Abstract
Deletion of COQ10 in Saccharomyces cerevisiae elicits a respiratory defect characterized by the absence of cytochrome c reduction, which is correctable by the addition of exogenous diffusible coenzyme Q2. Unlike other coq mutants with hampered coenzyme Q6 (Q6) synthesis, coq10 mutants have near wild-type concentrations of Q6. In the present study, we use Q-cycle inhibitors of the coenzyme QH2-cytochrome c reductase (bc1 complex) to assess electron transfer properties of coq10 cells. Our results show that coq10 mutants respond to antimycin A, indicating an active Q cycle in these mutants, even though they are unable to transport electrons through cytochrome c and are not responsive to myxothiazol. EPR spectroscopic analysis also suggests that wild type and coq10 mitochondria accumulate similar amounts of Q6 semiquinone, despite a lower steady state level of bc1 complex in the coq10 cells. Confirming the reduced respiratory chain state in coq10 cells, we found that the expression of the Aspergillus fumigatus alternative oxidase in these cells leads to a decrease in antimycin-dependent H2O2 release and improves their respiratory growth.
Keywords: Saccharomyces cerevisiae, mitochondria, coenzyme Q
Introduction
Coenzyme Q (ubiquinone) is an essential electron carrier of the mitochondrial respiratory chain whose main function is to transfer electrons from the NADH- and succinate-coenzyme Q reductases to the bc1 complex [1]. Electron transfer in the bc1 complex occurs through the “Q-cycle” [2–4], in which electrons from reduced coenzyme Q (QH2) follow a branched path to the iron-sulfur protein and to cytochrome bL [4].
Biosynthesis of coenzyme Q in eukaryotes occurs in mitochondria. In Saccharomyces cerevisiae, the benzene ring of coenzyme Q6 (Q6) has a polyprenyl side chain with 6 isoprenoid units [5]. The size of the isoprenoid chain varies among species and affects coenzyme Q diffusion through cell membranes [6]. On the other hand, at least nine yeast nuclear genes [7–9] have been shown to be involved in the synthesis of Q6. COQ10, however, is not involved in the synthesis of Q6 but, interestingly, respective mutants have Q6 respiratory deficiency [10–12]. All products of COQ genes, including Coq10p, are located in the mitochondrial inner membrane [1]. There are genetic and physical evidences that enzymes of Q6 biosynthesis, but not Coq10p, are part of a multi-subunit complex [13–15].
Coq10p is a member of the START domain super family [10,12]. Members of this family were shown to bind lipophilic compounds such as cholesterol [16]. When over-expressed in yeast, purified Coq10p contains bound Q6 [10,11]. The inability of Q6 in coq10 mutants to promote electron transfer to the bc1 complex suggests that Coq10p might function in the delivery of Q6 to its proper site in the respiratory chain. A direct role of Coq10p in electron transfer is not completely excluded, although it appears unlikely based on stoichiometric considerations [10]. The present studies were undertaken to assess the respiratory functionality of Q6 in the coq10 mutants which are defective in the reduction of cytochrome c. Using bc1 complex inhibitors, we observed that coq10 mitochondria were responsive to antimycin A but not to myxothiazol, indicating an active Q-cycle, and a defective transfer of QH2 to the bc1 Rieske protein. EPR spectroscopic analysis also suggests that wild type and coq10 mitochondria have similar amounts of Q6 semiquinone, even with a lower steady state level of bc1 complex. On the other hand, the expression of Aspergillus fumigatus alternative oxidase [17], which transports electrons directly from QH2 to oxygen, reduced the H2O2 release in coq10 cells and improved their respiratory growth.
Results
Effect of antimycin A and myxothiazol on semiquinone formation in the coq10 mutant
Antimycin A and myxothiazol are well known inhibitors of the bc1 complex acting, respectively, at the N and P sites of the Q cycle [18–21]. Both inhibitors enhance formation of oxygen radicals from the P site [20,21]. Antimycin A binds to the N site and blocks oxidation of cytochrome bH, resulting in a reverse flow of electrons from cytochrome bL to coenzyme Q to form the semiquinone (see Fig. 1). Myxothiazol, on the other hand, binds to the P site and prevents the reduction of cytochrome bL, but allows a slow reduction of the Rieske iron-sulfur protein [4,20]. An increase of myxothiazol- dependent semiquinone is thought to occur at the P site due to an incomplete inhibition of ubiquinone oxidation [20–22]. However, the existence of semiquinones at the P site is still controversial [20,23].
Fig. 1. Protonmotive Q cycle of electron transfer and proton translocation in the bc1 complex.
