Abstract
Nearly all DNA polymerases require processivity factors to ensure continuous incorporation of nucleotides. Processivity factors are specific for their cognate DNA polymerases. For this reason, the vaccinia DNA polymerase (E9) and the proteins associated with processivity (A20 and D4) are excellent therapeutic targets. In this study, we show the utility of stepwise rapid plate assays that i) screen for compounds that block vaccinia DNA synthesis, ii) eliminate trivial inhibitors, e.g. DNA intercalators, and iii) distinguish whether inhibitors are specific for blocking DNA polymerase activity or processivity. The sequential plate screening of 2,222 compounds from the NCI Diversity Set library yielded a DNA polymerase inhibitor (NSC 55636) and a processivity inhibitor (NSC 123526) that were capable of reducing vaccinia viral plaques with minimal cellular cytotoxicity. These compounds are predicted to block cellular infection by the smallpox virus, variola, based on the very high sequence identity between A20, D4 and E9 of vaccinia and the corresponding proteins of variola.
Keywords: Poxvirus; vaccinia virus; DNA polymerase; processivity factor, antiviral inhibitors; high throughput screening; rapid plate assay, smallpox
1. Introduction
The Centers for Disease Control has designated the smallpox virus, variola, as a class A agent of bioterrorism (Lane et al., 2001). Smallpox was globally eradicated in 1980 by vaccination with vaccinia virus. Currently, however, much of the human population is at risk since the vaccine has been discontinued. Moreover, the vaccine is contraindicated for the considerable portion of the population that is immunologically compromised (Lane and Millar, 1971; Lane et al., 1969; 1970). Therapeutics can protect these populations and serve as a rapid response to [MSOffice1]a smallpox outbreak.
New anti-poxvirus agents have recently emerged in the field, targeting the different points of the viral life cycle. Novel and promising agents include neutralizing antibodies that inhibit virus entry (Chen et al., 2006); nucleoside analogs such as cidofovir and ribavirin that terminate DNA replication upon incorporation into the growing DNA strand (Magee et al., 2005) or RNA transcript (Bougie and Bisaillon, 2004); ST-246 that targets the envelope protein production (Quenelle et al., 2007a; Yang et al., 2005) and Gleevec that blocks the tyrosine kinase activity (Reeves et al., 2005) required for motility, preventing the release of the extracellular enveloped virus (EEV). At present, cidofovir and ST-246 are the most effective therapeutics against poxvirus infection (De Clercq, 2002; Quenelle et al., 2007a; Sbrana et al., 2007).
The emergence and characterization of drug-resistant viruses (Beadle et al., 2002; Wodarz and Lloyd, 2004) including cidofovir-resistant poxviruses (Andrei et al., 2006; Kornbluth et al., 2006; Robbins et al., 2005; Smee et al., 2002) makes it compelling to discover new antiviral compounds. Additionally, therapeutics directed against a multiplicity of targets will further alleviate the problem of drug resistance. Indeed, synergistic inhibitory effect against vaccinia and cowpox viruses was recently demonstrated in vitro by combining ST-246 with hexadecyloxypropyl-cidofovir, CMX001, a derivative of cidofovir with increased bioavailability (Quenelle et Almost., 2007b).
The poxvirus replication cycle occurs entirely within the cytoplasm. The 192-kilobase vaccinia genome provides a multitude of potential targets that play distinct roles in vaccinia replication. The vaccinia proteins involved in DNA synthesis include a DNA polymerase (E9), nucleoside triphosphatase (D5), protein kinase (B1), putative DNA polymerase processivity factor (A20), uracil DNA glycosylase (D4), Holliday junction endonuclease (A22), DNA topoisomerase (H7), single-stranded DNA binding protein (I3), DNA ligase (A50), and enzymes involved in precursor metabolism, namely thymidine kinase (J2), thymidylate kinase (A48), ribonucleotide reductase (F4, I4) and dUTPase (F2) (Beaud, 1995; Moss, 2001).
An excellent target for therapeutics is the processivity complex. Almost all DNA polymerases from bacteriophages to mammals require processivity factors that enable them to replicate extended strands (Jeruzalmi et al., 2002; Kuriyan and O’Donnell, 1993). Processivity factors ensure that the rate of nucleotide incorporation by the DNA polymerase exceeds the rate of its dissociation from the template (Hingorani and O’Donnell, 2000; Weisshart et al., 1999). When the E9 DNA polymerase of vaccinia virus is presented alone to a DNA primer-template, it can incorporate approximately ten nucleotides (McDonald and Traktman, 1994). However, E9 complexed with processivity-associated proteins A20 and D4 incorporates thousands of nucleotides into the nascent DNA strand. Importantly, since A20, D4 and E9 vaccinia proteins share 98% sequence identity to the corresponding proteins of variola, they are excellent antiviral targets to prevent an outbreak of smallpox.
It is still unclear how A20 and D4 collaborate to enable E9 to synthesize DNA processively under physiological conditions. Through the use of recombinant virus and co-immunoprecipitation studies, A20 was shown to physically interact with E9 (Klemperer et al., 2001). Significantly, A20 has been shown to interact with D4 in vitro and by yeast-two hybrid assays (Ishii and Moss, 2002; McCraith et al., 2000; Stanitsa et al., 2006). On the other hand, D4 belongs to uracil DNA glycosylase (UDG) family (Scaramozzino et al., 2003), a class of enzymes which initiates the base excision and DNA repair pathway for misincorporated uracil through cleavage of the glycosidic bond. While D4 is required for vaccinia DNA replication (Millns et al., 1994), mutations of the D4 conserved catalytic region abolished the glycosylase activity but did not prevent viral DNA replication (De Silva and Moss, 2003; Ellison et al., 1996). Crystal structure of D4 recently revealed the possible formation of a tetrameric structure which consequently suggests its role as the sliding clamp associated with E9 DNA polymerase (Schormann et al., 2007). This model further implies the role of A20 as a scaffold for E9, D4 and possibly other replication proteins. Thus, desirable therapeutics include those that disrupt the interactions in the A20, D4 and E9 complex.
We have developed a rapid plate assay (Lin and Ricciardi, 2000) as a means of identifying novel antiviral therapeutics with lower toxicity. Previously, we used this assay to identify the promising compound, NSC 373989, that inhibits Kaposi’s sarcoma-associated herpes virus (KSHV) processive DNA synthesis in vitro and blocks lytic KSHV infection (Dorjsuren et al., 2006). In this study, we screened for potential therapeutics that block DNA synthesis of vaccinia virus. The experimental design employed a stepwise approach that subjects compounds to a series of distinct and increasingly stringent criteria to identify both polymerase and processivity inhibitors of vaccinia DNA synthesis. From a screen of 2,222 compounds, we identified both a polymerase inhibitor and a processivity inhibitor that block vaccinia DNA synthesis and viral infection. These inhibitory compounds have therapeutic potential due to their low cellular cytotoxicity.
2. Materials and Methods
2.1 Compound library
The NCI Training and Diversity Set libraries were obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, National Cancer Institute (http://dtp.nci.nih.gov/). The Training Set contains 230 anti-cancer compounds that are frequently used to confirm the reproducibility of high throughput screening assays (Shoemaker et al., 2002). The Diversity Set comprises 1,992 compounds selected from approximately 140,000 compounds using the Chem-X program (Accelrys, San Diego, CA). These compounds represent diversity in terms of three-dimensional pharmacophores (http://dtp.nci.nih.gov/branchers/dscb/diversity_explanation.html).
