Abstract
Background
The cytoadherence of Plasmodium falciparum is thought to be mediated by variant surface antigens (VSA), encoded by var, rif, stevor and pfmc-2tm genes. The last three families have rarely been studied in the context of cytoadherence. As most VSA genes are unique, the variability among sequences has impeded the functional study of VSA across different P. falciparum strains. However, many P. falciparum genomes have recently been sequenced, allowing the development of specific microarray probes for each VSA gene.
Methods
All VSA sequences from the HB3, Dd2 and IT/FCR3 genomes were extracted using HMMer software. Oligonucleotide probes were designed with OligoRankPick and added to the 3D7-based microarray chip. As a proof of concept, IT/R29 parasites were selected for and against rosette formation and the transcriptomes of isogenic rosetting and non-rosetting parasites were compared by microarray.
Results
From each parasite strain 50-56 var genes, 125-132 rif genes, 26-33 stevor genes and 3-8 pfmc-2tm genes were identified. Bioinformatic analysis of the new VSA sequences showed that 13 rif genes and five stevor genes were well-conserved across at least three strains (83-100% amino acid identity). The ability of the VSA-supplemented microarray chip to detect cytoadherence-related genes was assessed using P. falciparum clone IT/R29, in which rosetting is known to be mediated by PfEMP1 encoded by ITvar9. Whole transcriptome analysis showed that the most highly up-regulated gene in rosetting parasites was ITvar9 (19 to 429-fold up-regulated over six time points). Only one rif gene (IT4rifA_042) was up-regulated by more than four fold (five fold at 12 hours post-invasion), and no stevor or pfmc-2tm genes were up-regulated by more than two fold. 377 non-VSA genes were differentially expressed by three fold or more in rosetting parasites, although none was as markedly or consistently up-regulated as ITvar9.
Conclusions
Probes for the VSA of newly sequenced P. falciparum strains can be added to the 3D7-based microarray chip, allowing the analysis of the entire transcriptome of multiple strains. For the rosetting clone IT/R29, the striking transcriptional upregulation of ITvar9 was confirmed, and the data did not support the involvement of other VSA families in rosette formation.
Background
Plasmodium falciparum is the apicomplexan organism causing most malaria deaths. Clinical symptoms occur when the parasite enters the intraerythrocytic cycle. One feature characterising a red blood cell infected with P. falciparum at pigmented trophozoite stage is its ability to cytoadhere to human cells. The three main forms of cytoadherence are binding to microvascular endothelial cells, binding to uninfected red blood cells (rosetting) and binding to platelets ("platelet-mediated clumping"), (reviewed in [1]). Sequestration of infected red blood cells (iRBC) in the microvasculature, whether by direct attachment to microvessel walls or indirectly via platelets or red blood cells, is probably a way for parasites to avoid clearance by the spleen [2]. Sequestration in combination with high parasite burdens can, however, cause microvascular obstruction leading to acidosis, hypoxia and release of harmful inflammatory cytokines [3]. Rosetting in particular has been consistently associated with severe malaria cases in sub-Saharan Africa [4-6]. The importance of rosetting in virulent infections is underlined by the observation that human red blood cell polymorphisms that reduce the ability of P. falciparum to form rosettes offer substantial protection against life-threatening malaria [7,8].
The parasite molecules thought to mediate cytoadherence are the variant surface antigens (VSA). VSA genes, generally located in the subtelomeric regions, encode proteins exported to the surface of the iRBC. They include the var, rif, stevor and pfmc-2tm families, a total of 200 to 300 genes per genome. The var gene family, encoding P. falciparum Erythrocyte Membrane Protein 1 (PfEMP1), is the most-well studied VSA family (reviewed in [9]). The ~60 var genes per isolate can be subdivided into three main groups (A, B or C) based on the upstream regions [10]. The classification has functional and clinical relevance [11-15]. The var gene family undergoes mutually exclusive expression, so that a single PfEMP1 variant is found at the surface of the iRBC, although exceptions can occur [16]. Each PfEMP1 is composed of DBL (Duffy Binding-Like) and CIDR (Cysteine rich Inter-Domain Region) domains. Some of these domains have been shown to bind to human cell surface receptors, such as ICAM-1 (by DBLβ), CD36 (by CIDRα, found in group B and C PfEMP1) and red cells via CR1 (DBLα of group A PfEMP1) (reviewed in [1]).
By definition, VSA sequences show low similarity between paralogues, while orthologues cannot be found across strains (one notable exception is var2csa, a relatively well conserved var gene member) [17]. This implies that VSA sequences are unique to a particular P. falciparum strain/isolate. A function assigned to a particular VSA is thus difficult to extrapolate to other P. falciparum strains.
So far, virtually all studies investigating the parasite ligands for cytoadherence have focused on var genes only. Whether other VSA or non-VSA genes could be involved in adhesion is therefore untested. Moreover, the reference strain 3D7 is often used despite its poor ability to cytoadhere compared to other laboratory strains and field isolates. Recently, many P. falciparum genomes, including Dd2, HB3 and IT/FCR3 have been sequenced [18,19]. These three strains are selectable for various cytoadherence phenotypes [20,21] (and JA Rowe, unpublished data). Therefore, to study cytoadherence-related genes in these strains by microarray, it is possible to extract the VSA sequences from a sequenced genome and design oligonucleotide probes specific to each VSA gene. These oligos are added to the 3D7-based microarray chip, allowing the analysis of the full transcriptome of a new strain. As a proof of concept, the IT/R29 strain was selected for and against rosetting and the transcriptome of both populations was analysed with a "VSA-supplemented" microarray chip containing oligos specific for IT VSA. IT/R29 was chosen because it is a well-characterized strain whose rosetting ligand is the group A PfEMP1 variant encoded by the ITvar9 gene (also known as R29var1) [22,23]. Therefore, the aim of this work was to determine whether the VSA-supplemented microarray chip successfully identified ITvar9 as the major up-regulated gene in rosetting parasites, and to use whole transcriptome analysis to identify other candidate genes that could be accessory molecules in rosette formation.
Methods
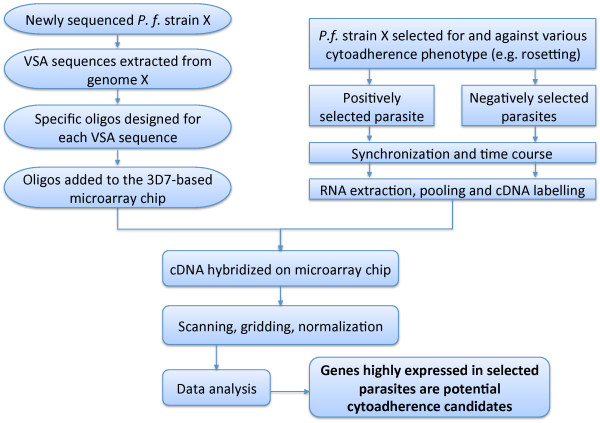
The overall approach used to design a VSA-supplemented microarray chip and investigate the transcriptional profiles of parasites selected for and against a particular cytoadherence phenotype is shown in Figure 1.
