Abstract
The differentiation of patient-derived induced pluripotent stem cells (iPSCs) to committed fates such as neurons, muscle and liver is a powerful approach for understanding key parameters of human development and disease1–6. Whether undifferentiated iPSCs themselves can be used to probe disease mechanisms is uncertain. Dyskeratosis congenita (DC) is characterized by defective maintenance of blood, pulmonary tissue, and epidermal tissues and is caused by mutations in genes controlling telomere homeostasis7,8. Short telomeres, a hallmark of DC, impairs tissue stem cell function in mouse models, suggesting that a tissue stem cell defect underlies the pathophysiology of DC9,10. Here, we show that even in the undifferentiated state, iPSCs from DC patients harbor the precise biochemical defects characteristic of each form of the disease and that the magnitude of the telomere maintenance defect in iPSCs correlates with clinical severity. In iPSCs from patients with heterozygous mutations in TERT, the telomerase reverse transcriptase, a 50% reduction in telomerase levels blunts the natural telomere elongation that accompanies reprogramming. In contrast, mutation of dyskerin (DKC1) in X-linked DC severely impairs telomerase activity by blocking telomerase assembly and disrupts telomere elongation during reprogramming. In iPSCs from a form of DC caused by mutations in TCAB1, telomerase catalytic activity is unperturbed, yet the ability of telomerase to lengthen telomeres is abrogated, because telomerase mislocalizes from Cajal bodies to nucleoli within the iPSCs. Extended culture of DKC1-mutant iPSCs leads to progressive telomere shortening and eventual loss of self-renewal, suggesting that a similar process occurs in tissue stem cells in DC patients. These findings in iPSCs from DC patients reveal that undifferentiated iPSCs accurately recapitulate features of a human stem cell disease and may serve as a cell culture-based system for the development of targeted therapeutics.
Patients with DC have high rates of bone marrow failure, pulmonary fibrosis and cancer, and a triad of epidermal findings, including oral leukoplakia, nail dystrophy and abnormal skin pigmentation7,11. The severity of DC and its age of onset vary widely; the reason for this range of phenotypes is unclear, but depends in part on the mode of inheritance and the specific genes involved. Patients with X-linked DC due to mutations in DKC1 typically present in early childhood with the classic manifestations of the disease11,12. In contrast, autosomal dominant DC due to mutations in TERT or TERC, the telomerase RNA component, presents later in life (i.e. in adolescence or young adulthood), and disease manifestations are often milder, with patients commonly lacking the epidermal triad. Patients with autosomal recessive DC due to TCAB1 mutations have the classic and severe form of the disease, with early age of onset and shortened life expectancy13. All forms of DC are associated with very short telomeres in peripheral blood lymphocytes14. Telomerase is restricted in its expression in many tissues to stem cells and progenitor cells, and the challenges in isolating and studying these rare cells have precluded a direct analysis of telomere maintenance mechanisms in stem cells from patients with DC. In skin fibroblasts, telomerase expression is silenced, but during reprogramming the TERT gene is reactivated and telomerase activity is reconstituted1,15–17. DC iPSCs have been used to study telomerase reactivation and TERC regulation during reprogramming, but thus far disease-specific iPSCs have not recapitulated telomere shortening15.
To study DC in patient-derived iPSCs, fibroblasts from five patients carrying different mutations in TERT (P704S and R979W), TCAB1 (H376Y/G435R) and DKC1 (DKC1_L54V and ΔL37) were transduced with retroviruses or lentiviruses expressing the reprogramming factors SOX2, c-Myc, KLF4 and OCT4 (Supplementary Tables 1, 2). DC fibroblasts were resistant to reprogramming in ambient oxygen, but successful reprogramming was achieved under low oxygen conditions (5% O2), a method that mitigates cellular stress responses18 (Supplementary Table 1). To generate isogenic iPSCs with the DKC1_ΔL37 mutation but with long telomeres, we reprogrammed DKC1_ΔL37 fibroblasts in which TERT and TERC were stably overexpressed, which bypasses the effects of the dyskerin mutation19 (DKC1_ΔL37_TT iPSCs). The resulting iPSCs from DC patients were morphologically indistinguishable from human embryonic stem cells (hESCs), were positive for all markers of pluripotency tested and gave rise to cells derived from all three germ layers (Supplementary Fig. 1–6).