2.2 In vitro translated proteins and vaccinia virus-infected cytoplasmic lysate
Vaccinia virus polymerase (E9), A20 and UDG (D4) proteins were expressed from pcDNA3.2/v5 (Invitrogen) in vitro using Promega TNT T7 coupled transcription/translation system. KSHV DNA polymerase-8 (Pol8) and processivity factor-8 (PF8) were translated in vitro from pTM1-Pol8 and pTM1-PF8, respectively (Lin et al., 1998). To confirm expression, an aliquot of the translation reactions was labeled with [35S]-methionine, fractionated on an SDS-10% polyacrylamide gel, and visualized by autoradiography. Throughout the study, a thymidine kinase (TK) deficient WR strain vaccinia virus was used to infect monolayers of BSC-1 cells. This virus was kindly provided by Drs. G. Cohen and R. Eisenberg. The vaccinia virus-infected cell lysate was prepared according to previously described methods (Kit et al., 1962). Briefly, the cells were infected at a multiplicity of infection (MOI) of 15. The vaccinia-infected cells were incubated at 37 °C for 6 h in the presence of hydroxyurea. The cells were harvested by scraping then pelleting at 500 rpm. The pellet was washed with phosphate-buffered saline (PBS) followed by hypotonic buffer (10 mM Hepes, 1.5 nM MgCl2, 10 mM KCl). After resuspension in hypotonic buffer, the cells were Dounce-homogenized and centrifuged at 15,000 rpm for 30 min. The suspension was passed through a 2 micron filter to remove the viral cores and nuclear particles. The vaccinia-infected cytoplasmic lysate was stored in -80 °C in the presence of 20% glycerol.
2.3 High-throughput screening for inhibitors of DNA synthesis using the rapid plate assay
A rapid plate DNA synthesis assay (Lin and Ricciardi, 2000) was performed using vaccinia-infected cell lysate. A 1.2:1 ratio of a 20-mer oligonucleotide primer (5′-GCGAATGAATGACCGCTGAC-3′) and a 5′-end biotinylated 100-mer oligonucleotide template (5′- Biotin — GCACTTATTGCATTCGCTAGTCCACCTTGGATCTCAGGCT ATTCGTAGCGAGCTACGCGTACGTTAGCTTCGGTCATCCCGTCAGCGGTCATT CATTGGC — 3′) were heated at 90 °C for 5 min and annealed by gradual cooling to room temperature. The annealed primer-template (P/T) was diluted with PBS to a concentration of 10 pmol/μL. The 96-well microtiter streptavidin-coated plates (Streptawell plates, Roche Applied Science, Indianapolis, IN) were coated with 5 pmol/well of the P/T solution and incubated at 37 °C for 90 min. The wells were washed with 100 μL PBS. Control (DMSO, acyclovir, azidothymidine (AZT) and ethylenediamine tetracetic acid (EDTA)) and test compounds were individually added to the wells to a final concentration of 167 μM. The 60 μL DNA synthesis reaction mixture contained 100 mM (NH4)2SO4, 20 mM Tris-HCl pH 7.4, 3 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 2% glycerol, 40 ug/mL BSA, 5 uM dNTPs, 1 uM digoxigenin-11-2′-deoxyuridine-5′-triphosphate (DIG-dUTP, Roche Applied Science) and 1 μL vaccinia lysate. The plates were incubated at 37 °C for 30 min. DNA synthesis was determined by quantitating incorporation of DIG-dUTP using a DIG detection ELISA kit (Roche Applied Science) using anti-digoxigenin-peroxidase (anti-DIG-POD) and its substrate 2, 2′-azino-bis(3-ethylbenzthiazoline)-sulfonate (ABTS), and by measuring the absorbance at 405 nm on a microplate reader (Tecan Genious Pro, Grodig, Austria). The inhibitory threshold was set at 50%.
2.4 Screen to eliminate trivial inhibitors
The selectivity screen was used to eliminate non-specific inhibitors of vaccinia DNA synthesis. A microplate assay was employed in which DNA synthesis was directed by KSHV Pol8/PF8 as previously described (Dorjsuren et al., 2006; Lin and Ricciardi, 2000).
2.5 Polymerase vs. processivity inhibition
Two different plate assays were used to distinguish polymerase versus processivity inhibitors. For the polymerase plate assay, dA was distributed in the template such that the DIG epitope could be uniformly incorporated throughout the newly synthesized strand. The DNA synthesis reaction was performed using a low salt buffer (20 mM Tris-HCl pH 7.4, 3 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, and 2 % glycerol) and 1 μL in vitro translated polymerase enzyme E9. For the processivity plate assay, a biotinylated primer/template (5′- GCCAATGAATGACCGCTGAC -3′) / (5′- Biotin — AGCACTATTGACATTACAGAGTCGCCTTGGCTCTCTGGCTGTTCGTTGCGGGC TCCGCGTGCGTTGGCTTCGGTCGTCCCGTCAGCGGTCATTCATTGGC — 3′) was designed so that the DIG epitope would be incorporated only at the distal end of the template. The processivity plate assay was conducted using a high salt buffer (100 mM (NH4)2SO4, 20 mM Tris-HCl pH 7.4, 3 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 2 % glycerol), to which one microliter each of the in vitro translated proteins A20, D4, and E9 was added in each well. All inhibitors were tested at a final concentration of 167 μM.
2.6 M13 DNA synthesis
In vitro DNA synthesis using an M13-primed template was performed with slight modifications as described (Klemperer et al., 2001; Lin and Ricciardi, 1998). Briefly, the DNA synthesis reaction mixtures (25 μL final volume), contained 10 mM Tris-HCl (pH 7.5), 40 mg/mL of BSA, 4% glycerol, 0.1 mM EDTA, 5 mM dithiothreitol (DTT), 8 mM MgCl2, 20 fmol of primed M13mp18 single-stranded (ss) DNA, 750 ng of E. coli SSB, 60 μM each dGTP, dTTP and dATP, and 20 μM [α-32P]dCTP. The mixtures were pre-incubated with 1 μL each of A20, D4 and E9 at 30 °C for 3 min and the reactions were initiated by the addition of radiolabeled dCTP, incubated for 30 min and stopped with an equal volume of 1% SDS-40 mM EDTA. The reaction products were fractionated on a 0.8% non-denaturing agarose gel at 60V and analyzed by a PhosphorImager (Amersham Biosciences).
2.7 Plaque reduction assay
African green monkey kidney BSC-1 cells (ATCC) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco BRL Life Technologies, Gaithersburg, MD) and 0.1% gentamicin antibiotic, at 37 °C in a humidified 5% CO2 environment. BSC-1 cells were infected in 48-well plates at an MOI of 0.001 with the WR strain of vaccinia virus. One hour post-infection, 400 μL of DNA synthesis inhibitors, ranging from 200 mM to 200 μM, were added per well and incubated at 37 °C overnight. All inhibitor and control compounds were dissolved in DMSO and diluted with DMEM. A 5% solution of formaldehyde in PBS was used to fix the cells. After washing twice with PBS, the plate was stained with 0.2% crystal violet in 50% ethanol.
For HSV-2, a black plaque assay, which enhances the sensitivity of viewing plaques, was performed under similar conditions as for the vaccinia plaque reduction assay. Confluent monolayers were infected with HSV-2 (MOI = 0.001) for 1 h. The inhibitors (200 nM to 200 μM) were added one hour post-infection, and incubated at 37 °C overnight. The wells were washed with PBS, fixed with 2:1 methanol/acetone for 1 h and dried completely. HSV anti-gB, gD and gC antibodies were added and incubated for two hours at RT. The wells were washed twice with PBS and analyzed by immunoperoxidase staining. Both plaque assays were performed in triplicate in at least three independent experiments.