Figure 1.
Summary of the microarray chip design and experimental protocol. A bioinformatic approach was used to extract VSA sequences and design oligonucleotide probes for a particular strain, while the parasites were selected for and against a cytoadherence phenotype (rosetting). cDNA from positively and negatively selected parasites was hybridized on the "VSA-supplemented" 3D7-based microarray chip in order to identify cytoadherence gene candidates that were up-regulated in rosetting parasites compared to isogenic non-rosetting parasites.
Genome sequences
The HB3 and Dd2 genomes (1189 and 2837 supercontigs, respectively) were downloaded from the Broad institute [18]. The IT/FCR3 genome (2185 supercontigs) was downloaded from the Sanger institute [19]. All other sequences were obtained from PlasmoDB [24].
Extracting sequences from a genome
The entire method used for extracting VSA sequences is outlined in Figure 2. The HMMer software, an implementation of profile Hidden Markov Models (HMM), was used with default parameters [25]. In this case, HMMer is used to search for VSA homologues in the HB3/Dd2/IT genomes using an "HMMer profile" based on an alignment of 3D7 sequences. In other words, the programme uses conserved motifs within a given protein family and finds similar motifs in a new genome.
Figure 2.
Flowchart of the process used for extracting VSA sequences from a genome.
To establish the HMMer profile, each translated exon of each VSA family (var, rif, stevor, pfmc-2tm) of the well-annotated 3D7 genome was aligned separately with ClustalW. Each resulting file was used to build a profile HMM specific to that exon. These profiles were used to search for homologues in the six frames of the translated HB3, Dd2 and IT genomes, as well as the Neurospora crassa (negative control, genome of similar size) and 3D7 genomes (positive control). The HMMer output file only shows the coordinates of the hits, thus Perl scripts were used to retrieve the actual amino acid sequences from the coordinates. Sequences with stop codon(s) and/or frameshift were annotated as "pseudogene" while shorter sequences with missing amino acids were annotated as "truncated". As a final control, each group of newly derived VSA sequences was used to build a HMM profile and searched against their original genome. If a comprehensive set of VSA had been extracted in the initial procedure, then no new sequences would be expected in the repeated search.
Designing specific oligos for a microarray chip
70-mer long oligonucleotide probes ("oligos") specific to the extracted VSA sequences were designed using OligoRankPick [26]. Oligos were added onto the 3D7-based microarray chip [27].
Parasite culture and rosette selection
Plasmodium falciparum strain R29 is a clone derived from the IT/FCR3 strain [21] and has been used to study the molecular basis of rosetting [22,28]. R29 was cultured at 2% haematocrit with group A erythrocytes (Scottish National Blood Transfusion Service, Edinburgh, UK) in supplemented RPMI as described elsewhere [29]. Parasitaemia was maintained at 5-10% and cultures were synchronized by sorbitol lysis [30]. Parasites were incubated at 37°C in the presence of 3% CO2, 1% O2 and 96% N2 and routinely screened to avoid mycloplasma contamination [31]. Parasites were replenished with media everyday and fresh erythrocytes every other day. The R29 strain was separated into rosette-positive (R+) and rosette-negative populations (R-) by centrifugation through 60% Percoll or gelatin flotation 2-3 times a week [32]. The rosette frequency of both rosette positive and negative populations was measured by counting the percentage of infected erythrocytes that form rosettes out of 300 infected erythrocytes. Wet preparations of culture suspension stained with 25 mg/ml of ethidium bromide were viewed using a combination of fluorescence and white light. A rosette was defined as an infected erythrocyte that bound two or more uninfected erythrocytes. R29R+ was at a rosette frequency of 73.2% and R29R- was at a rosette frequency of 1.3% at the pigmented trophozoite stage prior to RNA sample collection.
Synchronisation and time-course experiment
In the three weeks prior to RNA collection, sorbitol lysis was carried out twice at 12 hour intervals in every asexual cycle. For the time course, schizont stage cultures were examined hourly until the first ring was seen, then sorbitol lysis was carried out 12 hours later, giving ring stage parasites within an 12-hour time window. Samples were collected for RNA immediately after sorbitol lysis (time point 1), and then 8-hourly, resulting in 6 time points covering the entire intra-erythrocytic life cycle. For RNA, room temperature TRIzol reagent (Invitrogen 15596-026) was added (ten times the packed cell volume of the cell pellet), and after thorough mixing, stored at -80°C.
RNA extraction and cDNA synthesis
RNA was extracted as described [33] with minor modifications as follows. The tube with TRIzol solution was thawed on ice. Two volumes of chloroform (Sigma) were added per volume of packed cells. After mixing, the tube was incubated on ice for 5 min then centrifuged at 3600 g for 40 min at 4°C without brakes. The supernatant (aqueous layer) was carefully transferred into a fresh tube without disturbing the interface. The same volume of ice-cold isopropanol (Sigma) was added and the tube was incubated at 4°C overnight. The next day, the tube was centrifuged at 3600 g for 60 min at 4°C. The supernatant was discarded and the pellet was resuspended and washed with ice-cold 70% ethanol (Sigma). After another centrifugation at 3600 g and 4°C for 10 min, the supernatant was completely but carefully removed using a fine Pasteur pipette. The tube was left upside down to air dry for 15 to 60 min until no liquid was visible. The dried pellet was resuspended with 25 μl of warm DEPC-H2O then placed on ice. RNA concentration was measured using a spectrophotometer. 12 μg of RNA from the R29 non-rosetting parasites at each of the six time points was combined together to form the reference pool. The pool and 12 μg of each individual time point sample from both rosetting and non-rosetting parasites were then used for first-strand cDNA synthesis using an amino-allyl dye coupling protocol [34].
Printing the VSA-supplemented microarray chip
The microarray for the 3D7 P. falciparum genome designed by Hu et al [26], consists of 10,166 70-mer long oligonucleotide elements (oligos) for 5,363 genes, with one unique oligo every 2 kb per gene. To these were added oligos specific for the VSA from HB3, Dd2 and IT strains. Oligos were spotted onto polylysine-covered slides and post-processed as described in [35,36].
cDNA labelling and microarray hybridization
Microarray hybridizations were performed as previously described [35]. Briefly, each aminoallyl-cDNA sample was coupled to Cy5 (red dye) while Cy3 (green dye) was added to the pool. Cy5-labelled time point samples were mixed with the same amount of Cy3-labelled pool sample. The solution was loaded on a microarray slide and hybridized for 14-16 h using a Maui hybridization system (Bio Micro Systems) at 65°C. Microarrays were scanned with a GenePix 4000B scanner (Axon Instruments).