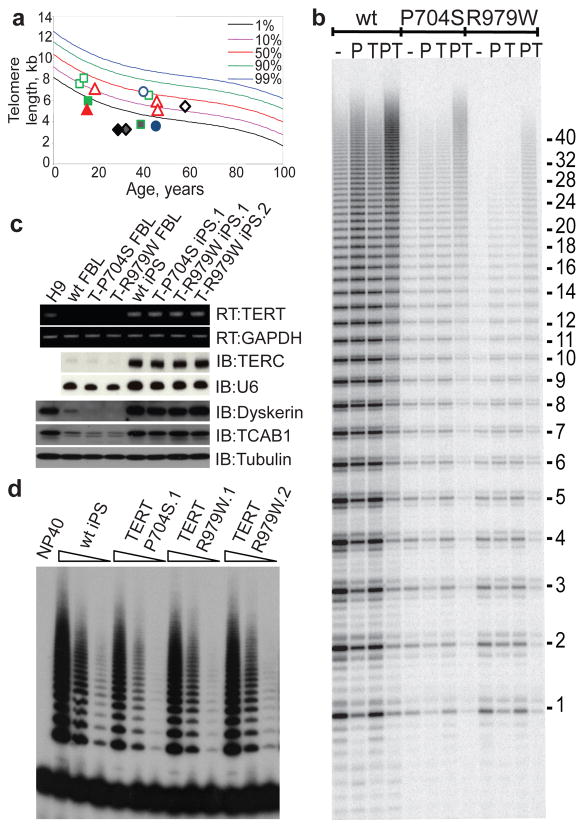
Both autosomal dominant TERT mutation-positive patients presented with bone marrow failure and short telomeres, but lacked the epidermal triad (Fig. 1a; Supplementary Fig. 7a, Table 2). To assess the effects of the mutations on telomerase catalytic activity, wild-type or mutant TERT proteins were assembled into telomerase in human 293T cells. Following immunopurification, telomerase activity of each reconstituted enzyme was analyzed using a quantitative direct enzymatic assay (Fig. 1b). For each mutant TERT, the enzymatic activity of reconstituted telomerase was reduced by 90%, and the defect was not suppressed by the telomere-binding proteins POT1 and TPP1, which enhance processivity20 (Supplementary Fig. 8). In the iPSCs derived from these patients, TERT mRNA and TERC were upregulated similarly compared with wild-type iPSCs by RT-PCR and Northern blot, respectively (Fig. 1c). Both dyskerin and TCAB1 were strongly upregulated by Western blot with reprogramming (Fig. 1c). Telomerase activity in both TERT-mutant iPSCs was reduced by approximately 50% compared to wild-type iPSCs, consistent with our findings that each mutant TERT protein retains only 10% residual activity, which when added to the activity from the wild-type allele would be predicted to yield 55% total activity in a heterozygote (Fig. 1d). Thus, our findings in TERT-mutant iPSCs are compatible with a mechanism of telomerase haploinsufficiency, whereby a 50% reduction in activity is the cause of disease in this form of DC21,22.
Figure 1. DC iPSCs with heterozygous TERT mutations show reduced telomerase levels.
a, Telomere lengths by Flow-FISH in peripheral blood lymphocytes from DC patients and their first-degree relatives. Squares, TERT_P704S family; Diamonds, TERT_R979W family; Triangles, TCAB1_H*Y/G*R family; Circles, DKC1_L54V family. Symbols: solid, probands; grey, carriers; Open, first-degree relatives. b, Direct telomerase assays on wild-type TERT, or TERT mutants, assembled with TERC + or − recombinant Pot1 (P), TPP1 (T) or T+P. c, Expression of TERT, TERC, DKC1, and TCAB1 with reprogramming. RT, RT-PCR; IB, immunoblot. GAPDH, U6 and Tubulin, loading controls. d, Telomerase activity by TRAP in wild-type, TERT_P704S and TERT_R979W iPSCs. Range of concentrations represent 4-fold serial dilutions. NP40, buffer control.
