renewal and proliferation of hematopoietic stem cells (Holst et al., 2010), muscle derived stem cells (Gilbert et al., 2010) and embryonic stem cells (ESCs) (Chowdhury et al., 2010). Note, earlier studies had shown increased proliferation on stiffer surfaces for lineage committed cells as mentioned above. In fact, Gilbert et al. observed within a mixed population enrichment of stem versus non-stem cells by increasing cell survival depending on substrate mechanics. Furthermore, addition of ROCK inhibitors such as Y27632 to freshly isolated ESCs in culture also increases survival and self-renewal (Watanabe et al., 2007; Harb et al., 2008), as well as increases the efficiency of reprogramming of dermal fibroblasts into induced pluripotent stem cells (iPSC) (Lai et al., 2010). These studies highlight the role of low contractility in the preservation of stemness, though the mechanistic basis for this effect remains to be defined.
Stiffness and contractility also modulate commitment of mesenchymal stem cells (MSCs) into different lineages. Depending on the soluble factors in medium, MSCs will adopt the adipogenic (McBeath et al., 2004; Sordella et al., 2003) or chondrogenic (Gao et al., 2010) phenotype when cell size is restricted or when contractility is reduced. Conversely, spreading and high contractility promotes osteogenic (McBeath et al., 2004; Kilian et al., 2010) and myogenic differentiation (Gao et al., 2010), albeit through different mechanotransduction pathways. Whereas the adipo/osteo switch is driven through RhoA/ROCK, the decision making at chondro/myo switch depends on the activation of Rac1. In analogy to previous studies, stiffness also dictates differentiation of MSCs towards a neurogenic or adipogenic phenotype on compliant substrates (<1kPa), myogenesis at medium stiffness (8-17kPa) and osteogenesis at high stiffness (>25kPa) (Engler et al., 2006; Fu et al., 2010). Early studies now also show that the lineage fates of neuronal stem cells are also regulated by substrate stiffness and cell spreading (Saha et al., 2008).
MSCs are a heterogeneous cell population; not all MSCs undergo lineage commitment with equal efficiency. Intriguingly, MSCs that ultimately differentiate in response to soluble osteogenic supplements display significantly enhanced contractility one day after initial stimulation, predicting osteogenic differentiation that only can be confirmed seven days later (Fu et al., 2010). In addition, microscopy-based quantitative metrics describing changes in cell shape and actin cytoskeletal structure have been able to discriminate osteogenically induced MSCs from untreated or adipogenically induced controls 24h after stimulation, demonstrating early changes in cytoskeletal integrity preceding osteogenic differentiation (Treiser et al., 2010). The notion that contractility and cytoskeletal responses can act as harbingers of a differentiation response suggests that biophysical measures of stem cells could one day have utility in selection of clones with differential response characteristics.
Mechanotransduction in embryogenesis
Two popular animal models to study embryogenesis are Drosophila melanogaster and Caenorhabditis elegans. Time lapse microscopy of developing Drosophila eggs reveals a remarkable sequence of mechanical events during early fly development. During gastrulation, the transcription factors Snail and Twist induce a pulsed actomyosin contraction in ventral midline cells which invaginate according a ratchet-like mechanism, forming the ventral furrow and presumptive mesoderm (Martin et al., 2010; Pouille et al., 2009). At the same time, cells flanking the ventral furrow undergo passive cell shape changes (Butler et al., 2009). Under control of polarized actomyosin dynamics, cellular convergence-extension movements and cell intercalation are regulated along the anterior-posterior axis (Irvine and Wieschaus, 1994; Bertet et al., 2004; Blankenship et al., 2006; Fernandez-Gonzalez et al., 2009). This process, also called germ band extension, elongates the axial body to 2.5 times of its original length. When fully extended, the caudal end of the germ band retracts and pulls the amnioserosa over the dorsal epidermal gap that becomes exposed. At this stage, dorsal closure is initiated by the formation of a supracellular contractile ring in the epidermal cells at the leading edges of the opening and the pulsed contractions of amnioserosa cells pulling the leading edges together (Kiehart et al., 2000; Solon et al., 2009; Gorfinkiel et al., 2009). At the time when the lateral flanks of the epidermis come together, the remaining amnioserosa cells undergo apoptosis and involute, apparently distorting the neighboring cells and exerting a pull on surrounding cells. This apoptotic force contributes to the total force generation and speed of dorsal closure (Toyama et al., 2008).
Most of aforementioned findings are based on careful observation of the dynamics of fluorescently tagged proteins with time-lapse laser scanning confocal or two-photon microscopy. Hypotheses are tested with genetic and biochemical manipulations, such as silencing myosin-regulating gene expression with siRNA, or introducing drugs to affect contractility. In addition, laser ablation has been utilized as a ‘mechanical manipulation’ to abolish long range force transduction between cells (Kiehart et al., 2000; Fernandez-Gonzalez et al., 2009). Such techniques fall short of quantitative characterization of the endogenous magnitudes of force experienced in these settings, but they do provide a clear demonstration that such processes are at least partially driven by actomyosin contractility (Mammoto and Ingber, 2010; Kasza and Zallen, 2011). In addition, it is known that application of forces can directly impact myosin itself (Fernandez-Gonzalez et al., 2009). Despite these limitations, these approaches have contributed to our understanding of how genetics and biochemical signaling initiate mechanical events, such as the regulation of invagination of the ventral furrow by Snail and Twist. Far fewer studies, however, have been able to address the question of whether force is critical for inducing biochemical signaling that is required for subsequent stages of development.