Pre-analysis and quality control
All arrays were visually inspected using GenePix and any poor quality spots (signal below background or dust on the chip) were flagged out. After gridding, the data were loaded onto the Acuity 4.0 software. Within this database, each spot was expressed as:
'F' is the foreground signal intensity, 'B' is the background signal intensity, '635' is the wavelength of the red channel, '532' is the wavelength of the green channel. Thus, the data for each spot is the ratio between red and green signal. Each array was then normalized with Lowess (locally weighted least squares regression). A dataset with all time points was created using the following parameters:
Cutoff: Discard flagged spots AND (% > B532+2SD > = 95 OR% > B635+2SD > = 95). In other words, only unflagged spots and spots with median intensities (green or red) greater than the local background plus two times the standard deviation of the background were used.
Microarray data analysis
Expression ratios correspond to the "red signal" (from a rosetting R+ or non-rosetting R- time point sample) divided by the "green signal" (from the pool). These ratios were used to visualize the timing of expression of a particular gene. To measure the change of expression in R+ compared to R-, the [rosetting/pool] ratios were divided by the [non-rosetting/pool] ratios to obtain [rosetting/non-rosetting] ratios. Values from oligos specific to the same genes were averaged using OligoAverage.pl [26]. Data analysis was carried out using Microsoft Excel, Cluster [37] and Jalview [38] for data visualization. All microarray data have been deposited in the GEO repository [39]. Genes showing a three-fold change between the rosetting R+ and non-rosetting R- parasites in at least one of the five paired time points (TP1-5) were subjected to K-means clustering of groups. Data from TP6 were not examined further due to synchronization differences between R+ and R- at this time point. Functional Enrichment Analysis was carried out to calculate the over-representation of genes belonging to functional groups for each cluster as compared to their respective frequency in the whole genome.
Results
All variant surface antigen sequences from HB3/Dd2/IT were extracted to design specific microarray probes (70-mer oligos)
The software HMMer was used to extract all VSA sequences from HB3, IT and Dd2 genomes (Table 1). Rif, stevor and pfmc-2tm sequences are available as supplementary data (Additional files 1, 2, 3, 4). The var gene sequences from these three strains have been published by other groups [40,41]. A total of 990 70-mer oligos corresponding to the var, rif and stevor gene families of HB3, Dd2 and IT (Additional file 5) were added to the 10,166 existing oligos on the P. falciparum microarray chip [26]. Strain-specific oligos to pfmc-2tm were not included because the hypervariable loop of this gene family is only about 50 nucleotides long [42]. Surfin genes were first described as a VSA family [43], however ClustalW alignments with 3D7, HB3 and Dd2 surfins indicate that each orthologue is well-conserved between strains. Thus surfin is not a VSA family and there is no need to design new oligos for microarray purposes.
Table 1.
Total number of gene sequences for each VSA family for each parasite strain
| 3D7 | HB3 | Dd2 | IT | ||
|---|---|---|---|---|---|
| var | genes | 62 | 38 | 39 | 49 |
| Pseudogenesa | 1 | 6 | 4 | 3 | |
| Truncated genesb | 0 | 6 | 7 | 4 | |
| Total | 63 | 50 | 50 | 56 | |
| rif | genes | 153 | 100 | 99 | 102 |
| Pseudogenes | 28 | 21 | 21 | 15 | |
| Truncated genes | 3 | 4 | 13 | 8 | |
| Total | 184 | 125 | 132 | 125 | |
| stevor | stevor genes | 30 | 17 | 24 | 25 |
| Pseudogenes | 7 | 9 | 7 | 8 | |
| Truncated genes | 0 | 0 | 0 | 0 | |
| Total | 37 | 26 | 31 | 33 | |
| pfmc-2tm | genes | 10 | 6 | 2 | 5 |
| Pseudogenes | 3 | 1 | 0 | 2 | |
| Truncated genes | 0 | 1 | 1 | 1 | |
| Total | 13 | 8 | 3 | 8 | |
aPseudogene: one or more stop codons or frameshifts within the sequence.
bTruncated gene: part of the gene sequence is missing, most likely due to incomplete sequencing.
Some VSA sequences are conserved amongst strains
The similarities between the VSA from different parasite strains were examined. The var genes are not described further here because a detailed examination of the HB3/Dd2/IT var gene repertoires was reported recently [40]. Bioinformatic analysis of rif and stevor genes revealed that several of these VSA family members are conserved across the four analyzed strains. Those that were found in at least three out of the four strains with >90% amino acid identity between at least one pair are shown in Table 2 (13 strain-transcendent Rifins) and Table 3 (5 strain-transcendent Stevors). For comparison, a typical pairwise alignment score for two random Rifins is 30 to 50% amino acid identity. The average pairwise alignment score of all Rifins is alike for each strain (~ 44%) as well as all strains together (43.2%). This is in agreement with the hypothesis that each member of a VSA family can recombine with each other, possibly through heterologous meiotic crossing-over [44]. Pairwise alignment scores are in general higher for Stevors (~60%).
Table 2.
Summary of strain-transcendent Rifins found in at least three out of 3D7, HB3, Dd2 and IT
| 3D7 | HB3 | Dd2 | IT | Pairwise alignment score1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Found in F.I.2 | Gene ID | Gene ID | Gene ID |
3D7- HB3 |
3D7- Dd2 |
HB3- Dd2 |
IT- 3D7 |
IT- HB3 |
IT- Dd2 |
| RifA2_PFD0070c | HB3rifA2_025 | Dd2rifA2_p1263 | IT4rifA2_125 | 92 | 83 | 85 | ||||
| / | HB3rifB1_050 | Dd2rifB1_027 | IT4rifB_114 | 100 | 100 | 100 | ||||
| RifB_MAL13P1.495 | HB3rifB_t0224 | Dd2rifB_t038 | IT4rifB_111 | 94 | 94 | 99 | 100 | 99 | 100 | |
| RifB_PFE1630w | yes | HB3rifB_079 | Dd2rifB_081 | / | 99 | 85 | 87 | |||
| RifA_PFL2585c | yes | HB3rifA_098 | IT4rifA_031 | 99 | 96 | 99 | ||||
| RifA_PFD0645w | / | Dd2rifA_086 | IT4rifA_021 | 98 | 94 | 95 | ||||
| RifA_p_PFD1020c | yes | HB3rifA_p112 | / | IT4rifA_050 | 97 | 90 | 98 | |||
| / | yes | HB3rifA_051 | Dd2rifA_p100 | IT4rifA_052 | 99 | 100 | 98 | |||
| RifB_p_PFA0710c | yes | HB3rifB_p116 | Dd2rifB_p111 | IT4rifB_104 | 97 | 98 | 99 | 98 | 99 | 98 |
| RifA_PFE1635w | yes | HB3rifA_p115 | Dd2rifA_p107 | IT4rifA_p084 | 98 | 94 | 95 | 86 | 86 | 92 |
| RifA_p_t_PFD0134c | HB3rifA_p101 | / | IT4rifA_t072 | 100 | 95 | 90 | ||||
| RifA_p_PF11_0022 | / | Dd2rifA_p112 | IT4rifA_p087 | 98 | 86 | 87 | ||||
| RifA_p_MAL7P1.52 | / | Dd2rifA_p098 | IT4rifA_p088 | 91 | 97 | 96 | ||||
1Rifins with >90% amino acid identity between at least one pair of strains plus >80% amino acid identity between at least 2 other pairs of strains are shown.