The patient with compound heterozygous mutations in TCAB1 presented with classical symptoms of DC, including very short telomeres (Fig. 1a; Supplementary Fig. 7b, Table 2). TERT, TERC, and dyskerin were each appropriately upregulated in TCAB1-mutant iPSCs, whereas TCAB1 protein levels were markedly reduced (Fig. 2a) 13. Although patients with mutations in TCAB1 have short telomeres, telomerase activity was unperturbed by TCAB1 mutations and indistinguishable from activity in wild-type iPSCs (Fig. 2b). TCAB1 is enriched in Cajal bodies, nuclear sites of ribonucleoprotein modification and assembly, and is required for trafficking of telomerase to Cajal bodies23,24. Whereas TCAB1 colocalized in discrete foci with the Cajal body marker p80-coilin in wild-type iPSCs, TCAB1-mutant iPSCs exhibited a dramatic reduction of TCAB1 accumulation in Cajal bodies (Fig. 2c; Supplementary Fig. 9a; p<0.0001). Dyskerin normally accumulates both in Cajal bodies, where it binds small Cajal Body RNAs (scaRNAs) and TERC, and in the nucleolus, where it binds H/ACA small nucleolar RNAs (snoRNAs). Efficient accumulation of dyskerin in Cajal bodies requires functional TCAB113. TCAB1-mutant iPSCs showed a significant reduction in dyskerin accumulation in Cajal bodies, whereas nucleolar localization of dyskerin was unperturbed (Fig. 2d; Supplementary Fig. 9b; p<0.0001). RNA fluorescent in situ hybridization (FISH) using probes complementary to TERC revealed that, whereas TERC localized to a single Cajal body focus in wild-type iPSCs, it dramatically mislocalized to nucleoli in TCAB1-mutant iPSCs (Fig. 2e; Supplementary Fig. 9c,10; p<0.0001). Together, these data show that TCAB1 mutations in patient-derived iPSCs result in mislocalization of the telomerase complex without affecting telomerase activity. Our results indicate that simple catalytic assays can falsely suggest that telomerase is active in a setting in which the telomerase enzyme is profoundly dysfunctional, results reminiscent of the first telomerase mutations in yeast25.
Figure 2. Preserved activity, but pronounced mislocalization of telomerase in TCAB1-mutant iPSCs.
a, Expression of TERT, TERC, DKC1, and TCAB1 with reprogramming. RT, RT-PCR; NB, northern blot; IB, immunoblot. GAPDH, U6 and Tubulin, loading controls. b, Telomerase activity by TRAP in wild-type and TCAB1_H*Y/G*R iPSCs. Range of concentrations represent 4-fold serial dilutions. NP40, buffer control. c, Immunofluorescence for TCAB1 (red) and p80-coilin (green) in wild-type and TCAB1_H*Y/G*R iPSCs. d, Co-staining for dyskerin (red) and p80-coilin (green) in wild-type and TCAB1_H*Y/G*R iPSCs. e, RNA FISH analysis for TERC (red) in wild-type and TCAB1_H*Y/G*R iPSCs. White arrows, Cajal bodies. Blue, DAPI.
Patients with X-linked DC included one with classic DC due to the DKC1_ΔL37 mutation12,15 and another who presented with bone marrow failure, the epidermal triad and very short telomeres due to a DKC1_L54V mutation (Fig. 1a; Supplementary Fig. 7c, Table 2). TERT mRNA, dyskerin protein and TCAB1 protein were upregulated appropriately following cellular reprogramming in DKC1-mutant iPSCs (Fig. 3a). Dyskerin serves a central role in assembling telomerase and other ribonucleoprotein complexes with RNAs containing H/ACA motifs12,26. The H/ACA motif within TERC is shared with scaRNAs and a subset of snoRNAs, involved in modification of splicing RNAs and ribosomal RNAs, respectively26. TERC was reduced in DKC1 fibroblasts by northern blot, consistent with previous studies12 (Fig. 3a). Despite an upregulation of TERC with reprogramming, TERC in DKC1-mutant iPSCs remained significantly suppressed compared with wild-type iPS controls. DKC1 point mutations selectively reduced TERC levels without affecting H/ACA snoRNAs and scaRNAs recapitulating results in lymphoblasts and fibroblasts12 (Supplementary Fig. 11). In marked contrast to TERT-mutant and TCAB1-mutant iPSCs, all DKC1-mutant iPS clones exhibited a severe reduction of telomerase activity, ranging from 5–15% of wild-type controls (Figs. 3b, 3c; Supplementary Fig. 12). Overexpression of TERT and TERC restored TERC levels by northern blot and rescued telomerase activity in DKC1_ΔL37_TT fibroblasts and DKC1_ΔL37_TT iPSCs by TRAP (Supplementary Fig. 12).
Figure 3. Diminished TERC levels, reduced activity and impaired assembly of mature telomerase in X-linked DC iPSCs.
a, Expression of TERT, TERC, DKC1, and TCAB1 with reprogramming. RT, RT-PCR; NB, northern blot; IB, immunoblot. GAPDH, U6 and Tubulin, loading controls. b–c, Telomerase activity by TRAP in (b) DKC1_L54V and (c) DKC1_ ΔL37 iPSCs. Range of concentrations represent 4-fold serial dilutions. NP40, buffer control. Internal PCR control band at bottom of gel. d, Analysis of mature telomerase in iPSCs. Immunoprecipitation of 1 mg of whole-cell extracts with IgG, anti-dyskerin antibodies, or anti-TCAB1 antibodies. Purified complexes were analyzed for the indicated proteins by IB and for TERC by NB. Recovery control, TERC fragment control for differential recovery of RNA.
