Summary
In parallel with evolutionary developments, the Hsp90 molecular chaperone system shifted from a simple prokaryotic factor into an expansive network that includes a variety of cochaperones. We have taken high-throughput genomic and proteomic approaches to better understand the abundant yeast p23 cochaperone Sba1. Our work revealed an unexpected p23 network that displayed considerable independence from known Hsp90 clients. Additionally, our data uncovered a broad nuclear role for p23, contrasting with the historical dogma of restricted cytosolic activities for molecular chaperones. Validation studies demonstrated that yeast p23 was required for proper Golgi function, ribosome biogenesis and was necessary for efficient DNA repair from a wide range of mutagens. Notably, mammalian p23 had conserved roles in these pathways as well as being necessary for proper cell mobility. Taken together, our work demonstrates that the p23 chaperone serves a broad physiological network and functions both in conjunction with and sovereign to Hsp90.
Introduction
Molecular chaperones generally display a promiscuous protein binding capacity enabled by an affinity for short hydrophobic amino acid motifs that likely accounts for the shared ability to suppress non-native protein aggregation in vitro (Hendrick and Hartl, 1993). To offset the broad binding, chaperones typically have short-lived, low affinity interactions to avoid interfering with the functional activity of a client protein. These characteristics fit well with the adopted molecular chaperone definition—a protein that has a functional effect on another protein/protein complex without becoming part of the final operative structure (Ellis, 1987). While these evolved characteristics are likely pivotal to the cellular operations of a chaperone, these properties have hindered the identification of client proteins and impeded our ability to gain an overall understanding of the physiological networks that are served by the various molecular chaperones.
Based upon high cellular abundances and wide-ranging influences on diverse pathways Hsp70 and Hsp90 and their associated cochaperones constitute the central eukaryotic molecular chaperone system (Wegele et al., 2004). While Hsp70 has been extensively studied (Mayer et al., 2009), the general in vivo roles of Hsp90 are just being revealed. Although the Hsp70 system is one of the most conserved, the Hsp90 machine is a recently evolved chaperone system. Escherichia coli contains a single Hsp90 homolog, HtpG, but does not express any apparent cochaperones that are common to eukaryotes. Nevertheless, Hsp90 has evolved into an essential eukaryotic protein, as have many of its cochaperones including Cdc37 and mammalian p23 (Johnson and Brown, 2009). Hence, Hsp90 and its associated cofactors form a chaperone system that has been adopted to support the unique challenges of higher order organisms.
Hsp90 was originally identified in stable association with signaling proteins (i.e., kinases and steroid receptors) and it has been argued that its primary function is to maintain metastable factors in activatable states (Pratt and Toft, 2003). However, signaling protein maintenance likely does not account for all duties of Hsp90. Recent studies indicate that Hsp90 serves in a broad range of cellular processes including protein transport, epigenetic status and cell cycle progression (Zhao et al., 2005; Millson et al., 2005; McClellan et al., 2007). Thus, Hsp90 has a central role in cell homeostasis, which is exemplified by its use as a therapeutic target for a variety of cancers and other diseases (Whitesell and Lindquist, 2005). Given the breadth of pathways affected by Hsp90, an efficient means to control and guide this chaperone is needed to insure proper function and avoid detrimental pleiotropic effects that might occur if this abundant chaperone was left unchecked.
To control Hsp90 numerous cochaperones including p23, Hop, Cdc37, Sgt1 and Aha1 have coevolved with the eukaryotic Hsp90s (Johnson and Brown, 2009). In a manner paralleling Hsp70 cochaperones, the Hsp90 cohorts can modulate Hsp90s' ATP hydrolysis activity and potentially direct substrate specificity. A number of biochemical and structural studies have dissected the ATPase cycle of Hsp90 along with the potential influence of cochaperones (Hessling et al., 2009; Wandinger et al., 2009). In addition, a mechanism by which a cochaperone might guide Hsp90 to select clients has been revealed, as Cdc37 forms a tripartite complex having contacts to both Hsp90 and the substrate protein (Vaughan et al., 2006). Besides Hsp90-dependent events, cochaperones might also regulate client proteins autonomously using their innate chaperone activities (Freeman et al., 1996). In general, however, independent cochaperone functions have been difficult to conclude since the known client proteins are shared with Hsp90.
We have investigated the physiological pathways served by the yeast p23 cochaperone Sba1 using high-throughput genomic and proteomic approaches. We focused on Sba1 since this cochaperone is broadly expressed in most eukaryotes, is an abundant cochaperone, forms co-complexes with Hsp90 alone and with client proteins and might also work autonomously. To comprehend the Sba1 interaction network and its relation to the yeast Hsp90 molecular chaperone Hsp82, we have incorporated in-depth primary and comparative bioinformatic analyses. In addition, we have assessed the Sba1-dependence of potential target pathways using various experimental techniques and we have determined whether the reliance is conserved in higher eukaryotic cells. Taken together, the presented studies establish a broad Sba1/p23 cellular network that includes a sizeable nuclear subnet.
Results
Sba1 genetic interaction network
We initiated our investigation on the Sba1 cellular network using Synthetic Genetic Array (SGA) analysis, which is a high-throughput procedure for identifying synthetic sick and lethal genetic interactions (SSL) (Tong et al., 2001). In our SGA screen sba1Δ yeast were systematically crossed with a deletion mutant array (4786 strains) comprised of knockout yeast for each non-essential open reading frame (ORF) in order to reveal parallel or compensatory pathways affected by Sba1. Double mutants were visually screened and scored as lethal, very sick or sick based upon colony size relative to control yeast that were processed in parallel. Of the 4786 strains assessed we found 234 ORFs that displayed a SSL phenotype with sba1Δ (Table S1). Of the 6 previously identified genes to have a synthetic genetic interaction with sba1Δ 2 genes, ACT1 and ELF1, were within our data set (Collins et al., 2007; Haarer et al., 2007). To better understand how Sba1 might intersect with the identified ORFs we performed a bioinformatic analysis of the data.
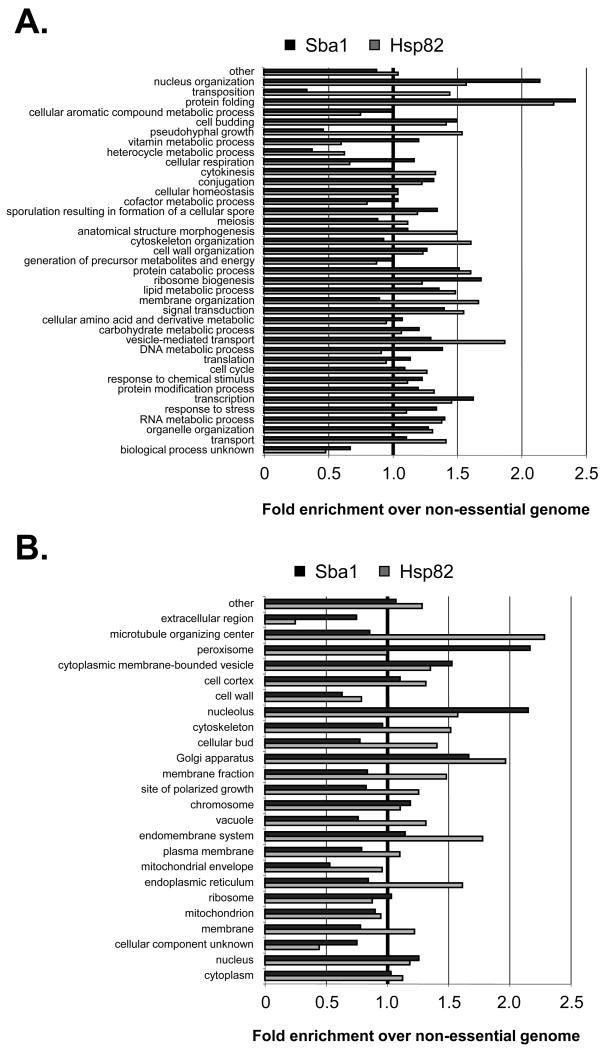
The SSL hits were partitioned into a variety of cellular process using a GO Slim analysis. For illustrative purposes, we displayed each hit in a single GO Slim category (Figure 1A). However, in determining whether the ORFs were favored within certain cellular processes relative to all non-essential genes we considered all potential functions of each ORF (Figure 1B). We found that the Sba1 interactors were enriched in signal transduction, transcription, protein catabolism, cell budding, DNA metabolism, cellular respiration and vesicle-mediated transport (Figure 1B).
