Abstract
Cells interact with the surrounding environment by making tens to hundreds of thousands of nanoscale interactions with extracellular signals and features. The goal of nanoscale tissue engineering is to harness the interactions through nanoscale biomaterials engineering in order to study and direct cellular behaviors. Here, we review the nanoscale tissue engineering technologies for both two- and three-dimensional studies (2- and 3D), and provide a holistic overview of the field. Techniques that can control the average spacing and clustering of cell adhesion ligands are well established and have been highly successful in describing cell adhesion and migration in 2D. Extension of these engineering tools to 3D biomaterials has created many new hydrogel and nanofiber scaffolds technologies that are being used to design in vitro experiments with more physiologically relevant conditions. Researchers are beginning to study complex cell functions in 3D, however, there is a need for biomaterials systems that provide fine control over the nanoscale presentation of bioactive ligands in 3D. Additionally, there is a need for 2- and 3D techniques that can control the nanoscale presentation of multiple bioactive ligands and the temporal changes in cellular microenvironment.
Introduction
Organs and tissues organize over multiple length-scales from the nanoscale to the macroscale. For example, centimeter high vertebrae stack to form a half-meter long column, actin and myosin filaments assemble into muscle fibers with micrometer striations, and lung bronchioles extend out into sub-milimeter alveoli. At the single cell level, interactions with the extracellular environment occur on a nanometer length scale; cell surface receptors that span the cell membrane bind ligands and induce cascades of intracellular biophysical and biochemical events that lead to changes in cellular states. In this way, cells receive, process, and respond to information presented in the surrounding environment. Fine control over the information, the molecular signals and physical cues, is essential to controlling cell behaviors. The goal of nanoscale tissue engineering is to create biomaterials that can direct the interactions between cells and the environment by engineering the nanoscale presentation of biologically relevant molecular signals. The ideal system is one in which a biologically inert background can be patterned with bioactive ligands in a controlled manner independently of the mechanical properties. Such systems enable parametric studies of controlled presentations of bioactive ligands on cellular functions.
The broad goal of the research efforts that we review herein is the development of biomaterials and biotechnologies to advance tissue engineering therapies and to help develop a better understanding of cellular biology. To this end, many different microscale techniques and synthetic polymer reaction schemes have been used to design biomaterials with controlled nanoscale presentations and surface densities of bioactive peptides and small molecules on two dimensional (2D) substrates.[1–13] Typically, a glass, gold, synthetic polymer, or other suitable substrate is modified so that the peptides, proteins, or small molecules of interest can be selectively grafted to the substrate in a controlled manner. Additionally, when peptide ligands are used to impart bioactivity control of the peptide sequence is also possible. Combined, these techniques provide the ability to engineer the spacing, spatial organization, and bioactivity at the nanoscale. Hydrogels and polymeric scaffolds decorated with pendent ligands[14–17] and self-assembled supramolecular structures[18, 19] have been used to study cells in 3D. Such experimental designs more closely mimic physiological conditions and can lead to experimental outcomes that can further direct our understanding of in vivo cell behaviors.
Extracellular matrix (ECM) proteins and glycosylaminoglycans, soluble factors and cytokines from autocrine, paracrine and endocrine signaling, and ligands on neighboring cells present a complex set of information in the environment surrounding a cell.[20] In combination with the physical and chemical properties of the environment, ECM proteins, neighboring cells and molecular signals define the cellular microenvironment, and the temporal, spatial and contextual presentation of the different aspects of the microenvironment directs cell behavior.[21, 22] Biological presentations, i.e. the spatial conformations that can induce bioactivity, of ECM-derived peptides and bioactive molecules can be engineered to direct cell behavior. For example, the presentation of cell adhesion ligands on (2D) substrates and in three-dimensional (3D) scaffolds affects cell morphology[23], and cell motility on substrates is dependent on the concentration of cell adhesion molecules[24], as is the migration of cells within 3D microenvironments[14]. Additionally, substrate mechanical properties can influence cell fate.[25] External cell signaling does not often occur in a straightforward binary manner and the induction of cellular pathways often requires multiple cell surface binding events to occur in concert.[26, 27] Complex signaling can be seen in the cellular responses to different spatial presentations of cell adhesion peptides. Examples of such responses include the effects of cell adhesion ligand clustering on cell morphology and adhesion[28], the effects of stem cell morphology on differentiation[29], and the effects of nanoscale presentation of adhesion ligands on DNA transfection efficiency.[30] Investigating cell functions such as adhesion and migration as well as differentiation requires accurate mimicking of the in vivo microenvironment. This mimicking of the natural ECM requires biomaterials that are tunable down to the nanometer length scale.
Here, we present an overview of the field of nanoscale tissue engineering focusing on the experimental techniques commonly used to control bioactive ligand presentation and the knowledge of cell behaviors derived from the outcomes of these experiments. We discuss the methods and techniques used to control the nanoscale presentation of cell adhesion ligands on substrates for cell culture and in 3D biomaterials scaffolds and hydrogels. We also describe some of the outcomes of experiments that employ these techniques, and review experimental and modeling efforts that explore the interplay between cell behavior and the nanoscale presentation of bioactive ligands. We discuss the experimental investigations and modeling of cell spreading and migration on surfaces as it represents some of the most advanced knowledge generated from nanoscale tissue engineering. We also discuss experimental investigations that explore changes in cellular behaviors in response to nanoscale presentations of adhesion molecules including stem cell differentiation, transfection efficiency, and protein expression among others. As background to understanding some of the fundamental biological aspects of cell-surface and cell-biomaterials interactions, we briefly discuss the structure and function of integrin surface receptors and cell adhesion ligands derived from ECM proteins. Additionally, we discuss the design, fabrication, and cell-biomaterials interactions of 3D systems in which the biochemical and biophysical conditions surrounding embedded or encapsulated cells are tunable down to the nanometer length scale. This area represents the future of the field and is less well defined then our understanding of 2D systems and cell behaviors in 2D. We also assess the needs of future technology development and biological studies to advance the field of nanoscale tissue engineering.
Integrins: mediating extracellular signals
Cells interact with nanoscale-engineered biomaterials through integrins and cell surface receptors; as such, we describe some of the fundamental aspects of integrins to provide context to the design and engineering of nanoscale biomaterials. Integrins are transmembrane proteins that mediate cell adhesion and cellular interactions to the extracellular microenvironment. Transmembrane proteins typically consist of a ~50 amino acid cytoplasmic domain that interacts with the intracellular space and a ~1000 amino acid domain that binds extracellular ligands.[31] Molecular binding at one end of an integrin results in conformational change at the opposite end, thus enabling a two-way exchange of information: outside-in and inside-out signaling.[32] Function is determined by the α/β domain pair that make up an integrin noncovalent heterodimer; however, only 24 of the possible pairs of the 18 α and 8 β domains have so far been observed.[33] The combination of α/β domains together with the different conformational states accessible to a single integrin results in a wide range of functions, thus allowing for interactions with many different biochemical signaling pathways. It is important to note that integrins are not fixed at specific locations in the cell membrane but are mobile. The mobility allows integrins to diffuse along the cell membrane and cluster at areas of high ligand density.
The basis for biological specificity in receptor-ligand binding arises from the complimentary structure of ligand and receptor binding pocket, and it is down to this scale that nanoscale tissue engineering can impart engineering design. Crystal structures of extracellular integrin domains have provided insights into the molecular structure of the ligand binding pocket as well as the interactions with cell adhesion ligands.[34] For example, it has been shown that the binding affinity of the cell adhesion ligand arginine-glycine-aspartic acid (RGD) is tunable with changes in the N-terminal residue.[35] In Figure 1 we schematically represent cell-biomaterials interactions through integrin binding and reproduce the x-ray crystal structure of the extracellular domain of αV/β3 integrin with the bound RGD ligand (1L5G).[34]
Figure 1.
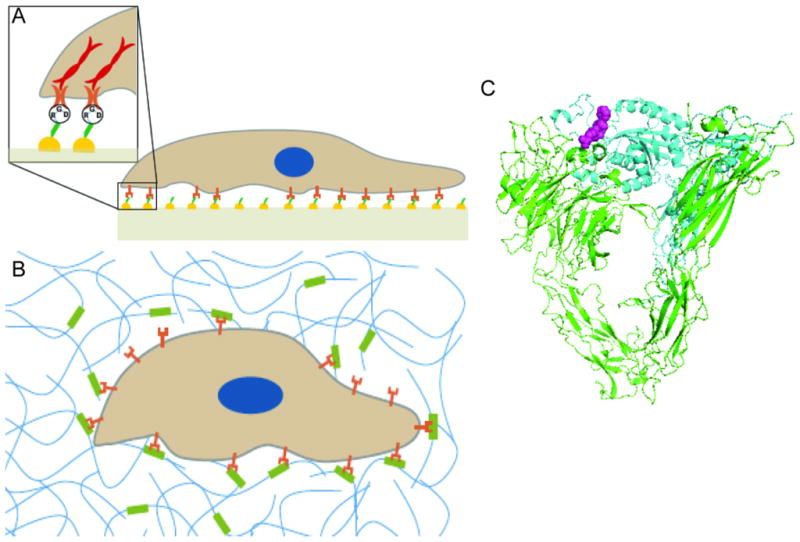
Cellular-ECM interaction. (A) Schematic of 2D cell-substrate interaction where cells acquire a non-physiologically flattened morphology. (B) Schematic of 3D cell-substrate interaction in which cells retain in vivo morphology. (C) The crystal structure of the extracellular domain of αV/β3 integrin (green/blue) with bound RGD ligand (purple) (1L5G).
Integrins couple to, and interact with, many different intracellular signaling pathways including calcium channels, kinases, phosphatases, and the Rho family of GTP binding proteins, among others.[33] For example, in focal adhesions (protein and macromolecule assemblies that form within a cell at adhesion sites) integrins not only bind to extracellular ligands in the ECM, but also recruit intracellular signaling agents such as the non-receptor tyrosine kinases Src and FAK.[36] Depending on the particular intermolecular interactions, Src and FAK can influence biochemical pathways responsible for cellular functions such as cell survival, membrane extension and cytoskeletal tension, cell motility and directional control, matrix assembly, and tissue invasion.[33] Engineering biomaterials and bioactive surfaces with controlled spacing and presentations of extracellular ligands enables parametric studies of these cellular functions.
Cell adhesion peptides from ECM proteins
One of the most successful strategies of patterning bioactivity is the modification of a biologically inert substrate with bioactive peptide ligands. This strategy requires peptides with bioactive sequences, and represents one level of control at the nanoscale, i.e. the primary sequence of a peptide and the peptide structure. In an effort to identify the structure-function relationships of ECM proteins many functional peptide sequences have been identified. Cell adhesion peptides have been identified in laminin including, RGD, YIGSR, LGTIPG, IKVAV, PDGSR, LRE, LRGDN and IKLLI (amino acid sequences given in single amino acid letter code).[37] Similarly, RGD and DGEA, and RGD, KQAGDV, REDV and PHSRN, have been identified from collagen I and fibronectin, respectively.[38–40] The incorporation of cell adhesive peptides into otherwise biologically inert substrates allows for a tunable platform for creating bioactive substrates and scaffolds through selective cell attachment.[41] Most often the RGD-containing peptides are used to impart cell adhesion properties. The other peptides sequences are listed here are also have cell adhesion properties and are under-explored relative to RGD.