2F.I: Brazilian field isolates [45]
3p=pseudogene: one or more stop codons or frameshifts within the sequence.
4t=truncated gene:part of the gene sequence is missing, possibly due to incomplete sequencing.
Table 3.
Summary of strain-transcendent Stevors found in at least three out of 3D7, HB3, Dd2 and IT
| 3D7 | HB3 | Dd2 | IT | Pairwise alignment score1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | Found in F.I.2 | Gene ID | Gene ID | Gene ID |
3D7- HB3 |
3D7- Dd2 |
HB3 - Dd2 |
IT- 3D7 |
IT- HB3 |
IT- Dd2 |
| SteA_MAL13P1.7 | HB3steA_14 | / | IT4steA_11 | 99 | 87 | 87 | ||||
| SteB_PFD0125c | HB3steB_p183 | Dd2steB_18 | IT4steB_01 | 99 | 93 | 91 | 99 | 93 | 92 | |
| SteB_t_PFB0955w4 | yes | HB3steB_p26 | / | IT4steB_14 | 98 | 99 | 99 | |||
| SteB_p_PF10_0009 | HB3steB_p20 | Dd2steB_p29 | IT4steB_p31 | 98 | 98 | 98 | 88 | 88 | 88 | |
| SteA_p_PFA0705c | yes | HB3steA_p23 | Dd2steA_p31 | / | 99 | 96 | 98 | |||
1Stevors with >90% amino acid identity between at least one pair of strains plus >80% amino acid identity between at least 2 other pairs of strains are shown.
2F.I: Brazilian field isolates [45]
3p=pseudogene: one or more stop codons or frameshifts within the sequence.
4t=truncated gene:part of the gene sequence is missing, possibly due to incomplete sequencing.
Comparison with data from field isolates in Brazil [45] showed that many of the conserved rif and stevor sequences are also found in field isolates (Tables 2 and 3). In several cases, conserved rif and stevor sequences are located in pairs next to each other in the subtelomeric regions (eg. chromosomes 1, 4, 10 and 13, Figure 3). When sufficient genomic data were available to allow examination of gene location, this synteny was conserved among strains. The internal non-coding DNA sequence in between such pairs of conserved genes was also conserved.
Figure 3.
Genomic location of strain-transcendent VSA in relation to drug-resistance genes. The 14 P. falciparum chromosomes are represented to scale. The location of six drug resistance-related genes, 22 rif and stevor genes that are conserved between 3D7 and at least one other strain, two conserved pfmc-2tm genes, var1csa and var2csa are indicated. "ψ" indicates a pseudogene.
The presence of conserved VSA cannot be explained by location next to known drug resistance genes
Previously, five var genes (varS1-varS5) were reported to be shared at relatively high frequency among field isolates from the West Pacific region [46]. Two of these genes, varS2 and varS3 were physically linked to the pyrimethamine-resistance pfdhfr locus (within 200 Kb), whereas another (varS4) was within 100 Kb of the chloroquine-resistance pfcrt locus. Thus, the fact that these var genes are conserved amongst isolates can be explained by a selective sweep due to heavy selection pressure for drug resistance in these parasites. A similar mechanism of VSA occurring adjacent to parasite genes that give a high survival advantage and undergo a selective sweep could in principle apply to the conserved VSA identified here. However, a chromosomal map reveals that few of the conserved VSA genes are located adjacent to known drug resistance genes (Figure 3). In addition, 3D7 and HB3 are sensitive to most of the commonly used anti-malarial drugs. Therefore, some other explanation is required to explain the presence of strain-transcendent rif and stevor genes.
Time-course experiment with IT/R29 rosetting (R+) and non-rosetting (R-) parasites
As proof of concept of the ability of the VSA-supplemented microarray chip to identify genes important in cytoadherence, we compared the whole transcriptome of IT/R29R+ parasites (rosette frequency 73.2%) with IT/R29R- parasites (rosette frequency 1.3%). RNA was collected from synchronized rosetting and non-rosetting parasites at six time points throughout the asexual blood stage cycle. The maturity of the two parasite populations was compared by Giemsa smear (Figure 4) and found to be similar throughout, apart from minor differences at time point six in which there were more rings and fewer schizonts in R+ parasites. In addition, statistical evaluation of gene transcript levels at each time point was carried out by Pearson correlation comparing data from all oligos in IT/R29R+ with all oligos at the same time point in IT/R29R-. A strong positive correlation was found (correlation coefficients above 0.88 for time points 1 to 5; 0.75 at time point 6), indicating that the two parasite populations were at similar levels of maturity. Therefore specific differences in gene expression detected in subsequent analyses are unlikely to be artefacts due to maturity differences between rosetting and non-rosetting parasites.
Figure 4.
Parasite maturity during the time-course experiment. Sorbitol lysis was carried out at 12 hours after the first ring invasion was seen (time point one), and samples were then taken 8-hourly throughout the asexual blood stage cycle. The maximum parasite maturity in terms of hours post invasion at each time point is shown in the second column. Samples for RNA extraction were taken from the culture at each time point, mixed with TRIzol reagent and frozen, and a Giemsa-stained thin blood smear was performed to record the developmental stage of rosetting (R29R+) and non-rosetting (R29R-) parasites.
