Abstract
Currently available PCR genotyping methods for point mutations in the mouse dystrophin gene can lead to false positives resulting in wasted time and money breeding or treating the wrong mice. Here we describe a simple and accurate method for sequencing the point mutations in mdx, mdx4cv and mdx5cv mice. This method clearly distinguishes between wild-type, heterozygous and mutant transcripts thereby saving time and money by avoiding false positives.
Introduction
Duchenne muscular dystrophy is caused by X-linked recessive mutations in the dystrophin gene. Our understanding of the pathogenesis of DMD and development of potential treatments has benefited enormously from the multiple mdx mouse models. Here we describe simple genotyping protocols for the most commonly used mdx, mdx4cv and mdx5cv strains. The original and naturally occurring mutation in mdx mice is a G to A transition that leads to a premature stop codon in exon 23 [1,2]. Four additonal mdx strains (2cv to 5cv) were generated with N-ethylnitrosourea chemical mutagenesis [3]. The mdx4cv mouse has a C- to T- transition in exon 53, creating a nonsense ochre codon [4]. The mdx5cv allele has an A to T transition in exon 10, creating a new splice donor site that generates a premature stop codon in RNA transcripts [4]. Each mutation leads to a loss of dystrophin protein expression in striated muscles. The mice vary in the number of revertant muscle fibers in which some muscle fibers express partially functional truncated dystrophins [5]. Both mdx4cv and mdx5cv have the fewest revertant fibers and each strain varies in which isoforms of dystrophin are expressed in non-muscle tissues [4,6]. Various methods have been published to genotype the different mdx mice ([7] and references within), but we and others find these methods difficult to apply on a routine basis as they can lead to false positives that waste large amounts of time and money breeding or treating the wrong mice. Here, we describe methods to genotype the mdx, mdx4cv and mdx5cv mice by sequence analyses to avoid false positives.
Materials and Methods
Mouse strains
The strains used in this study were mdx (C57BL/10ScSn-mdx/J), mdx4cv (B6Ros.Cg-Dmdmdx-4Cv/J), and mdx5cv (B6Ros.Cg-Dmdmdx-5Cv/J). The mice were originally obtained from the Jackson Labs (Bar Harbor, ME), but have been maintained via sibling matings at the University of Washington for more than 5 generations. All procedures were carried out in accordance with approved protocols by the Institutional Animal Care and Use Committee (IACUC) of the University of Washington.
Preparation of DNA
Approximately 1 cm of tail was clipped using a clean razor and the mice appropriately marked. The tails were digested with proteinase K in a 56°C water bath overnight and the DNA was isolated and dissolved in 100 μl of Tris-EDTA (TE) buffer using the spin column protocol from the Qiagen DNEasy blood and tissue extraction kit (Valencia, CA), as per manufacturers instructions.
Polymerase chain reaction
We prefer to utilize a proofreading Taq in a pre-made mix from Invitrogen called Accuprime (Carlsbad, CA), although other PCR reagents may suffice. We added 30–150 ng of genomic DNA and 20 pM of each primer to the Accuprime mix. The primers were mdx F1: AACTCATCAAATATGCGTGTTAGT mdx R1:5′ CTCAATCTCTTCAAATTCTG. mdx4cv F1 TCAAGAACAGCTGCAGAACAGGAGA. mdx4cv R1 GGATTGCATCTACTGTGTGAGGACC. mdx5cv F1 ATTTGGAAGCTCCCAGAGAC. mdx5cv R1 TGCTTTAGCTTCAGAAGTCA. The PCR conditions for each of the mdx strains is 94°C for 5′, followed by 35 cycles of 94°C for 30″, 60°C for 30″ and 72°C for 30″ followed by an extension of 72°C for 3′.
Isolation and sequencing of the amplified DNA
We isolated the amplified DNA using the Qiaquick PCR purification kit (Qiagen, Valencia, CA) into 50 μl of filter-sterilized water. The DNA concentration was quantified using a spectrophotometer (NanoDrop 1000; Thermo Scientific; Miami, OK). 5 μl of the cleaned DNA was run on a 1.25% agarose gel to verify the correct PCR product was amplified (179 bp for mdx, 157 bp for mdx4cv and 221 bp for mdx5cv) and no product was found in the negative control that had 1 μl TE instead of DNA.
Approximately 100 ng of the PCR product was required for sequencing (Biochemistry sequencing facility, University of Washington, WA). The facility utilizes the Applied Biosystems 3730XL sequencer (Applied Biosystems, USA). We used the forward primers for sequencing mdx and mdx5cv products and the reverse primer for sequencing the mdx4cv products.