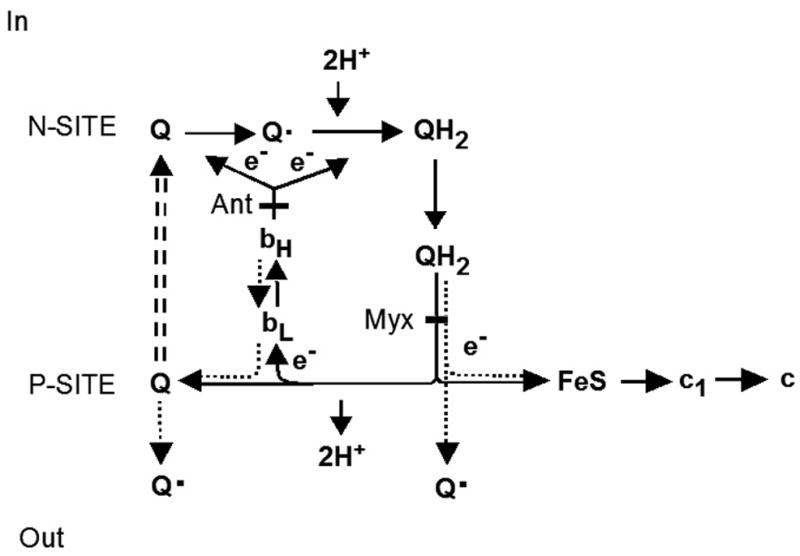
The Q cycle depicted schematically is based on Trumpower et al. and Snyder et al. [4, 32] showing the pathway of electron transfer from reduced Q6 (QH2) to cytochrome c. At the P site, two electrons are transferred in a concerted manner from QH2 to the iron-sulfur protein and to cytochrome bL. Myxothiazol (Myx) binds to the P site and prevents electron transfer to the Rieske protein. At the N site, coenzyme Q (Q) is reduced by cytochrome bH first to the semiquinone and then to QH2. This step is inhibited by antimycin (Ant), which binds to the N site. The stippled arrows show the pathway of reduction of coenzyme Q to the semiquinone at the P site in the presence of antimycin A or myxothiazol. The semiquinone formed in the presence of myxothiazol is the result of a slow leak of electrons to the iron sulfur protein [21].
The functionality of the P site in a coq10 mutant was studied by examining antimycin A or myxothiazol-dependent production of reactive oxygen species by assaying for H2O2 [21,22]. Yeast stains with different respiratory capacities were also used as controls. Therefore, the effect of the two inhibitors was also tested in the parental wild type strain, in a coq2 mutant lacking Q6 as a result of a deletion in the gene for p-hydroxybenzoate: polyprenyl transferase (that catalyzes the second step of coenzyme Q biosynthesis [24]), in a bcs1 mutant arrested in assembly of the bc1 complex [25], and in wild type and coq10 harboring the pYES2/AfAOX plasmid, expressing Aspergillus fumigatus alternative oxidase (AOX) under the control of the GAL10 promoter [17]. A. fumigattus AOX transfers electrons directly from QH2 to oxygen [17].
Antimycin A increased H2O2 release in wild type and coq10 mitochondria. However, a clear myxothiazol-dependent increment occurred only in the wild type. (Fig. 2A). On the other hand, the spontaneously high H2O2 release witnessed in the coq2 and bcs1 mutants suggests a greater accumulation of flavin free radicals at the NADH and/or succinate dehydrogenase sites. Under conditions of Q6 deficiency, when the oxidation of reduced Q6 is blocked as a result of a defective bc1 complex or respiratory inhibitor, keeping the the FMN flavin reduced, NADH-coenzyme Q reductase (complex I) of mammalian and other mitochondria, including those of most yeast, has been shown to produce reactive oxygen species (ROS) [26]. NADH-coenzyme Q reductase of S. cerevisiae also contains FMN but is evolutionarily distinct from complex I. Even so, conditions that prevent reduction of Q6 in S. cerevisiae may be expected to also favor increased production of H2O2 through accumulation of flavin semiquinones.
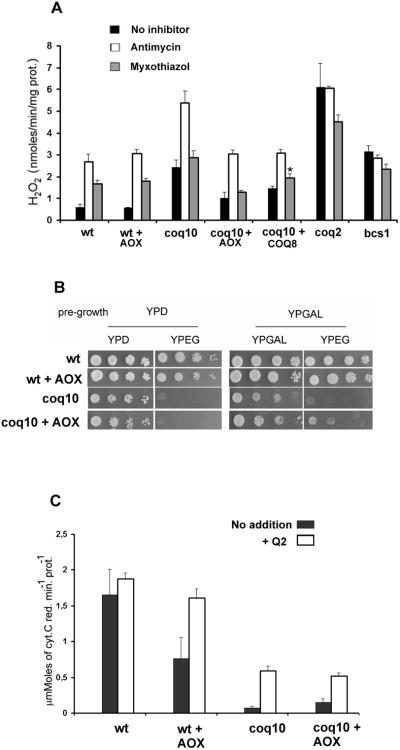
Fig. 2. Antimycin- and myxothiazol-dependent production of H2O2.