The canonical cell adhesion peptide is the tri-peptide RGD found in ECM proteins such as fibronectin, laminin, and collagen I, among others. The structure, function, and engineering of RGD and RGD containing peptide sequences have been reviewed in detail elsewhere.[35, 42] The structure of RGD ligands has also been investigated. Most notably, cyclic and linear RGD sequences have been compared,[43, 44] and changes in binding affinity with varying the N-terminal residue of RGDX been investigated.[35, 41] Techniques for presenting RGD and other cell adhesion peptides must allow for control over the accessibility of the ligand to integrin binding, and control of the spatial organization and effective density of the ligands. To this end, the effect of spacing between substrate and active integrin binding sequence has been explored by varying the number of amino acids C-terminal to RGD (XnRGD, X = any amino acid).[41] Other works have explored RGD presentations by grafting oligomer end-groups to the cell binding peptide.[41] These effects have been extensively studied [35, 41], and as such are not described in great detail here. Techniques for controlling the spatial presentation of RGD and other peptide ligands are described in the following section.
Engineering the nanoscale presentations of bioactive ligands in 2D
Many different techniques have been used to control the spatial presentation of bioactive ligands on planar substrates. Here, we review the most successful of these techniques as well as some successful techniques for creating nanoscale topographies. Micropatterning techniques with UV radiation[45, 46], electrochemical reactions[47], plasma polymerization[48, 49], microfluidic-based systems [50, 51], photolithography[52, 53], and capillary force lithography[54] have been used to produce surface-grafted patterns and gradients of bioactive ligands. These techniques use microscale technologies or bulk polymer modifications, therefore control at the nanoscale is indirect. For example, patterns with nanoscale spatial resolution of peptides and clusters (islands) of peptides can be created by controlling the addition of chemical functional groups to an inert substrate to which bioactive ligands can be selectively grafted. Reaction schemes for hydroxyl [1–4], carboxyl [5–9], amino [8, 55–57], aldehyde [10], acrylate [11], and thiol [12, 13] functional groups have been used to create patterns of peptides with nanoscale spacing. Similarly, co-polymer systems where one block contains active functional groups to allow for peptide linkage peptide clusters have been synthesized with controlled spacing and number of peptides per cluster. The desired arrangement of peptides within clusters and the spatial arrangement of these clusters can be tuned by mixing set ratios[58, 59] of peptide containing polymers with unmodified polymers. In this way, the number of ligands per cluster can be tuned. For example, with a PEG/PMMA co-polymer system modified with RGD peptide ligands, ligand clusters with an average of 1.7 to 5.4 ligands per cluster were created. The average ligand density was tuned, independently of the cluster size, over a range of 260 to 5200 ligands per μm2.[58] Additional control at the nanoscale is gained through the peptide sequence and structure. For example, Hsiong et al. use carbodiimide chemistry to modify alginate hydrogels with linear and cyclic RGD ligands. Using mixtures of modified and unmodified alginate polymer chains the average spacing between ligand clusters was tuned at discrete intervals between 36 and 121 nm. This system also enables the independent tuning of substrate stiffness and cell adhesion ligand density, which was demonstrated by creating alginate hydrogels with ~40 to ~120 nm spacing between RGD islands on hydrogels with an elastic modulus of 20, 60, 120 kPa.[59]
Nanolithography has also been used to create nanoscale patterns of bioactive peptides. One such method entails the use of metal or organometallic precursors embedded in block copolymer micelles to create patterns on a substrate. Lithographic treatment of targeted areas on the substrate form metal nanoparticles that are used as attachment sites for thiolated ligands with nanoscale resolution.[60, 61] With this technique it was possible to create features down to 4 nm in width and extent features of this width along 1 – 50 μm. This technique has been used in conjunction with other lithography methods to pattern complex shapes and gradients with nanoscale resolution[62] as well as hierarchal patterned substrates with micron sized patterning with embedded nanoscale patterns[63, 64]. Other techniques such as dip-pen nanolithography[65] and polystyrene nanosphere technique[66] have also been used to create nano-patterns of bioactive ligands. In Figure 2 we show two examples of nano-patterned substrates. One example demonstrates the fabrication of complex patterns of cell adhesion ligands and the second demonstrates nanoscale features fabricated from functionalized gold particles.
Figure 2.
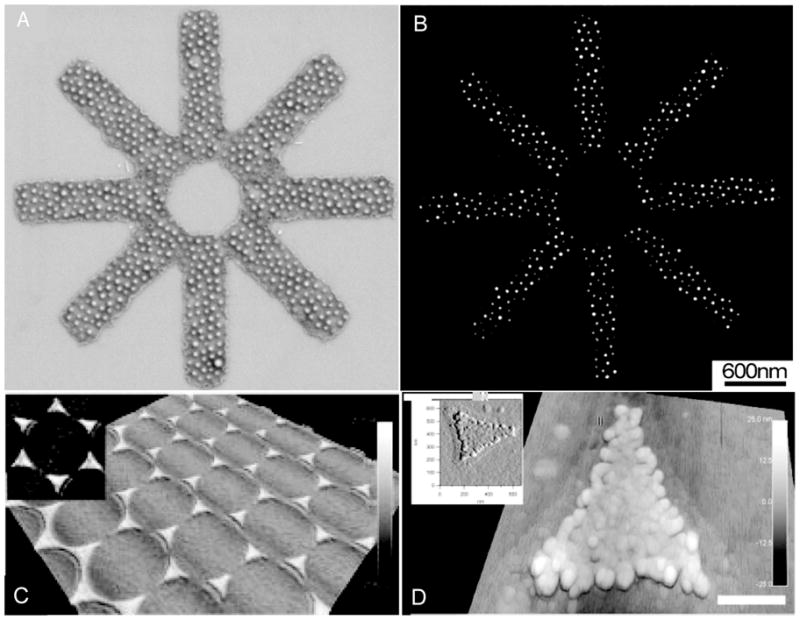
Examples of Nanoscale topographies for 2D cell-substrate investigations. (A, B) Hierarchal star structure is made by using a block copolymer templating technique where electron beam lithography is used to form 7 nm gold particles on substrate reproduced with permission from ref. 61 (doi: 10.1088/0957-4484/14/10/314 ). (C, D) Nano-sized polystyrene is used to template a functional gold surface to create nano-features. [66]
Nano-templating and nanolithography techniques have been devised to control cell behavior through defined nanoscale topographies. For example, patterning of surface topography on polymeric substrates by electron beam lithography has been used to direct stem cell fate[67, 68], and nanografted substrates have been used to spatially organize myocardial cells and control protein expression.[69] Readers are directed elsewhere for a more detailed review of nanofabrication techniques and cell interactions with nanoscale topographies.[70]
Controlling the average nanoscale spacing of bioactive ligands on 2D substrates has been highly successful, and these techniques has been used to extensively investigate cell adhesion, spreading, and migration on substrates with tuned parameters. While many different techniques have been successful in engineering the presentation of cell adhesion ligands there is a need to for techniques that can create controlled heterogeneous systems where multiple bioactive ligands are presented in a controlled manner. Additionally, substrates that change over time in a controlled manner would enable many interesting investigation of dynamic cellular processes.
Engineering cell adhesion and spreading in 2D
Cell-substrate interactions play a central role in regulation of cellular functions such as adhesion, locomotion, growth, proliferation, and differentiation.[71–75] The cell-substrate interactions are mediated by integrin receptors, and substrate properties including the chemical composition, ligand density and pattern, and ligand-receptor interaction binding energy.[39, 72, 76–85] Cell spreading is a dynamic process involving non-covalent association between membrane receptors on the cell surface and complementary ligands on the substrate. Two limiting regimes, reaction- and diffusion-controlled, can be defined for the displacement kinetics of the leading edge of an adhered cell.[86] In the reaction-controlled regime the formation and breaking of cell-substrate contacts are controlled by the rate of reversible reactions between ligands and cell surface receptors while in the diffusion-controlled regime, receptors are recruited from regions on the membrane far away from the adhesion zone.[87–89] At low receptor concentrations, receptor diffusion time is longer than that for the ligand-receptor reaction, and hence cell spreading is mediated by diffusion of mobile receptors. At high receptor concentrations, the rate of spreading of the adhesion zone is controlled by the rate of ligand-receptor association.
Cell spreading and attachment on 2D bioactive substrates has been the focus of many experimental investigations that have led to important finding of the adhesion of fibroblast on engineered substrates. For example, a number of studies have measured the spatial distribution of fibroblast adhesion on substrates with gradient density of fibronectin or RGD.[90, 91] The results of these studies demonstrate that the variation of cell distribution along the gradient is biphasic; cell density increases toward the increasing direction of gradient (Figure 3A), and above a certain ligand concentration it decreases or remains constant. The biphasic cell density distribution can be explained by the fact that above a certain ligand concentration, the mechanism of cell spreading switches from reaction-controlled to diffusion-controlled regime. Therefore, increasing the ligand concentration reduces the rate of cell-substrate association that in turn can inhibit cell spreading. Similar reasoning can be used to explain the biphasic dependence of the cell adhesion ligand density and motility of fibroblasts on substrates coated with different densities of fibronectin or collagen IV.[74, 92]
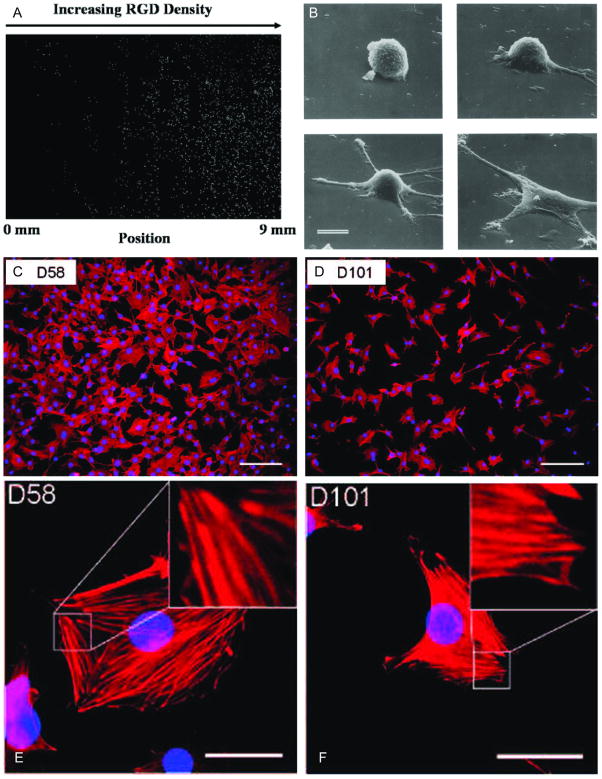
Figure 3.