2.8 Cytotoxicity assay
BSC-1 cells were grown to confluency in white 96-well plates at 37 °C in DMEM containing 10% FBS and 0.1% gentamicin in the presence or absence of inhibitor. Cytotoxicity of the inhibitors (200 nM — 200 μM) was assayed using the aCella-TOX bioluminescence cytotoxicity kit (Cell Technology Inc., Mountain View, CA) to follow the release of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). As a positive control, cells were completely lysed to cause maximum release of GAPDH. At least three independent assays were performed in triplicate for reproducibility testing.
2.9 Therapeutic Index (TI)
For each compound, the concentration of inhibitor needed to produce half maximal cell cytotoxicity (CC50) and to reduce 50% of the plaques (IC50) was determined. The therapeutics index was established as the ratio of the CC50 to IC50.
3.0 Compound confirmation
The molecular weights of the hit compounds were confirmed using low resolution liquid chromatography/mass spectrometry, electrospray ionization mode (ES+, Micromass LC, Opus software system, Department of Chemistry, University of Pennsylvania).
3.1 Preliminary evaluation of analogs
Analogs of the parental inhibitors (hit) compounds listed in Table 1 were obtained through bioinformatics compound mining. This was accomplished by performing structural similarity searches using the Bit Vector Structural Map online (http://spheroid.ncifcrf.gov/spheroid/) developed by the Developmental Therapeutic Program of the National Cancer Institute to aid in drug discovery.
Table I.
The data are reported as an average of at least three independent experiments.
| Inhibitor | Structure | Targeted Activity |
CC50 on BSC-1a (μM) |
IC50b (μM) | TIc | ||
|---|---|---|---|---|---|---|---|
| VV | HSV-2 | VV | HSV-2 | ||||
| 123526 |

|
Processivity | > 200 | 47.5± 2.6 | 34.8± 1.4 | > 4.2 | > 5.7 |
| 124808 |
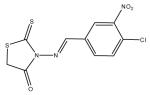
|
Processivity | > 200 | > 200 | 33.1± 1.4 | - | > 6 |
|
55636 Fentichlor |
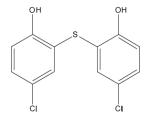
|
DNA Polymerase |
> 200 | 5.7± 0.3 | 2.4± 1.7 | > 40 | > 100 |
|
69343 Tetracycline |
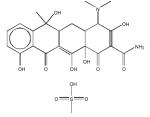
|
DNA Polymerase |
> 200 | > 200 | 47.5± 1.3 | - | > 4.2 |
| Cidofovir |
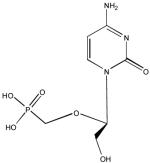
|
DNA Polymerase |
> 200 | 4 | 1.1± 2.1 | > 50 | > 182 |
Concentration at which compound is 50% cytotoxic
Concentration at which compound reduced plaques by 50%
Ratio of CC50 to IC50
3. Results
The rapid plate assay was initially used in a primary screen that led to the identification of an inhibitor of KSHV processive DNA synthesis (Dorjsuren et al., 2006). In this study, we expanded the utility of the rapid plate assay by incorporating sequential steps to select for inhibitors that block vaccinia virus DNA synthesis. A flowchart depicting the stepwise screening protocol is presented in Figure 1.
Figure 1.
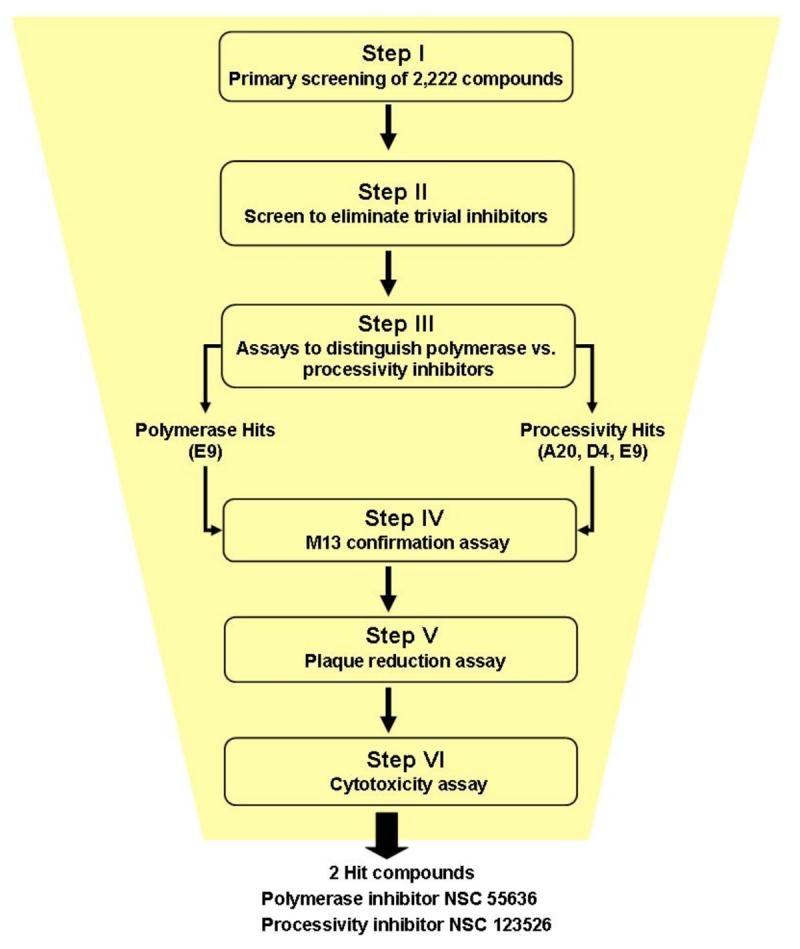
Flowchart of stepwise screening protocol to identify inhibitors of vaccinia virus DNA synthesis. The two most effective compounds targeted the DNA polymerase and processivity activity, respectively.
Step I. Primary high-throughput screen
For this study, the rapid plate assay was performed in a microtiter plate in which a 5′-biotinylated template was immobilized onto streptavidin-coated wells. In the presence of vaccinia lysate proteins, four dNTPs as well as digoxigenin-dUTP were incorporated. A peroxidase-conjugated anti-digoxigenin antibody generated a colorimetric reaction that was read at 405 nm.
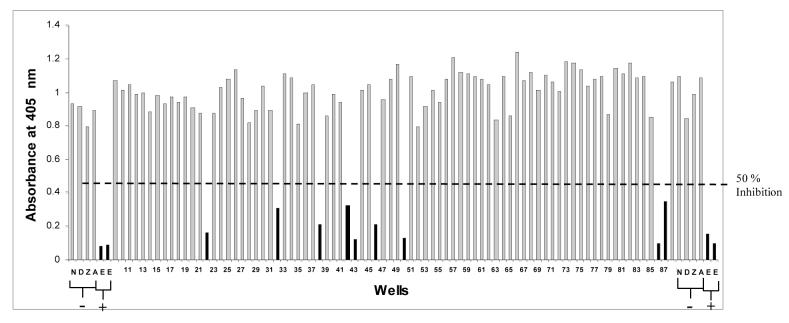
We used the rapid plate assay to screen the NCI Diversity and Training Set library of 2,222 compounds. This library contains diverse three-dimensional structures representative of approximately 140,000 compounds from the NCI-DTP library. For each compound of the diversity set, 1 μL was added to the reaction mix resulting in a final concentration 167 μM. It is noted that we used a thymidine kinase (TK) deficient vaccinia virus (derived from WR strain) in order to disfavor obtaining nucleoside inhibitors that require phosphorylation. As negative controls we used AZT and acyclovir, which do not inhibit vaccinia virus (Holzer et al., 2005; St. Clair et al., 1980). A total of 169 compounds decreased DNA synthesis by 50% or greater, giving a hit rate of 7.6% (Figure 2).
Figure 2.