Mitochondria were isolated from the following strains: wild type W303-1A; the coq mutants aW303ΔCOQ2 (coq2) and aW303 ΔCOQ10 (coq10); the bc1 deficient mutant aW303 ΔBCS1 (bcs1); wild type and coq10 mutant transformed with pYES2-AfAOX (wt+AOX and coq10 + AOX) and YEp352-COQ8 [10] (coq10 + COQ8). A) Mitochondria (100 μg protein) were assayed as described in the Materials and Methods section for H2O2 release before and after the addition of 0.5 μg/ml antimycin A or myxothiazol at a final concentration of 0.5 μM. Both inhibitors increase the basal rate of monoelectronic reduction of oxygen, which generates the superoxide radical O2. [21] that then dismutates to H2O2 [30]. The vertical bars indicate ranges of four independent experiments. * p< 0.01 vs absence of inhibitor, statistical analysis and comparison were performed using unpaired Student’s t test conducted by graphPad Prism software. B) Respiratory growth properties of wild type cells, coq10 mutants, and respective transformants with pYES2/AfAOX (wt+AOX, coq10+AOX) after pre-growth on glucose (YPD), or galactose (YPGal). C) Measurements of NADH cytochrome c reductase activity in isolated mitochondria from wild type cells and coq10 mutants and respective transformants with pYES2/AfAOX (wt+AOX, coq10+AOX), with or not the addition of 1 μM of synthetic Q2. The vertical bars indicate ranges of four independent experiments.
We reasoned that the presence of a bypass for reduced coenzyme Q might alleviate the production of ROS in the coq10 mitochondria, and, indeed we did observe less H2O2 in the mutant expressing the alternative oxidase (AOX) of Aspergillus fumigatus.
Indeed, the ROS production in the coq10 mutant was enhanced by a factor of 4–6 (Fig. 2A), while in the coq10/AOX transformant the H2O2 release was only two times that observed in the wild type cells. There was also a decrement in the antimycin A-dependent release in the mutant strain expressing AOX. Antimycin A stimulation in the coq10 mutants, however, was qualitatively different from that seen in the coq2 or bcs1 mutant. Antimycin A elicited a three fold increase in ROS formation in the coq10 mutant when normalized to the rate measured in the absence of inhibitor. In agreement with a previous report [21], antimycin A increased the rate of H2O2 release in wild type and AOX transformants, but had no effect in the coq2 and bcs1 mutants over and above the rate seen without the inhibitor (Fig. 2B). The ability of antimycin A to stimulate ROS formation in the coq10 mutant suggests that electron transfer from the low potential cytochrome bL to Q6 at the P site does not depend on Coq10p. Myxothiazol also increased H2O2 production in wild type mitochondria, although the increment over the basal rate was less pronounced (3-fold). However, in the coq10 mutant and in the coq10/AOX transformant, there were no significant effects on H2O2 release due to the addition of myxothiazol. Overexpression of COQ8 partially suppresses the coq10 mutant respiratory defect [10]. Accordingly, we found that the presence of extra COQ8 in these experiments decreased the rate of H2O2 release, whereas antimycin A treatment promoted H2O2 levels similar to the wild type strains and coq10+AOX transformant. On the other hand, we also observe that the COQ8 overexpressing strain presents a mild, but statistically significant increase in H2O2 when in the presence of myxothiazol.
The expression of the GAL10/AfAOX fusion in coq10 cells also improved their respiratory growth when pre-incubated in media containing galactose (Fig. 2B). However the specific enzymatic activity of NADH cytochrome c reductase of coq10/AOX transformants did not change significantly (Fig. 2C). Curiously, wild type cells harboring the AOX plasmid had less NADH cytochrome c reductase activity when compared to the untransformed cells, but the addition of synthetic Q2 to wild type/AOX mitochondria reestablished the enzymatic activity to wild type levels, indicating that the AOX electronic bypass is responsible for this decrement.
Detection of semiquinones by EPR spectroscopy and the steady state level of bc1 complex in the coq10 mutants
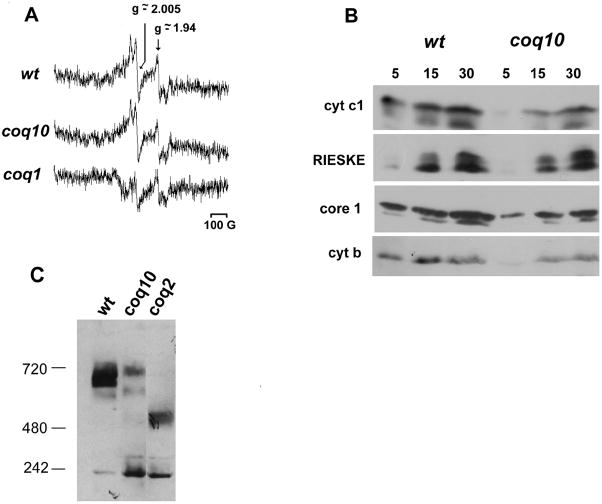
The presence of Q6 semiquinones in coq10 mutants was checked by low temperature EPR spectroscopy of mitochondria from wild type, coq10 and coq1 mutants. coq1 mitochondria are completely devoid of Q6 whereas coq10 organelles have near wild type levels of Q6 [10]. Spectra were obtained from mitochondria with membrane potentials maintained at 65 mV by the addition of extramitochondrial KCl [27] and using succinate as a respiratory substrate, to minimize the contribution of flavins to the semiquinone signal at g ~2.005 [28,29]. Under these conditions, the magnitude of the g ~2.005 signal was comparable in wild type and coq10 mutant mitochondria but was significantly lower in the coq1 mutant (Fig. 3). Because of the absence of Q6 in the coq1 mutant, this signal is most likely derived from flavin semiquinones (Fig. 3a). Semiquinone concentrations in these samples were estimated by double integration of the EPR spectrum and comparison to the standard 