ITvar9 is the only VSA highly up-regulated in IT/R29 rosetting parasites
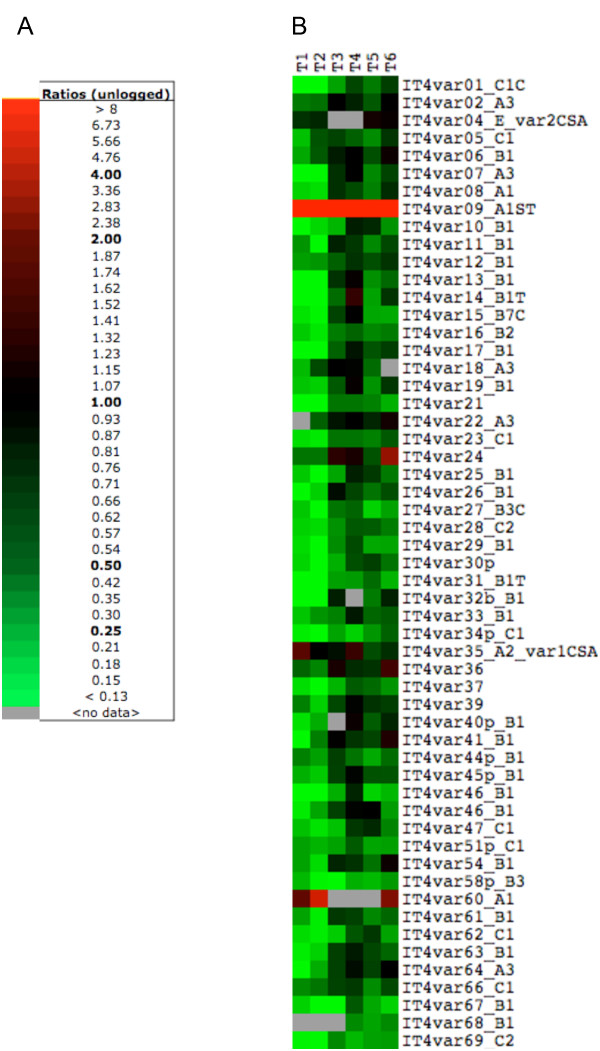
As the parasite ligand(s) mediating rosetting should be located on the surface of infected red cells, VSA are prime candidates for this function. For IT/R29R+, PfEMP1 encoded by ITvar9 (also called R29var1) is known to be the parasite rosetting ligand [22,23], although the existence of accessory molecules for rosetting has not been excluded. Examination of the var gene data from the microarray (Figure 5) showed that 55 out of the 56 annotated var genes in IT were transcribed, but only one, ITvar9, was highly up-regulated in IT/R29 rosetting parasites compared to non-rosetting parasites (19 to 439 fold up-regulated, Figure 5. By "up-regulated " we mean that the amount of mRNA for a particular gene is increased in IT/R29R+ compared to IT/R29R- parasites). This striking upregulation of ITvar9 was seen at all six time points. In the var gene family, the only other member to be up-regulated by more than two fold in rosetting parasites was ITvar60 (up to five fold up-regulated at time point 2), which is also a rosette-mediating variant in the IT strain (Ghumra and Rowe, in preparation). Virtually all other var genes were downregulated (i.e. expressed at a lower level in R29R+ than in R29R-, Figure 5). Therefore these data are consistent with previously published results indicating that ITvar9 (R29var1) encodes the ligand mediating rosetting in IT/R29 parasites [22,23], and authenticate the use of the VSA-supplemented microarray chip to detect important cytoadherence-associated gene candidates.
Figure 5.
IT/R29 var gene transcription in rosetting versus non-rosetting parasites analysed by VSA-supplemented microarray. (A) Colour legend used for the microarray data. (B) Each row represents one var gene and values from multiple oligos specific to the same gene were averaged. Each var gene is represented by 2 to 5 oligos on the chip, with an oligo approximately every 2 kb for each gene. Each column is a time point with 8-hour intervals. Var gene names and groups (A, B or C) are as described previously [40,41]. Data represented are the expression ratio of data from IT/R29R+ over IT/R29R-, thus red squares indicate higher amount of cDNA in the IT/R29R+ compared to the IT/R29R- population, and green squares indicate less cDNA in IT/R29R+ than IT/R29R-. A grey square indicates the lack of data (expression intensity below background level). Var genes with expression intensity below background level at all time points are not shown. ITvar9 is up-regulated in IT/R29R+ by 19, 21, 115, 57, 109 and 439 fold from time point 1 to 6, respectively.
Whole transcriptome analysis with the VSA-supplemented chip also has the potential to identify accessory molecules for cytoadherence phenotypes. Analysis of the other VSA families in R29R+ and R29R- showed that only 45 rif and six stevor genes showed expression above background level in at least one time point, out of a total of 125 and 33 genes in the IT genome, respectively (Figure 6). This contrasts with the var gene family where almost all variants were transcribed (Figure 5). In the rif family, only IT4rifA_042 showed upregulation in rosetting parasites by more than four fold in a single time point (five fold at time point 1). Interestingly, IT4rifA_054 (alias Rif13-1), located upstream of ITvar9 in a "head to head" manner [47], is up-regulated by up to three fold in time point 3 (Figure 6A). This upregulation may result from the high transcription level of its neighbouring gene ITvar9. No stevor or pfmc-2tm variant was up-regulated by more than two fold in IT/R29R+ (Figure 6B).
Figure 6.
IT/R29 rif and stevor gene transcription in rosetting versus non-rosetting parasites analysed by VSA-supplemented microarray. (A) Rif genes and (B) stevor genes. The microarray data are as described in the legend for Figure 5. There was only one oligonucleotide probe per gene for rif and stevor genes due to their small size.
Taking all VSA data together, ITvar9 is by far the most highly up-regulated VSA gene in R29 rosetting parasites, and these data do not support the involvement of other VSA families in rosette formation in this strain.
Some non-VSA genes are up-regulated in IT/R29R+ parasites
Although only surface proteins could act as a direct rosetting ligand, other proteins could contribute to the phenotype, for example by influencing the trafficking of PfEMP1. Such non-VSA genes could also be up-regulated after rosette selection. As rosetting begins at the late ring/early trophozoite stage, genes expressed at time points 1 and 2 are likely to be the most relevant, although the possibility of relevant genes expressed later in the cycle cannot be excluded. Examination of the dataset showed that 377 genes (7.8%) were differentially expressed by at least three fold in rosetting compared to non-rosetting parasites (a full list of the differentially regulated genes is shown in Additional file 6). 47 of the 377 differentially regulated genes in rosetting parasites had a PEXEL motif, indicating export to the infected red cell cytoplasm [48]. Furthermore, a total of 17 genes belonging to the PHIST (Plasmodium helical interspersed subtelomeric) family were up-regulated. This gene family could be involved indirectly in the process of rosetting, as it is linked to export to the erythrocyte and host cell remodelling.
K-means cluster analysis of the differentially regulated genes resulted in five distinct clusters in relation to timing of expression (Figure 7 and Additional file 6). Gene functional groups that were significantly enriched in rosetting parasites within each cluster were examined using three types of pathway, namely Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Malaria Parasite Metabolic Pathways (MPMP). At time points 1 and 2 (clusters 2 and 3, Figure 7), enriched gene groups included mitochondrial transporters, invasion-related genes and protein kinases (the enriched genes within each cluster are shown in Additional file 6). A different set of invasion-related genes were up-regulated at time point 4 (cluster 5, Figure 7). Some cytoadherence-related genes were up-regulated at time point 4 (cluster 5, Figure 7), such as the cytoadherence linked asexual proteins 3.1, 3.2 and 2. At time point 5 (cluster 1, Figure 7), a significant enrichment of genes coding for established and putative Maurer's clefts proteins was observed. These include, MAHRP, Skeletal-binding protein, and several etramps. However, in all cases, the upregulation of the above genes enriched in rosetting parasites was modest in comparison to the upregulation of ITvar9 encoding the known rosetting ligand.
Figure 7.
Differentially expressed genes in rosetting and non-rosetting IT/R29 parasites. The k-means clusters derived from the 377 genes that were differentially expressed by three-fold or more in rosetting compared to non-rosetting parasites are shown (colours as in Figure 5A). The corresponding graphs represent the enriched functional groups (p < 0.05) showing the over-representation of genes belonging to functional groups for each cluster as compared to their respective frequency in the genome as a whole.