A representative primary rapid plate assay. Compounds that inhibited DNA synthesis by more than 50% are indicated by solid bars, and those that inhibit DNA synthesis by less than 50% are indicated by shaded bars. Indicated are wells with the positive (+) control inhibitor EDTA (E) in duplicate and the negative (-) control inhibitors: unphosphorylated AZT (Z), unphosphorylated acyclovir (A), DMSO alone (D) and no compound added (N).
Step II. Screen to eliminate trivial inhibitors
We next tested the 169 hit compounds obtained from the primary screen (Step I) to determine if these vaccinia inhibitors were also able to block DNA synthesis conducted by a DNA polymerase and processivity complex from a different virus. Unlike vaccinia which requires three proteins for processive DNA synthesis, KSHV relies on only two proteins, a DNA polymerase (Pol8) and processivity factor (PF8) (Chen et al., 2004, 2005; Lin et al., 1998). Compounds (at a concentration of 167 μM) that block DNA synthesis of both vaccinia virus and KSHV could, as one reason, be acting as trivial inhibitors, e.g. by intercalating DNA, and therefore were not pursued. The result of this step yielded 47 compounds, giving a hit rate of 2.1% from Step I (2,222 compounds) (Figure 3).
Figure 3.
Assay to eliminate trivial inhibitors. Hit compounds from the primary screen using vaccinia lysates were futher tested in a KSHV rapid plate assay for their abilities to inhibit DNA synthesis directed by Pol8/PF8. Compounds that inhibited Pol8/PF8 DNA synthesis (solid bars) were categorized as potentially trivial inhibitors. Compounds that failed to block Pol8/PF8 DNA synthesis (shaded bars) were considered to be bona fide inhibitors of vaccinia DNA synthesis. Indicated are wells with the positive (+) control inhibitor EDTA (E) in duplicate and the negative (-) control inhibitor DMSO alone (D) and no compound added (N).
Step III. Screen to distinguish polymerase vs. processivity inhibitors
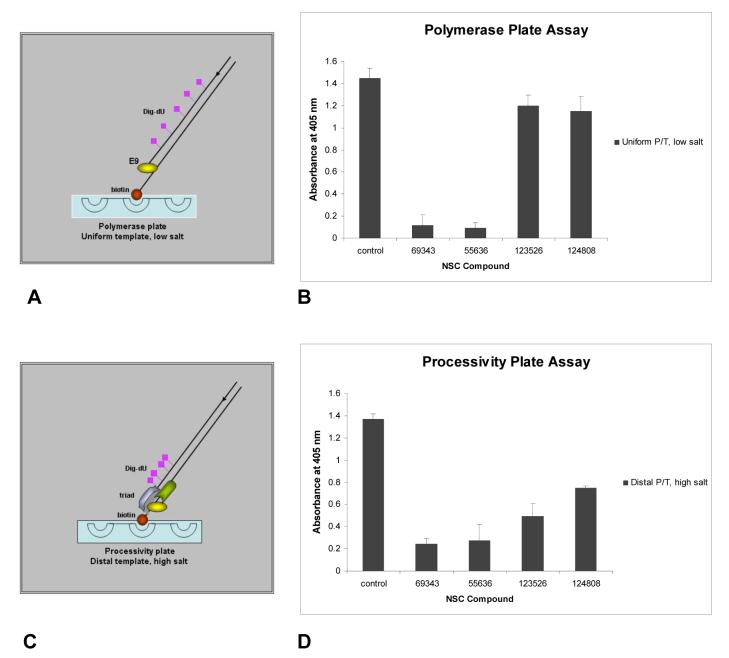
We individually synthesized the vaccinia A20, D4, E9 proteins in vitro in order to distinguish inhibitors that target nucleotide incorporation by E9 alone from inhibitors that target processive incorporation by the triad, A20, D4 and E9. These translated proteins were analyzed in two distinct plate assays.
To define polymerase inhibitors, E9 activity was conducted in low salt, in which DNA polymerases are able to synthesize fully extended strands in the absence of processivity factors (Chaudhuri and Parris, 2002; Williams et al., 1993). DNA synthesis by E9 was tested on a uniform template, which contains evenly distributed sites for DIG-dU incorporation (Figure 4A). Two compounds (69343 and 55636) were designated as polymerase inhibitors since they prevented E9 from incorporating nucleotides on the uniform template (Figure 4B). As expected, these polymerase inhibitors also blocked incorporation on the distal template (Figure 4D).
Figure 4.
Assay to distinguish polymerase and processive inhibitors of vaccinia DNA synthesis. (A) The model depicts the uniform incorporation of the DIG-dU on a template by E9 polymerase under low salt conditions. Under low salt conditions, E9 incorporates dNTPs along the DNA template. (B) NCI hit compounds were analyzed on the uniform tempate in the presence of E9 alone under low salt conditions to identify polymerase inhibitors. (C) The model depicts incorporation of the label DIG-dUTP on the distal end of the template by the triad (A20, D4, E9) under high salt conditions. This template was designed with all the adenines near the biotinylated end to direct incorporation of the DIG label towards the 3′ of the growing strand. Under high salt conditions, E9 requires A20 and D4 to accomplish processive DNA synthesis. (D) NCI hit compounds were analyzed on the distal tempate in the presence of the A20, D4 and E9 triad under high salt conditions to identify processivity inhibitors. Compounds that block E9 polymerase activity also blocked processivity. For both A and C, the template 5′ end is biotinylated for attachment to the streptavidin-coated plates.
To define processivity inhibitors, high salt was used which limits nucleotide incorporation by E9 alone but allows processive DNA synthesis by the A20, D4, E9 triad to occur. For the processivity assay, a distal template was used in which DIG-dU is incorporated only at the 3′ end of the nascent DNA by virtue of dA being positioned solely at the 5′ end of the template (Figure 4C). Two compounds (123526 and 124808) were designated as processivity inhibitors since they only prevented DNA synthesis by the triad on the distal template (Figure 4D) but not by E9 on the uniform template (Figure 4B).
It is noted that of the 47 compounds that passed Step II, 43 compounds did not prove inhibitory in either the vaccinia polymerase or processivity assay. The use of infected cell lysate in Step I vs. in vitro translated proteins in Step III may account for this difference. We have in fact noted the ability of in vitro translated proteins to support a more robust DNA synthesis as opposed to infected cell lysates (data not shown). Another reason could be due to a possible difference in sensitivity between KSHV vs. vaccinia proteins. Regardless, four compounds emerged from these sequential steps (I — III) that involved passing a rigorous screen employing different protein sources and viral systems.
Step IV. M13 DNA Synthesis Assay
The ability of compounds to inhibit DNA synthesis in the rapid plate assay was confirmed using the M13 DNA synthesis assay. In the M13 assay, full-length DNA strands of 7,249 nucleotides are produced and visualized by autoradiography. Because the length of the M13 product is extensive, this assay serves as a rigorous test for processive DNA synthesis. As shown in Figure 5, lane 2, full-length double stranded M13 DNA was successfully synthesized in the presence of the A20, D4, E9 triad. The brackets shown in Figure 5 indicate the typical formation of greater than unit length products (Hamilton et al., 2007; Willer et al., 1999; Zhang and Evans, 1993). The synthesis of M13 DNA was blocked by increasing concentrations of the two polymerase inhibitors, NSC 69343 (lanes 3-7) and NSC 55636 (lanes 8-12), and the two processivity inhibitors, NSC 123526 (lanes 13-17) and NSC 124808 (lanes 18-22). Most notably, the polymerase inhibitor NSC 69343 completely inhibited M13 DNA synthesis at 100 μM (lane 6). All of the other compounds inhibited at a concentration between 100 μM and 1 mM.
Figure 5.