Examples of cellular morphology on 2D substrate with nanoscale presentation of bioactive ligands. (A) Image of nuclear stained cells on RGD density gradient. Cell density increase with increasing RGD density. [91] (B) Scanning electron microscope image of cell-substrate adhesion with different degrees of cell spreading. (© Rockefeller University Press, 1991. Originally published in J. Cell Biol. 114: 1089–1100). [28] (C, D) Attachment of osteoblasts on disordered RGD modified substrate. Decoupled RGD density from spacing demonstrated a significant decrease in cell attachment for RGD spacing >70 nm. (E, F) Higher magnification shows actin filament distribution caused by cell-substrate traction forces. Acin filament (red) nucleus (blue); numbers on top-left represent average RGD spacing. [96]
There have been a number of efforts made to quantify the nanoscale effects in cell adhesion. For example, the concentration of GRGDY peptides on a substrate was varied to achieve nanometer scale spacing between RGD peptides.[28] By varying the surface concentration of RGD peptides (0.1–100 fmol cm−2) grafted onto a non-adherent polymeric thin film, a systematic study was conducted to determine the minimum RGD coating required for effective cell attachment (Figure 3B). The study showed that a peptide spacing of 440 nm promotes maximal cellular spreading while a spacing of 140 nm is required to promote focal contact formation (1 fmol cm−2 and 10 fmol cm−2 respectively). In another study, poly(ethylene glycol) (PEG) based semi-interpenetrating network hydrogels modified with RGD peptides were used to investigate the same phenomenon.[93] Concentrations of 66 and 110 pmol cm−2 provided substrates with well-adhered and flattened cells with cytoplasmic extensions. A number of other investigations using similarly modified substrates have reported a range of cell binding ligand surface densities apt for cell adhesion and spreading.[94, 95] Experimentally observed differences in optimum and minimum binding ligand spacing may be in part due to differences in experimental parameters (peptide sequence and substrate material) in these studies that lead to differences in the nanoscale presentation of cell adhesion ligands. Collectively, these studies have helped develop a strong understanding of the minimum requirements for cell adhesion and have informed the design criteria of many new microscale cell engineering techniques.
In many experimental systems peptides are covalently grafted to compliant polymeric substrates, an alternative method is to graft the RGD peptides to nano-patterned gold-islands.[62–64] The experimental design of these studies entailed grafting individual cyclic-(RGDK) peptides to gold patterned substrates with 6–8 nm resolution.[62, 63] Initially, adhesion of osteoblast cells on the nano-patterned substrates resulted in the conclusion that 28–58 nm cyclic-RGD spacing promotes cell attachment and proliferation, while larger peptide spacing substantially reduced the effects.[63] The result was supported by a subsequent investigation that also concluded that a cyclic-RGDK spacing greater than 70 nm does not support cell adhesion.[64, 96] Additionally, the effect of order and disorder of cyclic-(RGDK) nanopatterns on a substrate was investigated for cell attachment. It was observed that cell adhesion showed a significant increase for disordered RGD patterns compared to ordered patterns with the same average spacing, shown in Figure 3C–F.[96] Two parallel studies using this same technique investigated the minimal gradient strength (Δ15 nm mm−1) required to polarize cells as well as demonstrating an increase in cellular polarization along a gradient strength of Δ15 nm mm−1.[62, 97] By devising precisely arranged nano-patterns on a substrate, the two studies were able to conclude that osteoblasts are capable of detecting a ligand spacing difference of ~1 nm from the front to the back of the cell.
Directing cell migration on 2D substrates
Cell migration is central to many biological events and pathological processes.[98–101] The regulatory effect of substrates on cell motility has been used to design novel biomaterials and bioactive substrates with the ability to direct cell migration, proliferation, and morphogenesis.[102, 103] Guided migration of progenitor cells from a host tissue surrounding a scaffold and differentiation and proliferation of the migrating cells are central to the success of tissue engineered implants. Therefore, understanding cell-substrate and intracellular forces responsible for cell migration not only allows for the investigation of the underlying mechanisms of many pathological processes but also holds promise for designing improved engineered constructs for tissue regeneration. Cell migration on engineered substrates has also been the focus of many experimental efforts. The data generated in many of these works has formed the basis for the development of migration models in 1- and 2D.
At the cellular scale, migration is described by cell locomotion in which, 1) the cell extends lamellipod by polymerization of the actin microfilaments in the front edge of the cell and reversible bonding of the cell-surface receptors to the substrate ligands (protrusion); 2) contraction of the cytoskeletal network, mediated by protein motors (myosin-II), to generate a rear detachment force; and, 3) relaxation of the cytoskeletal network to reach a new cell configuration. The cycle repeats to produce cell migration. Surface density of cell adhesion ligands has been shown to influence the migration of cells on substrates. In particular, substrates with nanoscale gradient patterns of ligand density are widely used for guiding cell migration on biomaterials, and studies have demonstrated that cells preferentially move toward regions of increasing adhesiveness.[91, 104–107] Cellular migration on a substrate requires a balance between the generation of a contraction force by adhesion of the extending lamellae at the leading edge and lamellae retraction from the trailing end. Substrates with weak cellular adhesion exhibit slow migration as cells do not have sufficient traction to generate force to detach the trailing lamellae, while substrates with strong adhesion hinder detachment of lamellae altogether.[108]
Experimental studies have demonstrated that cell migration increases with ligand density up to a critical value, above which cell density and speed of migration reach either a plateau value or decrease.[74, 92, 109] Similarly, cell adhesion experiments with fibroblasts on substrates with RGD ligand density gradients show higher cell adhesion in the direction of increasing RGD density; however, a saturation effect is observed above a density of adhesive ligands where increasing the RGD density no longer improves cell adhesion.[91] Cell migration has also been investigated on substrates with nanopatterned clusters of cell adhesion ligands. In one study, a systematic variation of nanometer RGD spacing was conducted between 6–300 nm for randomly placed RGD peptides and clustered islands.[24] Substrates with tighter peptide spacing and with at least five peptides per cluster resulted in well-formed actin stress fibers with higher cell motility. The results of the study demonstrate that cell motility across a substrate is dependent not only on the adhesion ligand spacing but also on the number and spacing of ligands within a cluster.
Experimental data shows that cells preferentially move towards the direction of higher adhesiveness on substrates with increasing density of ligands, with a velocity which is tunable by the slope of ligand gradient [51, 110, 111]. Single cell locomotion can be modeled as time-dependent changes in the cell boundaries, which is governed by interactions with the adhering substrate and the density fluctuations of cytoplasmic proteins [112]. One dimensional (1D) models have proven useful in elucidating the underlying mechanism of cell locomotion on substrates with uniform ligand density [109, 113, 114]. Since cells are polarized and migrate along the gradient direction [111], 1D models can also be used to predict the speed of migration and the time-dependent variations of cell length on ligand gradient substrates. Figure 4 shows the schematic representation of a 1D model for cell locomotion on a ligand gradient substrate. The cell has position-dependent elasticity due to the variation of actin network density. Cell-substrate interaction is characterized by a frictional force, controlled by the density of ligand-receptor pairs. The generation of contractile stresses is described in terms of reactions between actins, myosins, and guanine nucleotide regulatory proteins. The model predicts a biphasic dependence between locomotion speed and ligand gradient slope with the maximum speed occurring at an intermediate gradient slope and more pronounced for higher ligand-receptor affinities. The model predictions can be utilized in the design of biomimetic substrates for guided tissue regeneration as it predicts an optimum range for the slope of ligand gradient with respect to the speed of cell migration.
Figure 4.
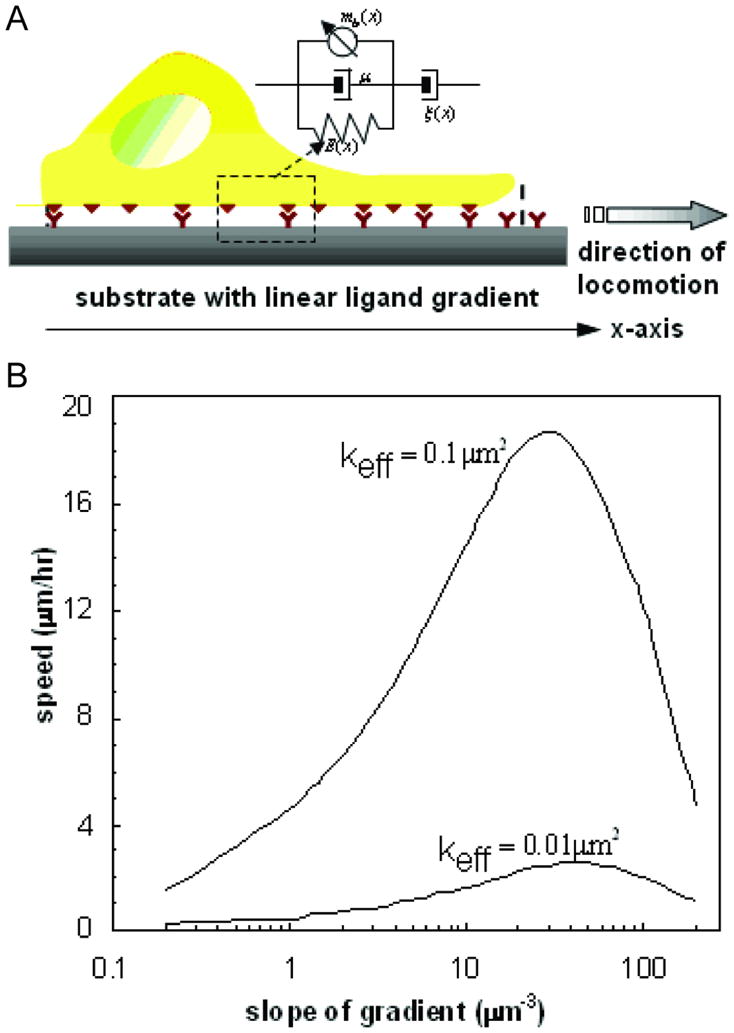
Modeling 2D cell migration. (A) Representation of forces involved in cell locomotion. ξ (x) is the position-dependant friction coefficient due to repetitive attachment-detachment interactions between the cell surface receptors and ligands on the substrate. E(x) and μ represent the elasticity and viscosity of cytoskeleton and cytosol, respectively. The myosin dynamics is described by the density of myosin molecules bound to actin microfilaments and generating contractile stress mb (x). (B) The speed of cell migration as a function of the slope of the gradient of cell adhesion ligands shows a biphasic distribution of cell migration. The speed of cell migration increases to a maximum prior to decreasing with increasing cell adhesion ligands.[112]
For motile cells that show appreciable polarization through gradients in ligand density, such as sprouting of endothelial cells in angiogenesis, the model can be used to predict the effect of substrate factors (type and density of ligand, slope of the gradient, interaction energy) and cell biomolecular factors (signal transduction pathways and proteins, and proteases activated by ligand-receptor interactions) on migration. This model and other 1D models [75, 109] are potentially useful for analyzing cell migration in confined channels, like in microfluidic devices, where the substrate width is of the same order of magnitude as the cell size [115], and the cell is physically restricted to migrate along the direction of gradient. More importantly, the predictions of the model can be used to design novel microdevices to determine biophysical properties of the cell (cytoskeletal elasticity, interaction energy per ligand-receptor pair, stress generated by one actin-myosin pair, regulatory proteins coupling parameter).