Results
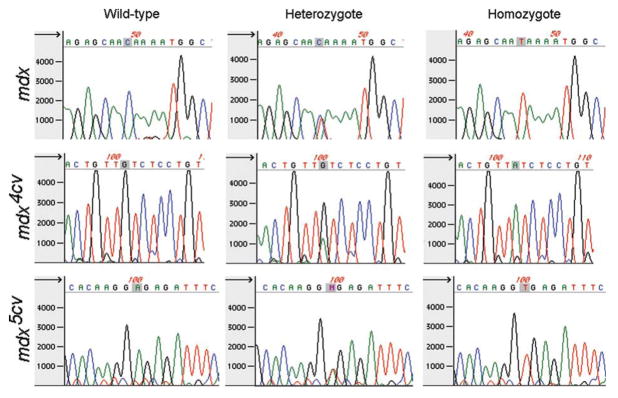
The chromatogram clearly showed a single peak for wild-type, mdx, mdx4cv and mdx5cv mice, and two peaks at the appropriate nucleotide for heterozygous mice (example of mdx in Fig. 1). Only the chromatogram was used to analyze the sequence, because the nucleotide in the letter file for heterozygous mice usually reads as a wild-type or N (unable to decipher) (Fig. 1). As expected, heterozygous mice were only found in the DNA samples from female mice. We successfully retrieved the sequence data from 92% of samples (232 readable sequences from 253 mice) on the first attempt from mdx, mdx4cv and mdx5cv mice. All but one of the sequences showed the mouse genotype on the second attempt (20 readable sequences from 21 mice). The second attempt for sequencing the amplified DNA utilized the same PCR product as the first. The single failure resulted from problems extracting the genomic DNA from the tail sample. Thus, the genotype of only one mouse was not positively identified out of 253 samples, and we have not had a single false positive in over a year of subsequent analyses of the genotyped mice.
Figure 1.
Representative chromatograms from wildtype, heterozygous, and homozygous mice for the point mutation in mdx, mdx4cv, and mdx5cv mice (shaded). Note that the heterozygous mouse samples have two peaks, but the nucleotide sequence reads as a wildtype nucleotide or “N,” necessitating manual analysis of chromatograms for genotyping these mice.
Discussion
In the present study we developed a sequencing method for genotyping the point mutations in mdx, mdx4cv and mdx5cv mice. We chose to sequence the PCR amplified DNA encompassing the point mutations, which gave a high level of success. Previous methods developed by our lab and others for genotyping mdx mice using PCR alone are typically faster and cheaper than sequencing the DNA as described in this study ([7] and references within). However, despite years of methodological optimization we find that PCR alone can lead to false positives and difficulty in obtaining reproducible results, which can be a constant source of frustration when trying to breed various mdx mice to different strains or examining mice that have been treated with expensive reagents. The methods described here did not lead to any false positives thereby saving time and money for housing, breeding, treating and examining the mice.
Acknowledgments
We would like to thank the staff at the Biochemistry sequencing facility, University of Washington. GBB was supported by the CJ Martin postdoctoral fellowship and an MDA development grant. This work was also supported by NIH grants AR044533 and PO1N5046788 to JSC.
References
- 1.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci U S A. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, et al. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 3.Chapman VM, Miller DR, Armstrong D, Caskey CT. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proc Natl Acad Sci U S A. 1989;86:1292–1296. doi: 10.1073/pnas.86.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Im WB, Phelps SF, Copen EH, Adams EG, Slightom JL, et al. Differential expression of dystrophin isoforms in strains of mdx mice with different mutations. Hum Mol Genet. 1996;5:1149–1153. doi: 10.1093/hmg/5.8.1149. [DOI] [PubMed] [Google Scholar]
- 5.Danko I, Chapman V, Wolff JA. The frequency of revertants in mdx mouse genetic models for Duchenne muscular dystrophy. Pediatr Res. 1992;32:128–131. doi: 10.1203/00006450-199207000-00025. [DOI] [PubMed] [Google Scholar]
- 6.Haenggi T, Fritschy JM. Role of dystrophin and utrophin for assembly and function of the dystrophin glycoprotein complex in non-muscle tissue. Cell Mol Life Sci. 2006;63:1614–1631. doi: 10.1007/s00018-005-5461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trebbin AL, Hoey AJ. A novel and simple method for genotyping the mdx mouse using high-resolution melt polymerase chain reaction. Muscle Nerve. 2009;39:603–608. doi: 10.1002/mus.21215. [DOI] [PubMed] [Google Scholar]