Discussion
VSA sequences from HB3, Dd2 and IT genomes were extracted using HMMer software and Perl scripts. The multiple controls carried out using HMMer (repeating the approach with the output sequences and extraction of the original 3D7 VSA) give confidence that no VSA sequences were missed. The number of var genes sequences identified here matches or exceeds those identified previously using other data extraction methods [40,41]. Some rif and stevor sequences have been described previously for HB3 and IT [49], but not the entire repertoire. The possibility that some VSA genes are missing altogether from these sequenced genomes cannot be excluded, although the good sequence coverage (>9X) suggests that the gaps are likely to be small. Recently, Joannin et al released rif and stevor sequences from HB3 and Dd2 (not IT) on varDB [50]. They described more rif genes than this study, however, these extra sequences are truncated duplicates of other rif genes, typically found on short contigs. Considering the HB3 and Dd2 genomes are not fully assembled, these short duplicated sequences may well be assembly errors.
The VSA sequences were extracted in order to design unique oligos for each sequence to generate a VSA-supplemented microarray chip. However, further examination of the sequences revealed that several rif and stevor genes are well-conserved across strains (Tables 2 and 3). Almost half of these "conserved" VSA are predicted pseudogenes. Strain-transcendent stevor genes have been described previously [42,45,51], however, with the exception of rifA2 [49,50], this is the first comprehensive description of strain-transcendent rif genes. Two well-conserved strain-transcendent var genes have been described previously, var2CSA involved in placental malaria [52] and var1CSA of unknown function [53,54]. Interestingly, two of the conserved rif genes occur adjacent to var1CSA (within 6kb, Figure 3). Furthermore, the intervening non-coding sequence between these genes is also well-conserved, suggested that specific chromosomal regions in the subtelomeres may be conserved between strains. A previous example of conserved var genes has been described in P. falciparum isolates from the western pacific [46]. These conserved var genes are physically linked to drug resistance genes (within 100-200 Kb) and may, therefore, have hitch-hiked along with the drug resistance genes during a selective sweep under high drug pressure [46]. Examination of the location of the conserved rif and stevor genes in relation to known drug resistance loci (Figure 3) does not support a similar explanation in this case. The reason why these rif and stevor genes (some of them pseudogenes) appear well-conserved across strains is unknown. It is possible that these conserved VSA are linked to other genes that are highly benefical for parasite fitness and have undergone a selective sweep. Further work is needed to investigate this intriguing observation.
The original P. falciparum microarray chip [27] is only useful for the analysis of VSA from the 3D7 parasite strain on whose genome it is based. To allow whole transcriptome analysis (including VSA) of additional strains, we supplemented the 3D7-based chip with probes for the VSA of three strains (IT, HB3 and Dd2). As proof that this VSA-supplemented chip could be used to identify candidate genes of importance in cytoadherence, the transcriptomes of IT/R29 rosetting (R+) and IT/R29 non-rosetting (R-) parasites were analysed in a time-course experiment over the intraerythrocytic cycle. By far the most highly up-regulated gene in rosetting parasites was ITvar9 (alias R29var1), part of the group A family of var genes, which was up to 429-fold increased in R+ compared to R- parasites (Figure 5). This was expected from previous data showing that DBL1α of the PfEMP1 variant encoded by ITvar9 binds red cells to mediate rosetting [22], and antibodies to ITvar9 inhibit rosetting with high potency [23]. Transcription of the ITvar9 gene in rosetting parasites was up-regulated at all six time points, suggesting either that the mRNA for this gene is produced throughout the intraerythrocytic cycle, or that it is maintained without degradation right through to schizogony. The only other consistently up-regulated var gene in R29R+ parasites was ITvar60 (up to five fold up-regulated, Figure 5), which encodes another rosette-mediating PfEMP1 variant in IT-derived parasites (Ghumra and Rowe, in preparation). The VSA-supplemented microarray chip therefore successfully identified the known rosetting ligand in IT/R29 as the leading candidate gene, and also identified another IT rosette-mediating variant.
Whether other VSA such as Rifins or Stevor are also involved in IT/R29 rosetting has not been investigated previously. The VSA-supplemented chip allowed us to determine whether any rif or stevor genes are highly up-regulated after selection for rosetting. Although a few rif genes were found expressed at a higher level in IT/R29R+ than in IT/R29R- parasites, the slight upregulation (two to five fold for IT4rifA_042) does not correlate with the large difference in rosetting frequency between the two populations and is minor compared to the upregulation of ITvar9. For stevor, no gene was even two-fold up-regulated in rosetting parasites. These data do not support the hypothesis that rif or stevor genes play a role in rosette formation in IT/R29.
One interesting observation regarding rif genes was that the gene "head to head" with ITvar9 (IT4rifA_054, alias Rif13-1), was up-regulated up to three fold. The association in expression between group A var genes and their upstream rif gene has already been shown in 3D7 parasites using the PfSir2 knockout line [55] as well as in 3D7 and IT/FCR3 parasites selected for group A var gene expression with children's serum from a malaria endemic area [49]. This co-regulation may be because the neighbouring var and rif genes are under the control of a common promoter. In fact, it has recently been shown that a titratable factor activates the transcription of all VSA families [56].
The analysis of the microarray data of non-VSA genes revealed 377 genes that were differentially expressed in rosetting and non-rosetting parasites by three fold or more (Figure 7). Many of these genes have PEXEL motifs [48] and could potentially be involved indirectly in cytoadherence, for example via PfEMP1 trafficking to the infected red cell surface. Replicates of the microarray experiment and further investigations would be needed before drawing any conclusion regarding the role of these genes. Parasites selected for other adhesion phenotypes including binding to brain endothelial cells and platelet-mediated clumping are currently being investigated using similar techniques, and will provide further information on non-VSA genes up-regulated after selection. Mok and colleagues previously performed a microarray analysis, comparing the transcriptome of rosetting versus CD36-selected 3D7 parasites [34]. Apart from the strain-specific VSA genes, they identified six non-VSA genes up-regulated by at least five fold in rosetting parasites. Only three of them (SERA-5, RESA-2 and PFI1445w) were up-regulated in R29 rosetting parasites (by two fold in a single time point), hence there is little overlap between their data set and the one reported here.
Conclusions
New technologies allow the sequencing of parasite genomes at unprecedented scales and many additional P. falciparum laboratory strains and field isolates are expected to be sequenced in the near future. The method presented here, VSA sequence extraction with HMMer and oligo design with OligoRankPick, followed by microarray analysis of parasites selected for a cytoadherence phenotype, is straightforward and could be applied to any new Plasmodium genome. One advantage of this approach over other methods for studying VSA and cytoadherence such as real-time PCR, is that the entire transcriptome can be examined in each experiment. This allows for the possible identification of novel cytoadherence-related gene candidates that could not be predicted on the basis of previous knowledge. The VSA-supplemented microarray chip therefore has the potential to shed new light on the role of VSA and non-VSA genes in cytoadherence mechanisms.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AG performed the rosetting selection and time course. AC performed the bioinformatics analysis, the microarray hybridizations and the data analysis. SM and APG assisted with the microarray experiment and microarray analysis. ZB provided advice and expertise on microarray analysis. AR conceived the study. AG, AC and AR drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Rif gene sequences from HB3, Dd2 and IT.