Investigating cell functions on 2D nano-patterns
As the field of nanoscale tissue engineering matures and a more comprehensive understanding of cellular adhesion is produced, experimenters are beginning to explore more diverse cell functions including gene and protein expression as well as stem cell fate in response to nanoscale presentations of cell adhesion ligands. This is an area that is highly under-explored in comparison to the abundance of data describing cell adhesion and migration in 2D. However, there are interesting works that demonstrate the importance of cell adhesion properties on cellular functions. For example, Kong et al. investigated DNA transfection (the exogenous uptake of DNA) in MC3T3 preosteoblast cells attached to nanopatterned RGD substrates.[30] RGD presentation on the substrate was independently tuned with respect to the overall density of RGD peptides (3×109 to 60×109 mm−2) and peptide island spacing (36 to 120 nm). An exponential decrease in expression of the transfected gene was observed for peptide spacing of 36–120 nm on a substrate with statistically similar peptide densities. It was hypothesized that the increase in proliferation of cells on lower RGD densities, resulting in an increase in cell mitosis, altered transfection efficiency. A recent study by the same group investigated the effect of RGD nano-island arrangement on the proliferation and differentiation of stem cell cultures including, preosteoblasts (MC3T3-E1), clonally derived bone marrow stromal cells (D1), and human bone marrow stromal cells (hBMSC).[59] The investigation demonstrates that proliferation and differentiation of stem cells can be guided by engineering microenvironments mimicking the complex feedback of the native ECM environment. Together, these works demonstrate that changes in the underlying substrate can have significant effect on different cellular functions.
Another example considered the effects of external stress caused by a micropatterend substrate on the differentiation of human mesynchemal stem cells (MSCs).[29] Polydimethylsilane (PDMS) micropatterned substrates were used to create corresponding MSC micropatterns constrained to different geometries as to produce cultures with varying external stresses (Figure 5A). Under contractile stress of a concave curvature human MSCs preferentially underwent adiopogenesis while a convex curvature promoted osteogenesis.[29] It is also known that substrate elasticity affects stem cell fate. Two studies that investigated the effects of stiffness demonstrate that the fate of MSCs[25] and preosteoblasts[116] can be directed by tuning substrate elastic modulus without changing the nanoscale presentation of adhesion ligands. These examples demonstrate the wide range of effects that substrates can have on cell functions, and that there is a need for similar studies that employ engineered substrates to investigate stem cell differentiation and changes in gene expression.
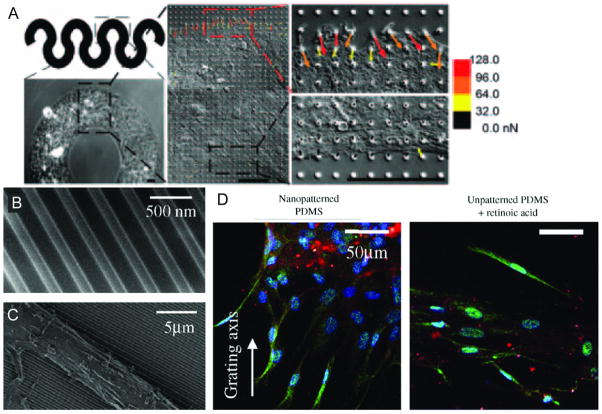
Figure 5.
Complex cell behavior on nanopatterned substrates. (A) Geometric related tension on cells directs mesenchymal stem cell fate. Vector map on right panels sow traction forces imposed on cells. [29] (B) Scanning electron images of nanoscale graftings on 2D substrate. (C) Cellular elongation along axis of nanografting. (D) Cells grown on nanopatterned gratings shown to the left and planar substrate containing neuronal stimulator (retinoic acid0 are stained for cell nucleus (blue), nestin (red), MAP2 (green). [67]
The effects of nanoscale topographies have also been investigated. For example, Dalby et al. investigated the level of order/disorder of nanopits on a substrate on the differentiation of MSCs.[68] To investigate the effect of order/disorder of a nanopatterned substrate the expression of two bone-specific proteins, osteopontin and osteocalcin, where monitored. Interestingly, MSCs on both highly ordered and highly disordered substrates exhibited low expression of both proteins; however, substrates with slightly irregular patterning significantly upregulated the expression of both proteins. Additionally, to study the expression of osteospecific genes via messenger RNA microarray, MSCs were cultured on planar and ordered nanopit substrates, with and without dexamethasone (DEX), a corticosteroid shown to induce bone formation, as well as on a slightly ordered nanoptit. MSCs cultured on the nanopit array expressed a comparable level of osteoblast-specific genes compared to the flat substrate in the presence of DEX. Additionally, some genes were specifically upregulated when MSCs were cultured on the nanopit compared to the flat substrate with DEX. Another example demonstrates that nanotopography alone can induce an upregulation of neuronal markers of MSCs. In this example, nanopatterned substrates (350 nm wide ridges with varying pitch between 250 nm and 10 mm shown in Figures 5B and 5C) induced cytoskeletal rearrangement and nuclei elongation, producing significant changes in signal transduction for transdifferentiation.[67] Analyzing MAP2 expression on nanopatterned collagen coated PDMS with and without retinoic acid showed significant upregulation (as per quantitative real-time PCR analysis) compared with unpatterned controls after 7 days of incubation (Figure 5D). These studies suggest the potential of nanotopography on substrates to direct stem cell fate. There is a need for more experimental works such as those described here. In comparison to the volume of data on the adhesion and migration of cells on controlled nanoscale substrates, there is a relatively little information on the effects of varying cell-substrate interactions on more complex tissue and cell engineering. New experiments are required that employ the many existing strategies of controlling cell adhesion ligand density, pattern, and clustering. There is also a need to extend these technologies to new bioactive peptide ligands know to signal specific pathways.
Nanoscale tissue engineering in three-dimensions
While 2D nanoscale tissue engineering techniques have provided many important insights into cellular functions, there is an inherent asymmetry in such experimental systems. On 2D substrates, flattened cells spread along the plane of the substrate while the non-adhered side of the cell is exposed to liquid media. Such a situation approximates the conditions of the vasculature endothelial lining and of epithelial cell sheets; however, 2D experimental designs result in asymmetric signaling that can lead to misinformed findings as cells adapt to the artificial microenvironment.[23, 117] The complex presentation of polysaccharides, ECM proteins, and neighboring cells creates a unique set of inputs to cells in vivo. Recapitulating these conditions in vitro is necessary for parametric studies of cells in controlled 3D microenvironments needed to develop a more complete understanding of cellular functions.[20, 118, 119] In addition to expanding 2D investigations to include a variety of bioactive peptides, there is an important need to develop new biomaterials technologies that can control 3D nanoscale presentations of bioactive ligands.
Often 3D cell culture experiments are conducted with biomaterial scaffolds and hydrogels made with naturally derived ECM components such as decellulized tissue[120], Matrigel (a commercially available ECM protein mixture secreted by Engelbreth-Holm-Swarm (EHS) mouse sarcoma cells)[121], as well as collagen[122] and gelatin[123]. Such biomaterials have proven to be highly useful for studying 3D cell cultures as they contain naturally occurring integrin binding ligands and are often susceptible to proteolytic cleavage, but such materials are not easily adapted to controlled studies for varying the nanoscale presentations of bioactive ligands. Similar to 2D biomaterials design, a strategy for controlling the nanoscale presentations of bioactive ligands in 3D is the modification of an otherwise biologically inert material with bioactive ligands in a controllable and scalable manner. This strategy has been used to create 3D biomaterials with controlled microenvironments from micro- and nanofiber scaffolds[124] as well as hydrogels made from synthetic and natural polymers[125]. Pseudo-3D microenvironments where cells are cultured in microwells have also used such a strategy to investigate single cells and cell aggregates.[126]
3D biomaterials systems for nanoscale tissue engineering must 1) use mild fabrication conditions such that cell viability is not adversely affected during cell embedding or encapsulation; and, 2) allow for tunable biochemical and biophysical parameters of the cellular microenvironment. Additionally, 3D biomaterials should, 3) be susceptible to cell invasion and allow for cell migration within the biomaterial; and, 4) be sufficiently porous or otherwise accessible to diffusion of nutrients and dissolved gases throughout the material. The complexity of 3D environments is such that, thus far, there are no 3D techniques that can replicate the high level of nanoscale control that is possible with the 2D systems described in the previous sections. In 3D systems the nanoscale presentation of cell adhesion ligands and other bioactive peptides is an average property, and the systems are largely disordered. This level of control represents the state-of-the-art in 3D nanoscale tissue engineering and reveals an area where more work is required in the field—the development of techniques that can directly control bioactive ligand presentation at the nanoscale. Here, we review the most common techniques used to create 3D microenvironments that address criteria 1 through 4 including, natural and synthetic polymer hydrogels, self-assembled peptide nanofibers and hydrogels, as well as synthetic and natural polymer nanofiber scaffolds.
Engineering hydrogels and nanoscale scaffolds with controlled 3D nanoscale presentations
Hydrogels are attractive biomaterials for 3D cell culture as they form highly swollen structures that can approximate the conditions of the ECM.[119, 127] Many synthetic or natural polymer hydrogels can form mechanically robust systems from 5 wt%, or less, polymer with the remaining 95 wt% comprised of water. Such conditions allow for diffusion of nutrients and dissolved gases throughout the material and are sufficiently porous to allow for encapsulation of high cell densities. Hydrogels are also attractive for nanoscale tissue engineering as many different chemistries have been developed to encapsulate cells in situ under mild, non-cytotoxic conditions. For example, cell-laden hydrogels have been made by covalently crosslinking polymer networks via Michael-type addition[16, 128], photo-initiated free radical chain polymerization[129, 130], and enzymatic reactions[131]. Self-assembled networks of peptide nanofibers [132–134], and polysaccharides, such as alginate that gels in the presence of divalent cations, have also been used to form hydrogels, allowing for simple and non-cytotoxic methods for cell encapsulation.[59]
Non-cell adhesive hydrogels such as a PEG, PEG derivatives, and alginate have been used to create 3D microenvironments with controlled adhesion properties. For example, the number of RGD ligands per alginate chain can be altered to control the nanoscale spacing between ligands.[135] Similarly, diacrylated PEG hydrogels have been fabricated by photopolymerization in the presence of linear and cyclic RGD ligands, and pendent cell adhesion and bioactive ligands have been incorporated into PEG hydrogels by Michael-type addition[16, 128] and NHS peptide conjugation chemistries.[136] Click chemistry has also been used to synthesize 3D microenvironments.[17] In these systems, the quantitative translation of bulk ligand densities to nanometer scale spatial presentations is not straightforward. To address this issue, a multiscale predictive model was created to characterize the presentations of ligand spacing and quantify the fraction of ligands accessible to integrin binding.[59]
Click chemistry has been used to create ‘click hydrogels’ with independently tunable mechanical and chemical properties. PEG-based click hydrogels have been used to create hydrogels with tuned biochemical gradients.[137] Microscale gradient generation devices have also been used to create hydrogels with controlled gradients of chemical properties, thus allowing for the fabrication of gradients of 3D microenvironments.[138] Such devices have also been used in fabrication of hydrogels with gradients of mechanical properties.[139]
It is known that substrate stiffness can effect cellular functions[25], therefore it is necessary to independently tune the biochemical and biophysical properties of the hydrogel to effectively control 3D microenvironments and design parametric studies of nanoscale presentations. Decoupling chemical and physical properties has been demonstrated with alginate and PEG-based hydrogels with tunable moduli and RGD ligand densities.[16, 135, 137] In such systems, it is possible to control the physical properties in terms of elastic modulus, porosity and crosslink density, as well as the biochemical properties by modification with pendent bioactive peptides. In many of these examples, peptide sequences susceptible to proteolytic cleavage are incorporated in the polymer networks to allow for cell invasion.[16, 17, 128]
The ECM is a meshwork of fibers, tens to thousands of nanometers in diameter, which form a cell scaffold with nanoscale porosity. Cells embedded in the fibrous network are presented with structurally complex 3D environment with nanoscale features. Electrospun fibers of collagen [140–142] and hyaluronic acid (HA) [143–145] as well as poly(D,L-lactide-co-glycolide) [146], poly(ε-caprolactone) (PLA) [147], poly(L-lactic acid) (PLLA) [148], polyanaline [149], and polyamides[150] have been used to mimic the structure of the ECM. The fabrication of such fibers has been the focus of much research and the control of fiber diameter, scaffold porosity, and materials composition has been demonstrated. A more comprehensive overview of electrospun nanofibers can be found elsewhere[151].