4-hydroxy-2,2,6,6-tetramethyl-1-piperidinyloxy solution scanned under the same conditions. The calculated value for the wild type mitochondria was 1.3 nmoles/mg protein while the coq10 mutant was 1.7 nmoles/mg protein. The semiquinone concentration in the coq1 mutant was not calculated because the spectrum obtained for this mutant contained a depression close to the semiquinone signal precluding the quantification by double integration. The signals detected at g ~ 1.94, corresponding to the iron-sulfur centers, were similar in the two mutants. Approximately half of coq10ρ+ cells and one fifth of coq1ρ+ were converted to ρ− and ρ0 after cell growth for mitochondrial preparation. There are a number of cellular events that lead to mitochondrial DNA instability in yeast [30]. We can speculate that changes in the mitochondrial redox state may trigger the observed instability in these coq mutants. Nevertheless, this fact could also explain their lower iron-sulfur signal compared to wild type mitochondria. In order to evaluate the presence of the bc1 complex in the coq10 mutant mitochondria, the steady state concentrations of some bc1 subunits was checked and compared to wild type mitochondria using different amounts of mitochondrial proteins for quantitative evaluation (Fig. 3B). Western analyses with subunit-specific antibodies revealed six times less cytochrome b, and half to two orders of magnitude decrements in the amounts of cytochrome c1, Rieske iron-sulfur and core 1 proteins in the coq10 mitochondria, probably as a consequence of the coq10 mitochondrial DNA instability. On the other hand, in a coq2 mutant, the steady state levels of these bc1 complex proteins were one forth lower that of the wild type (not shown). Accordingly, the addition of diffusible Q2 to the coq10 mitochondria restored less than half of the NADH-cytochrome c reductase activity of the wild type (Fig. 2C), which is also observed in other coq mutants [9, 14, 24]. In agreement with this lower concentration of bc1 complex subunits in the coq10 mutant, Fig. 3C shows one-dimensional BN-PAGE of wild type, coq10 and coq2 mutants mitochondria digitonin extracts, immuno-detected with apocytochrome b. The predominant signal indicates the presence of high molecular mass complexes in the wild type and in the coq10 mitochondria digitonin extracts but with altered size in the coq2 extract as detected before in a coq4 point mutant [31]. These high molecular mass complexes correspond to respiratory super complexes, which in yeast should involve the association of cytochrome c oxidase and bc1 complex dimer [32]. Immuno-detection using Cox4p antibodies also revealed the same high molecular mass complexes at the same size and intensity (not shown). It is noteworthy that coq10 mitochondria extracts revealed complexes apparently at the same size of the wild type, but much less abundant. Altogether, the EPR spectra and bc1 complex steady state levels suggest that even with less active bc1 complex in the coq10 mitochondria, it accumulates semiquinones concentrations similar to the wild type.
Fig. 3. Detection of semiquinone by EPR spectroscopy and bc1 steady state level.
A) Representative low temperature EPR spectra of mitochondria isolated from W303 wild type cells (wt), coq10 and coq1 mutants maintained at 65 mV by the addition of KCl and succinate. The experimental conditions were as described in Materials and Methods. Spectra were obtained at a microwave power of 10 mW, modulation amplitude of 5G, time constant of 81.920 ms, and a scan rate of 5.96 G. s−1. The receiver gain was 1.12 × 105. Arrows correspond to the expected signal peaks for semiquinones (g ~2.004) and iron-sulfur centers (g ~ 1.94). B) Western blot of bc1 complex subunit polypeptides. 5, 15 and 30 μg of mitochondrial proteins from wild type (wt) and coq10 mutants were separated on a 12% polyacrylamide gel as indicated. The proteins were transferred to nitrocellulose and separately probed with Rieske iron-sulfur protein, core1, cytochrome c1, and cytochrome b antiserum. C) Mitochondria from wild type (wt), coq10, and coq2 mutants were isolated with 2% digitonin and samples representing 250 mg of starting mitochondrial protein were analyzed by BN-PAGE, which immuno-blot was probed with cytochrome b antisera. Estimated molecular masses are indicated and were based on the migration of Fo-F1 ATPase dimmers and monomers [42].
Superoxide anion formation and redox state of coq mutants
Leakage of electrons emanating from NADH and succinate reduce oxygen to the superoxide anion (O2·−), which is dismutated to H2O2 [33]. As already noted, the H2O2 assays indicated substantially higher rates of superoxide production in coq10 and in the coq2 mutant (lacking Q6) (Fig. 3B). Measurements of cellular glutathione, a natural ROS scavenger, were used to further assess the redox state of mutants blocked in electron transfer at the level of the bc1 complex. The increased oxidant production in coq10 and coq2 mutants was supported by their significantly greater content of oxidized glutathione relative to reduced and total glutathione (Fig. 4).
Fig. 4. Whole cell glutathione in wild type and coq10 mutants.