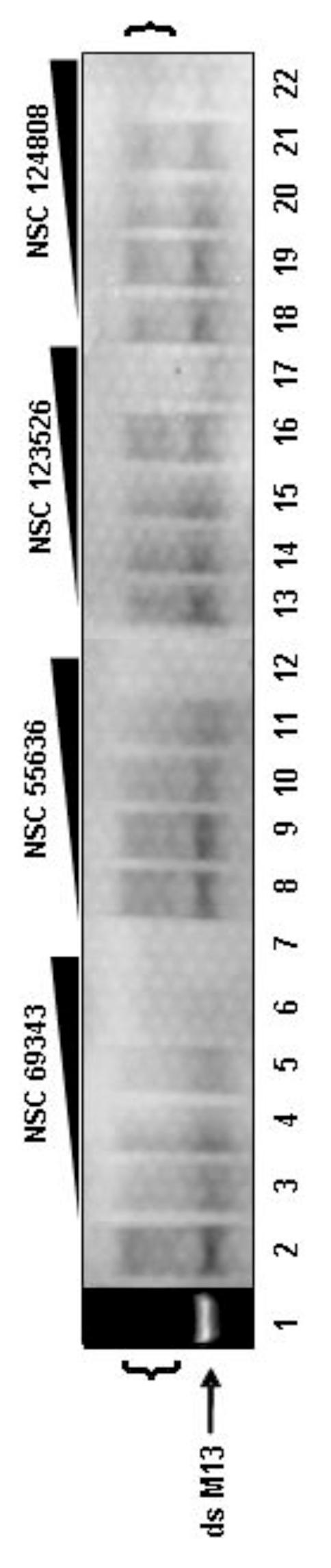
Confirmation of inhibitors using the M13 DNA synthesis assay. Full-length ds DNA products labeled with [α-32P]dCTP were synthesized from the M13 DNA primer/template in the presence of the vaccinia triad (A20, D4, E9) and examined on a non-denaturing gel. The ability of each polymerase and processivity inhibitor to block the M13 DNA synthesis was examined by adding increasing concentrations (100 nM, 1 μM, 10 μM, 100 μM and 1 mM) of each compound: NSC 69343, lanes 3-7; NSC 55636, lanes 8-12; NSC 123526, lanes 13-17; NSC 124808, lanes 18-22. Lane 1 shows the position of the full-length ds M13 DNA marker (arrow) as detected by cybergold. Lane 2 is the control reaction containing DMSO, the solvent for each of the compounds. The position of greater than unit length products are indicated by brackets.
Step V. Plaque reduction assays
A plaque reduction assay was used to evaluate the antiviral activity of the two polymerase and the two processivity inhibitors that were selected from Step III. For comparison, we also performed plaque reduction assays on HSV-2. The results are summarized in Table I. Surprisingly, NSC 69343, the more potent polymerase inhibitor based on in vitro assays, was less remarkable in its ability to reduce vaccinia viral plaques when compared to NSC 55636, the less potent polymerase inhibitor. Interestingly, NSC 69343 was more effective in inhibiting HSV-2 plaques than vaccinia plaques whereas NSC 55636 was equally effective in inhibiting both viruses. With respect to the two processivity inhibitors, NSC 123526 was significantly more effective than NSC 124808 in inhibiting vaccinia virus. However, both of these processivity inhibitors were comparable in their abilities to block HSV-2. Overall, NSC 55636 was the strongest viral inhibitor.
Step VI. Cytotoxicity assay
Cytotoxicity of the four inhibitors of vaccinia DNA synthesis was determined using the aCella-TOX assay. This assay quantifies the cellular release of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is essential for the production of ATP in the glycolysis pathway. GAPDH released into the cell media is used in a coupled reaction to generate ATP, which is detected by luciferase/luciferin bioluminescence. As indicated in Table I, all of the inhibitors had CC50 of greater than 200 μM in BSC-1 cells based on GAPDH release.
Cellular therapeutic indices for polymerase inhibitor NSC 55636 and processivity inhibitor NSC 123526
The polymerase inhibitor NSC 55636 and processivity inhibitor NSC 123526 were considered to be the most significant compounds based on their efficacy at reducing vaccinia virus plaques with the least cytotoxicity. NSC 55636 gave a plaque reduction IC50 of 5.7 μM and CC50 of greater than 200 μM, resulting in a cellular TI of greater than 40. NSC 123526 gave a plaque reduction IC50 of 47.5 μM and CC50 of greater than 200 μM, resulting in a cellular TI of greater than four.
Evaluation of analogs of polymerase and processivity inhibitors
Bioinformatic mining of[MSOffice2] the four inhibitors listed in Table I generated 118 compounds that were similar in structure, spatial conformation (3D), and chemical properties. Each of these compounds was subjected to the stepwise screening procedure. Two compounds related to NSC 55636 and one compound related to NSC 69343 emerged as hits (Figure 6). The two NSC 55636 analogs (NSC 646023 and 406932) had similar IC50 values in the vaccinia plaque assays (7.5 μM and 6.6 μM, respectively). By contrast, the NSC 69343 analog (NSC 159628) had an IC50 value (1.6 μM) that was dramatically improved over that of the parental compound NSC 69343 (IC50 > 200).
Figure 6.
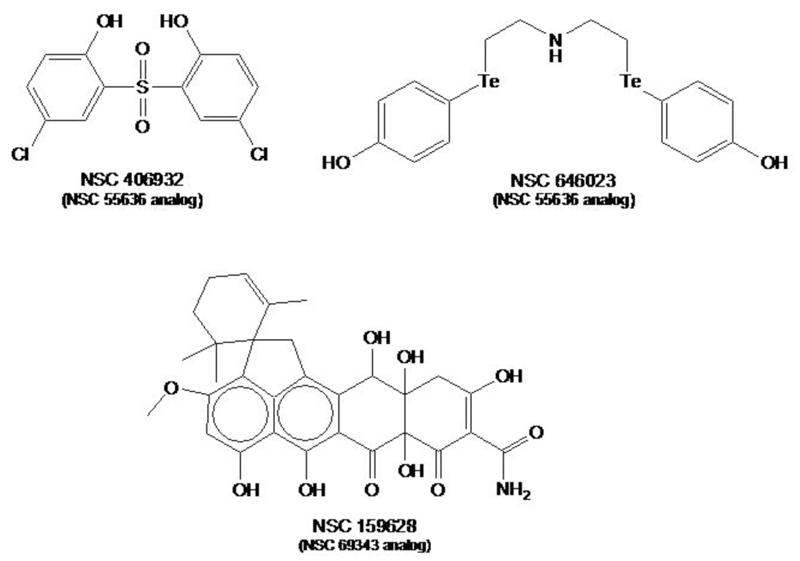
Analogs of the indicated hit compounds that inhibit vaccinia DNA synthesis.
4. Discussion
The availability of effective new therapeutics against variola virus, the causative agent of smallpox, is essential to combat a potential smallpox outbreak. Vaccinia, a prototypic member of the poxviridae family, serves as an ideal model for variola, as these two viruses exhibit a high degree of genomic sequence identity (Afonso et al., 2002). For the purpose of identifying new therapeutics against smallpox, we screened for compounds that target the polymerase and processivity complex of vaccinia virus. We reasoned that compounds that target the DNA synthesis proteins of vaccinia would likely target the corresponding proteins of variola.
In order to identify inhibitors of the polymerase/processivity complex, we utilized a sequence of rapid plate assays. The first screening step selects for functional inhibitors that block vaccinia DNA synthesis. This step, which uses infected cell lysate, was designed for poxviruses, which are unique amongst the DNA viruses in that they replicate in the cytoplasm. This provides a great advantage since there are no competing nuclear polymerases in the cytoplasmic lysate that could contribute to anomalous results. The second screening step ideally eliminates trivial inhibitors, e.g. DNA intercalators and DNA groove binders. The third screening step distinguishes whether inhibition of the polymerase or processive mechanism is targeted. Interestingly, this step could be applied to defining inhibitors of other viruses and eukaryotes that engage processivity complexes in their DNA synthesis.