Elegant experiments from the Farge group have given insight in to how compression forces between two cell types during Drosophila germ band elongation can induce the expression of Twist, a transcription factor that regulates the differentiation of the anterior mid-gut. Using a custom-built microscope (two-photon microscope with a third harmonics generation module and a femto-second titanium/sapphire oscillator), dorsal cells of the germ band were laser ablated, preventing the germ band from pushing the stomodeal primordium at the anterior side of the embryo (Supatto et al., 2005). To restore deformation, anterior pole stomodeal cells were indented with a micropipette, or a ferrofluid was injected and concentrated with an electromagnet in approximately 50 cells surrounding the stomodeal primordium. Then magnetic microtweezers were used to push the magnetized patch against the stomodeal cells with a force of 60 ± 20 nN. These mechanical manipulations rescued Armadillo/Beta Catenin translocation from the cell junctions to the nucleus which restored transcription of Twist (Desprat et al., 2008).
A slightly different approach was ventured by Zhang et al. to reveal a role for force in the maturation of hemidesmosomes and its impact on embryonic elongation in C. elegans (Zhang et al., 2011). Embryonic elongation is regulated by both the epidermis and muscles as muscle defective mutants arrest midway through elongation. C. elegans hemidesmosomes (CeHD) are anchoring structures found at the apical and basal plasma membrane of the epidermis, connecting the exoskeleton (apical side) with intermediate filaments to the ECM at the muscle-epidermis interface (basal side). Zhang and colleagues identified p21 activated kinase (PAK1), G-protein-coupled receptor kinase interactor (GIT1), and PAK interacting exchange factor (PIX1) to be three key signaling proteins involved in CeHD biogenesis and intermediate filament phosphorylation. Interestingly, during growth arrest in defective muscle mutants, PAK1 and PIX1 remained localized around the CeHDs, whereas GIT1 gradually disappeared from the CeHDs as embryos stopped elongation. By compressing muscle defective mutant embryos between a blunted microneedle tip and a programmable microscope stage, the authors were able to rescue GIT1 signaling and CeHD maturation, thus providing a link between muscle-generated tension and longitudinal growth in C. elegans. Hence, CeHDs might be mechanosensors, in the same way as FAs and AJs are.
Similar to Drosophila and C. elegans, zebrafish (Danio rerio) and Xenopus laevis embryos can be easily manipulated and monitored under a microscope making these lower vertebrate animals attractive for mechanotransduction research. Indeed, scientists have focused on the role of myosin II driven cortical forces in sorting out progenitor cells into three germ layers in zebrafish (Krieg et al., 2008; Schotz et al., 2008), and regulation of convergence-extension cellular movement (Skoglund et al., 2008) or fibronectin fibril assembly in the blastocoel roof in Xenopus embryos (Dzamba et al., 2009). Despite the fact that zebrafish and Xenopus are vertebrates, surprisingly little data is available addressing mechanoregulation of skeletal development in these models. In chicken, however, abolishment of muscle contractions by injecting a neuromuscular blocker reagent during limb development impairs cartilage synthesis and tenascin-c expression, suggesting a role for force in developmental skeletogenesis (Mikic et al., 2000; Mikic et al., 2004).
The function of myosin II and its isoforms has been explored to some extent in mouse development. Because direct manipulation is experimentally difficult, mechanotransduction studies in mice often restricted to examination of phenotypic abnormalities observed in knockout or transgenic mice. Deletion of myosin IIB in mice results in death from cardiac malformation and brain defects at 14 days post coitus (dpc) (Tullio et al., 2001), a surprising finding considering its role during gastrulation in other species (Skoglund et al., 2008). Moreover, myosin IIA knockout mice are also embryonic lethal but at 6.5/7.5 dpc because they fail to develop a normal visceral endoderm associated with mislocation of E-cadherin and beta catenin and are therefore unable to assemble AJs (Conti et al., 2004). Interestingly, knocking in myosin IIB driven by a myosin IIA promoter can rescue this phenotype. However, death occurs in the knock-in mice at 12.5-14.5 dpc due to abnormal angiogenesis and migration defects in the placenta which may indicate a unique role for myosin IIA in placenta development (Wang et al., 2010). Despite the lack of technology to apply forces directly on cells in utero, knockout/knock-in strategy, which controls for the total amount of myosin II, might suggest that localization of beta catenin and E-cadherin in presumptive visceral endodermal cells may be driven by contractile forces, independent of the myosin isoform producing the force. As conditional knockouts are introduced for these myosin isoforms, we will begin to better define their roles more completely in tissue-specific physiologic or pathologic responses to force.