To assess the composition of the fully assembled telomerase holoenzyme in DKC1-mutant iPSCs, dyskerin and TCAB1 were immunoprecipitated from whole-cell extracts using antibodies directed against each protein. TERC was readily detected by northern blot in dyskerin and TCAB1 complexes from wild-type iPSCs. In contrast, the amount of TERC assembled with either dyskerin or TCAB1 was markedly reduced in DKC1_ΔL37 iPSCs (Fig. 3d, Supplementary Fig. 13). Overexpression of TERT and TERC in DKC1_ΔL37_TT iPSCs rescued the assembly defect and led to an amount of TERC in the mature holoenzyme that exceeded wild-type levels. Overall the amount of TERC in dyskerin and TCAB1 complexes in DKC1-mutant, wild-type and DKC1_ΔL37_TT iPSCs correlated directly with telomerase enzymatic activity in X-linked DC IPSCs. Thus, the reduction in both TERC and telomerase activity in DKC1-mutant iPSCs is consistent with a defect in a dyskerin-mediated assembly step, impairing the maturation of the active telomerase complex.
Upregulation of telomerase leads to significant telomere lengthening during reprogramming of wild-type fibroblasts1,15,16(Fig. 4a–d). However, in TERT-mutant iPSCs, telomere elongation during reprogramming was blunted, with telomeres in TERT-mutant iPSCs always remaining significantly shorter than in wild-type iPSCs (Fig. 4a; Supplementary Fig. 14). In marked contrast, telomere elongation failed in all TCAB1-mutant iPSCs and DKC1-mutant iPSCS. For both TCAB1-mutant iPSCs and DKC1-mutant iPSCs, telomeres were shorter than in their parental fibroblasts and telomeres continued to shorten as cells divided in culture (Fig. 4b–d and Supplementary Fig. 14,15). In DKC1_ΔL37_TT fibroblasts and iPSCs, telomerase overexpression fully restored telomere elongation, with telomere lengths increasing significantly beyond those of their wild-type counterparts (Fig. 4d). These data show that telomerase mutations can severely impair telomere maintenance in DC iPSCs, providing evidence for a defect in maintaining telomeres in DC stem cells.
Figure 4. Impaired telomere maintenance and loss of self-renewal in DC iPSCs.
a–d, Telomere lengths by Southern blot using genomic DNA from fibroblasts and iPSCs. (a) TERT_P704S and TERT_R979W iPSCs (b) TCAB1_H*Y/G*R (c) DKC1_L54V and (d) DKC1_ΔL37 iPSCs and DKC1_ΔL37_TT iPSCs at indicated passages after reprogramming. Molecular weight, kilobases. Black line in a, membrane cut for hybridization. e–h, Telomere FISH on metaphase chromosomes from: e, wild-type iPSCs f, DKC1_ΔL37 iPSC clone 1 and g, DKC1_ΔL37_TT iPSC at p22. White arrows, SFEs. High magnification, inset. h, Quantification of SFEs per metaphase. i, Western blot for p53 and p21 at p9 and p36 in DKC1_ΔL37 iPSCs. DOX, doxorubicin treated. Tubulin, loading control.
With extended growth in cell culture of DKC1_ΔL37 iPSCs, telomeres continued to shorten through passage 19 and the bulk population of telomeres reached a plateau at passage 26 by Southern blot (Supplementary Fig. 15b). Using telomere-FISH, we found that telomere signals were readily detected at all chromosome ends in wild-type iPSCs and in DKC1_ΔL37_TT iPSCs. In contrast, average telomere intensity was greatly reduced in DKC1_ΔL37 iPSCs, which also showed an increase in the number of signal-free ends (SFEs), chromosome ends lacking detectable telomere repeats (Fig. 4e–h p<0.01). Continued passage of DKC1_ΔL37 iPSCs resulted in an abrupt increase in spontaneous differentiation within iPSC colonies and the culture could no longer be maintained as undifferentiated iPSCs after passage 36. Critical telomere shortening leads to a loss of telomere capping function, which triggers a DNA damage response that activates the p53 tumor suppressor protein. The p53 pathway was strongly activated in DKC1_ΔL37 iPSCs at passage 36, as evidenced by p53 protein stabilization and induction of its downstream target p21 by western blot. No such activation of p53 was seen at passage 9, or in late passage human ES cells (Fig. 4i). Taken together, these data show that impaired telomere maintenance in DC iPSCs can ultimately compromise self-renewal, resulting in a finite cellular lifespan.