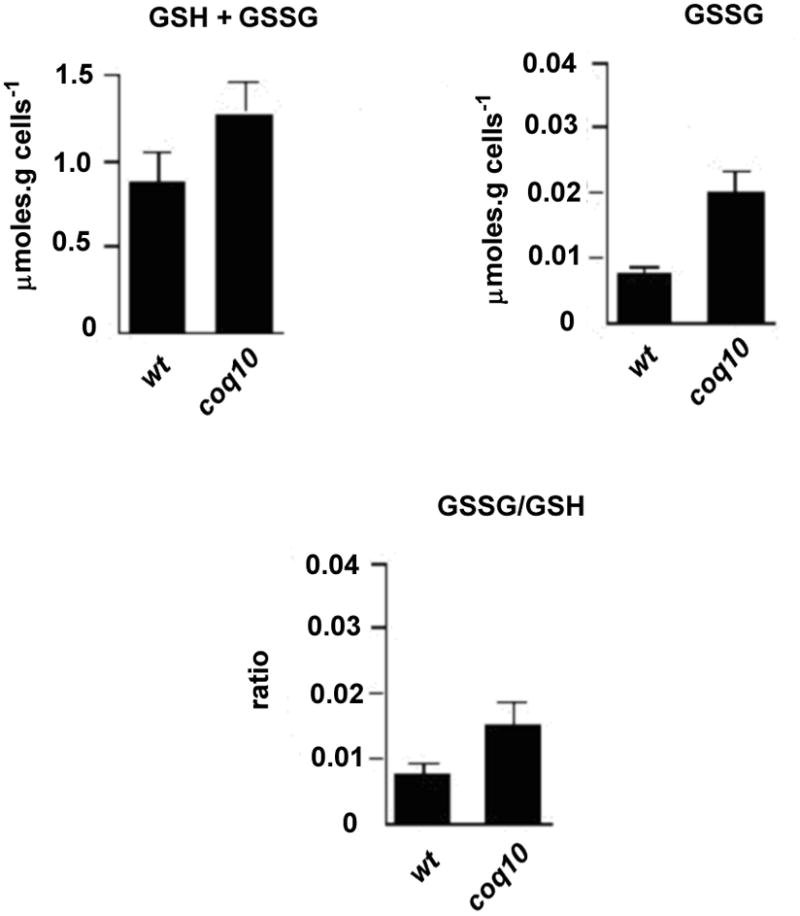
A) Oxidized (GSSG) and total glutathione were assayed in whole cells as previously described [33]. Briefly, total glutathione was determined with 76 μM DTNB in the presence of 0.27 mM NADPH and 0.12 U/ml glutathione reductase. GSSG was estimated by incubation of cells for 1h in the presence of 5 mM N-ethylmaleimide at pH to 7. The concentration of reduced glutathione (GSH) was calculated from the difference between total glutathione and GSSG and used to express the ratio GSSG/GSH. The values reported are averages of three independent measurements with the ranges indicated by the vertical bars.
Discussion
The yeast COQ10 gene codes for a mitochondrial inner membrane protein that binds Q6 and is essential for respiration [10–12] Unlike coq1-9 mutants that fail to synthesize Q6 [7–9], yeast coq10 mutants have normal amounts of Q6, but respiration is completely restored by the addition of the more diffusible Q2 [10,12].
The ability of Coq10p to bind Q6 suggested that one of its functions might be the delivery/exchange of Q6 between the bc1 complex and the large pool of free Q6 during electron transport [10]. This idea was supported by the homology of Coq10p to the reading frame CC1736 of Caulobacter crescentus, which codes for a member of the START superfamily [10, 12] implicated in the delivery of polycyclic compounds such as cholesterol. These compounds bind to a hydrophobic tunnel that is a structural hallmark of this protein family. Another possible function of Coq10p was proposed to be in the transport of Q6 from its site of synthesis to its active sites in the bc1 complex, which would also require Coq10p binding to Q6.
To better understand the function of Coq10p we tested the reducibility of Q6 in a coq10 null mutant in the presence of inhibitors that block Q6 binding to the P (o) and N (i) sites of the bc1 complex. Reduction of Q6 was also examined by comparing the EPR signals associated with semiquinone radicals in wild type and mutant mitochondria and by measuring their concentration of oxidized and reduced glutathione. Since glutathione is an effective scavenger of ROS, the ratio of oxidized to reduced glutathione serves as an index of redox state.
Inhibition of respiration in mammalian and yeast mitochondria with antimycin A has previously been shown to increase the rate of Q reduction to oxygen radicals [20, 21]. In agreement with these data, addition of antimycin A and myxothiazol to respiratory competent yeast mitochondria was found to stimulate oxygen radicals formation by 6 and 3-fold, respectively, as inferred by the rate of H2O2 released. A significant (3-fold) antimycin A-dependent increment in ROS was also observed in the coq10 mutant. The stimulation by antimycin A was not observed in a bc1 mutant or in mutants lacking Q6, and was much lower in the coq10 mutant when myxothiazol was used. The increase in ROS production in the presence of antimycin A indicates that the mutant is capable of transferring an electron from cytochrome bL to Q6 at the P site. Coq10p, therefore, is not required for the accessibility of Q6 to the bL center at the P site. Moreover, the presence of the Aspergillus fumigatus alternative oxidase [17] as a bypass for reduced Q alleviates H2O2 release from the coq10 mutant and even improved the respective respiratory growth. These results are also supported by EPR spectroscopy of mitochondria. The signal at g~2.005 corresponds to semiquinones and presented a lower magnitude in coq1 mitochondria. Since this mutant lacks Q6, the residual signal at g~2.005 is most likely contributed by flavin semiquinone. Because of the lower steady state level of bc1 complex in the coq10 mitochondria, the real magnitude of the EPR signal should be larger in the mutant than in wild type cells.