A major challenge in fibrous scaffold design is mimicking the 3D nanoscale architecture of the natural ECM. Often, in scaffolds made from microscale fibers, cells adhere in a manner resembling cell adhesion on 2D substrates. Similar effects occur in fibrous scaffolds with pore diameters significantly larger than the cell diameter, thus causing cells to acquire morphologies as though on a 2D substrate. Such effects have been demonstrated with photodegradable polymers [152, 153] and templating [154, 155] techniques that allow for the engineering of microchannels and pores where cellular behavior can be closely monitored.
Self-assembled peptides have also been used to create nanofibrous hydrogels. The self-assembly approach is bottom-up in that peptide and peptide-polymer building blocks assemble into hierarchical structures.[132–134] These techniques allow for the controlled placement of bioactive peptide ligands along the length of the nanofibers. Additionally, it is possible to incorporate different bioactive ligands within a single fiber.[132, 156]
Investigating cell behaviors in 3D
Hydrogels and 3D scaffolds have been used in many experimental studies to investigate microenvironmental factors including cell-cell, cell-matrix, and cell-soluble factor interactions. Here, we discuss hydrogel and nanofiber scaffolds that have been used to explore cell behaviors in controlled microenvironments. Each of the discussed systems demonstrates control of the nanoscale presentation of cell adhesion ligands and/or other bioactive ligands and molecules. Additionally, many of these systems control the physical properties of the biomaterials. We focus our discussion on the biological outcomes from the studies and contrast the outcomes with cell behaviors observed in 2D. For example, there are a number of works that describe and compare cell migration in 2- and 3D. In one example, a novel photodegradable hydrogel technology was used to investigate real time cellular spreading of hMSCs encapsulated in 3D scaffolds with bioactive and non-active background.[152] This technology allows for temporal changes in the cellular microenvironment and consequently in the nanoscale presentation of bioactive ligands, and represents an important advancement in hydrogel technologies. In another example, Tayalia et al. engineered a polymeric scaffold with controlled macroporosity via two-photon polymerization to investigate 3D cell migration.[153]. By altering the average pore diameter from less than a cell diameter to several times larger (12 um to 110 um), comprehensive tracking of cellular migration in the scaffold was made possible. This is a highly interesting area of research that is beginning to reveal key differences between the behavior of cells in 2- and 3D. Together, these examples demonstrate two important challenges in 3D cell studies: the analysis of the outcomes, for example cell migration, is highly complicated in comparison to analysis in 2D, and controlling the micro- and nano-architecture over time is important as cell can change and remodel their environment over time.
It has also been demonstrated that the inhibitory effect of high ligand densities in 2D is not observed with fibroblast migration in 3D.[23] A recent study directly addressed the issue of cellular migration in 2- and 3D. By using microscale photopatterning Doyle et al. created 1D fibrillar patterns, 2D planes, and 3D microenvironments (Figure 6A).[157] The study demonstrated that cell migration and morphology in 1D and 3D are similar and distinctly different than migration and morphology in 2D. The study also demonstrated that fibroblast migration is independent of ligand density in 1D and 3D systems but is more likely to depend on microenvironment topography.
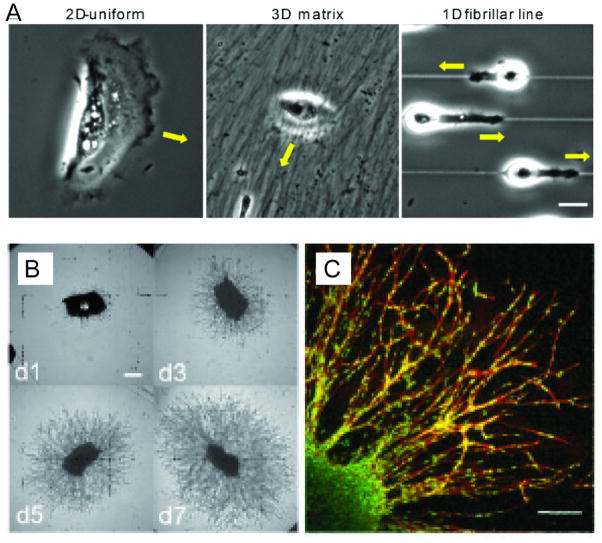
Figure 6.
Cell migration in 3D. (A) A comparison of cell migration in 1D, 2D, and 3D(© Doyle et al., 2009. J. Cell Biol. doi:10.1083/jcb.200810041).[157] The images show fibroblast morphology and migration on microengineered fibronectin substrates including 2D (left) and 3D (middle) fibronectin substrates, and along 1D fibrillar lines. (B, C) Fibroblasts invading a PEG-based hydrogel that is sensitive to proteolytic cleavage from cell-secreted protease.[128]
Translation of in vitro experimental studies to in vivo therapies will require 3D biomaterials systems that can be invaded by proliferating cells and in vivo progenitor cells. Additionally, the biomaterial for cell-based therapies should be susceptible to remodeling by the encapsulated and surrounding cells. To this end, a PEG-based hydrogel containing proteolytically cleavable sequences and controlled concentrations of cell adhesion peptides have been designed (Figure 6B,C). This PEG-hydrogel technology was also demonstrated as a potential bone regeneration therapy. Hydrogels loaded with bone morphogenic protein-2 applied to critical defects in rat craniums were completely invaded over a four-week period. Additionally, significant remolding of the hydrogels into bony tissue was observed.[128] Similar PEG-based hydrogel technologies has also been used to investigate cardioprogenitor differentiation,[158] scar tissue formation,[159] as well as to create hydrogels with pro-angiogenic properties.[160]
To decouple the effects of physical and biochemical properties, it is necessary to control materials stiffness over time. For example, alginate hydrogels modified with RGD ligands have been used to study MSCs in controlled 3D microenvironments.[135] This study demonstrated that osetogenic commitment of MSCs is not correlated with morphology, in opposition to MSCs behaviors on 2D substrates of controlled stiffness. Rather, matrix stiffness dictates integrin binding and adhesion ligand reorganization at the nanoscale, both of which correlate with commitment to osteogenesis.
Differentiation of MSCs has also been explored with nanofiber scaffolds. In one example, tunable nanofiber scaffolds were developed to study MSCs to osteoblast differentiation. By tuning the pore structure and mechanical properties of the biodegradable nanofibers scaffold Yoshimoto et al. demonstrated that microenviroments can be created to support mineralization required for bone tissue engineering.[147] Other investigations have considered hyaluronic acid and collagen nanofibers, both of which have structural functions in the native tissue, as compositional choices for tissue engineering. Nesti et al. has reported interesting results for intervertebral disc tissue engineering.[145] Long term culturing of MSCs in hyaluronic acid nanofibrous scaffolds resulted in high expression of target ECM proteins as well as the development of constructs similar to the target native tissue.
Nanofiber scaffolds have also been used to study neurons and neural tissue. For example, one investigation into neural tissue engineering found that nanofibers compared to microfibers increased the differentiation rate of neural stem cells and improved neurite outgrowth [148]. Alternatively, Li et al. synthesized a bioactive and conductive nanofibrous scaffold for neural tissue engineering through the use of polyanaline-gelatin mixture. This was a first step in developing a scaffold where structural (mechanical cues), bioactive, and electro-active signals could be presented to neural cells for intelligent tissue culturing [149]. Interested readers are referred to an excellent review on conductive polymers in tissue engineering [161].
Self-assembling peptide amphiphiles has been used to create bioactive hydrogels for tissue regenerative therapies (Figure 7, top).[162] This technology was used to present the neurite promoting peptide sequence IKVAV to encapsulated neural progenitor cells.[163] Peptide amphiphiles have also been used to deliver growth factors in controlled 3D microenvironments. In one example, RGD containing peptide nanofibers with bound basic fibroblast growth factor (bFGF) were subcutaneously injected into mice. The peptide amphiphiles formed a clear hydrogel in vivo with a sustained release of bFGF that induced significant angiogenesis at the injection site (Figure 7, bottom).[156] Self-assembled peptide nanofibers have also been used to create hydrogels with controlled presentations of RGD and other cell adhesion ligands.[134] Such materials have been used to study bone regeneration therapies[164], culturing of synthetic dermis[165], and tubulogenesis of endothelial cells[166], among other biomedical applications[167].
Figure 7.
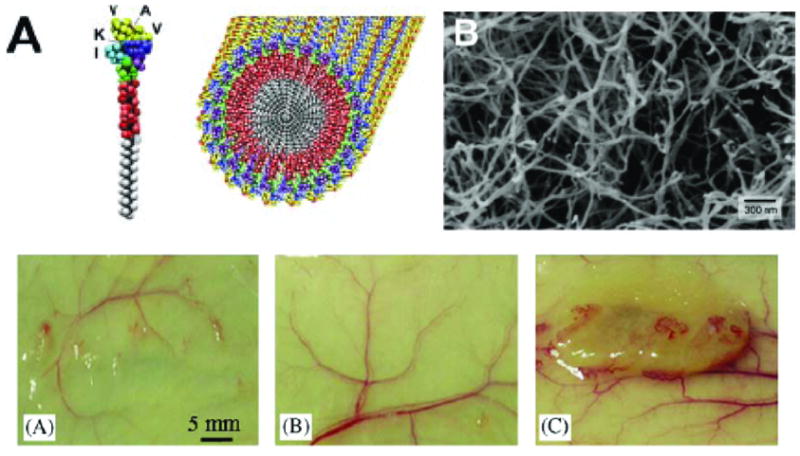
Nanofiber hydrogels from peptide amphphiles. Top: (A) Cartoon of the single peptide amphiphile and a self-assembled nanofiber.[132] (B) A scanning electron micrograph of a nanofiber network.[132] Bottom: (A) Representative photographs of tissue samples with injected angiogenetic peptide amphiphile hydrogels, (A) control nanofiber hydrogel, (B) control bFGF solution, and (C) bFBF containing nanofiber hydrogel.[156]
The relative lack of data focused towards specific cellular functions, such as migration, cell spreading or directed differentiation, described here for 3D systems, as compared to the data presented for 2D systems is a reflection of the maturity of these systems. Comparatively, 3D biomaterials with nanoscale control of cell adhesion and bioactive properties are new. These systems have not yet evolved to be able to directly control the 3D patterning and organization of bioactivity that is possible in 2D. Such systems would be highly useful for the investigation of interesting biological questions that explore the importance of order and disorder in the presentation of cell adhesion ligands. In some 2D systems it has been shown that disordered presentation increases cell adhesion[68], comparison to similarly designed 3D studies would produce highly interesting results.