Figure 1.
SGA analysis revealed a broad genetic interaction network for Sba1 that included factors involved in vesicle-mediated protein transport. The genes that produced an SSL phenotype with sba1Δ were categorized by a GO Slim analysis. (A) Each hit was assigned to an initial cellular process and displayed or (B) all potential processes were considered for each gene and the enrichments in each category were determined relative to the distribution of all non-essential yeast ORFs. (C) To test if Golgi function was Sba1-dependent parental (WT) and sba1Δ transformants carrying either an empty or an Sba1-expression vector (+ Sba1) were treated with the Golgi transport inhibitor Brefeldin A (BFA) (100 μg mL-1). (D) To check if mammalian Golgi function is p23-dependent, exponentially growing parental or p23 null MEFs were cultured in unsupplemented or BFA-supplemented (50 μg mL-1) media. Error bars represent the standard error of the mean.
To better understand how the SGA hits might connect with Sba1 we tested 59 targets for direct Sba1 interactions using the yeast 2-hybrid assay and observed 11 positive results (Table S1). We had not anticipated this relatively high level (∼20%) of contact between Sba1 and the SGA hits. For comparison, an SGA and 2-hybrid investigation of Hsp82 targets yielded <1% overlap (Zhao et al., 2005). While it is plausible that the positive correlation between our SGA and 2-hybrid results is skewed since we did not test all of the hits by 2-hybrid, we suspect that our results are representative of the entire set since the tested SGA interactors were selected randomly. Hence, Sba1 likely utilizes more direct contacts to influence cellular pathways relative to the mechanisms used by Hsp82.
Though the majority of the identified SSL hits had not been previously revealed, the favored cell processes fit within the known regulatory network for the Hsp90 molecular chaperone system and suggest that our screen identified valid Sba1 interactors. For instance, the signal transduction and protein catabolism categories were anticipated given the historical roles of Hsp90 and more recent studies support an Sba1 involvement in transcription and DNA metabolic events (DeZwaan and Freeman, 2008). The enrichment in the vesicle-mediated transport process and predicted enhanced Sba1 localization to cytoplasmic membrane bound vesicles and the Golgi apparatus (Figure S1) agree with a prior high-throughput screen in which an Hsp82 role in protein transport was revealed (McClellan et al., 2007). In general, the SSL interactors are within cellular processes expected for Hsp90.
Sba1 and p23 affect Golgi organelle function
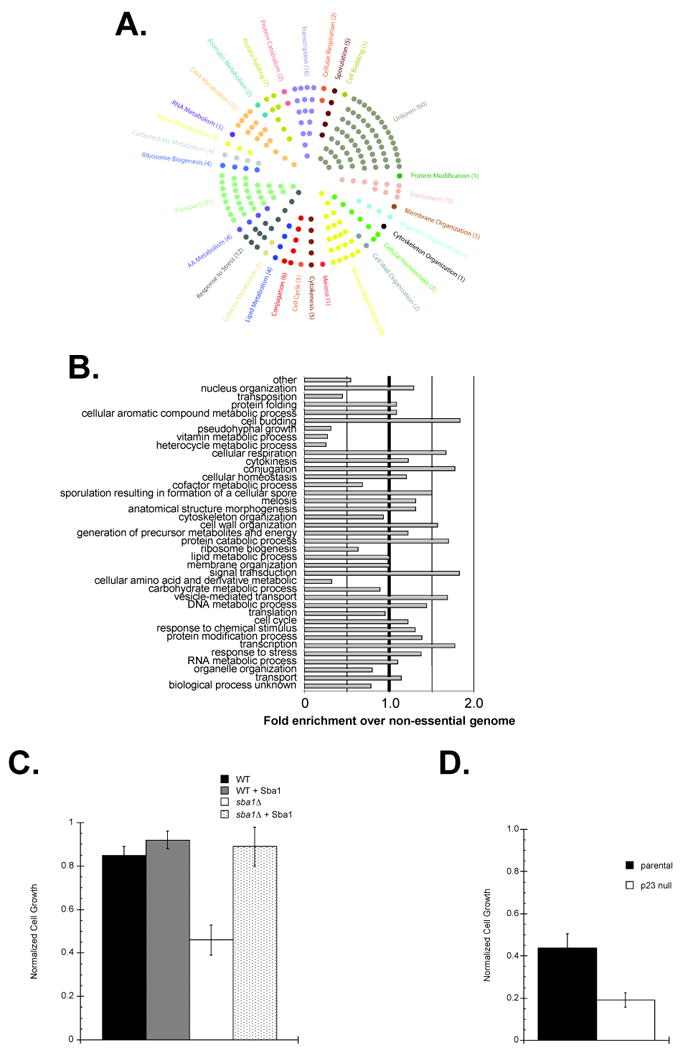
To begin to understand how Sba1 might contribute to vesicle-mediated protein transport, we created an in silico map of the relative locale of the identified Sba1 and curated Hsp82 hits along the Endoplasmic Reticulum (ER) and Golgi organelles (Figure S2). While the Hsp82 targets were evenly distributed between the ER and Golgi, the Sba1 interactors were typically downstream of the ER lumen. To assess whether a p23 protein can influence transport events beyond the ER, we took advantage of the small-molecule inhibitor Brefeldin A (BFA). BFA is a heterocyclic lactone that blocks protein trafficking through disassembly of the Golgi apparatus and trans-Golgi network. Although normal yeast are relatively resistant to BFA (Shah and Klausner, 1993), we found that the sba1Δ cells were sensitive to BFA treatment and that ectopic expression of Sba1 restored BFA resistance (Figure 1C). Notably, we also observed that mouse embryonic fibroblast (MEF) cells derived from a p23 null mouse were more sensitive to BFA relative to parental MEFs (Figure 1D). Since BFA blocks recruitment of coat proteins that are responsible for the formation of transport vesicles in the Golgi apparatus, we suggest that Sba1 and p23 actively participate in membrane-vesicle protein transport.
To further support a role for mouse p23 in Golgi function we examined the cellular localization of p23 in parental MEFs. Overall, p23 was found throughout the cell with enrichment at the Golgi as determined by colocalization with the established Golgi marker Giantin (Figure 2A); a computational assessment of the fluorescent signals indicated a ∼98% overlap. In contrast, the ER protein Calnexin IFF-signal demonstrated a ∼50% coincidence with p23's (Figure S2D). Thus, p23 appears to preferentially localize to the Golgi organelle.
Figure 2.