The severe phenotypes observed in knockout mice lacking the genes encoding for members of the Kinesin Family 3 and Polycystic Kidney Disease have directed the interest to explore mechanotransduction in cilia, motile cilia, and primary cilia in mouse development. Cilia and primary cilia are microtubule-based structures enveloped in a specialized membrane that originate from the cell body into the extracellular space (Figure 1B). They act as mechanosensors steering the development of organs and tissues, such as kidney, pancreas, liver, cartilage, and bone among others. Although the molecular basis for cilia mechanosensing is still elusive, cilia appear to transduce mechanical stimuli in part by gating polycystin-based ion channels and modulating Sonic Hedgehog and Wnt signaling. The specific mechanosensory roles for cilia have been reviewed in detail elsewhere (Wallingford, 2010; Wallingford and Mitchell, 2011; Bisgrove and Yost, 2006; Berbari et al., 2009).
Thus, one can appreciate that despite the ubiquitous presence of forces driving developmental processes, only a small handful of studies have begun to shed light on these mechanical events. As genetic and biophysical tools continue to mature, we anxiously anticipate a substantial increase in the pace of discovery in this space.
Parting Perspectives
Identifying, characterizing, and understanding how mechanical forces contribute to cell and developmental biology depends on the establishment of tools to both measure and manipulate forces in cells and tissues. Over recent decades, substantial progress has been made in establishing tools, such as methods to measure traction forces, to apply forces to cells, and to modulate ECM stiffness. Although these techniques have enabled much of the current understanding of cellular mechanotransduction, much of the current work is limited to a few cell types in a few contexts, so our appreciation of when and where forces might be dissimilar is still quite limited. In addition to expanding the application of existing techniques to a wider range of biological settings, significant innovation is still needed to build a more powerful toolkit for the field. For example, while forces can be measured for cells cultured on specialized surfaces, they are only beginning to be observed in 3D hydrogels. We still cannot measure forces on natural ECMs such as collagen owing to their complex mechanical properties. In vivo, methods are even more limited and as such the force distribution between cells and tissues in each stage of development is still unknown. To begin to address this question, Brodland and colleagues combined computational finite element analysis with time-lapse movies obtained with two-photon microscopy to estimate forces during Drosophila development (Brodland et al., 2010). The finite element method is a general, widely used approach in engineering to model the mechanics of complex structures by examining the mechanics of each individual voxel (finite element). Although done for the first time in a living organism, the combination of measuring deformation with finite element analysis is still an indirect method to estimate force vectors. This shortcoming may be overcome by the development of molecular force sensors. Recently, a calibrated force sensor that undergoes efficient FRET upon deformation has been developed, allowing the measurement of traction forces across vinculin (Grashoff et al., 2010) and α-actinin (Meng and Sachs, 2011) in living cells. When applicable to other force transmitting molecules, this type of force beacons might provide a platform for ‘visualizing’ forces in vitro and in vivo.
In addition to force mapping, the field would benefit from additional techniques to mechanically manipulate cells in vivo. Such techniques could involve the use of caged proteins to control drug release, targeting mechanotransduction mechanisms to specific sites within the cell. Optogenetics approaches involving transgenic expression of light-gated ion channels are also being developed to manipulate myosin contractility. For example, Arrenberg and colleagues modulated the beating rate of a zebrafish heart by emitting light pulses (Arrenberg et al., 2010). A possible approach to circumventing the challenges of in vivo models is to focus more effort on organotypic in vitro models that might mimic in vivo mechanotransduction events. For lung tissue, this biomimetic approach has been successful for gaining insight into nanoparticle uptake of lung epithelial and underlying endothelial cells under cyclic stretch, mirroring alveolar expansion during breathing (Huh et al., 2010). Such models offer the advantages of in vitro accessibility and control, with the potential for capturing more complex tissue behaviors.
In conclusion, experimental evidence is mounting to suggest that forces are present and evolving in virtually every biological setting, whether that setting involves the forces of physical activity, remodeling in multicellular and ECM structures, or even simple changes in cell-cell and cell-ECM adhesion. These forces also appear to regulate many basic cellular processes from cell adhesion itself to cellular signaling, proliferation, and differentiation. As such, it may be best not to regard mechanics as applicable only to specialized circumstances, but instead as an ever-present physical dimension of cell and developmental biology. Similar to studying biochemical signaling, exploring mechanotransduction relies on the development of enabling technologies. Over the past thirty years, the development of PCR, antibodies, and fluorescent molecules have drastically changed the way scientists investigate biochemical signaling pathways. Now, several laboratories are developing tools such as traction force microscopy, micropost array detectors, patterned substrates and tunable hydrogels to facilitate mechanotransduction research. It will only be a matter of time before derivatives of these tools will be part of a standard biology laboratory tool kit.