Conclusions
Engineering biomaterials to interact with cells at the nanoscale has led to many insights into important biological questions. Investigators have produced a large body of knowledge describing cell adhesion and migration in 2D with engineered bioactive substrates. Extension of the design concepts used to create controlled bioactive substrates to 3D has produced important hydrogel and nanofiber technologies that can mimic many different aspects of the ECM. The outcomes of these experiments demonstrate that cell behavior is markedly different in 3D as compared to 2D. Advances in tissue engineering therapies and the development of a better understanding of in vivo cell behaviors will come with advances in 3D biomaterials technologies that can recapitulate physiological conditions in vitro. Designing 3D biomaterials with controlled nanoscale features including the physical and biochemical features of the ECM is paramount to the successful transition of tissue engineering therapies from experimental investigations to clinical applications.
Acknowledgments
This paper was supported by the National Institutes of Health (EB008392 and DE019024 to AK; DE19180 to EJ), National Science Foundation (DMR0847287 to AK; CBET0756394 and CBET0931998 to EJ), the Institute for Soldier Nanotechnology, the Office of Naval Research and the US Army Corps of Engineers. AB is supported by MSTP grant 2T32GM007753-29 from the NIH.
References
- 1.Massia SP, Hubbell JA. Human endothelial cell interactions with surface-coupled adhesion peptides on a nonadhesive glass substrate and two polymeric biomaterials. J Biomed Mat Res. 1991;25(2):223–242. doi: 10.1002/jbm.820250209. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee P, et al. Polymer latexes for cell-resistant and cell-interactive surfaces. J Biomed Mat Res. 2000;50(3):331–339. doi: 10.1002/(sici)1097-4636(20000605)50:3<331::aid-jbm6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.Tong YW, Shoichet MS. Peptide surface modification of poly(tetrafluoroethylene-co-hexafluoropropylene) enhances its interaction with central nervous system neurons. J of Biomed Mat Res. 1998;42(1):85–95. doi: 10.1002/(sici)1097-4636(199810)42:1<85::aid-jbm11>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson K, Mosbach K. [2] Immobilization of ligands with organic sulfonyl chlorides. In: William BJ, editor. Methods in Enzymology. Academic Press; 1984. pp. 56–69. [DOI] [PubMed] [Google Scholar]
- 5.Barrera DA, et al. Copolymerization and Degradation of Poly(lactic acid-co-lysine) Macromole. 1995;28(2):425–432. [Google Scholar]
- 6.Jo S, Engel PS, Mikos AG. Synthesis of poly(ethylene glycol)-tethered poly(propylene fumarate) and its modification with GRGD peptide. Polymer. 2000;41(21):7595–7604. [Google Scholar]
- 7.Besselink G, Beugeling T, Bantjes A. N-Hydroxysuccinimide-activated glycine-sepharose. App Biochem Biotechnol. 1993;43(3):227–246. [Google Scholar]
- 8.Morpurgo M, Bayer EA, Wilchek M. N-hydroxysuccinimide carbonates and carbamates are useful reactive reagents for coupling ligands to lysines on proteins. J Biochem Biophys Method. 1999;38(1):17–28. doi: 10.1016/s0165-022x(98)00027-x. [DOI] [PubMed] [Google Scholar]
- 9.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20(1):45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 10.Scheibler L, et al. Functional molecular thin films: Topological templates for the chemoselective ligation of antigenic peptides to self-assembled monolayers. Angew Chem- Int Ed. 1999;38(5):696–699. doi: 10.1002/(SICI)1521-3773(19990301)38:5<696::AID-ANIE696>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 11.Elbert DL, Hubbell JA. Conjugate Addition Reactions Combined with Free-Radical Cross-Linking for the Design of Materials for Tissue Engineering. Biomacromole. 2001;2(2):430–441. doi: 10.1021/bm0056299. [DOI] [PubMed] [Google Scholar]
- 12.Stile RA, Healy KE. Thermo-Responsive Peptide-Modified Hydrogels for Tissue Regeneration. Biomacromole. 2001;2(1):185–194. doi: 10.1021/bm0000945. [DOI] [PubMed] [Google Scholar]
- 13.Bearinger JP, Castner DG, Healy KE. Biomolecular modification of p(AAm-co-EG/AA) IPNs supports osteoblast adhesion and phenotypic expression. J Biomat Sci, Polymer Ed. 1998;9:629–652. doi: 10.1163/156856298x00064. [DOI] [PubMed] [Google Scholar]
- 14.Miller JS, et al. Bioactive hydrogels made from step-growth derived PEG-peptide macromers. Biomaterials. 31(13):3736–3743. doi: 10.1016/j.biomaterials.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 16.Lutolf MP, Hubbell JA. Synthesis and physicochemical characterization of end-linked poly(ethylene glycol)-co-peptide hydrogels formed by Michael-type addition. Biomacromole. 2003;4(3):713–722. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 17.DeForest CA, Polizzotti BD, Anseth KS. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat Mater. 2009;8(8):659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stupp SI, et al. Supramolecular materials: Self-organized nanostructures. Science. 1997;276(5311):384–389. doi: 10.1126/science.276.5311.384. [DOI] [PubMed] [Google Scholar]
- 19.Zhang SG, et al. Design of nanostructured biological materials through self-assembly of peptides and proteins. Curr Opin Chem Bio. 2002;6(6):865–871. doi: 10.1016/s1367-5931(02)00391-5. [DOI] [PubMed] [Google Scholar]
- 20.Stevens MM, George JH. Exploring and engineering the cell surface interface. Science. 2005;310(5751):1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 21.Scott J. Supramolecular organization of extracellular matrix glycosaminoglycans, in vitro and in the tissues. FASEB J. 1992;6(9):2639–2645. [PubMed] [Google Scholar]
- 22.Huang S, Ingber DE. The structural and mechanical complexity of cell-growth control. Nat Cell Biol. 1999;1(5) doi: 10.1038/13043. [DOI] [PubMed] [Google Scholar]
- 23.Cukierman E, et al. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 24.Maheshwari G, et al. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113(10):1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 25.Engler AJ, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 26.Discher DE, Mooney DJ, Zandstra PW. Growth Factors, Matrices, and Forces Combine and Control Stem Cells. Science. 2009;324(5935):1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7(3):211–224. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 28.Massia SP, Hubbell JA. An Rgd Spacing of 440nm Is Sufficient for Integrin Alpha-V-Beta-3-Mediated Fibroblast Spreading and 140nm for Focal Contact and Stress Fiber Formation. J Cell Biol. 1991;114(5):1089–1100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz SA, Chen CS. Emergence of Patterned Stem Cell Differentiation Within Multicellular Structures. Stem Cells. 2008;26(11):2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong HJ, Hsiong S, Mooney DJ. Nanoscale cell adhesion ligand presentation regulates nonviral gene delivery and expression. Nano Lett. 2007;7(1):161–6. doi: 10.1021/nl062485g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell ID. Studies of focal adhesion assembly. Biochem Soc Trans. 2008;36:263–266. doi: 10.1042/BST0360263. [DOI] [PubMed] [Google Scholar]
- 32.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 33.Romer LH, Birukov KG, Garcia JG. Focal adhesions: paradigm for a signaling nexus. Circ Res. 2006;98(5):606–616. doi: 10.1161/01.RES.0000207408.31270.db. [DOI] [PubMed] [Google Scholar]
- 34.Xiong J-P, et al. Crystal Structure of the Extracellular Segment of Integrin alpha Vbeta 3 in Complex with an Arg-Gly-Asp Ligand. Science. 2002;296(5565):151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 35.Perlin L, MacNeil S, Rimmer S. Production and performance of biomaterials containing RGD peptides. Soft Matt. 2008;4(12):2331–2349. [Google Scholar]
- 36.Hildebrand JD, Schaller MD, Parsons JT. Identification of sequences required for the efficient localization of the focal adhesion kinase, pp125FAK, to cellular focal adhesions. J Cell Biol. 1993;123(4):993–1005. doi: 10.1083/jcb.123.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J. Bioactive modification of poly(ethylene glycol) hydrogels for tissue engineering. Biomaterials. 2010 doi: 10.1016/j.biomaterials.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hynes RO, Yamada KM. Fibronectins: multifunctional modular glycoproteins. J Cell Biol. 1982;95(2 Pt 1):369–77. doi: 10.1083/jcb.95.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238(4826):491–7. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 40.Lauffenburger DA, Linderman JJ. Receptors: Models for Binding, Trafficking, and Signalling. 1993. [Google Scholar]
- 41.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24(24):4385–415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 42.Ratner BD, Bryant SJ. Biomaterials: Where We Have Been and Where We are Going. Annu Rev Biomed Eng. 2004;6(1):41–75. doi: 10.1146/annurev.bioeng.6.040803.140027. [DOI] [PubMed] [Google Scholar]
- 43.Porte-Durrieu MC, et al. Cyclo-(DfKRG) peptide grafting onto Ti-6Al-4V: physical characterization and interest towards human osteoprogenitor cells adhesion. Biomaterials. 2004;25(19):4837–4846. doi: 10.1016/j.biomaterials.2003.11.037. [DOI] [PubMed] [Google Scholar]
- 44.Verrier S, et al. Function of linear and cyclic RGD-containing peptides in osteoprogenitor cells adhesion process. Biomaterials. 2002;23(2):585–596. doi: 10.1016/s0142-9612(01)00145-4. [DOI] [PubMed] [Google Scholar]
- 45.Hypolite CL, et al. Formation of microscale gradients of protein using heterobifunctional photolinkers. Bioconjug Chem. 1997;8(5):658–63. doi: 10.1021/bc9701252. [DOI] [PubMed] [Google Scholar]
- 46.Herbert CB, et al. Micropatterning gradients and controlling surface densities of photoactivatable biomolecules on self-assembled monolayers of oligo(ethylene glycol) alkanethiolates. Chem Biol. 1997;4(10):731–7. doi: 10.1016/s1074-5521(97)90311-2. [DOI] [PubMed] [Google Scholar]
- 47.Plummer ST, Bohn PW. Spatial dispersion in electrochemically generated surface composition gradients visualized with covalently bound fluorescent nanospheres. Langmuir. 2002;18(10):4142–4149. [Google Scholar]
- 48.Haddow DB, et al. Comparison of proliferation and growth of human keratinocytes on plasma copolymers of acrylic acid/1,7-octadiene and self-assembled monolayers. J Biomed Mater Res. 1999;47(3):379–87. doi: 10.1002/(sici)1097-4636(19991205)47:3<379::aid-jbm13>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 49.Whittle JD, et al. A method for the deposition of controllable chemical gradients. Chem Comm. 2003;14:1766–1767. [Google Scholar]
- 50.Jeon NL, et al. Generation of solution and surface gradients using microfluidic systems. Langmuir. 2000;16(22):8311–8316. [Google Scholar]
- 51.Burdick JA, Khademhosseini A, Langer R. Fabrication of gradient hydrogels using a microfluidics/photopolymerization process. Langmuir. 2004;20(13):5153–6. doi: 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]
- 52.Buck CA, Horwitz AF. Cell surface receptors for extracellular matrix molecules. Annu Rev Cell Biol. 1987;3:179–205. doi: 10.1146/annurev.cb.03.110187.001143. [DOI] [PubMed] [Google Scholar]
- 53.Lanza RP, Langer RS, Chick WL. Principles of Tissue Engineering. Austin, TX: Academic Press; 1997. [Google Scholar]