Stevor gene sequences from HB3, Dd2 and IT.
Pfmc-2tm gene sequences from HB3, Dd2 and IT.
Rif and stevor gene names and supercontig location.
All VSA oligos added to the 3D7 microarray chip.
Non-VSA genes differentially expressed in rosetting versus non-rosetting parasites.
Contributor Information
Antoine Claessens, Email: antoineclaessens@gmail.com.
Ashfaq Ghumra, Email: aghumra@staffmail.ed.ac.uk.
Archna P Gupta, Email: archana@ntu.edu.sg.
Sachel Mok, Email: MOKS0007@ntu.edu.sg.
Zbynek Bozdech, Email: ZBozdech@ntu.edu.sg.
J Alexandra Rowe, Email: Alex.rowe@ed.ac.uk.
Acknowledgements and funding
We thank Ann Hedley for help in writing the Perl script, and Guangan Hu for VSA oligo design. This work was funded by the Wellcome Trust (4 year PhD studentship to AC and Senior Fellowship in Basic Biomedical Science to JAR, grant number 084226).
References
- Rowe JA, Claessens A, Corrigan RA, Arman M. Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: molecular mechanisms and therapeutic implications. Expert Rev Mol Med. 2009;11:e16. doi: 10.1017/S1462399409001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffet PA, Safeukui I, Milon G, Mercereau-Puijalon O, David PH. Retention of erythrocytes in the spleen: a double-edged process in human malaria. Curr Opin Hematol. 2009;16:157–164. doi: 10.1097/MOH.0b013e32832a1d4b. [DOI] [PubMed] [Google Scholar]
- Dondorp AM, Lee SJ, Faiz MA, Mishra S, Price R, Tjitra E, Than M, Htut Y, Mohanty S, Yunus EB, Rahmen R, Nosten F, Anstey NM, Day NP, White NJ. The relationship between age and the manifestations of and mortality associated with severe malaria. Clin Infect Dis. 2008;47:151–157. doi: 10.1086/589287. [DOI] [PubMed] [Google Scholar]
- Carlson J, Helmby H, Hill AV, Brewster D, Greenwood BM, Wahlgren M. Human cerebral malaria: association with erythrocyte rosetting and lack of anti-rosetting antibodies. Lancet. 1990;336:1457–1460. doi: 10.1016/0140-6736(90)93174-N. [DOI] [PubMed] [Google Scholar]
- Rowe A, Obeiro J, Newbold CI, Marsh K. Plasmodium falciparum rosetting is associated with malaria severity in Kenya. Infect Immun. 1995;63:2323–2326. doi: 10.1128/iai.63.6.2323-2326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumbo OK, Thera MA, Kone AK, Raza A, Tempest LJ, Lyke KE, Plowe CV, Rowe JA. High levels of Plasmodium falciparum rosetting in all clinical forms of severe malaria in African children. Am J Trop Med Hyg. 2009;81:987–993. doi: 10.4269/ajtmh.2009.09-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn IA, Mackinnon MJ, O'Donnell A, Allen SJ, Moulds JM, Baisor M, Bockarie M, Reeder JC, Rowe JA. A human complement receptor 1 polymorphism that reduces Plasmodium falciparum rosetting confers protection against severe malaria. Proc Natl Acad Sci USA. 2004;101:272–277. doi: 10.1073/pnas.0305306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JA, Handel IG, Thera MA, Deans AM, Lyke KE, Koné A, Diallo DA, Raza A, Kai O, Marsh K, Plowe CV, Doumbo OK, Moulds JM. Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc Natl Acad Sci USA. 2007;104:17471–17476. doi: 10.1073/pnas.0705390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tembo D, Montgomery J. Var gene expression and human Plasmodium pathogenesis. Future Microbiol. 2010;5:801–815. doi: 10.2217/fmb.10.33. [DOI] [PubMed] [Google Scholar]
- Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malar J. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, Thera MA, Kone AK, Doumbo OK, Plowe CV, Rowe JA. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol. 2006;150:211–218. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BA, Welch TL, Smith JD. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Mol Microbiol. 2003;47:1265–1278. doi: 10.1046/j.1365-2958.2003.03378.x. [DOI] [PubMed] [Google Scholar]
- Warimwe GM, Keane TM, Fegan G, Musyoki JN, Newton CR, Pain A, Berriman M, Marsh K, Bull PC. Plasmodium falciparum var gene expression is modified by host immunity. Proc Natl Acad Sci USA. 2009. [DOI] [PMC free article] [PubMed]
- Kaestli M, Cockburn IA, Cortes A, Baea K, Rowe JA, Beck HP. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J Infect Dis. 2006;193:1567–1574. doi: 10.1086/503776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, Chiucchiuini A, Salanti A, Vestergaard LS, Lusingu JP, Hermsen R, Sauerwein R, Christensen J, Nielsen MA, Hviid L, Sutherland C, Staalsoe T, Theander TG. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med. 2004;199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joergensen L, Bengtsson DC, Bengtsson A, Ronander E, Berger SS, Turner L, Dalgaard MB, Cham GK, Victor ME, Lavstsen T, Theander TG, Arnot DE, Jensen AT. Surface co-expression of two different PfEMP1 antigens on single Plasmodium falciparum-infected erythrocytes facilitates binding to ICAM1 and PECAM1. PLoS Pathog. p. e1001083. [DOI] [PMC free article] [PubMed]
- Templeton TJ. The varieties of gene amplification, diversification and hypervariability in the human malaria parasite, Plasmodium falciparum. Mol Biochem Parasitol. 2009;166:109–116. doi: 10.1016/j.molbiopara.2009.04.003. [DOI] [PubMed] [Google Scholar]
- BROAD Institute. http://www.broadinstitute.org/annotation/genome/plasmodium_falciparum_spp/Downloads.html
- Sanger Institute. ftp://ftp.sanger.ac.uk/pub/pathogens/Plasmodium/falciparum/IT_strain/
- Xiao L, Yang C, Dorovini-Zis K, Tandon NN, Ades EW, Lal AA, Udhayakumar V. Plasmodium falciparum: involvement of additional receptors in the cytoadherence of infected erythrocytes to microvascular endothelial cells. Exp Parasitol. 1996;84:42–55. doi: 10.1006/expr.1996.0088. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Craig AG, Berendt AR, Pinches R, Nash G, Marsh K, Newbold CI. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JA, Moulds JM, Newbold CI, Miller LH. Plasmodium falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature. 1997;388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- Ghumra A, Khunrae P, Ataide R, Raza A, Rogerson SJ, Higgins MK, Rowe JA. Immunisation with recombinant PfEMP1 domains elicits functional rosette-inhibiting and phagocytosis-inducing antibodies to Plasmodium falciparum. PLoS ONE. 2011;6:e16414. doi: 10.1371/journal.pone.0016414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ Jr, Treatman C, Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Multiple alignment using hidden Markov models. Proc Int Conf Intell Syst Mol Biol. 1995;3:114–120. [PubMed] [Google Scholar]