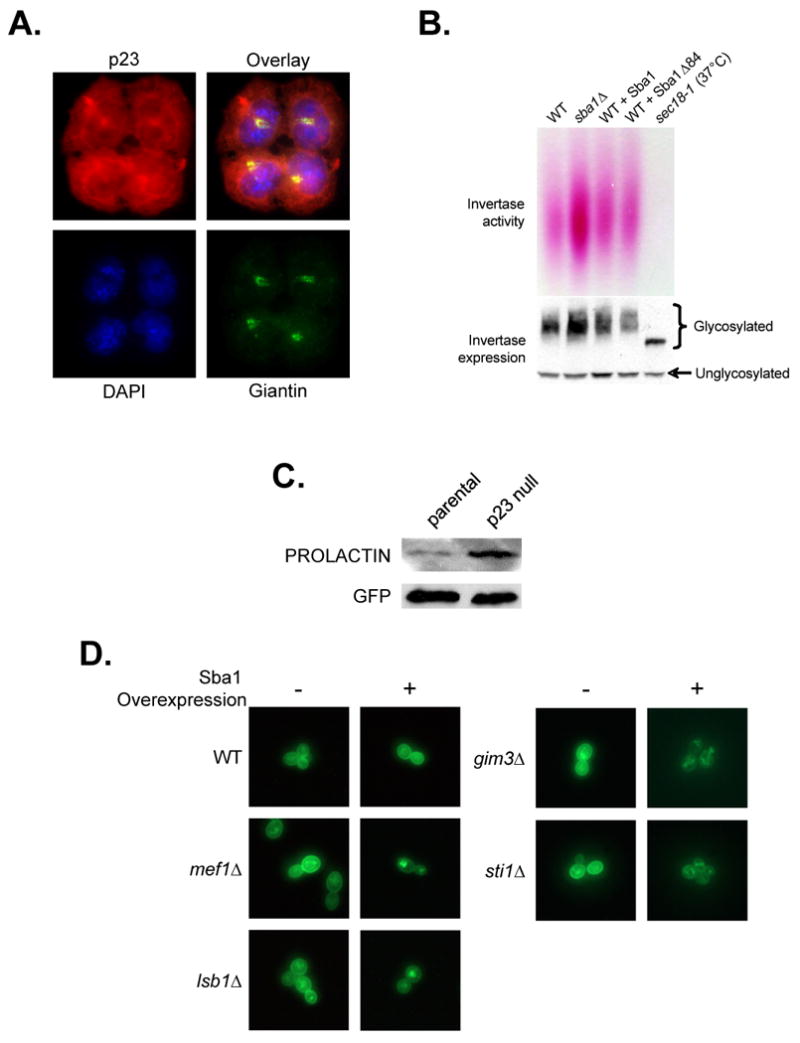
The yeast and mouse p23 proteins are negative modulators of protein transport. (A) Colocalization of mouse p23 (red) with the Golgi maker protein Giantin (green) was determined by IIF. The cells were counterstained with DAPI to detect the nuclei. (B) The impact of Sba1 levels on yeast Invertase secretion was determined. The influence of Sba1 loss (sba1Δ), Sba1 overexpression or Sba1Δ84 (chaperone mutant) expression was checked as well as the secretion defective mutant sec18-1. The levels of intracellular Myc-Invertase were detected by immunoblot analysis. (C) The efficiency of Preprolactin processing to Prolactin during transport in parental and p23 null MEF cells was determined. (D) The effect of Sba1 overexpression on Fus-Mid-GFP transport was examined by fluorescence microscopy in WT and secretion compromised yeast.
We used the classic yeast cargo-protein Invertase to assess the functional influence of Sba1 on the transport process (Poon et al., 1999). In sba1Δ cells, Invertase was more efficiently secreted suggesting that Sba1 is a negative regulator of protein transport (Figure 2B). In general, Sba1 overexpression had no apparent ill-effect indicating that Sba1 is not a limiting negative effector of Invertase secretion in wild type yeast dependent (+ Sba1) or independent (+ Sba1Δ84) of Sba1's innate chaperone activity. To test whether p23 also functions in mammalian protein transport we exploited the presecretory-protein Preprolactin, which is converted to Prolactin in the secretory pathway (Lakkaraju et al., 2008). We observed more efficient transport-dependent processing of Preprolactin to Prolactin in p23 null MEFS compared to parental (Figure 2C). Taken together, our data support the contention that Sba1/p23 serves as a negative protein transport regulator.
To identify the effected pathway step(s), we exploited the transport marker Fus-Mid-GFP. Fus-Mid-GFP is a GFP-tagged chimera of the Fus1 and Mid2 proteins that has been used to systematically screen for sorting defects within the secretory pathway, as Fus-Mid-GFP marks the transport pathway from the ER to cell surface including missorting to vacuoles (Proszynski et al., 2005). Of the 31 Sba1 genetic interactors known to have a role in protein secretion (Figure 1A), 13 were previously shown to be defective for Fus-Mid-GFP transport (Proszynski et al., 2005). Upon overexpression of Sba1, we observed phenotypic enhancement (PE) of the transport defects in 4 of the 13 single-gene knockout strains since the cell surface Fus-Mid-GFP signal was markedly decreased (Table S3; Figure 2D). Two of the hits occurred with strains containing the chaperone-related gene knockouts sti1Δ and gim3Δ. While a connection to Sti1 might be anticipated since it is a fellow Hsp90 cochaperone, Gim3 is a Prefoldin subunit and is generally considered a Chaperonin (GroEL) cofactor (Siegers et al., 1999). Perhaps during the protein transport process, Sba1 links the Hsp90 and Chaperonin systems. The other two PE hits were with mef1Δ and lsb1Δ, which encode a translation elongation factor and an Actin polymerizing protein, respectively. In addition to the PE hits, we found that Sba1 overexpression complemented the transport defects associated with two single-gene knockouts, mnn10Δ and mnn11Δ (Table S3; Figure S3A).
The Mnn proteins are subunits of Golgi mannosyltransferases that synthesize poly-mannose modifications with primarily α-1,6 linkages (Jungmann and Munro, 1998). The yeast mannosyltransferase complexes are distinguished by Van1- or Anp1-containing structures and Mnn10 and Mnn11 are responsible for the majority of the α-1,6 polymerizing activity of the Anp1-complex. Deletion of either MNN10 or MNN11 results in protein hyper-secretion (Bartkeviciūte and Sasnauskas, 2004). Upon Sba1 overexpression in mnn10Δ or mnn11Δ yeast we observed a reduction in cell surface Fus-Mid-GFP with a corresponding increase in the intracellular signal, which is comparable to the normal pattern (Figure S3A). Of note, the Sba1 complementation effect was particular to Anp1-mannosyltransferase since Fus-Mid-GFP transport in mnn9Δ yeast was unaffected (Figure S3A). Mnn9 is a component of both Van1- and Anp1-structures and mnn9Δ results in slower protein secretion rates (Jungmann and Munro, 1998). Thus, Sba1 affects the transport process by influencing protein Mannosylation, which might account for the elevated level of glycosylated Invertase in the sba1Δ yeast (Figure 2B). To determine if the relationship between a p23 protein and mannose glycosylation is conserved and functional, we checked the relative levels of α-1,6 mannose modified proteins in parental and p23 null MEFs. Using a biotinylated-lectin that selectively recognizes α-1,6 mannose (Vector Labs, Inc) we found that p23 null cells contain higher amounts of α-1,6 mannose modified proteins (Figure S3B).
Our protein transport investigation demonstrated that Sba1/p23 primarily functions at or near the Golgi as a negative regulator of vesicle-mediated protein transport events. Minimally, the influence involves modulation of protein α-1,6 mannose polymerization since both genetic and functional affects were apparent that correlate with secretion defficiencies. In conjunction with prior work (McClellan et al., 2007), a conserved role for the Hsp90 chaperone system in vesicle-mediated transport is supported. Notably, however, the impact of Sba1 and Hsp82 on secretion efficiency contrasts since Hsp82 positively supports the process whereas Sba1 suppresses it. While several plausible models might account for this difference, we suggest that Sba1 and Hsp82 function independently on separate clients to differentially modulate protein secretion.