Our data in patient-derived iPSCs provide evidence for severe defects in telomerase function and telomere maintenance in stem cells from DC patients. The spontaneous differentiation in DKC1-mutant iPSCs suggests that exhaustion of self-renewal in hematopoietic stem cells and other tissue stem cells may underlie the tissue defect in DC. Restoration of telomerase function through pharmacological or genetic means in stem cells from blood, lung or epidermal tissues may therefore provide a rational guide for therapy of DC. Data from these iPSCs provide an explanation for the longstanding clinical observation that X-linked DC is often more severe and presents at a younger age than autosomal dominant DC caused by mutations in TERT or TERC8. Our data indicate that effective telomerase activities in the 15%-50% range may represent a critical threshold in which telomere maintenance is particularly impaired. Thus, a reduction in telomerase activity to the 15–50% range may be necessary to yield severe phenotypes in a single generation, whereas genetic anticipation21,22 through inheritance of heterozygous mutations in TERT for several generations may be important in eliciting disease phenotypes for autosomal dominant patients with greater than 50% residual telomerase activity. Together, our data show that many important features of a human stem cell disease are accurately recapitulated in patient-derived iPSCs, providing an iPSC-based system that is not dependent upon differentiation to probe disease mechanisms or to identify potential therapeutics.
Methods summary
TERT (P704S; R979W), TCAB1 (H376Y/G435R) and DKC1 (L54V) fibroblasts were obtained from skin biopsies from patients in the National Cancer Institute’s IRB-approved study, “Etiologic Investigation of Cancer Susceptibility in Inherited Bone Marrow Failure Syndromes (IBMFS)” (http://marrowfailure.cancer.gov)8. Detailed medical record review, physical examination, comprehensive laboratory evaluation, measurement of telomere length, genetic counseling, and mutation analyses were conducted. DKC1_ΔL37 fibroblasts were purchased from Coriell Cell Repositories.
Methods
Cell culture
Human fibroblasts were cultured in fibroblast media (DMEM supplemented with 15% FBS) at 37°C, 5% CO2 and 5% O2. H9 human embryonic stem cells and iPSCs were cultured on γ-irradiated mouse embryonic fibroblasts in hES cell media consisting of DMEM F12-Glutamax, supplemented with 20% knockout serum, 0.1 mM non-essential amino-acids, 0.1 mM β-mercaptoethanol and 10 ng/ml recombinant human basic fibroblast growth factor (Invitrogen). Human ES and iPSCs were transferred to matrigel-coated plates (Invitrogen) and kept in mTeSR1 media (STEMCELL Technologies) prior to most experiments. In all culture conditions, hES and iPSCs culture media was changed daily and cells were treated with collagenase (1 mg/ml, 5 min) or manually passaged every 5–6 days.
Retroviral and lentiviral Production
pMXs retroviral plasmids encoding hSOX2, hOCT3/4, hc-MYC and hKLF4 as well as the packaging vectors pUMVC and pVSV-G were purchased from Addgene. Plasmids for the production of the single polycistronic lentiviral vector were donated by Dr. Gustavo Mostoslavsky. Retrovirus and lentivirus production for iPS cell reprogramming was performed as described27,28. For generation of DKC1 fibroblasts overexpressing TERT and TERC, retroviruses were generated by cotransfecting plasmids encoding RSV(Gag+Pol), VSV-G, and retroviral expression plasmids containing hTERT and hTERC into 293T cells using calcium phosphate precipitation. Approximately 105 cells were co-transduced with both genes and kept for 7 days under blasticidin and neomycin selection.
iPSC Generation
For retroviral and lentiviral transduction, 105 DC human fibroblasts were seeded per well of a 6-well plate 24 h pre-transduction. For retroviral transduction, cells were infected for 24 h with retroviruses encoding the 4 reprogramming factors (5x concentration for SOX2, KLF4 and c-MYC and 10x concentration for OCT3/4 in the presence of 8 ng/ml polybrene). Cells were then washed in PBS and kept in fibroblast growth media. DKC1 mutant fibroblasts required 3 rounds of viral infection on alternate days for efficient reprogramming. For lentiviral transduction, cells were kept for 24 h with single polycistronic lentiviral vectors encoding SOX2, KLF4, c-MYC and OCT3/4 (10x concentrarion, 8 ng/ml polybrene, one single round of infection), according to the protocol described in28. Three days after the final round of infection, approximately 2×105 cells were briefly trypsinized and transferred to 10cm dishes pre-plated with feeders. 24 h after passaging, cells were washed with PBS and hES media was added to the plate. Media was changed every other day, until background colonies emerged, after which media was changed daily. hES-like colonies appeared from large background colonies 20 days after viral transduction and were manually picked on days 24–30. Colonies that maintained their ES-like morphology were further passaged and analysed for pluripotency potential. During the entire reprogramming period, cells were kept under low oxygen conditions (5% O2). Wild-type iPS cells have been reprogrammed using both retroviral and lentiviral vectors. In all figures, wild-type cells shown were reprogrammed using the same strategy as the DC cells they are compared to. huF-4 (wild-type #1), huF-5 (wild-type #2) and huF-Q (wild-type #3) human dermal fibroblasts were reprogrammed and used as wild-type controls, either with retroviral (wild-type #1) or lentiviral vectors (wild-type #2 and #3).