- Hu G, Llinas M, Li J, Preiser PR, Bozdech Z. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinformatics. 2007;8:350. doi: 10.1186/1471-2105-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JA, Rogerson SJ, Raza A, Moulds JM, Kazatchkine MD, Marsh K, Newbold CI, Atkinson JP, Miller LH. Mapping of the region of complement receptor (CR) 1 required for Plasmodium falciparum rosetting and demonstration of the importance of CR1 in rosetting in field isolates. J Immunol. 2000;165:6341–6346. doi: 10.4049/jimmunol.165.11.6341. [DOI] [PubMed] [Google Scholar]
- Corrigan RA, Rowe JA. Strain variation in early innate cytokine induction by Plasmodium falciparum. Parasite Immunol. 2010;32:512–527. doi: 10.1111/j.1365-3024.2010.01225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. doi: 10.2307/3280287. [DOI] [PubMed] [Google Scholar]
- Rowe JA, Scragg IG, Kwiatkowski D, Ferguson DJ, Carucci DJ, Newbold CI. Implications of mycoplasma contamination in Plasmodium falciparum cultures and methods for its detection and eradication. Mol Biochem Parasitol. 1998;92:177–180. doi: 10.1016/S0166-6851(97)00237-5. [DOI] [PubMed] [Google Scholar]
- Handunnetti SM, van Schravendijk MR, Hasler T, Barnwell JW, Greenwalt DE, Howard RJ. Involvement of CD36 on erythrocytes as a rosetting receptor for Plasmodium falciparum-infected erythrocytes. Blood. 1992;80:2097–2104. [PubMed] [Google Scholar]
- Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/S0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- Mok BW, Ribacke U, Winter G, Yip BH, Tan CS, Fernandez V, Chen Q, Nilsson P, Wahlgren M. Comparative transcriptomal analysis of isogenic Plasmodium falciparum clones of distinct antigenic and adhesive phenotypes. Mol Biochem Parasitol. 2007;151:184–192. doi: 10.1016/j.molbiopara.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Zhu J, Joachimiak MP, Cohen FE, Pulliam B, DeRisi JL. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- Cluster software. http://bonsai.hgc.jp/~mdehoon/software/cluster/
- Jalview software. http://jtreeview.sourceforge.net/
- GEO repository number: GSE28990. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=thqzlmmgkawsuro&acc=GSE28990
- Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes--divide and conquer. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer SM, Kyes SA, Aggarwal G, Springer AL, Nelson SO, Christodoulou Z, Smith LM, Wang W, Levin E, Newbold CI, Myler PJ, Smith JD. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavazec C, Sanyal S, Templeton TJ. Hypervariability within the Rifin, Stevor and Pfmc-2TM superfamilies in Plasmodium falciparum. Nucleic Acids Res. 2006;34:6696–6707. doi: 10.1093/nar/gkl942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G, Kawai S, Haeggstrom M, Kaneko O, von Euler A, Kawazu S, Palm D, Fernandez V, Wahlgren M. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J Exp Med. 2005;201:1853–1863. doi: 10.1084/jem.20041392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Albrecht L, Merino EF, Hoffmann EH, Ferreira MU, de Mattos Ferreira RG, Osakabe AL, Dalla Martha RC, Ramharter M, Durham AM, Ferreira JE, Del Portillo HA, Wunderlich G. Extense variant gene family repertoire overlap in Western Amazon Plasmodium falciparum isolates. Mol Biochem Parasitol. 2006;150:157–165. doi: 10.1016/j.molbiopara.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Fowler EV, Chavchich M, Chen N, Peters JM, Kyle DE, Gatton ML, Cheng Q. Physical linkage to drug resistance genes results in conservation of var genes among West Pacific Plasmodium falciparum isolates. J Infect Dis. 2006;194:939–948. doi: 10.1086/506619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes S, Christodoulou Z, Pinches R, Kriek N, Horrocks P, Newbold C. Plasmodium falciparum var gene expression is developmentally controlled at the level of RNA polymerase II-mediated transcription initiation. Mol Microbiol. 2007;63:1237–1247. doi: 10.1111/j.1365-2958.2007.05587.x. [DOI] [PubMed] [Google Scholar]
- Sargeant TJ, Marti M, Caler E, Carlton JM, Simpson K, Speed TP, Cowman AF. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Magistrado PA, Nielsen MA, Theander TG, Lavstsen T. Preferential transcription of conserved rif genes in two phenotypically distinct Plasmodium falciparum parasite lines. Int J Parasitol. 2009;39:655–664. doi: 10.1016/j.ijpara.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Joannin N, Kallberg Y, Wahlgren M, Persson B. RSpred, a set of Hidden Markov Models to detect and classify the RIFIN and STEVOR proteins of Plasmodium falciparum. BMC Genomics. 2011;12:119. doi: 10.1186/1471-2164-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe JE, Niang M, Marsh K, Holder AA, Langhorne J, Preiser PR. Characterization of the repertoire diversity of the Plasmodium falciparum stevor multigene family in laboratory and field isolates. Malar J. 2009;8:140. doi: 10.1186/1475-2875-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanti A, Dahlback M, Turner L, Nielsen MA, Barfod L, Magistrado P, Jensen AT, Lavstsen T, Ofori MF, Marsh K, Hviid L, Theander TG. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JA, Kyes SA, Rogerson SJ, Babiker HA, Raza A. Identification of a conserved Plasmodium falciparum var gene implicated in malaria in pregnancy. J Infect Dis. 2002;185:1207–1211. doi: 10.1086/339684. [DOI] [PubMed] [Google Scholar]
- Kyes SA, Christodoulou Z, Raza A, Horrocks P, Pinches R, Rowe JA, Newbold CI. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol Microbiol. 2003;48:1339–1348. doi: 10.1046/j.1365-2958.2003.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham WH, Payne PD, Brown GV, Rogerson SJ. Identification of basic transcriptional elements required for rif gene transcription. Int J Parasitol. 2007;37:605–615. doi: 10.1016/j.ijpara.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Howitt CA, Wilinski D, Llinas M, Templeton TJ, Dzikowski R, Deitsch KW. Clonally variant gene families in Plasmodium falciparum share a common activation factor. Mol Microbiol. 2009;73:1171–1185. doi: 10.1111/j.1365-2958.2009.06846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rif gene sequences from HB3, Dd2 and IT.
Stevor gene sequences from HB3, Dd2 and IT.
Pfmc-2tm gene sequences from HB3, Dd2 and IT.
Rif and stevor gene names and supercontig location.
All VSA oligos added to the 3D7 microarray chip.
Non-VSA genes differentially expressed in rosetting versus non-rosetting parasites.