Cell motility is p23-dependent
Besides the Golgi localization, the p23 cellular IIF-pattern was consistent with a cytoskeleton association (Figure 2A). Given the connection to the Actin polymerizing factor Lsb1 and, more notably, that act1Δ has an SSL phenotype with sba1Δ and Actin scored positive for a physical Sba1 interaction by 2-hybrid (Table S1), we further explored the potential p23 relationship with the cytoskeleton. Unfortunately, in vitro experiments testing the impact of purified Sba1/p23 on Actin polymerization failed to reveal any significant effects besides confirming an ability to physically interact (Echtenkamp, Freeman and Brieher, unpublished). However, experiments using a standard cell-wounding assay revealed a declined p23 null MEF motility (Figure 3A and 3B). Correlating with the decreased mobility was a reduced (∼25% of parental) Vinculin signal as detected by IIF (Figure 3C). Vinculin is a subunit of focal adhesions, which link the outer cell membrane with the actin-cytoskeleton and are necessary for cell movements (Ziegler et al., 2006). Immunoblot analysis showed that Vinculin was selectively destabilized in p23 null cells, as the focal adhesion proteins α-Actinin and Talin appear unaffected (Figure 3C; Figure S4). Given the prominent reduction in the Vinculin IIF signal, we suggest that the declined p23 null MEF cell mobility results from both a Vinculin destabilization and a reduced Vinculin incorporation into focal adhesions.
Figure 3.

Cell motility is p23-dependent. (A) The abilities of parental and p23 null MEFs to migrate into an artificial wound was determined and (B) the results were quantified. Error bars represent the standard error of the mean. (C) The relative levels of the focal adhesion proteins Vinculin and α-Actinin were determined in parental and p23 null cells by IIF.
Although cell motility also has been shown to be Hsp90-dependent, the mechanism occurs through an extracellular, cell surface Hsp90 function (Tsutsumi et al., 2008). Together, the work suggests that p23 and Hsp90 can converge on a cell process by targeting distinct client proteins. Importantly, the p23 role in Vinculin processing and on cell motility likely accounts for the impact of p23 in cancer cell invasion events (Simpson et al., 2010).
Sba1 physical interaction network
To expand our understanding of the cellular factors directly contacted by Sba1, which would include client and partner proteins, we exploited high-density protein microarrays (Yeast ProtoArray Invitrogen Inc.). A ProtoArray contains 4,088 S. cerevisiae ORFs expressed as 5′-GST fusions that have been purified and spotted in duplicate on nitrocellulose-coated slides. Associations with the GST fusion proteins were detected using biotin-labeled Sba1 coated with streptavidin-conjugated Alexa Fluor 647. Of note, the biotinylated-Sba1 used to screen the arrays displayed comparable in vitro chaperone activity as unlabeled Sba1 (data not shown). Of the 4,088 proteins, we found that Sba1 reproducibly interacted with 80 of the spotted fusion proteins (Table S2). We further explored these hits using an independent interaction assay and bioinformatic analysis.
We substantiated 70 of the potential interactions using the yeast 2-hybrid assay and found that 59 of the 70 tested positive (Table S2). Given the relatively high validation rate of ∼84%, we evaluated the entire ProtoArray data set to obtain the broadest possible view of the Sba1 interactors. A GO Slim analysis revealed that the hits fall into a wide spectrum of cellular process (Figure 4A) and a comparison to all annotated yeast proteins showed an Sba1 target enrichment in nuclear organization, ribosome biogenesis, RNA metabolism, vitamin and cofactor metabolism (Figure 4B).
Figure 4.
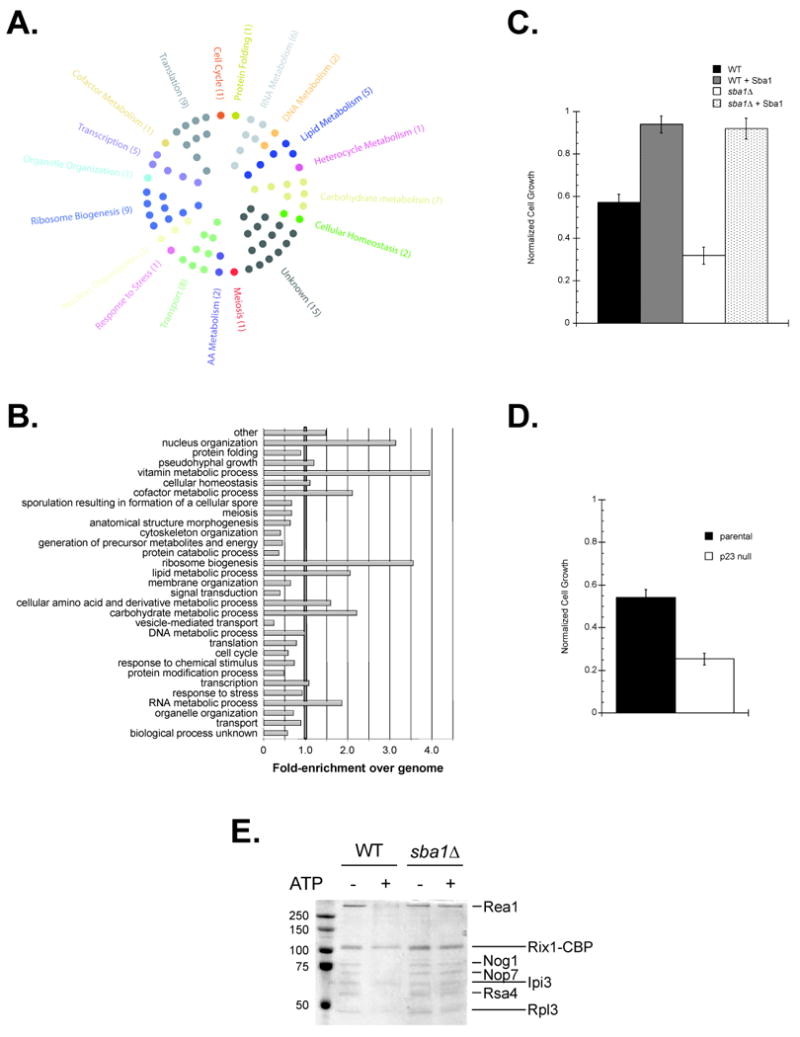
Sba1 physically interacts with proteins functioning in a wide variety of cellular process including ribosome biogenesis. Yeast ProtoArrays were used to detect proteins bound by Sba1. (A) Each hit was assigned to an initial cellular process using a GO Slim analysis and displayed or (B) all potential processes were considered for each gene and the enrichments in each category were determined relative to the distribution for all yeast ORFs. (C) The sensitivity of yeast to the ribosome inhibitor Hygromycin B (300 μg mL-1) fluctuates with Sba1 levels. (D) The Hygromycin B (100 μg mL-1) sensitivity of exponentially growing parental and p23 null MEFs was determined. Error bars represent the standard error of the mean. (E) Sba1 facilitates ATP-dependent release of ribosome biogenesis factors from Rix1-associated, nascent 60S particles. TAP purifications were performed using extracts prepared from wild type and sba1Δ yeast expressing Rix1-TAP, during the Calmodulin affinity step ATP (2 mM) was included, as indicated, and the EGTA eluates were analyzed by SDS-PAGE and Coomassie staining. The maturation factors known to dissociate in an ATP-dependent manner are demarked (short lines) along with the ATP-insensitive components (long lines) (Ulbrich et al., 2009).
An Osprey bioinformatic analysis of the ProtoArray interactors highlighted a possible Sba1 role in ribosome biogenesis (Figure S5A). Supporting this potential, we found that the sba1Δ cells were more sensitive to Hygromycin B, the vulnerability was alleviated upon plasmid-borne expression of Sba1 in the sba1Δ yeast and resistance was enhanced in the parental cells upon Sba1 overexpression (Figure 4D). As Hygromycin B induces misreading of aminoacyl-tRNA by distorting the ribosomal A site, the fluctuating Hygromycin B sensitivities with Sba1 levels supports an Sba1 role in yeast ribosome biogenesis.