Our screening method identified four compounds that inhibit vaccinia DNA synthesis. NSC 55636 and NSC 69343 prevented the catalytic activity of E9 DNA polymerase. By contrast, NSC 123526 and NSC 124808 inhibited the processive activity of the triad A20, D4 and E9.
NSC 55636 (Fentichlor)
Despite the fact that NSC 69343 is a more potent inhibitor of E9 in the rapid plate assay, it did not reduce vaccinia plaques efficiently. On the other hand, the compound of greatest interest is NSC 55636, which reduced vaccinia plaques at a lower IC50 (5.7 μM) in the rapid plate assay. This polymerase inhibitor is also known as Fentichlor, an antibacterial, anthelmintic and antifungal agent (Hugo and Bloomfield, 1971a,b,c; Watson, 1973) that has been used as an additive in cosmetic products (Schmahl and Hieke, 1980; Schmahl and Matissek, 1981). However, due to its photoallergenic nature, the use of Fentichlor in recent years seems to be limited to agrochemicals. Specifically, Fentichlor has been shown to undergo photodehalogenation generating aryl radicals (Delahanty et al., 1989; Li and Chignell, 1987) and byproducts that may act as antigens (Hindsén et al., 2006). Nevertheless, Fentichlor was reported to have no detectable teratogenic effects in pregnant rats that received Fentichlor intragastrically (Veselova et al., 1980), and the Ames test has shown that it is non-mutagenic (Serafimova et al., 2007). Interestingly, in silico modeling suggested that Fentichlor is inactive against HIV-1 integrase, protease, reverse transcriptase and virus uncoating (Vilar et al., 2006) (See supporting information in this reference).
In this study, we determined that Fentichlor inhibits the vaccinia viral DNA polymerases. Furthermore, we found that Fentichlor is similarly effective in reducing plaques in both HSV-2 and vaccinia-infected BSC-1 cells. Possibly, Fentichlor interacts with structural motifs common to the DNA polymerases of both viruses.
Fentichlor’s high cellular therapeutic index of greater than forty against vaccinia reflects its ability to effectively block vaccinia infection with minimal cytotoxicity. The potency of this compound is also reiterated by its analogs (NSC 406932 and NSC 646023). Our studies indicate that Fentichlor is a potent antipoxvirus candidate and that analogs which do not cause photosensitization should be pursued.
NSC 123526
Little is known about the processivity inhibitor NSC 123526, an S-fluorenylcysteine compound. Since NSC 123526 does not inhibit E9 polymerase activity, it is interesting to speculate that it perturbs the interactions of A20 and D4 with E9 to prevent continuous DNA strand synthesis. Interestingly, a structurally related compound, S-trityl-L-cysteine, is a potent tumor growth inhibitor of human Eg5 (Brier et al., 2004; Skoufias et al., 2006). The cysteine moiety of S-trityl-L-cysteine renders the compound cell permeable and likely delivers the flourenyl group to the interaction site, inducing local conformational changes. In a study of S-tritylcysteine-related compounds, NSC 123526 was found to be non-toxic on the leukemia cell line L-1210 (Zee-Cheng and Cheng, 1970). In our study, NSC 123526 had a cellular therapeutic index of greater than four, making it a modest antipoxvirus agent.
NSC 69343 and NSC 124808
This study yielded two other compounds, NSC 69343 and NSC 124808, that were effective in blocking vaccinia DNA synthesis in the in vitro rapid plate assays but were not as effective in blocking vaccinia infection. This was not due to poor cellular uptake, as these two compounds were able to reduce HSV-2 plaques. It remains to be determined whether these two inhibitors function by different mechanisms in inhibiting vaccinia and HSV-2.
The polymerase inhibitor NSC 69343 is in fact tetracycline, an antibiotic that inhibits the prokaryotic 30S ribosome. It is thus fascinating that tetracycline can inhibit vaccinia DNA synthesis. Our preliminary studies show that a more complex tetracycline-related compound, viridicatumtoxin, had a greater inhibitory activity (IC50 = 1.6 μM) than all parental hits and analogs.
SUMMARY
Our pilot screen of 2,222 compounds yielded two compounds, NSC 55636 and NSC 123526, which effectively block vaccinia virus DNA synthesis and infection. Future experiments will determine whether chemical modification of these compounds will further improve their antiviral potency. Based on our stepwise approach, it is anticipated that an expanded high throughput screen will identify additional new antiviral compounds that inhibit poxvirus DNA synthesis.
Acknowledgements
The authors thank Drs. G. H. Cohen and R. J. Eisenberg (University of Pennsylvania) for kindly providing the viral stocks and HSV antibodies, Dr. S. N. Isaac (University of Pennsylvania) for helpful advice on the plaque assay and Dr. M. J. Currens (NCI) for helping to obtain compounds from the NCI Diversity Set library. A.M.D.S. was supported by an NIH EID T32 training grant. This study was supported by NIAID U54-AI057168 MARCE grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonso CL, Tulman ER, Lu Z, Zsak L, Sandybaev NT, Kerembekova UZ, Zaitsev VL, Kutish GF, Rock DL. The genome of camelpox virus. Virology. 2002;295:1–9. doi: 10.1006/viro.2001.1343. [DOI] [PubMed] [Google Scholar]
- Andrei G, Gammon DB, Fiten P, De Clercq E, Opdenakker G, Snoeck R, Evans DH. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J. Virol. 2006;80:9391–9401. doi: 10.1128/JVI.00605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle JR, Hartline C, Aldern KA, Rodriguez N, Harden E, Kern ER, Hostetler KY. Alkoxyalkyl esters of cidofovir and cyclic cidofovir exhibit multiple-log enhancement of antiviral activity against cytomegalovirus and herpesvirus replication in vitro. Antimicrob. Agents and Chemother. 2002;46:2381–2386. doi: 10.1128/AAC.46.8.2381-2386.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaud G. Vaccinia virus DNA replication: a short review. Biochimie. 1995;77:774–779. doi: 10.1016/0300-9084(96)88195-8. [DOI] [PubMed] [Google Scholar]
- Bougie I, Bisaillon M. The broad spectrum antiviral nucleoside ribavirin as a substrate for a viral RNA capping enzyme. J. Biol. Chem. 2004;279:22124–22130. doi: 10.1074/jbc.M400908200. [DOI] [PubMed] [Google Scholar]
- Brier S, Lemaire D, Debonis S, Forest E, Kozielski F. Identification of the protein binding region of S-trityl-L-cysteine, a new potent inhibitor of the mitotic kinesin Eg5. Biochemistry. 2004;43:13072–13082. doi: 10.1021/bi049264e. [DOI] [PubMed] [Google Scholar]