Immunofluorescence
iPSCs cells were grown on feeders in 48 well plates, fixed with 4% paraformaldehyde/PBS, washed three times with PBS and blocked with 4% goat serum for 1 hr. For Oct4 and Nanog, cells were permeabilized with 1% Triton-X/PBS for 1 h at room temperature prior to blocking. After blocking, primary antibodies were diluted in PBS and cells were incubated overnight at 4°C. The following antibodies were used: SSEA3 (1:200), SSEA4 (1:200), TRA-1–60 (1:200), TRA-1–81 (1:200), all from Millipore; Nanog (1:100, Abcam), and Oct4 (1:200, Santa Cruz). Following incubation, cells were washed twice with PBS and incubated with secondary antibodies for 1 hr at room temperature in the dark. Secondary antibodies used were the Alexa Fluor Series from Invitrogen (all at 1:1,000). Cells where then washed three times with PBS and stained with DAPI for nuclei labeling. Images were taken using a Leica DM5000B microscope coupled to a Leica DFC360FX camera.
Telomere length analysis
Genomic DNA was collected from human fibroblasts, H9 human ESCs and iPSCs at different passages. The isolated genomic DNA was then digested with RsaI and HinfI and fractionated as described previously29. Membranes were prepared by Southern transfer and hybridized to a radioactively end-labeled (TTAGGG)4 oligonucleotide probe as described previously30.
Detection of telomerase activity by TRAP
Human fibroblasts and fully undifferentiated H9 ES cells and iPSCs (grown in matrigel) were lysed in NP40 buffer (25 mM HEPES-KOH, 150 mM KCl, 1.5 mM MgCl2, 10% glycerol, 0.5% NP40, and 5 mM 2ME [pH 7.5] supplemented with protease inhibitors) for 15–30 min on ice. Extracts clarified by centrifugation at 16,000 × g for 10 min were quantified by Bradford assay. TRAP reaction was performed using 2.0 μg, 0.5 μg and 0.125 μg of protein, following a modified protocol from the manufacturer (TRAPeze kit, Chemicon), where the TS extension was carried out prior to PCR amplification.
Direct telomerase activity assays
Activity of each human telomerase complex expressed in HEK 293T cells was determined by a direct assay modified from a published protocol31. Each hTERT contained an N-terminal 3xFLAG tag, and telomerase was immunopurified from cell extracts using anti-FLAG M2 affinity gel (Sigma). The reaction mixture (20 μL) contained 1X human telomerase assay buffer (50 mM Tris-HCl, pH 8.0, 50 mM KCl, 1 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM spermidine), 0.1 μM telomeric DNA primer, 0.5 mM dATP, 0.5 mM dTTP, 2.92 μM dGTP, and 0.33 μM 32P-dGTP (3000 Ci/mmole, 1 Ci = 37GBq) with 6 mL of immunopurified telomerase complex on beads. Reactions were performed with telomerase alone or supplemented with hPOT1, hTPP1-N (amino acids 89–334), or both (0.5μM each)32,33. Reactions were incubated at 30°C for 1 hour, then stopped with the addition of 100 μL of 3.6 M NH4Oac, 20 μg of glycogen, and ethanol (500 mL). After incubating at −80°C for 1 hour, samples were centrifuged at 4°C for 30 min. Pellets were washed with 70% ethanol and resuspended in 10 μL of H2O followed by 10 μL of 2X loading buffer (94% formamide, 0.1X TBE, 0.1% bromophenol blue, 0.1% xylene cyanol). The heat-denatured samples were loaded onto a 10% polyacrylamide/7 M urea/1X TBE denaturing gel for electrophoresis. After electrophoresis, the gel was dried and total radioactivity incorporated into telomerase products was quantified using a Phosphorimager (GE Healthcare). A portion of each immunopurified telomerase was subjected to electrophoresis on an SDS-polyacrylamide gel and Western blotting with an anti-FLAG antibody to confirm equal amounts of hTERT protein in each reaction.
