Abstract
Background
Inflammatory bowel disease (IBD) is used to describe a state of idiopathic, chronic inflammation of the gastrointestinal tract. The two main phenotypes of IBD are Crohn's disease (CD) and ulcerative colitis (UC). The major cause of IBD-associated mortality is colorectal cancer. Although both host-genetic and exogenous factors have been found to be involved, the aetiology of IBD is still not well understood. In this study we characterized thirteen Escherichia coli strains from patients with IBD by comparative genomic hybridization employing a microarray based on 31 sequenced E. coli genomes from a wide range of commensal and pathogenic isolates.
Results
The IBD isolates, obtained from patients with UC and CD, displayed remarkably heterogeneous genomic profiles with little or no evidence of group-specific determinants. No IBD-specific genes were evident when compared with the prototypic CD isolate, LF82, suggesting that the IBD-inducing effect of the strains is multifactorial. Several of the IBD isolates carried a number of extraintestinal pathogenic E. coli (ExPEC)-related virulence determinants such as the pap, sfa, cdt and hly genes. The isolates were also found to carry genes of ExPEC-associated genomic islands.
Conclusions
Combined, these data suggest that E. coli isolates obtained from UC and CD patients represents a heterogeneous population of strains, with genomic profiles that are indistinguishable to those of ExPEC isolates. Our findings indicate that IBD-induction from E. coli strains is multifactorial and that a range of gene products may be involved in triggering the disease.
Keywords: Inflammatory bowel disease, Escherichia coli, Urinary tract infections, LF82
Background
The term inflammatory bowel disease (IBD) is used to describe a state of idiopathic, chronic inflammation of the gastrointestinal tract. The two main phenotypes of IBD are Crohn's disease (CD) and ulcerative colitis (UC). CD is characterized by granulomatous inflammation and may affect any part of the gastrointestinal tract but is particularly prevalent in the ileocaecal area. UC, on the other hand, which is associated with extensive epithelial damage, crypt abscesses and significant neutrophil infiltration, is colon-specific. Patients suffering from extensive ulcerative colitis or colonic CD have a 10-fold increased risk of developing colorectal cancer, the major cause of IBD-associated mortality. The prevalence of IBD is 1-20 cases per 100,000 individuals and up to two million people are estimated to be affected by IBD worldwide [1,2]. IBD is more prevalent in developed countries than in developing countries, which has given rise to a number of theories regarding the significance of dietary preferences for the development and exacerbation of IBD [3-5].
Although the aetiology of IBD is still not well understood, it is generally believed that both host, environmental and microbial factors are involved. Several host-genetic factors (e.g. NOD2/CARD15) and exogenous factors (e.g. diet and smoking) have been identified [6]. Growing evidence also indicates that IBD results from an inappropriate immune response to the intestinal flora in genetically susceptible individuals. Animal models have shown that colitis does not occur in germ-free environment, but can be induced by the addition of bacteria [7,8]. The large intestine and colon are heavily colonized by microorganisms. While the bacteria associated with the intestinal flora are usually associated with a commensal lifestyle and form symbiotic multi-species communities, these bacteria may in some cases cause disease. There is mounting evidence that the composition of the microbiota in patients suffering from IBD is abnormal, and that the biodiversity is lower than in healthy subjects [9]. A range of bacterial species have been implicated in IBD, including Escherichia coli. Several studies have reported increased levels of E. coli in IBD tissues [10-12]. IBD patients have also been found to display increased serum immune-reactivity against E. coli as compared to healthy control subjects [13]. The identification of specific E. coli isolates with adherent and invasive capabilities in relation to CD patients has led to the coining of a new E. coli pathotype, the adherent-invasive E. coli (AIEC) [14]. The complete genome sequences of the prototypic CD E. coli isolate, LF82, as well as one other CD isolate, NRG857c, were recently published [15,16] and may pave the way for an increased understanding of the involvement of AIEC in IBD. Interestingly, studies with various probiotic microorganisms, including E. coli Nissle 1917, have shown that some probiotics may have a positive effect on inflammatory bowel disease [17-19]. This effect might be due to bacterial competition [20].