- 54.Jeong HE, et al. UV-assisted capillary force lithography for engineering biomimetic multiscale hierarchical structures: From lotus leaf to gecko foot hairs. Nanoscale. 2009;1(3):331–338. doi: 10.1039/b9nr00106a. [DOI] [PubMed] [Google Scholar]
- 55.Kondoh A, Makino K, Matsuda T. Two-dimensional artificial extracellular matrix: Bioadhesive peptide-immobilized surface design. J App Polym Sci. 1993;47(11):1983–1988. [Google Scholar]
- 56.Kim MR, Jeong JH, Park TG. Swelling Induced Detachment of Chondrocytes Using RGD-Modified Poly(N-isopropylacrylamide) Hydrogel Beads. Biotechnol Prog. 2002;18(3):495–500. doi: 10.1021/bp020287z. [DOI] [PubMed] [Google Scholar]
- 57.Carlisle ES, et al. Enhancing hepatocyte adhesion by pulsed plasma deposition and polyethylene glycol coupling. Tissue Eng. 2000;6(1):45–52. doi: 10.1089/107632700320883. [DOI] [PubMed] [Google Scholar]
- 58.Koo LY, et al. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J Cell Sci. 2002;115(7):1423–1433. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- 59.Hsiong SX, et al. Differentiation stage alters matrix control of stem cells. J Biomed Mater Res A. 2008;85(1):145–56. doi: 10.1002/jbm.a.31521. [DOI] [PubMed] [Google Scholar]
- 60.Glass R, et al. Micro-nanostructured interfaces fabricated by the use of inorganic block copolymer micellar monolayers as negative resist for electron-beam lithography. Adv Func Mat. 2003;13(7):569–575. [Google Scholar]
- 61.Glass R, Moller M, Spatz JP. Block copolymer micelle nanolithography. Nanotechnology. 2003;14(10):1153–1160. [Google Scholar]
- 62.Arnold M, et al. Induction of cell polarization and migration by a gradient of nanoscale variations in adhesive ligand spacing. Nano Lett. 2008;8(7):2063–2069. doi: 10.1021/nl801483w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnold M, et al. Activation of integrin function by nanopatterned adhesive interfaces. Chemphyschem. 2004;5(3):383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 64.Cavalcanti-Adam EA, et al. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur J Cell Biol. 2006;85(3–4):219–224. doi: 10.1016/j.ejcb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 65.Lee K-B, et al. Protein Nanoarrays Generated By Dip-Pen Nanolithography. Science. 2002;295(5560):1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 66.Slater JH, Frey W. Nanopatterning of fibronectin and the influence of integrin clustering on endothelial cell spreading and proliferation. J Biomed Mater Res A. 2008;87(1):176–95. doi: 10.1002/jbm.a.31725. [DOI] [PubMed] [Google Scholar]
- 67.Yim EKF, Pang SW, Leong KW. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp Cell Res. 2007;313(9):1820–1829. doi: 10.1016/j.yexcr.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dalby MJ, et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater. 2007;6(12):997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 69.Kim DH, et al. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc Natl Acad Sci U S A. 2010;107(2):565–70. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bettinger CJ, Langer R, Borenstein JT. Engineering Substrate Topography at the Micro- and Nanoscale to Control Cell Function. Angew Chem-Int Ed. 2009;48(30):5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Streuli CH, Bissell MJ. Expression of extracellular matrix components is regulated by substratum. J Cell Biol. 1990;110(4):1405–15. doi: 10.1083/jcb.110.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69(1):11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 73.Clark EA, Brugge JS. Integrins and signal transduction pathways: the road taken. Science. 1995;268(5208):233–9. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 74.Huttenlocher A, Ginsberg MH, Horwitz AF. Modulation of cell migration by integrin-mediated cytoskeletal linkages and ligand-binding affinity. J Cell Biol. 1996;134(6):1551–62. doi: 10.1083/jcb.134.6.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palecek SP, et al. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385(6616):537–40. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 76.Grinnell F, Feld MK. Adsorption characteristics of plasma fibronectin in relationship to biological activity. J Biomed Mater Res. 1981;15(3):363–81. doi: 10.1002/jbm.820150308. [DOI] [PubMed] [Google Scholar]
- 77.Hubbell JA. Bioactive biomaterials. Curr Opin Biotechnol. 1999;10(2):123–9. doi: 10.1016/s0958-1669(99)80021-4. [DOI] [PubMed] [Google Scholar]
- 78.Lewandowska K, et al. Cell-type-specific adhesion mechanisms mediated by fibronectin adsorbed to chemically derivatized substrata. J Biomed Mater Res. 1992;26(10):1343–63. doi: 10.1002/jbm.820261007. [DOI] [PubMed] [Google Scholar]
- 79.Martin JY, et al. Effect of titanium surface roughness on proliferation, differentiation, and protein synthesis of human osteoblast-like cells (MG63) J Biomed Mater Res. 1995;29(3):389–401. doi: 10.1002/jbm.820290314. [DOI] [PubMed] [Google Scholar]
- 80.Iuliano DJ, Saavedra SS, Truskey GA. Effect of the conformation and orientation of adsorbed fibronectin on endothelial cell spreading and the strength of adhesion. J Biomed Mater Res. 1993;27(8):1103–13. doi: 10.1002/jbm.820270816. [DOI] [PubMed] [Google Scholar]
- 81.Kao WJ, Lee D. In vivo modulation of host response and macrophage behavior by polymer networks grafted with fibronectin-derived biomimetic oligopeptides: the role of RGD and PHSRN domains. Biomaterials. 2001;22(21):2901–9. doi: 10.1016/s0142-9612(01)00037-0. [DOI] [PubMed] [Google Scholar]
- 82.Pettit DK, Hoffman AS, Horbett TA. Correlation between corneal epithelial cell outgrowth and monoclonal antibody binding to the cell binding domain of adsorbed fibronectin. J Biomed Mater Res. 1994;28(6):685–91. doi: 10.1002/jbm.820280605. [DOI] [PubMed] [Google Scholar]
- 83.Prime KL, Whitesides GM. Self-assembled organic monolayers: model systems for studying adsorption of proteins at surfaces. Science. 1991;252(5010):1164–7. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 84.Thomas CH, et al. The role of vitronectin in the attachment and spatial distribution of bone-derived cells on materials with patterned surface chemistry. J Biomed Mater Res. 1997;37(1):81–93. doi: 10.1002/(sici)1097-4636(199710)37:1<81::aid-jbm10>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 85.Tegoulia VA, Cooper SL. Leukocyte adhesion on model surfaces under flow: effects of surface chemistry, protein adsorption, and shear rate. J Biomed Mater Res. 2000;50(3):291–301. doi: 10.1002/(sici)1097-4636(20000605)50:3<291::aid-jbm2>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 86.Boulbitch A, Guttenberg Z, Sackmann E. Kinetics of membrane adhesion mediated by ligand-receptor interaction studied with a biomimetic system. Biophys J. 2001;81(5):2743–51. doi: 10.1016/S0006-3495(01)75917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brochard-Wyart F, de Gennes PG. Adhesion induced by mobile binders: dynamics. Proc Natl Acad Sci USA. 2002;99(12):7854–9. doi: 10.1073/pnas.112221299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Freund LB, Lin Y. The role of binder mobility in spontaneous adhesive contact and implications for cell adhesion. J Mechan Phy Sol. 2004;52(11):2455–2472. [Google Scholar]
- 89.Shenoy VB, Freund LB. Growth and shape stability of a biological membrane adhesion complex in the diffusion-mediated regime. Proc Natl Acad Sci USA. 2005;102(9):3213–8. doi: 10.1073/pnas.0500368102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mei Y, et al. Tuning cell adhesion on gradient poly(2-hydroxyethyl methacrylate)-grafted surfaces. Langmuir. 2005;21(26):12309–14. doi: 10.1021/la050668x. [DOI] [PubMed] [Google Scholar]
- 91.Harris BP, et al. Photopatterned polymer brushes promoting cell adhesion gradients. Langmuir. 2006;22(10):4467–71. doi: 10.1021/la053417x. [DOI] [PubMed] [Google Scholar]
- 92.Maheshwari G, et al. Biophysical integration of effects of epidermal growth factor and fibronectin on fibroblast migration. Biophys J. 1999;76(5):2814–23. doi: 10.1016/S0006-3495(99)77435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Drumheller PD, Hubbell JA. Polymer Networks with Grafted Cell Adhesion Peptides for Highly Biospecific Cell Adhesive Substrates. Analyt Biochem. 1994;222(2):380–388. doi: 10.1006/abio.1994.1506. [DOI] [PubMed] [Google Scholar]
- 94.Ostuni E, Yan L, Whitesides GM. The interaction of proteins and cells with self-assembled monolayers of alkanethiolates on gold and silver. Colloids and Surfaces B: Biointerfaces. 1999;15(1):3–30. [Google Scholar]
- 95.Pakalns T, et al. Cellular recognition of synthetic peptide amphiphiles in self-assembled monolayer films. Biomaterials. 1999;20(23–24):2265–2279. doi: 10.1016/s0142-9612(99)00157-x. [DOI] [PubMed] [Google Scholar]
- 96.Huang JH, et al. Impact of Order and Disorder in RGD Nanopatterns on Cell Adhesion. Nano Lett. 2009;9(3):1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hirschfeld-Warneken VC, et al. Cell adhesion and polarisation on molecularly defined spacing gradient surfaces of cyclic RGDfK peptide patches. Euro J Cell Biol. 2008;87(8–9):743–750. doi: 10.1016/j.ejcb.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dickinson RB, Tranquillo RT. Transport-Equations and Indexes for Random and Biased Cell-Migration Based on Single-Cell Properties. Siam J Appl Math. 1995;55(5):1419–1454. [Google Scholar]
- 99.Bray D. Cell Movements. New York, NY: Garland Publishing, Inc; 1992. [Google Scholar]
- 100.Lackie JM. Cell Movement and Cell Behaviour. London: Allen and Unwin; 1986. [Google Scholar]
- 101.Stossel TP. On the crawling of animal cells. Science. 1993;260(5111):1086–94. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 102.Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84(3):345–57. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 103.Howe A, et al. Integrin signaling and cell growth control. Curr Opin Cell Biol. 1998;10(2):220–31. doi: 10.1016/s0955-0674(98)80144-0. [DOI] [PubMed] [Google Scholar]
- 104.Kang CE, Gemeinhart EJ, Gemeinhart RA. Cellular alignment by grafted adhesion peptide surface density gradients. J Biomed Mater Res A. 2004;71(3):403–11. doi: 10.1002/jbm.a.30137. [DOI] [PubMed] [Google Scholar]
- 105.Bhat RR, et al. Tailoring cell adhesion using surface-grafted polymer gradient assemblies. Adv Mater. 2005;17(23):2802. [Google Scholar]
- 106.DeLong SA, Gobin AS, West JL. Covalent immobilization of RGDS on hydrogel surfaces to direct cell alignment and migration. J Control Rel. 2005;109(1–3):139–148. doi: 10.1016/j.jconrel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 107.Simon KA, et al. Enhancing cell adhesion and confinement by gradient nanotopography. J Am Chem Soc. 2007;129(16):4892–3. doi: 10.1021/ja0653472. [DOI] [PubMed] [Google Scholar]
- 108.Reinhart-King CA, Dembo M, Hammer DA. Endothelial Cell Traction Forces on RGD-Derivatized Polyacrylamide Substrata. Langmuir. 2002;19(5):1573–1579. [Google Scholar]
- 109.DiMilla PA, Barbee K, Lauffenburger DA. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys J. 1991;60(1):15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith JT, et al. Measurement of cell migration on surface-bound fibronectin gradients. Langmuir. 2004;20(19):8279–86. doi: 10.1021/la0489763. [DOI] [PubMed] [Google Scholar]