Significantly, p23 null MEFs were more vulnerable to Hygromycin B relative to parental cells (Figure 4D). Unexpectedly, p23 localization was not enhanced at nucleoli but rather it was disfavored, as determined by IIF (Figure S5B). While it is possible that a low level of p23 shuttles into the nucleoli, we suspect that the conserved Hygromycin B sensitivity results from a declined efficiency in a p23/Sba1-dependent post-nucleolar step of the ribosome pathway.
Within the ribosome-linked hits were several factors, including Rsa4, that promote the biogenesis of the 60S particle as it transitions away from the nucleolus (Figure S5A). Rsa4 mediates the ATP-dependent release of ribosomal maturation factors from Rix1-associated pre-60S ribosomal complexes (Ulbrich et al., 2009). We tested whether Sba1 influenced this process and found that the ATP-triggered release of Rea1, Nog1, Nop7 and Rsa4 was inefficient when the Rix1-complexes were isolated from sba1Δ yeast (Figure 4E). Hence, Sba1 minimally facilitates ribosome biogenesis by enhancing the Rsa4-mediated, ATP-dependent release of maturation factors from pre-60S complexes.
Composite Sba1 interaction network
To gain a better appreciation of the cellular pathways affected by Sba1 we compiled the SGA (234) and Protoarray (80) data sets along with all the curated (34) Sba1-associated genes and performed a bioinformatic analysis on the unified Sba1 interactor catalog (348 unique total hits) (Figure 5A). In general, the composite Sba1 net does not show significant enrichments in any particular category other than nuclear organization and protein folding (Figure 5B). Rather, we suspect, Sba1 acts like a typical molecular chaperone and serves a broad range of protein clientele. Still, several processes were mildly favored (>1.25-fold enrichment) including ribosome biogenesis, transcription, DNA metabolism, and RNA metabolism (Figure 5A and 5B). Given the general cellular locale of these functions, Sba1 has an apparent bias for nuclear processes. In particular, we found that many of the potential Sba1 targets function at or near DNA in a number of distinct pathways (Figure 5C). Since prior studies have shown roles for Sba1 in transcription and telomere maintenance pathways (DeZwaan and Freeman, 2008), we wanted to explore the functional relevance of Sba1 on a nuclear activity revealed in our screens. Thus, we tested the impact of Sba1 on yeast DNA repair function.
Figure 5.
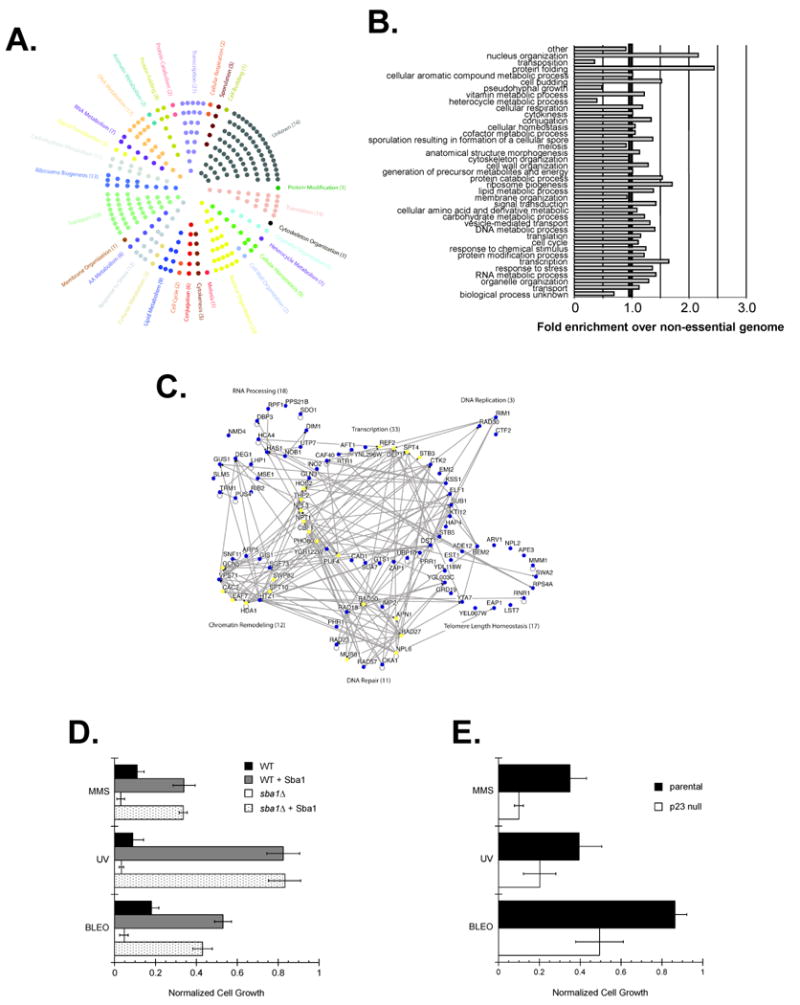
The Sba1 network has a significant nuclear component that includes DNA repair activities. The SGA, ProtoArray and all curated Sba1 interactors were pooled and analyzed using GO Slim. (A) Each hit was assigned to a cellular process and displayed or (B) all potential processes were considered for each gene and the enrichments in each category were determined relative to the distribution for all non-essential yeast ORFs. (C) The nuclear-associated Sba1 interactors were analyzed using Osprey. Genes marked in yellow are associated with multiple activities but for clarity each hit was placed only once in the presented network. (D) Wild type (WT) or sba1Δ yeast transformants carrying either an empty vector or SBA1 expression vector (+ Sba1) were grown in the presence of the DNA mutagens MMS (300 μg mL-1) or BLEO (20 mU). Alternatively, the transformants were exposed to UV light (200 J/m2) and then cultured in selective glucose media. (E) The sensitivities of parental or p23 null MEFs to DNA mutagens was tested by exposing the cells either to MMS (500 μg mL-1), BLEO (200 mU) or UV light (7.5 J/m2). Error bars represent the standard error of the mean.
Sba1 is a determinant in DNA repair processes
To investigate whether Sba1 has a general role in DNA repair pathways we determined the susceptibility of parental and sba1Δ yeast to Methyl methanesulfonate (MMS), bleomycin (BLEO) and ultraviolet light (UV). MMS causes methylated DNA bases that are repaired by the base excision repair (BER) pathway, bleomycin treatment creates single- and double-stranded breaks that are fixed by the recombination and post-replication DNA repair pathways and UV light induces pyrimidine dimers that are corrected by the nucleotide excision repair (NER) pathway (Nagy and Soutoglou, 2009). We found that the sba1Δ yeast were more susceptible to standard MMS, BLEO and UV treatments used to assess vulnerabilities to DNA mutagens (Figure 5D). The increased sensitivities of the sba1Δ yeast were suppressed by Sba1 expression and Sba1 overexpression in the parental yeast led to an enhanced resistance to all three mutagens (Figure 5D). Importantly, we found that the role of p23 cochaperones in DNA repair pathways was conserved as p23 null MEFs were more sensitive to MMS, BLEO and UV treatments relative to the parental cells (Figure 5E). Overall, our data show that the influence of p23 proteins in DNA repair pathways is conserved.
Relationships of the Sba1 and Hsp82 interactors
Cochaperones are typically considered obligate accessories proteins for the central molecular chaperone of a given system and the perceived relationship between Sba1 and Hsp82 is no exception (Wegele et al., 2004). To better understand the connection between Sba1 and Hsp82 we compared the composite networks for Sba1 and Hsp82—the Hsp82 net was formulated using a GO Slim analysis and all curated Hsp82-interactors (Figure 6). The Sba1 and Hsp82 subnets contained 348 and 1178 interactors, respectively. Unexpectedly, only 86 hits are common between the data sets, which corresponds to ∼25% of the Sba1 interactors (Figure 7A). While we had suspected that Sba1 might function independently, we had anticipated a greater overlap. Intriguingly, despite the relatively low coincidence with discrete ORF targets, Sba1 and Hsp82 hits are associated with similar cellular processes and have comparable cellular localization patterns based upon a GO Slim analysis (Figure 6).