Acknowledgements
We apologize to our colleagues whose work could not be cited due to space limitations. This work was supported by NIH (GM60692, EB 00262). J.E. is a post doctoral fellow of the Research Foundation - Flanders (FWO). X.Y. acknowledges support from the American Heart Association postdoctoral fellowship program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
Reference List
- Alenghat FJ, Fabry B, Tsai KY, Goldmann WH, Ingber DE. Analysis of cell mechanics in single vinculin-deficient cells using a magnetic tweezer. Biochem. Biophys. Res. Commun. 2000;277:93–99. doi: 10.1006/bbrc.2000.3636. [DOI] [PubMed] [Google Scholar]
- Altman D, Sweeney HL, Spudich JA. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- Arrenberg AB, Stainier DY, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330:971–974. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky A, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- Bancroft GN, Sikavitsas VI, van den DJ, Sheffield TL, Ambrose CG, Jansen JA, Mikos AG. Fluid flow increases mineralized matrix deposition in 3D perfusion culture of marrow stromal osteoblasts in a dose-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12600–12605. doi: 10.1073/pnas.202296599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr. Biol. 2009;19:R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C, Sulak L, Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429:667–671. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Yost HJ. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev. Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Brodland GW, Conte V, Cranston PG, Veldhuis J, Narasimhan S, Hutson MS, Jacinto A, Ulrich F, Baum B, Miodownik M. Video force microscopy reveals the mechanics of ventral furrow invagination in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22111–22116. doi: 10.1073/pnas.1006591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- Butler LC, Blanchard GB, Kabla AJ, Lawrence NJ, Welchman DP, Mahadevan L, Adams RJ, Sanson B. Cell shape changes indicate a role for extrinsic tensile forces in Drosophila germ-band extension. Natl. Cell Biol. 2009;11:859–864. doi: 10.1038/ncb1894. [DOI] [PubMed] [Google Scholar]
- Byron A, Humphries JD, Bass MD, Knight D, Humphries MJ. Proteomic analysis of integrin adhesion complexes. Sci. Signal. 2011;4:pt2. doi: 10.1126/scisignal.2001827. [DOI] [PubMed] [Google Scholar]
- Chancellor TJ, Lee J, Thodeti CK, Lele T. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys. J. 2010;99:115–123. doi: 10.1016/j.bpj.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AA, Khetani SR, Lee S, Bhatia SN, Van Vliet KJ. Modulation of hepatocyte phenotype in vitro via chemomechanical tuning of polyelectrolyte multilayers. Biomaterials. 2009;30:1113–1120. doi: 10.1016/j.biomaterials.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Chien S, Li S, Shyy YJ. Effects of mechanical forces on signal transduction and gene expression in endothelial cells. Hypertension. 1998;31:162–169. doi: 10.1161/01.hyp.31.1.162. [DOI] [PubMed] [Google Scholar]
- Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88:39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- Chowdhury F, Na S, Li D, Poh YC, Tanaka TS, Wang F, Wang N. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater. 2010;9:82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Scholey JM. Mitotic force generators and chromosome segregation. Cell Mol. Life Sci. 2010;67:2231–2250. doi: 10.1007/s00018-010-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti MA, Even-Ram S, Liu C, Yamada KM, Adelstein RS. Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J. Biol. Chem. 2004;279:41263–41266. doi: 10.1074/jbc.C400352200. [DOI] [PubMed] [Google Scholar]
- Crouzier T, Fourel L, Boudou T, Albiges-Rizo C, Picart C. Presentation of BMP-2 from a Soft Biopolymeric Film Unveils its Activity on Cell Adhesion and Migration. Adv. Mater. 2011;23:H111–H118. doi: 10.1002/adma.201004637. [DOI] [PubMed] [Google Scholar]
- David V, Martin A, Lafage-Proust MH, Malaval L, Peyroche S, Jones DB, Vico L, Guignandon A. Mechanical loading down-regulates peroxisome proliferator-activated receptor gamma in bone marrow stromal cells and favors osteoblastogenesis at the expense of adipogenesis. Endocrinology. 2007;148:2553–2562. doi: 10.1210/en.2006-1704. [DOI] [PubMed] [Google Scholar]
- Davies PF, Dewey CF, Jr., Bussolari SR, Gordon EJ, Gimbrone MA., Jr. Influence of hemodynamic forces on vascular endothelial function. In vitro studies of shear stress and pinocytosis in bovine aortic cells. J. Clin. Invest. 1984;73:1121–1129. doi: 10.1172/JCI111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PF, Remuzzi A, Gordon EJ, Dewey CF, Jr., Gimbrone MA., Jr. Turbulent fluid shear stress induces vascular endothelial cell turnover in vitro. Proc. Natl. Acad. Sci. U. S. A. 1986;83:2114–2117. doi: 10.1073/pnas.83.7.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J, Kerstens A, Danuser G, Schwartz MA, Waterman-Storer CM. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J. Cell Biol. 2005;171:153–164. doi: 10.1083/jcb.200506152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoe-Ayari H, Rieu JP, Sano M. 4D traction force microscopy reveals asymmetric cortical forces in migrating Dictyostelium cells. Phys. Rev. Lett. 2010;105:248103. doi: 10.1103/PhysRevLett.105.248103. [DOI] [PubMed] [Google Scholar]
- Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprat N, Supatto W, Pouille PA, Beaurepaire E, Farge E. Tissue deformation modulates twist expression to determine anterior midgut differentiation in Drosophila embryos. Dev. Cell. 2008;15:470–477. doi: 10.1016/j.devcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M, Kerssemakers JW, Romet-Lemonne G, Janson ME. Force generation by dynamic microtubules. Curr. Opin. Cell Biol. 2005;17:67–74. doi: 10.1016/j.ceb.2004.12.011. [DOI] [PubMed] [Google Scholar]
- du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Force mapping in epithelial cell migration. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzamba BJ, Jakab KR, Marsden M, Schwartz MA, DeSimone DW. Cadherin adhesion, tissue tension, and noncanonical Wnt signaling regulate fibronectin matrix organization. Dev. Cell. 2009;16:421–432. doi: 10.1016/j.devcel.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Haj AJ, Minter SL, Rawlinson SC, Suswillo R, Lanyon LE. Cellular responses to mechanical loading in vitro. J. Bone Miner. Res. 1990;5:923–932. doi: 10.1002/jbmr.5650050905. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Simoes SM, Roper JC, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev. Cell. 2009;17:736–743. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan LA, Ju YE, Marg B, Osterfield M, Janmey PA. Neurite branching on deformable substrates. Neuroreport. 2002;13:2411–2415. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed LE, Vunjak-Novakovic G, Langer R. Cultivation of cell-polymer cartilage implants in bioreactors. J. Cell Biochem. 1993;51:257–264. doi: 10.1002/jcb.240510304. [DOI] [PubMed] [Google Scholar]
- Frieboes LR, Gupta R. An in-vitro traumatic model to evaluate the response of myelinated cultures to sustained hydrostatic compression injury. J. Neurotrauma. 2009;26:2245–2256. doi: 10.1089/neu.2009.0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, McBeath R, Chen CS. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells. 2010;28:564–572. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin J, Qi J, Maloney M, Banes AJ. Novel system for engineering bioartificial tendons and application of mechanical load. Tissue Eng. 2003;9:967–979. doi: 10.1089/107632703322495619. [DOI] [PubMed] [Google Scholar]
- Georges PC, Janmey PA. Cell type-specific response to growth on soft materials. J. Appl. Physiol. 2005;98:1547–1553. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KE, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girton TS, Oegema TR, Tranquillo RT. Exploiting glycation to stiffen and strengthen tissue equivalents for tissue engineering. J. Biomed. Mater. Res. 1999;46:87–92. doi: 10.1002/(sici)1097-4636(199907)46:1<87::aid-jbm10>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Gjorevski N, Nelson CM. Endogenous patterns of mechanical stress are required for branching morphogenesis. Integr. Biol. (Camb. ) 2010;2:424–434. doi: 10.1039/c0ib00040j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N, Blanchard GB, Adams RJ, Martinez AA. Mechanical control of global cell behaviour during dorsal closure in Drosophila. Development. 2009;136:1889–1898. doi: 10.1242/dev.030866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SP, Welte MA, Block SM, Wieschaus EF. Dynein-mediated cargo transport in vivo. A switch controls travel distance. J. Cell Biol. 2000;148:945–956. doi: 10.1083/jcb.148.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- Harb N, Archer TK, Sato N. The Rho-Rock-Myosin signaling axis determines cell-cell integrity of self-renewing pluripotent stem cells. PLoS. One. 2008;3:e3001. doi: 10.1371/journal.pone.0003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AK, Stopak D, Wild P. Fibroblast traction as a mechanism for collagen morphogenesis. Nature. 1981;290:249–251. doi: 10.1038/290249a0. [DOI] [PubMed] [Google Scholar]
- Harris AK, Wild P, Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208:177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- Haudenschild CC, Grunwald J, Chobanian AV. Effects of hypertension on migration and proliferation of smooth muscle in culture. Hypertension. 1985;7:I101–I104. doi: 10.1161/01.hyp.7.3_pt_2.i101. [DOI] [PubMed] [Google Scholar]
- Helenius J, Heisenberg CP, Gaub HE, Muller DJ. Single-cell force spectroscopy. J. Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- Holst J, Watson S, Lord MS, Eamegdool SS, Bax DV, Nivison-Smith LB, Kondyurin A, Ma L, Oberhauser AF, Weiss AS, Rasko JE. Substrate elasticity provides mechanical signals for the expansion of hemopoietic stem and progenitor cells. Nat. Biotechnol. 2010;28:1123–1128. doi: 10.1038/nbt.1687. [DOI] [PubMed] [Google Scholar]
- Houseman BT, Mrksich M. The microenvironment of immobilized Arg-Gly-Asp peptides is an important determinant of cell adhesion. Biomaterials. 2001;22:943–955. doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]
- Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur SS, Zhao Y, Li YS, Botvinick E, Chien S. Live Cells Exert 3-Dimensional Traction Forces on Their Substrata. Cell Mol. Bioeng. 2009;2:425–436. doi: 10.1007/s12195-009-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber DE, Folkman J. Mechanochemical switching between growth and differentiation during fibroblast growth factor-stimulated angiogenesis in vitro: role of extracellular matrix. J. Cell Biol. 1989;109:317–330. doi: 10.1083/jcb.109.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber DE, Prusty D, Sun Z, Betensky H, Wang N. Cell shape, cytoskeletal mechanics, and cell cycle control in angiogenesis. J. Biomech. 1995;28:1471–1484. doi: 10.1016/0021-9290(95)00095-x. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Wieschaus E. Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development. 1994;120:827–841. doi: 10.1242/dev.120.4.827. [DOI] [PubMed] [Google Scholar]
- Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasza KE, Zallen JA. Dynamics and regulation of contractile actin-myosin networks in morphogenesis. Curr. Opin. Cell Biol. 2011;23:30–38. doi: 10.1016/j.ceb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keselowsky BG, Collard DM, Garcia AJ. Integrin binding specificity regulates biomaterial surface chemistry effects on cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5953–5957. doi: 10.1073/pnas.0407356102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J. Cell Biol. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirfel G, Rigort A, Borm B, Herzog V. Cell migration: mechanisms of rear detachment and the formation of migration tracks. Eur. J. Cell Biol. 2004;83:717–724. doi: 10.1078/0171-9335-00421. [DOI] [PubMed] [Google Scholar]
- Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr. Biol. 2009;19:1511–1518. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Forced unfolding of the fibronectin type III module reveals a tensile molecular recognition switch. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1351–1356. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg M, Arboleda-Estudillo Y, Puech PH, Kafer J, Graner F, Muller DJ, Heisenberg CP. Tensile forces govern germ-layer organization in zebrafish. Nat. Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- Kuo JC, Han X, Hsiao CT, Yates JR, III, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WH, Ho JC, Lee YK, Ng KM, Au KW, Chan YC, Lau CP, Tse HF, Siu CW. ROCK inhibition facilitates the generation of human-induced pluripotent stem cells in a defined, feeder-, and serum-free system. Cell Reprogram. 2010;12:641–653. doi: 10.1089/cell.2010.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansman JB, Hallam TJ, Rink TJ. Single stretch-activated ion channels in vascular endothelial cells as mechanotransducers? Nature. 1987;325:811–813. doi: 10.1038/325811a0. [DOI] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J. Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Noble F, Moyon D, Pardanaud L, Yuan L, Djonov V, Matthijsen R, Breant C, Fleury V, Eichmann A. Flow regulates arterial-venous differentiation in the chick embryo yolk sac. Development. 2004;131:361–375. doi: 10.1242/dev.00929. [DOI] [PubMed] [Google Scholar]
- Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, Chen CS. Mechanical tugging force regulates the size of cell-cell junctions. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9944–9949. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137:1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF. Integration of contractile forces during tissue invagination. J. Cell Biol. 2010;188:735–749. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruthamuthu V, Sabass B, Schwarz US, Gardel ML. Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4708–4713. doi: 10.1073/pnas.1011123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskarinec SA, Franck C, Tirrell DA, Ravichandran G. Quantifying cellular traction forces in three dimensions. Proc. Natl. Acad. Sci. U. S. A. 2009;106:22108–22113. doi: 10.1073/pnas.0904565106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BD, Overby DR, Alenghat FJ, Karavitis J, Numaguchi Y, Allen PG, Ingber DE. Mechanical properties of individual focal adhesions probed with a magnetic microneedle. Biochem. Biophys. Res. Commun. 2004;313:758–764. doi: 10.1016/j.bbrc.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Matthews BD, Thodeti CK, Tytell JD, Mammoto A, Overby DR, Ingber DE. Ultra-rapid activation of TRPV4 ion channels by mechanical forces applied to cell surface beta1 integrins. Integr. Biol. (Camb.) 2010;2:435–442. doi: 10.1039/c0ib00034e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- Meng F, Sachs F. Visualizing dynamic cytoplasmic forces with a compliance-matched FRET sensor. J. Cell Sci. 2011;124:261–269. doi: 10.1242/jcs.071928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikic B, Isenstein AL, Chhabra A. Mechanical modulation of cartilage structure and function during embryogenesis in the chick. Ann. Biomed. Eng. 2004;32:18–25. doi: 10.1023/b:abme.0000007787.39262.a7. [DOI] [PubMed] [Google Scholar]
- Mikic B, Wong M, Chiquet M, Hunziker EB. Mechanical modulation of tenascin-C and collagen-XII expression during avian synovial joint formation. J. Orthop. Res. 2000;18:406–415. doi: 10.1002/jor.1100180312. [DOI] [PubMed] [Google Scholar]