- 111.Smith JT, Elkin JT, Reichert WM. Directed cell migration on fibronectin gradients: effect of gradient slope. Exp Cell Res. 2006;312(13):2424–32. doi: 10.1016/j.yexcr.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 112.Sarvestani AS, Jabbari E. Analysis of cell locomotion on ligand gradient substrates. Biotechnol Bioeng. 2009;103:424–429. doi: 10.1002/bit.22273. [DOI] [PubMed] [Google Scholar]
- 113.Gracheva ME, Othmer HG. A continuum model of motility in ameboid cells. Bull Math Biol. 2004;66(1):167–93. doi: 10.1016/j.bulm.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 114.Larripa K, Mogilner A. Transport of a 1D viscoelastic actin-myosin strip of gel as a model of a crawling cell. Phys Stat Mech Appl. 2006;372(1):113–123. doi: 10.1016/j.physa.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chaw KC, et al. A quantitative observation and imaging of single tumor cell migration and deformation using a multi-gap microfluidic device representing the blood vessel. Microvas Res. 2006;72:153–160. doi: 10.1016/j.mvr.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 116.Kong HJ, et al. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc Nat Acad Sci USA. 2005;102(12):4300–4305. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trend Cell Biol. 2003;13(5):264–269. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- 118.Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462(7272):433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Slaughter BV, et al. Hydrogels in Regenerative Medicine. Adv Mater. 2009;21(32–33):3307–3329. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang Q, et al. A cartilage ECM-derived 3-D porous acellular matrix scaffold for in vivo cartilage tissue engineering with PKH26-labeled chondrogenic bone marrow-derived mesenchymal stem cells. Biomaterials. 2008;29(15):2378–87. doi: 10.1016/j.biomaterials.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 121.Levenberg S, et al. Differentiation of human embryonic stem cells on three-dimensional polymer scaffolds. Proc Nat Acad Sci USA. 2003;100(22):12741–12746. doi: 10.1073/pnas.1735463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mann BK, et al. Smooth muscle cell growth in photopolymerized hydrogels with cell adhesive and proteolytically degradable domains: synthetic ECM analogs for tissue engineering. Biomaterials. 2001;22(22):3045–51. doi: 10.1016/s0142-9612(01)00051-5. [DOI] [PubMed] [Google Scholar]
- 123.Nichol JW, et al. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials. 2010;31(21):5536–44. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Khademhosseini A, et al. Microscale technologies for tissue engineering and biology. Proc Nat Acad Sci USA. 2006;103(8):2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Drury JL, Mooney DJ. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials. 2003;24(24):4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 126.Charnley M, et al. Integration column: microwell arrays for mammalian cell culture. Integrat Biol. 2009;1(11–12):625–634. doi: 10.1039/b918172p. [DOI] [PubMed] [Google Scholar]
- 127.Peppas NA, et al. Hydrogels in biology and medicine: From molecular principles to bionanotechnology. Adv Mater. 2006;18(11):1345–1360. [Google Scholar]
- 128.Lutolf MP, et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: Engineering cell-invasion characteristics. Proc Nat Acad Sci USA. 2003;100(9):5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mann BK, et al. Smooth muscle cell growth in photopolymerized hydrogels with cell adhesive and proteolytically degradable domains: Synthetic ECM analogs for tissue engineering. Biomaterials. 2001;22(22):3045–3051. doi: 10.1016/s0142-9612(01)00051-5. [DOI] [PubMed] [Google Scholar]
- 130.Khademhosseini A, et al. Micromolding of photocrosslinkable hyaluronic acid for cell encapsulation and entrapment. J Biomed Mater Res A. 2006;79A(3):522–532. doi: 10.1002/jbm.a.30821. [DOI] [PubMed] [Google Scholar]
- 131.Mammoto A, et al. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457(7233):1103–U57. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;294(5547):1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 133.Hartgerink JD, Beniash E, Stupp SI. Peptide-amphiphile nanofibers: A versatile scaffold for the preparation of self-assembling materials. Proc Nat Acad Sci USA. 2002;99(8):5133–5138. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yokoi H, Kinoshita T, Zhang SG. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc Nat Acad Sci USA. 2005;102(24):8414–8419. doi: 10.1073/pnas.0407843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 9(6):518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhu JM, et al. Design and Synthesis of Biomimetic Hydrogel Scaffolds with Controlled Organization of Cyclic RGD Peptides. Bioconjug Chem. 2009;20(2):333–339. doi: 10.1021/bc800441v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.DeForest CA, Sims EA, Anseth KS. Peptide-Functionalized Click Hydrogels with Independently Tunable Mechanics and Chemical Functionality for 3D Cell Culture. Chem Mater. doi: 10.1021/cm101391y. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Du Y, et al. Convection-driven generation of long-range material gradients. Biomaterials. 31(9):2686–2694. doi: 10.1016/j.biomaterials.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.He J, et al. Rapid generation of biologically-relevant hydrogels containing long-range chemical and mechanical gradients. Adv Func Mat. 2009;20(1):131–137. doi: 10.1002/adfm.200901311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Matthews JA, et al. Electrospinning of Collagen Nanofibers. Biomacromole. 2002;3(2):232–238. doi: 10.1021/bm015533u. [DOI] [PubMed] [Google Scholar]
- 141.Teng S-H, et al. Collagen/hydroxyapatite composite nanofibers by electrospinning. Mater Lett. 2008;62(17–18):3055–3058. [Google Scholar]
- 142.Chen ZG, et al. Electrospun collagen-chitosan nanofiber: A biomimetic extracellular matrix for endothelial cell and smooth muscle cell. Acta Biomater. 2010;6(2):372–382. doi: 10.1016/j.actbio.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 143.Ji Y, et al. Electrospun three-dimensional hyaluronic acid nanofibrous scaffolds. Biomaterials. 2006;27(20):3782–92. doi: 10.1016/j.biomaterials.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 144.Kim TG, Chung HJ, Park TG. Macroporous and nanofibrous hyaluronic acid/collagen hybrid scaffold fabricated by concurrent electrospinning and deposition/leaching of salt particles. Acta Biomater. 2008;4(6):1611–9. doi: 10.1016/j.actbio.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 145.Nesti LJ, et al. Intervertebral disc tissue engineering using a novel hyaluronic acid-nanofibrous scaffold (HANFS) amalgam. Tissue Eng Part A. 2008;14(9):1527–37. doi: 10.1089/ten.tea.2008.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li W-J, et al. Electrospun nanofibrous structure: A novel scaffold for tissue engineering. Journal of Biomedical Materials Research. 2002;60(4):613–621. doi: 10.1002/jbm.10167. [DOI] [PubMed] [Google Scholar]
- 147.Yoshimoto H, et al. A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering. Biomaterials. 2003;24(12):2077–2082. doi: 10.1016/s0142-9612(02)00635-x. [DOI] [PubMed] [Google Scholar]
- 148.Yang F, et al. Electrospinning of nano/micro scale poly(L-lactic acid) aligned fibers and their potential in neural tissue engineering. Biomaterials. 2005;26(15):2603–2610. doi: 10.1016/j.biomaterials.2004.06.051. [DOI] [PubMed] [Google Scholar]
- 149.Li M, et al. Electrospinning polyaniline-contained gelatin nanofibers for tissue engineering applications. Biomaterials. 2006;27(13):2705–2715. doi: 10.1016/j.biomaterials.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 150.Schindler M, et al. A synthetic nanofibrillar matrix promotes in vivo-like organization and morphogenesis for cells in culture. Biomaterials. 2005;26(28):5624–5631. doi: 10.1016/j.biomaterials.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 151.Cao H, Liu T, Chew SY. The application of nanofibrous scaffolds in neural tissue engineering. Adv Drug Del Rev. 2009;61(12):1055–1064. doi: 10.1016/j.addr.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 152.Kloxin AM, et al. Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science. 2009;324(5923):59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tayalia P, et al. 3D Cell-Migration Studies using Two-Photon Engineered Polymer Scaffolds. Adv Mater. 2008;20(23):4494–4498. [Google Scholar]
- 154.Lu H, et al. Cartilage tissue engineering using funnel-like collagen sponges prepared with embossing ice particulate templates. Biomaterials. 2010;31(22):5825–5835. doi: 10.1016/j.biomaterials.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 155.Wei G, Ma PX. Partially nanofibrous architecture of 3D tissue engineering scaffolds. Biomaterials. 2009;30(32):6426–6434. doi: 10.1016/j.biomaterials.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hosseinkhani H, et al. Enhanced angiogenesis through controlled release of basic fibroblast growth factor from peptide amphiphile for tissue regeneration. Biomaterials. 2006;27(34):5836–5844. doi: 10.1016/j.biomaterials.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 157.Doyle AD, et al. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184(4):481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kraehenbuehl TP, et al. Three-dimensional extracellular matrix-directed cardioprogenitor differentiation: Systematic modulation of a synthetic cell-responsive PEG-hydrogel. Biomaterials. 2008;29(18):2757–2766. doi: 10.1016/j.biomaterials.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 159.Adeloew C, et al. The effect of enzymatically degradable poly(ethylene glycol) hydrogels on smooth muscle cell phenotype. Biomaterials. 2008;29(3):314–326. doi: 10.1016/j.biomaterials.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 160.Moon JJ, et al. Biomimetic hydrogels with pro-angiogenic properties. Biomaterials. 2010;31(14):3840–3847. doi: 10.1016/j.biomaterials.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Guimard NK, Gomez N, Schmidt CE. Conducting polymers in biomedical engineering. Progr Polym Sci. 32(8–9):876–921. [Google Scholar]
- 162.Shah RN, et al. Supramolecular design of self-assembling nanofibers for cartilage regeneration. Proc Nat Acad Sci USA. 107(8):3293–3298. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Silva GA, et al. Selective Differentiation of Neural Progenitor Cells by High-Epitope Density Nanofibers. Science. 2004;303(5662):1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 164.Yoshimi R, et al. Self-Assembling Peptide Nanofiber Scaffolds, Platelet-Rich Plasma, and Mesenchymal Stem Cells for Injectable Bone Regeneration With Tissue Engineering. J Craniofac Surg. 2009;20(5):1523–1530. doi: 10.1097/SCS.0b013e3181b09b7e. [DOI] [PubMed] [Google Scholar]
- 165.Kao B, Kadomatsu K, Hosaka Y. Construction of Synthetic Dermis and Skin Based on a Self-Assembled Peptide Hydrogel Scaffold. Tissue Eng Part A. 2009;15(9):2385–2396. doi: 10.1089/ten.tea.2008.0362. [DOI] [PubMed] [Google Scholar]
- 166.Wang XM, Horii A, Zhang SG. Designer functionalized self-assembling peptide nanofiber scaffolds for growth, migration, and tubulogenesis of human umbilical vein endothelial cells. Soft Matt. 2008;4(12):2388–2395. [Google Scholar]
- 167.Chow D, et al. Peptide-based biopolymers in biomedicine and biotechnology. Mater Sci Eng Rep. 2008;62(4):125–155. doi: 10.1016/j.mser.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]