The possible myxothiazol-dependent reduction of Q6 to the semiquinone at the P site has been proposed to result from an incomplete inhibition of electron transfer to the iron-sulfur protein [19, 20, 34]. In the strains tested, the presence of myxothiazol elevated the H2O2 release only in the wild type cells and in the coq10 mutant overexpressing COQ8.
The Q6 deficient mitochondria of the coq2 mutant had a higher basal rate of ROS production than the wild type. The source of the extra ROS is probably NADH and succinate dehydrogenase-associated flavins. Similar results were reported for a Q6 deficient coq7 mutant, but only when the mitochondria were assayed at 42°C [35]. Since the assays in the present study were done at 30°C, the difference in ROS production may stem from the genetic background of the W303 strain used in the present study, which could engender a feebler oxidative stress response [36]. Our experiments do not distinguish between flavin and Q6 as the source of the increased free radicals in the bcs1 mutant. It is worth emphasizing that even though the coq2 and bcs1 mutants both displayed higher basal rates of ROS production, these were not further enhanced by the addition of antimcyin A, as was the case with wild type and coq10 mutant mitochondria.
Experimental procedures
Yeast strains and growth media
The genotypes and sources of the yeast strains used in this study are listed in Table I. The compositions of YPD, YPEG and minimal glucose medium have been described elsewhere [10].
Table 1.
Genotypes and Sources of Saccharomyces cerevisiae Strains
| Strain | Genotype | Source |
|---|---|---|
| W303-1A |
MATa ade2-1, trp1-1, his3-115, leu2-3,112 ura3-1 ρ+, canR |
Rothstein, R. Columbia University |
| aW303 ΔCOQ1 |
MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coq1::LEU2 |
[14] |
| aW303 ΔCOQ2 |
MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coq2::HIS3 |
[23] |
| aW303 ΔCOQ10 |
MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coq10::HIS3 |
[10] |
| aW303 ΔBCS1 |
MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 bcs1::HIS3 |
[24] |
Dr. R. Rothstein, Department of Human Genetics, Columbia University, New York, NY
O2 consumption
Mitochondrial and spheroplast oxygen consumption was monitored on a computer-interfaced Clark-type electrode at 30°C with 1 mM malate/glutamate, 2% ethanol or 1 μmol of NADH as substrates in the presence of mitochondria at 400 μg/ml of protein concentration, or spheroplasts at 600 μg/ml of total cell protein. All measurements were carried out in the presence of 0.002% digitonin. In order to block cytochrome c oxidase respiration, 1 mM KCN was added at the end of the trace.
H2O2 production
H2O2 formation in mitochondria was monitored for 10 min at 30°C in a buffer containing 50 μM Amplex Red (Molecular Probes), 0.5 U/ml horse radish peroxidase (Sigma), 2% ethanol, 1 mM malate, 6 mM glutamate and 100 μg/ml of mitochondrial protein. Resorufin production was recorded using a fluorescence spectrophotometer at 563 nm excitation and 587 nm emission wavelengths. A calibration curve of known amounts of H2O2 was used to convert fluorescence to concentration of H2O2. Antimycin A and myxothiazol were added to a final concentration of 0.5 μg/ml and 0.5 μM, respectively.
Glutathione assays
Oxidized glutathione (GSSG), reduced glutathione (GSH), and total glutathione were determined in late stationary phase using the DTNB (5,5′-dithiobis-(2-nitrobenzoic acid) colorimetric assay [37].
EPR spectroscopy
EPR spectra were recorded at 77 K with a Bruker EMX spectrometer equipped with an ER4122 SHQ 9807 high sensitivity cavity. For these experiments, 8 mg of mitochondrial protein suspended in 0.6 M sorbitol, 10 mM Tris-Cl pH 7.5 and 1 mM EDTA, were maintained at 65 mV by incubation for 2 min with KCl (12.4 mM), valinomycin (0.1 μg/ml) and succinate (1 mM final) [27]. The samples were immediately transferred to a 1 ml disposable syringe, frozen and stored in liquid nitrogen until analysis. Spectra were acquired by extrusion of the samples from the syringe into a finger-tip Dewar flask containing liquid nitrogen and were examined at 77 K in the region of g~2.000 [38]. The spectra shown here were corrected by baseline subtractions. The spectrum of 1,1-diphenyl-2-picrylhydrazyl (DPPH) (g=2.004), and those of known concentrations of 4-hydroxy-2,2,6,6-tetramethyl-1-piperidinyloxy, acquired under the same conditions, were used as standards for determining the g values and semiquinone concentrations, respectively.
solutions acquired under the same conditions, was used as a standard for determining the g values.