Signal-free ends quantification by quantitative fluorescence in situ hybridrization (Telomere-FISH)
Metaphase chromosomes were prepared from human iPSCs by treatment with Colcemid 0.03 μg/ml KaryoMAX Colcemid Solution (Invitrogen), followed by hypotonic KCl and fixation in cold methanol-acetic acid. Chromosomes hybridization with a Cy3-conjugated (CCCTAA)3 peptide-nucleic acid probe has been described previously30. At least 10 metaphases per sample were analyzed to determine the amount of signal-free ends in each sample. Differences between samples were compared by the two-tailed Fisher’s exact test.
Immunoprecipitations, western blots, and northern blots
Protocols for immunoprecipitation, western-blotting and northern-blotting have been previously described23,34. For western-blotting, primary antibodies included Dyskerin (1:30,000- serum35), SHQ1 (20 ng/ml), TCAB1 (50 ng/ml), NAF1 (20 ng/ml), Pontin (10 ng/ml), Reptin (300 ng/ml), p21 (1:400; Santa Cruz), phospho-p53-Ser15 (1:1,000; Cell Signaling), Tubulin (1:50,000; Sigma) and Lamin-B (1:1,000; Santa Cruz). Generation of polyclonal antibodies has been previously described23,34. For northern-blot analysis, total RNA was purified with Trizol reagent (Invitrogen) and treated with Turbo DNA-free kit (Ambion) to remove genomic DNA contamination. The Northern-blot probes and recovery control used have been described previously23.
In vitro and in vivo differentiation
For spontaneous in vitro differentiation towards endoderm, mesoderm and ectoderm fates, undifferentiated iPSCs were transferred to matrigel plates. 24 h after attaching, cells were washed with PBS and cultured thereafter with differentiation media (hES media depleted of b-FGF), changed daily. When cells reached 90% confluency, they were transferred by trypsinization to gelatin-coated 6-well plates. 21 days after b-FGF depletion cells were fixed with 4% paraformaldehyde/PBS, permeabilized with 1% Triton-X and incubated overnight at 4°C with primary antibodies. The antibodies used were neuronal class III β-tubulin (TUJ1, 1:200, Covance), pan-Cytokeratin (Lu-5, 1:200, Abcam) for ectoderm staining I;α-smooth muscle actin (1:200, Abcam), Desmin (1:200, Lab Vision) for mesoderm staining; and human alpha-fetoprotein (1:200, R&D Biosystems) for endoderm staining. Immunofluorescence was performed as described above. In vivo differentiation through teratoma formation was done as described1. Briefly, approximately 106 iPSCs were injected subcutaneously into dorsal flanks of immunocompromised mice (a/a Foxn1nu/Foxn1nu; The Jackson Laboratory). Tumors were collected after 8–10 weeks, fixed and embedded into paraffin blocks.
Genomic DNA sequencing
For genetic identification of our TERT, TCAB1, and DKC1 cells, total genomic DNA of huF4, huF5, TERT_P704S, TERT_R979W, TCAB1_H*Y/G*R, DKC1_L54V, DKC1_ΔL37 and DKC1_DL37_TT fibroblasts, as well as their respective iPSCs was extracted by using lysis buffer34. 100 ng of isolated genomic DNA was used for PCR with primers flanking the specific mutations:
| TERT exon 5: | Fwd: 5′-GTGGCATGAGGATCCCGTGTGC-3′ |
| Res: 5′-CACAGTCGGCCCCATGTGCTG-3′ | |
| TERT exon 12: | Fwd: 5′-GGCCGTGCGAGGTTTGGATACAC-3′ |
| Res: 5′-GTGTATCCAAACCTCGCACGGCC-3′ | |
| TCAB1 exon 7: | Fwd: 5′-GGTCCTTTGGGAGGATAGATGTGG-3′ |
| Res: 5′-GGAACAGGACCTGGAGTCACCC-3′ | |
| TCAB1 exon 9: | Fwd: 5′-GTGCTGGGATCTCCGGCAGTC-3′ |
| Res: 5′-CTGACCGGAGGCAGTGGCC-3′ | |
| DKC1 exon 3: | Fwd: 5′-GTTCAAAATCGGGTGGGAAG-3′ |
| Res: 5′-CCAAAGTCAAGGATGCCAG-3′ |
Following gel extraction, PCR products were sequenced. Resulting sequences were aligned by Clustal-W2 (EMBL-EBI).
G-band analysis
After mitotic arrest, monolayer cell cultures in log-phase growth were harvested by standard cytogenetic methods of trypsin dispersal, hypotonic shock and fixation. Mitotic cell slide preparations were analyzed by the GTW (G-banding with trypsin and Wright’s stain) banding method36.