- Miyamoto S, Teramoto H, Coso OA, Gutkind JS, Burbelo PD, Akiyama SK, Yamada KM. Integrin function: molecular hierarchies of cytoskeletal and signaling molecules. J. Cell Biol. 1995;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar G, Schroedl NA, Gonda SR, Hartzell CR. Skeletal muscle satellite cells cultured in simulated microgravity. In Vitro Cell Dev. Biol. Anim. 1997;33:386–391. doi: 10.1007/s11626-997-0010-9. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Fukata M, Yamaga M, Itoh N, Kaibuchi K. Recruitment and activation of Rac1 by the formation of E-cadherin-mediated cell-cell adhesion sites. J. Cell Sci. 2001;114:1829–1838. doi: 10.1242/jcs.114.10.1829. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Chen CS. Cell-cell signaling by direct contact increases cell proliferation via a PI3K-dependent signal. FEBS Lett. 2002;514:238–242. doi: 10.1016/s0014-5793(02)02370-0. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Emergent patterns of growth controlled by multicellular form and mechanics. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Pirone DM, Tan JL, Chen CS. Vascular endothelial-cadherin regulates cytoskeletal tension, cell spreading, and focal adhesions by stimulating RhoA. Mol. Biol. Cell. 2004;15:2943–2953. doi: 10.1091/mbc.E03-10-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314:298, 300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Weaver VM. The tension mounts: mechanics meets morphogenesis and malignancy. J. Mammary. Gland. Biol. Neoplasia. 2004;9:325–342. doi: 10.1007/s10911-004-1404-x. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Jr., Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- Pirone DM, Liu WF, Ruiz SA, Gao L, Raghavan S, Lemmon CA, Romer LH, Chen CS. An inhibitory role for FAK in regulating proliferation: a link between limited adhesion and RhoA-ROCK signaling. J. Cell Biol. 2006;174:277–288. doi: 10.1083/jcb.200510062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille PA, Ahmadi P, Brunet AC, Farge E. Mechanical signals trigger Myosin II redistribution and mesoderm invagination in Drosophila embryos. Sci. Signal. 2009;2:ra16. doi: 10.1126/scisignal.2000098. [DOI] [PubMed] [Google Scholar]
- Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schafer LV, Brandmeier B, Grater F, Grubmuller H, Gaub HE, Gautel M. Mechanoenzymatics of titin kinase. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puech PH, Poole K, Knebel D, Muller DJ. A new technical approach to quantify cell-cell adhesion forces by AFM. Ultramicroscopy. 2006;106:637–644. doi: 10.1016/j.ultramic.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Raab-Cullen DM, Thiede MA, Petersen DN, Kimmel DB, Recker RR. Mechanical loading stimulates rapid changes in periosteal gene expression. Calcif. Tissue Int. 1994;55:473–478. doi: 10.1007/BF00298562. [DOI] [PubMed] [Google Scholar]
- Renkawitz J, Sixt M. Mechanisms of force generation and force transmission during interstitial leukocyte migration. EMBO Rep. 2010;11:744–750. doi: 10.1038/embor.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rid R, Schiefermeier N, Grigoriev I, Small JV, Kaverina I. The last but not the least: the origin and significance of trailing adhesions in fibroblastic cells. Cell Motil. Cytoskeleton. 2005;61:161–171. doi: 10.1002/cm.20076. [DOI] [PubMed] [Google Scholar]
- Rief M, Pascual J, Saraste M, Gaub HE. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- Roux W. Gesammelte Abhandlungen über Entwicklungsmechanik. Leipzig: 1895. [Google Scholar]
- Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoshima J, Takahashi T, Jahn L, Izumo S. Roles of mechano-sensitive ion channels, cytoskeleton, and contractile activity in stretch-induced immediate-early gene expression and hypertrophy of cardiac myocytes. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9905–9909. doi: 10.1073/pnas.89.20.9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Substrate modulus directs neural stem cell behavior. Biophys. J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Sheetz MP. Force transduction by Triton cytoskeletons. J. Cell Biol. 2002;156:609–615. doi: 10.1083/jcb.200110068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotz EM, Burdine RD, Julicher F, Steinberg MS, Heisenberg CP, Foty RA. Quantitative differences in tissue surface tension influence zebrafish germ layer positioning. HFSP. J. 2008;2:42–56. doi: 10.2976/1.2834817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler EJ, Lancin PA, Dasgupta A, Moghe PV. Engineering hepatocellular morphogenesis and function via ligand-presenting hydrogels with graded mechanical compliance. Biotechnol. Bioeng. 2005;89:296, 307. doi: 10.1002/bit.20328. [DOI] [PubMed] [Google Scholar]
- Sheetz MP. Microtubule motor complexes moving membranous organelles. Cell Struct. Funct. 1996;21:369–373. doi: 10.1247/csf.21.369. [DOI] [PubMed] [Google Scholar]
- Shyy YJ, Hsieh HJ, Usami S, Chien S. Fluid shear stress induces a biphasic response of human monocyte chemotactic protein 1 gene expression in vascular endothelium. Proc. Natl. Acad. Sci. U. S. A. 1994;91:4678–4682. doi: 10.1073/pnas.91.11.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JR, Karp S, Ingber DE. Altering the cellular mechanical force balance results in integrated changes in cell, cytoskeletal and nuclear shape. J. Cell Sci. 1992;103(Pt 4):1215–1222. doi: 10.1242/jcs.103.4.1215. [DOI] [PubMed] [Google Scholar]