Miscellaneous procedures
Measurements of respiratory enzymes were performed as described previously [39]. Mitochondria were prepared from yeast grown in rich media containing galactose as a carbon source [40]. Western blot quantifications were performed by 1Dscan EX software (Scanalytics, Inc.) For BN-PAGE, mitochondrial proteins were extracted with 2% final concentration of digitonin and separated on a 4–13% linear polyacrylamide gel [41]. Proteins were transferred to a PVDF membrane and probed with rabbit polyclonal antibodies against yeast cytochrome b. The antibody-antigen complexes were visualized using the SuperSignal chemiluminescent substrate kit (Pierce).
Acknowledgments
We thank Dr. C.F. Clarke, University of California, for providing yeast strains, and T Magnanini and S.A. Uyemura (Universidade de Sao Paulo) for the Aspergillus fumigatus alternative oxidase plasmid. We are indebt to Edlaine Linares (IQ-USP) and Fernando Gomes (ICB-USP) for technical assistance. This work was supported by grants and fellowships from the Fundação de Amparo a Pesquisa de São Paulo (FAPESP – 2007/01092-5; 2006/03713-4), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 470058/2007-2), INCT de Processos Redox em Biomedicina-Redoxoma (CNPq-FAPESP/CAPES) and Research Grant HL022174 from the National Institutes of Health.
References
- 1.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P. The protonmotive Q cycle: a general formulation. FEBS Lett. 1975;59:137–139. doi: 10.1016/0014-5793(75)80359-0. [DOI] [PubMed] [Google Scholar]
- 3.Trumpower BL. The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. J Biol Chem. 1990;265:11409–11412. [PubMed] [Google Scholar]
- 4.Trumpower BL. A concerted, alternating sites mechanism of ubiquinol oxidation by the dimeric cytochrome bc(1) complex. Biochim Biophys Acta. 2002;1555:166–173. doi: 10.1016/s0005-2728(02)00273-6. [DOI] [PubMed] [Google Scholar]
- 5.Gloor U, Wiss O. The biosynthesis of ubiquinone. Experientia. 1958;14:410–411. doi: 10.1007/BF02160434. [DOI] [PubMed] [Google Scholar]
- 6.Marchal D, Boireau W, Laval JM, Moiroux J, Bourdillon C. Electrochemical measurement of lateral diffusion coefficients of ubiquinones and plastoquinones of various isoprenoid chain lengths incorporated in model bilayers. Biophys J. 1998;74:1937–1948. doi: 10.1016/S0006-3495(98)77902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. icrobiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7S:S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson A, Gin P, Marbois BN, Hsieh EJ, Wu M, Barros MH, Clarke CF, Tzagoloff A. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J Biol Chem. 2005;280:31397–31404. doi: 10.1074/jbc.M503277200. [DOI] [PubMed] [Google Scholar]
- 10.Barros MH, Johnson A, Gin P, Marbois BN, Clarke CF, Tzagoloff A. The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J Biol Chem. 2005;280:42627–42635. doi: 10.1074/jbc.M510768200. [DOI] [PubMed] [Google Scholar]
- 11.Cui TZ, Kawamukai M. Coq10, a mitochondrial coenzyme Q binding protein, is required for proper respiration in Schizosaccharomyces pombe. FEBS J. 2009;276:748–759. doi: 10.1111/j.1742-4658.2008.06821.x. [DOI] [PubMed] [Google Scholar]
- 12.Busso C, Bleicher L, Ferreira-Junior JR, Barros MH. Site-directed mutagenesis and structural modeling of Coq10p indicate the presence of a tunnel for coenzyme Q6 binding. FEBS Letters. 2010;584:1609–1614. doi: 10.1016/j.febslet.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh EJ, Gin P, Gulmezian M, Tran UC, Saiki R, Marbois BN, Clarke CF. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch Biochem Biophys. 2007;463:19–26. doi: 10.1016/j.abb.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gin P, Clarke CF. Genetic evidence for a multi-subunit complex in Coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase. J Biol Chem. 2005;280:2676–2681. doi: 10.1074/jbc.M411527200. [DOI] [PubMed] [Google Scholar]
- 15.Tauche A, Krause-Buchholz U, Rödel G. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 2008;8:1263–1275. doi: 10.1111/j.1567-1364.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 16.Soccio RE, Adams RM, Romanowski MJ, Sehayek, Burley SK, Breslow JL. The cholesterol-regulated StarD4 gene encodes a StAR-related lipid transfer protein with two closely related homologues, StarD5 and StarD6. Proc Natl Acad Sci U S A. 2002;99:6943–6948. doi: 10.1073/pnas.052143799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnani T, Soriani FM, Martins VP, Nascimento AM, Tudella VG, Curti C, Uyemura SA. Cloning and functional expression of the mitochondrial alternative oxidase of Aspergillus fumigatus and its induction by oxidative stess. FEMS Microbiol Lett. 2007;271:230–238. doi: 10.1111/j.1574-6968.2007.00716.x. [DOI] [PubMed] [Google Scholar]
- 18.Wikström MK, Berden JA. Oxidoreduction of cytochrome b in the presence of Antimycin. Biochim Biophys Acta. 1972;283:403–420. doi: 10.1016/0005-2728(72)90258-7. [DOI] [PubMed] [Google Scholar]