Figure 6.
Sba1 and Hsp82 associate with factors functioning in common cellular processes and locales. The composite Sba1 catalog and all curated Hsp82 interactors were categorized by a GO Slim analysis and the relative enrichments in either cellular processes (A) or compartments (B) were determined relative to the distribution of all non-essential yeast ORFs.
Figure 7.
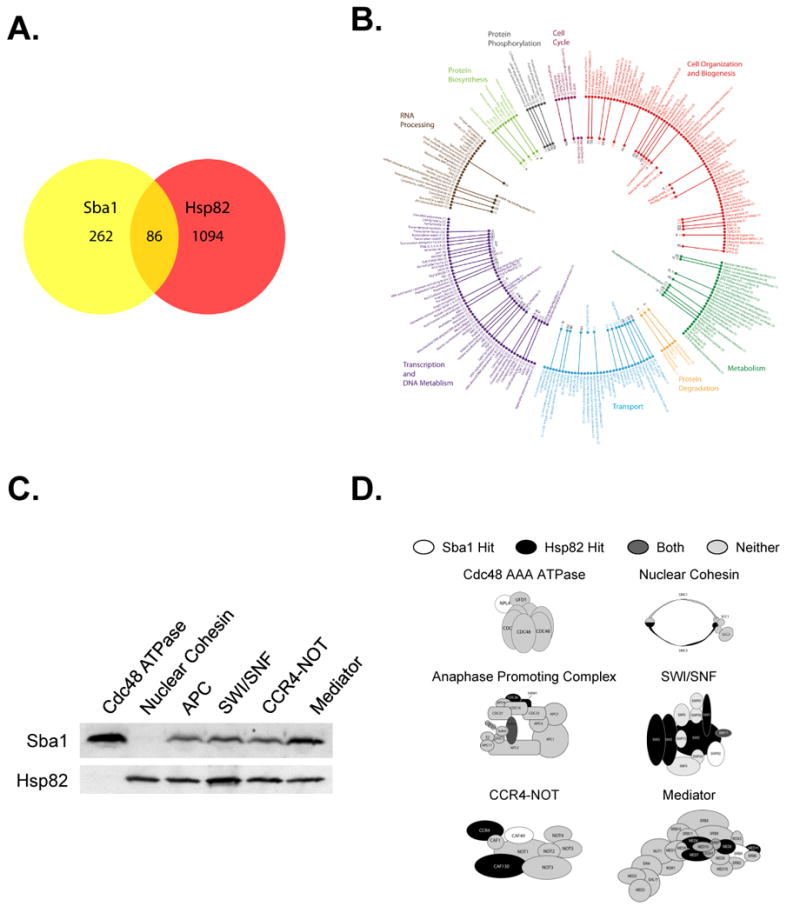
The Sba1 and Hsp82 networks share few mutual direct hits yet are connected to common cellular processes. (A) The relative overlap in discrete ORFs between the Sba1 and Hsp82 interactors was established and displayed in a Venn diagram.(B) The stable protein complexes that have Sba1 and Hsp82 interactors were determined. The inner circle represents complexes with Sba1 hits, the outer circle has Hsp82-linked structures and the lines connect complexes that are associated with both chaperones. (C) The ability of Sba1 and Hsp82 to interact with different protein complexes was determined using the TAP-tag pull-down assay. The indicated protein complexes were precipitated using a TAP-fusion subunit that was not predicted to be a chaperone target. Sba1 and Hsp82 association with the various complexes was detected by immunoblot analysis. (D) Organizational models for chaperone-associated complexes are shown. The subunits predicted to interact with Sba1 (white), Hsp82 (Black), both (dark grey) or neither (light grey) are marked.
Though the overall patterns were quite analogous, several variances were apparent including enhanced Sba1 involvement in nuclear organization, ribosome biogenesis and DNA metabolism whereas Hsp82 hits were increased in protein transport, membrane organization and cytoskeleton organization (Figure 6A). Several processes showed mild increases with one chaperone and a decline for the other including vitamin metabolism and cell respiration where Sba1 was favored and Hsp82 was disfavored and pseudohyphal growth and transposition were enriched for Hsp82 and reduced for Sba1 interactors. In contrast to the apparent processes that differentially rely on Sba1 and Hsp82, classic molecular chaperone processes including protein folding, protein catabolism and signal transduction were comparably enriched. Given the relatively low overlap in actual ORF targets, how are Sba1 and Hsp82 effectively serving many of the same cellular processes?
Typically, cellular processes function through protein assemblies rather than individual proteins. Thus, we investigated whether Sba1 and Hsp82 might associate with common protein targets by interacting with different subunits of the same complex. A GO Slim analysis revealed 86 stable protein complexes with shared Sba1/Hsp82 interactors (Figure 7B). Of the 86, 52 were with the same protein and 34 were through distinct subunits. Using the TAP-tag pull-down assay we tested whether several of the putative target complexes did indeed associate with Sba1 and/or Hsp82.
We isolated various protein assemblies using TAP-tagged subunits that were not predicted to be directly associated with either Sba1 or Hsp82. Therefore, if chaperone interactions are detected, the binding is likely occurring with the amassed structure rather than with an expected target. As initial controls, we examined the Cdc48 AAA ATPase and nuclear Cohesin complexes, which were predicted to interact with just Sba1 or Hsp82, respectively, but not both. Importantly, only Sba1 was isolated with the Cdc48 AAA ATPase complex and only Hsp82 was associated with the nuclear Cohesin structure (Figure 7C and 7D). Hence, Sba1 and Hsp82 can form autonomous interactions with protein complexes. Next, we tested the chaperone binding to structures that have an anticipated shared protein subunit, the anaphase-promoting complex and the SWI/SNF structure, and found that both Sba1 and Hsp82 were bound (Figure 7C and 7D). Finally, we examined Sba1 and Hsp82 associations with protein structures that were predicted to contain different clients for Sba1 and Hsp82—the CCR4-NOT and Mediator complexes. Significantly, both chaperones were associated, which supports the contention that Sba1 and Hsp82 can interact with a common protein complex through separate protein subunits.
With the addition of the common complexes, the shared space for the Sba1 and Hsp82-networks is comprised of 120 total interactions (i.e., 86 common ORFs and 34 mutual complexes). Thus, ∼31% of the Sba1 net involves Hsp82 while ∼69% appears to be independent Sba1 targets. Since Sba1 and Hsp82 had not previously been shown to affect protein complexes through distinct subunits, we wished to rationalize how this might occur. Given the involvement of molecular chaperones in protein complex assembly/disassembly (Ellis, 1987), we reasoned that the chaperone-associated subunits might be juxtaposed, which would permit cooperative chaperone actions in amassing and/or dissociating the common structures. Of the 86 identified shared complexes, subunit-organizational data was available for 10. An examination of these structures revealed that 7 of the 10 had neighboring chaperone-associated proteins (Figure 7D; data not shown). While this is a limited set, the data support the contention that Sba1 and Hsp82 affect complexes by targeting adjacent subunits.
A GO Slim analysis of the shared complexes revealed an enhanced joint role for Sba1 and Hsp82 in translation, cellular respiration, protein catabolism, nuclear organization and ribosome biogenesis (Figure S6). Hence, the combined Sba1 and Hsp82 activities appear to be concentrated on certain cellular activities. Taken together, Sba1 appears to expand the overall reach of the Hsp82 molecular chaperone system to autonomous targets in addition to affecting select clients in conjunction with Hsp82.