Bisulfite sequencing
Genomic DNA (1 μg) was treated with MethylEasy Xceed (Human Genetic Signatures) according to the manufacturer’s recommendations. The human OCT3/4 promoter was PCR-amplified and TOPO-cloned, with at least ten clones from each sample sequenced, following the protocol described37.
PCR
Reverse transcription was carried with Superscript II (Invitrogen). PCR was performed using Taq DNA-polymerase (Qiagen) in a GeneAmp PCR System 9700 machine (Applied Biosystems). Primers for pluripotency (TERT, endo-OCT3/4, endo-SOX2, endo-c-MYC, NANOG, REX1, FGF4, Nodal, GDF3 and ESG1) were described1.
Supplementary Material
Acknowledgments
L.F.Z.B. is the recipient of a Pew Fellowship. M.F.P. and K.T.X are the recipients of NSF Graduate Research Fellowships. F.L.Z. was supported by A*STAR, Singapore. We thank the patients for their valuable contributions and Lisa Leathwood, Westat, Inc., for excellent study support. This work was supported, in part, by the intramural research program of the Division of Cancer Epidemiology and Genetics, NCI, NIH; by a CIRM Shared Research Laboratory grant to R.R.P.; and by grants from the NCI, NIA, NHLBI and CIRM to S.E.A.
Footnotes
Contributions
L.F.Z.B., M.F.P., F.L.Z and S.E.A. designed the experiments and analyzed the data; L.F.Z.B., M.F.P., F.L.Z., H.N.N., K.T.X., A.J.Z., S.M.C., J.C., V.S. and A.C. performed the experiments; N.G., M.W., B.P.A., T.R.C., S.A.S. and R.R.P. analyzed the data; N.G., B.P.A and S.A.S. collected patients material; L.F.Z.B. and S.E.A. wrote the manuscript.
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Rashid ST, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu GH, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011 doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soldner F, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 6.Maehr R, et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci U S A. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walne AJ, Dokal I. Advances in the understanding of dyskeratosis congenita. Br J Haematol. 2009;145:164–172. doi: 10.1111/j.1365-2141.2009.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alter BP, et al. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol. 2010;150:179–188. doi: 10.1111/j.1365-2141.2010.08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HW, et al. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 10.Allsopp RC, Morin GB, DePinho R, Harley CB, Weissman IL. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 2003;102:517–520. doi: 10.1182/blood-2002-07-2334. [DOI] [PubMed] [Google Scholar]
- 11.Bessler M, Wilson DB, Mason PJ. Dyskeratosis congenita. FEBS Lett. 2010;584:3831–3838. doi: 10.1016/j.febslet.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 13.Zhong F, et al. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alter BP, et al. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110:1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal S, et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010 doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marion RM, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell stem cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell stem cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell stem cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Wong JM, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006;20:2848–2858. doi: 10.1101/gad.1476206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vulliamy T, et al. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet. 2004;36:447–449. doi: 10.1038/ng1346. [DOI] [PubMed] [Google Scholar]
- 22.Armanios M, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci U S A. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venteicher AS, et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644–648. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell. 2009;34:47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 26.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 27.Byrne JA, Nguyen HN, Reijo Pera RA. Enhanced generation of induced pluripotent stem cells from a subpopulation of human fibroblasts. PLoS One. 2009;4:e7118. doi: 10.1371/journal.pone.0007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sommer CA, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomlinson RL, et al. Telomerase reverse transcriptase is required for the localization of telomerase RNA to Cajal bodies and telomeres in human cancer cells. Molecular Biology of the Cell. 2008;19:3793–3800. doi: 10.1091/mbc.E08-02-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middleman EJ, Choi J, Venteicher AS, Cheung P, Artandi SE. Regulation of cellular immortalization and steady-state levels of the telomerase reverse transcriptase through its carboxy-terminal domain. Mol Cell Biol. 2006;26:2146–2159. doi: 10.1128/MCB.26.6.2146-2159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. Embo J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- 33.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 34.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mochizuki Y, He J, Kulkarni S, Bessler M, Mason PJ. Mouse dyskerin mutations affect accumulation of telomerase RNA and small nucleolar RNA, telomerase activity, and ribosomal RNA processing. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10756–10761. doi: 10.1073/pnas.0402560101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seabrigh M. Rapid Banding Technique for Human Chromosomes. Lancet. 1971;2:971. doi: 10.1016/s0140-6736(71)90287-x. [DOI] [PubMed] [Google Scholar]
- 37.Deb-Rinker P, Ly D, Jezierski A, Sikorska M, Walker PR. Sequential DNA methylation of the Nanog and Oct-4 upstream regions in human NT2 cells during neuronal differentiation. J Biol Chem. 2005;280:6257–6260. doi: 10.1074/jbc.C400479200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.