- 19.von Jagow G, Ljungdahl PO, Graf P, Ohnishi T, Trumpower BL. An inhibitor of mitochondrial respiration which binds to cytochrome b and displaces quinone from the iron–sulfur protein of the cytochrome bc1 complex. J Biol Chem. 1984;259:6318–6326. [PubMed] [Google Scholar]
- 20.Starkov AA, Fiskum G. Myxothiazol induces H2O2 production from mitochondrial respiratory chain. Biochem Biophys Res Commun. 2001;281:645–650. doi: 10.1006/bbrc.2001.4409. [DOI] [PubMed] [Google Scholar]
- 21.Dröse S, Brandt U. The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. J Biol Chem. 2008;283:21649–21654. doi: 10.1074/jbc.M803236200. [DOI] [PubMed] [Google Scholar]
- 22.Muller F, Crofts AR, Kramer DM. Multiple Q-cycle bypass reactions at the Qo site of the cytochrome bc1 complex. Biochemistry. 2002;41:7866–7874. doi: 10.1021/bi025581e. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Chobot SE, Osyczka A, Wraight CA, Dutton PL, Moser CC. Quinone and non-quinone redox couples in Complex III. J Bioenerg Biomembr. 2008;40:493–499. doi: 10.1007/s10863-008-9174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashby MN, Kutsunai SY, Ackerman S, Tzagoloff A, Edwards PA. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate: polyprenyltransferase. J Biol Chem. 1992;267:4128–4136. [PubMed] [Google Scholar]
- 25.Nobrega FG, Nobrega MP, Tzagoloff A. BCS1, a novel gene required for the expression of functional Rieske iron-sulfur protein in Saccharomyces cerevisiae. EMBO J. 1992;11:3821–3829. doi: 10.1002/j.1460-2075.1992.tb05474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.St-Pierre J, Buckingham JA, Roebuck SJ, Brand DM. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;47:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 27.Kowaltowski AJ, Cosso RG, Campos CB, Fiskum G. Effect of Bcl-2 overexpression on mitochondrial structure and function. J Biol Chem. 2002;277:42802–4807. doi: 10.1074/jbc.M207765200. [DOI] [PubMed] [Google Scholar]
- 28.Ruzicka FJ, Beinert H, Schepler KL, Dunham WR, Sands RH. Interaction of ubisemiquinone with a paramagnetic component in heart tissue. Proc Natl Acad Sci U S A. 1975;72:2886–2890. doi: 10.1073/pnas.72.8.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seddiki N, Meunier B, Lemesle-Meunier D, Brasseur G. Is cytochrome b glutamic acid 272 a quinol binding residue in the bc1 complex of Saccharomyces cerevisiae? Biochemistry. 2008;47:2357–2368. doi: 10.1021/bi701905a. [DOI] [PubMed] [Google Scholar]
- 30.Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marbois B, Gin P, Gulmezian M, Clarke CF. The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim Biophys Acta. 2009;1791:69–75. doi: 10.1016/j.bbalip.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boveris A, Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem J. 1973;134:707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder CH, Gutierrez-Cirlos EB, Trumpower BL. Evidence for a concerted mechanism of ubiquinol oxidation by the cytochrome bc1 complex. J Biol Chem. 2000;275:13535–13541. doi: 10.1074/jbc.275.18.13535. [DOI] [PubMed] [Google Scholar]
- 35.Davidson JF, Schiestl RH. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol Cell Biol. 2001;24:8483–8489. doi: 10.1128/MCB.21.24.8483-8489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veal EA, Ross SJ, Malakasi P, Peacock E, Morgan BA. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J Biol Chem. 2003;278:30896–30904. doi: 10.1074/jbc.M303542200. [DOI] [PubMed] [Google Scholar]
- 37.Monteiro G, Kowaltowski AJ, Barros MH, Netto LE. Glutathione and thioredoxin peroxidases mediate susceptibility of yeast mitochondria to Ca(2+)-induced damage. Arch Biochem Biophys. 2004;425:14–24. doi: 10.1016/j.abb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Giorgio S, Linares E, Ischiropoulos H, Von Zuben FJ, Yamada A, Augusto O. In vivo formation of electron paramagnetic resonance-detectable nitric oxide and of nitrotyrosine is not impaired during murine leishmaniasis. Infect Immun. 1998;66:807–814. doi: 10.1128/iai.66.2.807-814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tzagoloff A, Akai A, Needleman RB, Zulch G. Assembly of the mitochondrial membrane system. Cytoplasmic mutants of Saccharomyces cerevisiae with lesions in enzymes of the respiratory chain and in the mitochondrial ATPase. J Biol Chem. 1975;250:8236–8242. [PubMed] [Google Scholar]
- 40.Herrmann JM, Foelsch H, Neupert W, Stuart RA. Isolation of yeast mitochondria and study of mitochondrial protein translation. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. I. Academic Press; San Diego, CA: 1994. pp. 538–544. [Google Scholar]
- 41.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 42.Rak M, Tzagoloff A. F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc Natl Acad Sci U S A. 2009;106:18509–18514. doi: 10.1073/pnas.0910351106. [DOI] [PMC free article] [PubMed] [Google Scholar]