Discussion
Expansion of the eukaryotic molecular chaperone network
We have exploited genomic and proteomic high-throughput approaches to create the first extensive functional net for an Hsp90 cochaperone. A priori, a knowledge gap existed in our understanding of cellular chaperone networks since the broad activities of any cochaperone had yet to be resolved. Typically, molecular chaperone systems are described in terms of one or two focal-point chaperones (e.g., Hsp90 and Hsp70) that are affected by a variety of cochaperones (e.g., p23 and Hsp40) (Wegele et al., 2004). It has been suggested that cochaperones primarily function to guide particular client proteins to a central chaperone and then modulate the ATPase activity of that chaperone to achieve a desired effect on the client. Based on this limited paradigm, cochaperone subnets should be fully contained within the network of its cognate chaperone. Unexpectedly, we found that the p23 network is not superimposable on the Hsp90 net. Our work supports a model in which the cellular chaperone system expands beyond a few focal-point chaperones and includes the numerous cochaperones that can directly regulate client proteins. For simplicity, we will refer to Hsp90 and its associated cochaperones, including p23, as the Hsp90 molecular chaperone machine, which would constitute one significant branch of the eukaryotic chaperone system.
Our study has revealed an unanticipatedly extensive cast for p23 that both coincides with and is sovereign to known Hsp90 associated-targets. While it has been reasoned that the innate chaperoning activities for the cochaperones, which are often comparable to the central chaperones (Freeman et al., 1996), might be used to direct Hsp90 or Hsp70 to select clients, our data suggest that this is a minor feature for Sba1 since only a subset (∼25%) of the 348 total Sba1 interactors coincide with known Hsp82 targets (Figure 7A). Yet despite the relatively low overlap, the Sba1 and Hsp82 targets appear to work within common biological processes (Figure 6). Remarkably, the convergence minimally involves Sba1/Hsp82 interactions with different subunits of the same protein complex (Figure 7). It is plausible that Sba1 and Hsp82 cooperate to assemble and/or disassemble protein complexes by associating with neighboring subunits. Alternatively, by interacting with more than one component of a structure a greater level of chaperone-mediated regulation might be gained (i.e., cooperative effect). While additional work is required to determine the function relevance, the idea of Sba1 and Hsp82 interacting with neighboring subunits to affect a protein complex reveals a distinct avenue for chaperone action within the cell.
In addition to joint interactions with protein complexes, Sba1 and Hsp82 appear to merge onto common pathways by intersecting at different points along biological paths. For example, modulation of vesicular-mediated protein transport by Sba1 appears to involve regulation of protein mannosylation in the Golgi (Figure 2) whereas Hsp82 does not alter glycosylation events but rather affects transport by interacting with vesicle-tethering complexes (McClellan et al., 2007). In addition to protein transport, our studies showed that cell mobility is dependent upon p23 (Figure 3), as it is on Hsp90 (Tsutsumi et al., 2008). In a manner comparable to transport, p23 and Hsp90 appear to target different client proteins in order to influence the motility pathway.
By intersecting at individual proteins, protein complexes and protein pathways, p23 forms a triaxial relationship with Hsp90 and cellular processes. These crossroads, along with the independent p23 clients, likely expand the reach of the Hsp90 chaperone machine beyond the limited hub of the individual Hsp90 protein. In future studies, it will be interesting to determine if interactions between p23 and Hsp90 are important for modulating the shared protein complexes or for propagating the multi-step pathways. We anticipate that other cochaperones (e.g., FKBP52) contribute to the Hsp90 machine in a similar manner. Thus, in addition to merely guiding Hsp90 to select proteins, cochaperones likely utilize their innate chaperone activities to distinctively modulate proteins and to create a more extensive cellular molecular chaperone system.
Nuclear molecular chaperone activities
Historically, the Hsp90 molecular chaperone system had been viewed as a strict cytosolic machine (Pratt, 1993). This erroneous model was put forth to help explain the cytoplasmic localization of unactivated steroid hormone receptors. Yet, Hsp90 and its associated cochaperones have since been shown to have nuclear roles in diverse activities including transcriptional regulation and telomere DNA maintenance (DeZwaan and Freeman, 2008). The presented p23 data and established Hsp90 studies indicate a significant number of nuclear interactors for these chaperones that are involved in a wide-range of functions including chromatin, transcription, RNA processing, DNA replication, telomere maintenance, and DNA repair (Figure 6). Hence, in addition to established cytosolic roles, p23 and Hsp90 appear to serve as general chaperones for nuclear processes (DeZwaan and Freeman, 2008).
Dissecting the Hsp90 molecular chaperone network
A better understanding of the Hsp90 chaperone machine has important implications for both basic cell functions and medicinal therapeutics. Hsp90 has become a worthy therapeutic target, as demonstrated by the large number of progressing clinical trials (Tsutsumi et al., 2009). To date, the clinical studies have utilized Hsp90 inhibitors. Given the broad spectrum and shear number of clients, it might be useful to develop more directed means of disrupting the Hsp90 chaperone machine.
Cochaperones are one promising avenue to partition the cellular chaperone system more narrowly than targeting Hsp90 given the apparently smaller p23 network. Yet, by targeting p23 many clinically relevant proteins might still be affected, which appears to be a beneficial feature of efficacious therapeutic agents. Significantly, p23 levels are generally elevated in cancerous cells and p23 appears to have an active role in cell invasion events (Simpson et al., 2010). Our study provides an in-depth evaluation of the p23 cochaperone, which is a highly conserved and abundant component of the Hsp90 machine (Johnson and Brown, 2009). Importantly, the presented work provides insights into the functional capacity of the cellular molecular chaperone system that is critical for achieving cellular proteostasis (Balch et al., 2008).
Experimental Procedures
Synthetic Genetic Array analysis
The sba1Δ SGA analysis was performed essentially as described in Tong et al. (2004). In brief, a sba1∷natMX query strain was systematically mated with a Deletion Mutant Array (DMA) and the genetic interactors with reproducible phenotypes (2 of the 3 replicas) were compiled and used for further analysis.
ProtoArray screen
The ProtoArray experiments were performed according to instructions (Invitrogen Inc.). In brief, biotinylated Sba1 was used to screen 4,088 yeast proteins expressed as glutathione-S-transferase fusion proteins and spotted on nitrocellulose membrane. The associated biotinylated Sba1 was detected using streptavidin-conjugated Alexa Fluor 647. Statistically significant positive hits were determined using 3-independent array hybridizations and the ProtoArray Prospector software (Invitrogen Inc.).
Yeast 2-hybrid and TAP-Tag experiments
Select SGA and ProtoArray interactors (Supplemental Tables 1 and 2) were analyzed using the yeast 2-hybrid assay. The candidate genes were expressed as Gal4 activation domain fusions (pGADT7), Sba1 was expressed as a LexA DBD fusion (pRS314-LexA) and positive interactions were detected using a LexA-regulated β-galactosidase reporter (pLacGus) (Invitrogen Inc.).
The ability of Sba1 and/or Hsp82 to associate with protein structures was determined using the TAP-tag method as previously described (Rigaut et al., 1999). In short, yeast expressing the indicated TAP-fusion proteins were utilized to isolate the various protein complexes by affinity for IgG sepharose, the bound protein was eluted with SDS sample buffer, resolved by SDS-PAGE and the associated Sba1 or Hsp82 was detected by immunoblot analysis. For the Rix1-TAP experiments we followed an established protocol (Ulbrich et al., 2009).
Yeast inhibitor assays
We used established protocols for testing the sensitivities of yeast cells to Methyl methanesulfonate (MMS), bleomycin (BLEO) and ultraviolet light (UV) to also assess the effect of the ribosome inhibitor Hygromycin B (Hyg B) and the Golgi organelle inhibitor Brefeldin A (BFA) on yeast growth (Toussaint and Conconi, 2006). Basically, exponentially growing parental or sba1Δ yeast transformants (pRS315-GPD or pRS315-GPD-SBA1) were diluted into selective media unsupplemented or supplemented with the indicated levels of inhibitor or mutagen and the optical densities were determined following growth at 30°C. The relative growth rates were normalized to the optical densities of the unsupplemented transformants.