- Chaudhuri M, Parris DS. Evidence against a simple tethering model for enhancement of herpes simplex virus DNA polymerase processivity by accessory protein UL42. J. Virol. 2002;76:10270–10281. doi: 10.1128/JVI.76.20.10270-10281.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lin K, Ricciardi RP. Human Kaposi’s sarcoma herpesvirus processivity factor-8 functions as a dimer in DNA synthesis. J. Biol. Chem. 2004;279:28375–28386. doi: 10.1074/jbc.M400032200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ciustea M, Ricciardi RP. Processivity factor of KSHV contains a nuclear localization signal and binding domains for transporting viral DNA polymerase into the nucleus. Virology. 2005;340:183–191. doi: 10.1016/j.virol.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Chen Z, Earl P, Americo J, Damon I, Smith SK, Zhou YH, Yu F, Sebrell A, Emerson S, Cohen G, Eisenberg RJ, Svitel J, Schuck P, Satterfield W, Moss B, Purcell R. Chimpanzee/human mAbs to vaccinia virus B5 protein neutralize vaccinia and smallpox viruses and protect mice against vaccinia virus. Proc. Natl. Acad. Sci. USA. 2006;103:1882–1887. doi: 10.1073/pnas.0510598103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Cidofovir in the treatment of poxvirus infections. Antiviral Res. 2002;55:1–13. doi: 10.1016/S0166-3542(02)00008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Moss B. Vaccinia virus uracil DNA glycosylase has an essential role in DNA synthesis that is independent of its glycosylase activity: catalytic site mutations reduce virulence but not virus replication in cultured Cells. J. Virol. 2003;77:159–166. doi: 10.1128/JVI.77.1.159-166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahanty JN, Evans JC, Rowlands CC, Barratt MD, Pendlington RU. Radicals involved in photoallergen/protein interactions. Free Radic. Biol. Med. 1989;7:231–236. doi: 10.1016/0891-5849(89)90129-9. [DOI] [PubMed] [Google Scholar]
- Dorjsuren D, Burnette A, Gray GN, Chen X, Zhu W, Roberts PE, Currens MJ, Shoemaker RH, Ricciardi RP, Sei S. Chemical library screen for novel inhibitors of Kaposi’s sarcoma-associated herpesvirus processive DNA synthesis. Antiviral Res. 2006;69:9–23. doi: 10.1016/j.antiviral.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Ellison KS, Peng W, McFadden G. Mutations in active-site residues of the uracil-DNA glycosylase encoded by vaccinia virus are incompatible with virus viability. J. Virol. 1996;70:7965–7973. doi: 10.1128/jvi.70.11.7965-7973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MD, Nuara AA, Gammon DB, Buller RM, Evans DH. Duplex strand joining reactions catalyzed by vaccinia virus DNA polymerase. Nucleic Acids Res. 2007;35:143–151. doi: 10.1093/nar/gkl1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindsén M, Zimerson E, Bruze M. Photoallergic contact dermatitis from ketoprofen in southern Sweden. Contact Dermatitis. 2006;54:150–157. doi: 10.1111/j.0105-1873.2005.00778.x. [DOI] [PubMed] [Google Scholar]
- Hingorani MM, O’Donnell M. DNA polymerase structure and mechanisms of action. Curr. Org. Chem. 2000;4:887–913. [Google Scholar]
- Holzer GW, Mayrhofer J, Gritschenberger W, Falkner FG. Dominant negative selection of vaccinia virus using a thymidine kinase/thymidylate kinase fusion gene and the prodrug azidothymidine. Virology. 2005;337:235–241. doi: 10.1016/j.virol.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Hugo WB, Bloomfield SF. Studies on the mode of action of the phenolic antibacterial agent fentichlor against Staphylococcus aureus and escherichia coli.II. The effects of fentichlor on the bacterial membrane and cytoplasmic constituents of the cell. J. Appl. Bacteriol. 1971a;34:569–578. doi: 10.1111/j.1365-2672.1971.tb02319.x. [DOI] [PubMed] [Google Scholar]
- Hugo WB, Bloomfield SF. Studies on the mode of action of the phenolic antibacterial agent fentichlor against Staphylococcus aureus and escherichia coli.I. The adsorption of fentichlor by bacterial cell and its antibacterial activity. J. Appl. Bacteriol. 1971b;34:557–567. doi: 10.1111/j.1365-2672.1971.tb02318.x. [DOI] [PubMed] [Google Scholar]
- Hugo WB, Bloomfield SF. Studies on the mode of action of the phenolic antibacterial agent fentichlor against Staphylococcus aureus and Escherichia Coli.III. The effect of fentichlor on the metabolic activities of Staphylococcus areus and escherichia coli. J. Appl. Bacteriol. 1971c;34:579–591. doi: 10.1111/j.1365-2672.1971.tb02320.x. [DOI] [PubMed] [Google Scholar]
- Ishii K, Moss B. Mapping interaction sites of the A20R protein component of the vaccinia virus DNA replication complex. Virology. 2002;303:232–239. doi: 10.1006/viro.2002.1721. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, O’Donnell M, Kuriyan J. Clamp loaders and sliding clamps. Curr. Opin. Struc. Biol. 2002;12:217–224. doi: 10.1016/s0959-440x(02)00313-5. [DOI] [PubMed] [Google Scholar]
- Kit S, Dubbs DR, Piekarski LJ. Enhanced thymidine phosphorylating activity of mouse fibroblasts (strain LM) following vaccinia infection. Biochem. Biophys. Res. Commun. 1962;8:72–75. doi: 10.1016/0006-291x(62)90238-3. [DOI] [PubMed] [Google Scholar]
- Klemperer N, McDonald W, Boyle K, Unger B, Traktman P. The A20R protein is a stoichiometric component of the processive form of vaccinia virus DNA polymerase. J. Virol. 2001;75:12298–12307. doi: 10.1128/JVI.75.24.12298-12307.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth RS, Smee DF, Sidwell RW, Snarsky V, Evans DH, Hostetler KY. Mutations in the E9L polymerase gene of cidofovir-resistant vaccinia virus strain WR are associated with the drug resistance phenotype. Antimicrob. Agents Chemother. 2006;50:4038–4043. doi: 10.1128/AAC.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyan J, O’Donnell M. Sliding clamps of DNA polymerases. J. Mol. Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- Lane HC, Montagne JL, Fauci AS. Bioterrorism: a clear and present danger. Nat. Med. 2001;7:1271–1273. doi: 10.1038/nm1201-1271. [DOI] [PubMed] [Google Scholar]
- Lane JM, Millar JD. Risks of smallpox vaccination complications in the United States. Am. J. Epidemiol. 1971;93:238–240. doi: 10.1093/oxfordjournals.aje.a121252. [DOI] [PubMed] [Google Scholar]
- Lane JM, Ruben FL, Neff JM, Millar JD. Complications of smallpox vaccination, 1968. N. Engl. J. Med. 1969;281:1201–1208. doi: 10.1056/NEJM196911272812201. [DOI] [PubMed] [Google Scholar]
- Lane JM, Ruben FL, Neff JM, Millar JD. Complications of smallpox vaccination, 1968: results of ten statewide surveys. J. Infect. Dis. 1970;122:303–309. doi: 10.1093/infdis/122.4.303. [DOI] [PubMed] [Google Scholar]
- Li ASW, Chignell CF. Generation of free-radicals during the photolysis of the photoallergens bithionol and fentichlor - a spin trapping study. Environ. Health Perspect. 1987;75:131. [Google Scholar]
- Lin K, Dai CY, Ricciardi RP. Cloning and functional analysis of Kaposi’s sarcoma-associated herpesvirus DNA polymerase and its processivity factor. J. Virol. 1998;72:6228–6232. doi: 10.1128/jvi.72.7.6228-6232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Ricciardi RP. The 41-kDa protein of human herpesvirus 6 specifically binds to viral DNA polymerase and greatly increases DNA synthesis. Virology. 1998;250:210–219. doi: 10.1006/viro.1998.9348. [DOI] [PubMed] [Google Scholar]
- Lin K, Ricciardi RP. A rapid plate assay for the screening of inhibitors against herpesvirus DNA polymerases and processivity factors. J. Virol. Methods. 2000;88:219–225. doi: 10.1016/s0166-0934(00)00190-7. [DOI] [PubMed] [Google Scholar]