- Skoglund P, Rolo A, Chen X, Gumbiner BM, Keller R. Convergence and extension at gastrulation require a myosin IIB-dependent cortical actin network. Development. 2008;135:2435–2444. doi: 10.1242/dev.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smutny M, Cox HL, Leerberg JM, Kovacs EM, Conti MA, Ferguson C, Hamilton NA, Parton RG, Adelstein RS, Yap AS. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat. Cell Biol. 2010;12:696–702. doi: 10.1038/ncb2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon J, Kaya-Copur A, Colombelli J, Brunner D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell. 2009;137:1331–1342. doi: 10.1016/j.cell.2009.03.050. [DOI] [PubMed] [Google Scholar]
- Sordella R, Jiang W, Chen GC, Curto M, Settleman J. Modulation of Rho GTPase signaling regulates a switch between adipogenesis and myogenesis. Cell. 2003;113:147–158. doi: 10.1016/s0092-8674(03)00271-x. [DOI] [PubMed] [Google Scholar]
- Sumpio BE, Banes AJ, Levin LG, Johnson G., Jr. Mechanical stress stimulates aortic endothelial cells to proliferate. J. Vasc. Surg. 1987;6:252–256. [PubMed] [Google Scholar]
- Supatto W, Debarre D, Moulia B, Brouzes E, Martin JL, Farge E, Beaurepaire E. In vivo modulation of morphogenetic movements in Drosophila embryos with femtosecond laser pulses. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1047–1052. doi: 10.1073/pnas.0405316102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DW. On Growth and Form. Cambridge University Press; Cambridge, UK: 1917. [Google Scholar]
- Thompson MT, Berg MC, Tobias IS, Rubner MF, Van Vliet KJ. Tuning compliance of nanoscale polyelectrolyte multilayers to modulate cell adhesion. Biomaterials. 2005;26:6836–6845. doi: 10.1016/j.biomaterials.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Toyama Y, Peralta XG, Wells AR, Kiehart DP, Edwards GS. Apoptotic force and tissue dynamics during Drosophila embryogenesis. Science. 2008;321:1683–1686. doi: 10.1126/science.1157052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiser MD, Yang EH, Gordonov S, Cohen DM, Androulakis IP, Kohn J, Chen CS, Moghe PV. Cytoskeleton-based forecasting of stem cell lineage fates. Proc. Natl. Acad. Sci. U. S. A. 2010;107:610–615. doi: 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullio AN, Bridgman PC, Tresser NJ, Chan CC, Conti MA, Adelstein RS, Hara Y. Structural abnormalities develop in the brain after ablation of the gene encoding nonmuscle myosin II-B heavy chain. J. Comp Neurol. 2001;433:62–74. doi: 10.1002/cne.1125. [DOI] [PubMed] [Google Scholar]
- Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, Cao G, DeLisser H, Schwartz MA. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Koach MA, Whitmore L, Lamers ML, Horwitz AF. Segregation and activation of myosin IIB creates a rear in migrating cells. J. Cell Biol. 2008;183:543–554. doi: 10.1083/jcb.200806030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB. Planar cell polarity signaling, cilia and polarized ciliary beating. Curr. Opin. Cell Biol. 2010;22:597–604. doi: 10.1016/j.ceb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Mitchell B. Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes Dev. 2011;25:201–213. doi: 10.1101/gad.2008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Ma X, Conti MA, Liu C, Kawamoto S, Adelstein RS. Nonmuscle myosin II isoform and domain specificity during early mouse development. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14645–14650. doi: 10.1073/pnas.1004023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S, Muguruma K, Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Weber GF, Bjerke MA, DeSimone DW. Integrins and cadherins join forces to form adhesive networks. J. Cell Sci. 2011;124:1183–1193. doi: 10.1242/jcs.064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster KD, Crow A, Fletcher DA. An AFM-Based Stiffness Clamp for Dynamic Control of Rigidity. PLoS. One. 2011;6:e17807. doi: 10.1371/journal.pone.0017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, Lobdell NA, Settleman J, Reynolds AB. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–1039. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
- Willits RK, Skornia SL. Effect of collagen gel stiffness on neurite extension. J. Biomater. Sci. Polym. Ed. 2004;15:1521–1531. doi: 10.1163/1568562042459698. [DOI] [PubMed] [Google Scholar]
- Wipff PJ, Rifkin DB, Meister JJ, Hinz B. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J. Cell Biol. 2007;179:1311–1323. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff Y. Das Gesetz der Transformation der Knochen. Verlag von August Hirschwald; Berlin: 1892. [Google Scholar]
- Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol. 2003;163:583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki H, Pan S, Lee RT, Berk BC. Fluid shear stress inhibits vascular inflammation by decreasing thioredoxin-interacting protein in endothelial cells. J. Clin. Invest. 2005;115:733–738. doi: 10.1172/JCI200523001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lin JS, Chen J, Wang JH. Determining substrate displacement and cell traction fields--a new approach. J. Theor. Biol. 2006;242:607–616. doi: 10.1016/j.jtbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R, Milo R, Kam Z, Geiger B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 2007;120:137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471:99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- Zimerman B, Volberg T, Geiger B. Early molecular events in the assembly of the focal adhesion-stress fiber complex during fibroblast spreading. Cell Motil. Cytoskeleton. 2004;58:143–159. doi: 10.1002/cm.20005. [DOI] [PubMed] [Google Scholar]