MEF inhibitor studies
For mammalian cell survival assays, parental and p23 null MEFs (Grad et al., 2006) were plated at 104 cells/well in 24-well plates and exposed either to UV light, BLEO, BFA, MMS or Hygromycin B at indicated concentrations. Cell viabilities were determined following 1 (UV, MMS, BFA) or 2 (BLEO, Hygromycin B) days of incubation at 37°C. Data represent averages of nine experiments.
Protein transport protocols
The Invertase assays were performed as described (McClellan et al., 2007). In brief, the levels of secreted Invertase in the growth media of the indicated, low glucose-induced yeast was detected using 2,3,5-Triphenyltetrazolium chloride following resolution by native gel electrophoresis. Intracellular levels of Myc-Invertase were detected by immunoblot analysis using an anti-Myc antibody. The ts mutant sec18-1 was a control for Golgi glycosylation affects since this mutation blocks Invertase passage out of the ER. To monitor Fus-Mid-GFP transport, yeast transformants (pTPQ55 and either pRS425GPD or pRS425GPD-SBA1) were grown exponentially and the locale of Fus-Mid-GFP was determined using conventional GFP fluorescence micrographs (Proszynski et al., 2005).
The processing efficiency of Preprolactin to Prolactin during transport was determined as previously described (Lakkaraju et al., 2008). MEF cells were transiently transfected with a Flag-Preprolactin expression construct (pPrl3f) and the levels of Prolactin were detected by immunoblot analysis using an anti-Flag antibody. To control for transfection efficiencies a GFP expression vector was included and GFP levels were determined by immunoblot analysis with an anti-GFP antibody.
Mammalian in vitro wound assay
The abilities of parental and p23 null MEFs to migrate into a predefined wound were determined, as described (Rodriguez et al., 2005). Upon reaching confluency, cell layers were scratched and cell migration into the wound was determined by manual cell counting using conventional light microscopy following a 14 h incubation at 37°C.
Mammalian indirect immunofluorescence
Antibodies directed against p23 (JJ3), Giantin (ab24586; Abcam), Vinculin (V4319; Sigma) or α-Actinin (985; Brieher, unpublished) were used to detect the indicated proteins in formaldehyde-fixed, triton-permeabilized parental or p23 null MEFs. The p23, Vinculin and α-Actinin antibodies were visualized using Alexa Fluor 488-conjugated anti-rabbit antibody and Giantin antibody was visualized using Alexa Fluor 694-conjugated anti-rabbit antibody.
Supplementary Material
Highlights.
The p23 chaperone works in conjunction with and sovereign to Hsp90.
p23 modulates protein transport by regulating Golgi-linked Mannosylation activity.
p23 is required for both ribosome biogenesis and cell mobility.
p23 serves a broad, nuclear-associated protein network.
Acknowledgments
We thank Natalie Simpson (NYU) for helpful comments on the manuscript. We are grateful to William Brieher (UIUC) and Kizhakke Sathyan (UIUC) for all their technical assistance. B.C.F. was supported by the Public Service grant DK074270 and B.J.A. was supported by CIHR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Bartkeviciūte D, Sasnauskas K. Disruption of the MNN10 gene enhances protein secretion in Kluyveromyces lactis and Saccharomyces cerevisiae. FEMS Yeast Res. 2004;4:833–840. doi: 10.1016/j.femsyr.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- DeZwaan DC, Freeman BC. Hsp90: The Rosetta Stone of cellular protein dynamics? Cell Cycle. 2008;7:1006–1012. doi: 10.4161/cc.7.8.5723. [DOI] [PubMed] [Google Scholar]
- Ellis J. Proteins as molecular chaperones. Nature. 1987;328:378–379. doi: 10.1038/328378a0. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Toft TO, Morimoto RI. Molecular chaperone machines: Chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor associated protein, p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- Grad I, McKee TA, Ludwig SM, Hoyle GW, Ruiz P, Wurst W, Floss T, Miller CA, 3rd, Picard D. The Hsp90 cochaperone p23 is essential for perinatal survival. Mol Cell Biol. 2006;26:8976–8983. doi: 10.1128/MCB.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B, Viggiano S, Hibbs MA, Troyanskaya OG, Amberg DC. Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev. 2007;21:148–159. doi: 10.1101/gad.1477507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick JP, Hartl FU. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Brown C. Plasticity of the Hsp90 chaperone machine in divergent eukaryotic organisms. Cell Stress Chaperones. 2009;14:83–94. doi: 10.1007/s12192-008-0058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Prodromou C, Frydman J. The Hsp90 mosaic: a picture emerges. Nat Struct Mol Biol. 2009;16:2–6. doi: 10.1038/nsmb0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- Millson SH, Truman AW, King V, Prodromou C, Pearl LH, Piper PW. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p) Eukaryot Cell. 2005;4:849–860. doi: 10.1128/EC.4.5.849-860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Soutoglou E. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol. 2009;19:617–629. doi: 10.1016/j.tcb.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Poon PP, Cassel D, Spang A, Rotman M, Pick E, Singer RA, Johnston GC. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 1999;18:555–564. doi: 10.1093/emboj/18.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB. The role of heat shock proteins in regulating the function, folding, and trafficking of the glucocorticoid receptor. J Biol Chem. 1993;268:21455–21458. [PubMed] [Google Scholar]
- Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Proszynski TJ, Klemm RW, Gravert M, Hsu PP, Gloor Y, Wagner J, Kozak K, Grabner H, Walzer K, Bagnat M, Simons K, Walch-Solimena C. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc Nat l Acad Sci USA. 2005;102:17981–17986. doi: 10.1073/pnas.0509107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rodriguez LG, Wu X, Guan JL. Wound healing assay. Methods Mol Biol. 2005;294:23–29. doi: 10.1385/1-59259-860-9:023. [DOI] [PubMed] [Google Scholar]
- Shah N, Klausner RD. Brefeldin A reversibly inhibits secretion in Saccharomyces cerevisiae. J Biol Chem. 1993;268:5345–5348. [PubMed] [Google Scholar]
- Siegers K, Waldmann T, Leroux MR, Grein K, Shevchenko A, Schiebel E, Hartl FU. Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J. 1999;18:75–84. doi: 10.1093/emboj/18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson NE, Lambert WM, Watkins R, Giashuddin S, Huang SJ, Oxelmark E, Arju R, Hochman T, Goldberg JD, Schneider RJ, Reiz LF, Soares FA, Logan SK, Garabedian MJ. High levels of Hsp90 cochaperone p23 promote tumor progression and poor prognosis in breast cancer by increasing lymph node metastases and drug resistance. Cancer Res. 2010;70:8446–8456. doi: 10.1158/0008-5472.CAN-10-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Toussaint M, Conconi A. High-throughput and sensitive assay to measure yeast cell growth: a bench protocol for testing genotoxic agents. Nat Protoc. 2006;1:1922–1928. doi: 10.1038/nprot.2006.304. [DOI] [PubMed] [Google Scholar]
- Tsutsumi S, Scroggins B, Koga F, Lee MJ, Trepel J, Felts S, Carreras C, Neckers L. A small molecule cell-impermeant Hsp90 antagonist inhibits tumor cell motility and invasion. Oncogene. 2008;27:2478–2487. doi: 10.1038/sj.onc.1210897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi S, Beebe K, Neckers L. Impact of heat-shock protein 90 on cancer metastasis. Future Oncol. 2009;5:679–688. doi: 10.2217/fon.09.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, Böttcher B, Hurt E. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 2009;138:911–922. doi: 10.1016/j.cell.2009.06.045. [DOI] [PubMed] [Google Scholar]
- Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele H, Muller L, Buchner J. Hsp70 and Hsp90--a relay team for protein folding. Rev Physiol Biochem Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of Vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.