- Magee WC, Hostetler KY, Evans DH. Mechanism of inhibition of vaccinia virus DNA polymerase by cidofovir diphosphate. Antimicrob. Agents Chemother. 2005;49:3153–3162. doi: 10.1128/AAC.49.8.3153-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Nat. Acad. Sci. USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WF, Traktman P. Vaccinia virus DNA polymerase. In vitro analysis of parameters affecting processivity. J. Biol. Chem. 1994;269:31190–31197. [PubMed] [Google Scholar]
- Millns AK, Carpenter MS, DeLange AM. The vaccinia virus-encoded uracil DNA glycosylase has an essential role in viral DNA replication. Virology. 1994;198:504–513. doi: 10.1006/viro.1994.1061. [DOI] [PubMed] [Google Scholar]
- Moss B. Poxviridae: the viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. Lippincott-Raven; Philadelphia: 2001. pp. 2849–2883. [Google Scholar]
- Quenelle DC, Buller RM, Parker S, Keith KA, Hruby DE, Jordan R, Kern ER. Efficacy of delayed treatment with ST-246 given orally against systemic orthopoxvirus infections in mice. Antimicrob. Agents Chemother. 2007a;51:689–695. doi: 10.1128/AAC.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenelle DC, Prichard MN, Keith KA, Hruby DE, Jordan R, Painter GR, Robertson A, Kern ER. Synergistic efficacy of the combination of ST-246 with CMX001 against orthopoxviruses. Antimicrob. Agents Chemother. 2007b;51:4118–4124. doi: 10.1128/AAC.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PM, Bommarius B, Lebeis S, McNulty S, Christensen J, Swimm A, Chahroudi A, Chavan R, Feinberg MB, Veach D, Bornmann W, Sherman M, Kalman D. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat. Med. 2005;11:731–739. doi: 10.1038/nm1265. [DOI] [PubMed] [Google Scholar]
- Robbins SJ, Jackson RJ, Fenner F, Beaton S, Medveczky J, Ramshaw IA, Ramsay AJ. The efficacy of cidofovir treatment of mice infected with ectromelia (mousepox) virus encoding interleukin-4. Antiviral Res. 2005;66:1–7. doi: 10.1016/j.antiviral.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Sbrana E, Jordan R, Hruby DE, Mateo RI, Xiao SY, Siirin M, Newman PC, Da Rosa AP, Tesh RB. Efficacy of the antipoxvirus compound ST-246 for treatment of severe orthopoxvirus infection. Am. J. Trop. Med. Hyg. 2007;76:768–773. [PubMed] [Google Scholar]
- Scaramozzino N, Sanz G, Crance JM, Saparbaev M, Drillien R, Laval J, Kavli B, Garin D. Characterisation of the substrate specificity of homogeneous vaccinia virus uracil-DNA glycosylase. Nucleic Acids Res. 2003;31:4950–4957. doi: 10.1093/nar/gkg672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahl HJ, Hieke E. Separation and identification of some anti-microbials used also in cosmetic products by means of thin-layer chromatography. Fresenius Zeitschrift Fur Analytische Chemie. 1980;304:398–404. [Google Scholar]
- Schmahl HJ, Matissek R. Separation and determination of phenolic antimicrobials from non-emulsified cosmetics containing surfactants. Fresenius Zeitschrift Fur Analytische Chemie. 1981;307:392–399. [Google Scholar]
- Schormann N, Grigorian A, Samal A, Krishnan R, DeLucas L, Chattopadhyay D. Crystal structure of vaccinia virus uracil-DNA glycosylase reveals dimeric assembly. BMC Struct. Biol. 2007;7:45. doi: 10.1186/1472-6807-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafimova R, Todorov M, Pavlov T, Kotov S, Jacob E, Aptula A, Mekenyan O. Identification of the structural requirements for mutagencitiy, by incorporating molecular flexibility and metabolic activation of chemicals. II. General Ames mutagenicity model. Chem. Res. Toxicol. 2007;20:662–676. doi: 10.1021/tx6003369. [DOI] [PubMed] [Google Scholar]
- Shoemaker RH, Scudiero DA, Melillo G, Currens MJ, Monks AP, Rabow AA, Covell DG, Sausville EA. Application of high-throughput, molecular-targeted screening to anticancer drug discovery. Curr. Top Med. Chem. 2002;2:229–246. doi: 10.2174/1568026023394317. [DOI] [PubMed] [Google Scholar]
- Skoufias DA, DeBonis S, Saoudi Y, Lebeau L, Crevel I, Cross R, Wade RH, Hackney D, Kozielski F. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J. Biol. Chem. 2006;281:17559–17569. doi: 10.1074/jbc.M511735200. [DOI] [PubMed] [Google Scholar]
- Smee DF, Sidwell RW, Kefauver D, Bray M, Huggins JW. Characterization of wild-type and cidofovir-resistant strains of camelpox, cowpox, monkeypox, and vaccinia viruses. Antimicrob. Agents Chemother. 2002;46:1329–1335. doi: 10.1128/AAC.46.5.1329-1335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clair MH, Furman PA, Lubbers CM, Elion GB. Inhibition of cellular alpha and virally induced deoxyribonucleic acid polymerases by the triphosphate of acyclovir. Antimicrob. Agents Chemother. 1980;18:741–745. doi: 10.1128/aac.18.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanitsa ES, Arps L, Traktman P. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J. Biol. Chem. 2006;281:3439–3451. doi: 10.1074/jbc.M511239200. [DOI] [PubMed] [Google Scholar]
- Veselova TP, Lapteva LA, Khrustaleva LI. Embryotoxic and teratogenic effects of oksid. Veterinariya. 1980;8:56. [Google Scholar]
- Vilar S, Santana L, Uriarte E. Probabilistic neural network model for the in silico evaluation of anti-HIV activity and mechanism of action. J. Med. Chem. 2006;49:1118–1124. doi: 10.1021/jm050932j. [DOI] [PubMed] [Google Scholar]
- Watson G. Biocidal materials containing fentichlor. GB Patent 1308511 19730228 1973
- Weisshart K, Chow CS, Coen DM. Herpes simplex virus processivity factor UL42 imparts increased DNA-binding specificity to the viral DNA polymerase and decreased dissociation from primer-template without reducing the elongation rate. J. Virol. 1999;73:55–66. doi: 10.1128/jvi.73.1.55-66.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer DO, Mann MJ, Zhang W, Evans DH. Vaccinia virus DNA polymerase promotes DNA pairing and strand-transfer reactions. Virology. 1999;257:511–523. doi: 10.1006/viro.1999.9705. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Wernette CM, Kaguni LS. Processivity of mitochondrial DNA polymerase from Drosophila embryos. Effects of reaction conditions and enzyme purity. J. Biol. Chem. 1993;268:24855–24862. [PubMed] [Google Scholar]
- Wodarz D, Lloyd AL. Immune responses and the emergence of drug-resistant virus strains in vivo. Proc. Biol. Sci. 2004;271:1101–1109. doi: 10.1098/rspb.2003.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Pevear DC, Davies MH, Collett MS, Bailey T, Rippen S, Barone L, Burns C, Rhodes G, Tohan S, Huggins JW, Baker RO, Buller RL, Touchette E, Waller K, Schriewer J, Neyts J, DeClercq E, Jones K, Hruby D, Jordan R. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus Challenge. J. Virol. 2005;79:13139–13149. doi: 10.1128/JVI.79.20.13139-13149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee-Cheng KY, Cheng CC. Experimental antileukemic agents. Preparation and structure-activity study of S-tritylcysteine and related compounds. J. Med. Chem. 1970;13:414–418. doi: 10.1021/jm00297a019. [DOI] [PubMed] [Google Scholar]
- Zhang WD, Evans DH. DNA strand exchange catalyzed by proteins from vaccinia virus-infected cells. J. Virol. 1993;67:204–212. doi: 10.1128/jvi.67.1.204-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]