We have previously described the design and application of a 31-genome CGH microarray of E. coli [21,22]. In this study we have used this array to examine the genomic profile of a range of IBD isolates. We recently characterized a set of E. coli strains isolated from IBD patients [23]. Here we present a detailed comparative genomics study of these strains. We show that IBD isolates represents a heterogeneous population of strains, which display a remarkable resemblance with ExPEC isolates. Given that most ExPEC isolates originate from the intestinal tract, where they normally are present only in low numbers, it is conceivable that in IBD patients these clones are overrepresented.
Results
Genomic profiling of IBD-related E. coli isolates
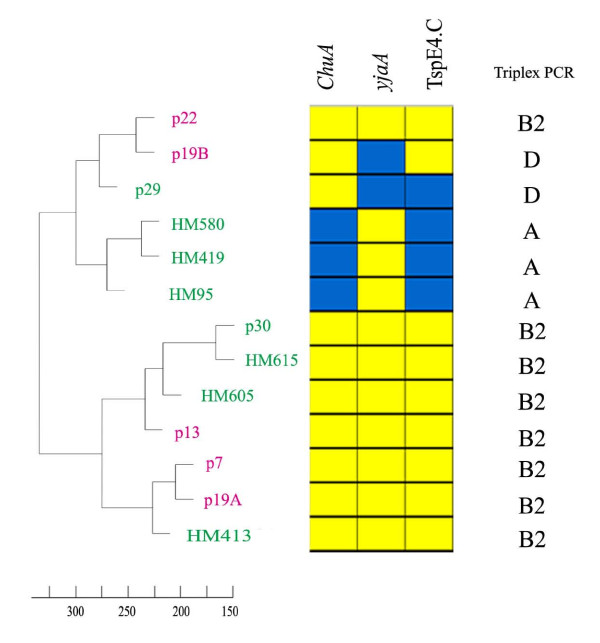
The comparative genomic analysis included five ulcerative colitis (UC) isolates, eight Crohn's disease (CD) isolates and the prototypic uropathogenic E. coli (UPEC) strain CFT073 (Table 1). Based on the CGH data, we initially sought to compare the IBD isolates with respect to their overall genomic profile. Interestingly, the comparison did not reveal the presence of two distinct subpopulations, i.e. UC and CD; rather it grouped the strains primarily according to their phylogenetic group (Figure 1). The only exception to this pattern was UC isolate p22, a group B2 isolate, which clustered most closely with UC isolate p19B, a group D strain. Therefore, based on the overall genotype, the UC and CD strains were indistinguishable. This clearly shows that the single most important determinant for the genomic profiles of IBD isolates is their phylogenetic group origin. With the exception of p22, this also suggests that the presence of horizontally acquired genetic segments does not generally obscure the phylogenetic signal. Within each phylogenetic group, some clustering according to disease was observed, although the relatively small number of isolates limits the delineation. Combined, the data shows that bacterial isolates from UC and CD display remarkably heterogeneous genomic profiles.
Table 1.
Strains used in this study
| E. coli strain | Relevant characteristics | Reference |
|---|---|---|
| Nissle 1917 | Nonpathogenic probiotic isolate (O6:K5:H1) | [38] |
| MG1655 | K-12 reference strain | [37] |
| LF82 | Prototypic Crohn's disease isolate | [12] |
| p7 | Ulcerative colitis isolate, active | [23] |
| p13 | Ulcerative colitis isolate, active | [23] |
| p19A | Ulcerative colitis isolate, active | [23] |
| p19B | Ulcerative colitis isolate, active | [23] |
| p22 | Ulcerative colitis isolate, active | [23] |
| p25 | Ulcerative colitis isolate, active | [23] |
| p29 | Crohn's disease isolate, active | [23] |
| p30 | Crohn's disease isolate, active | [23] |
| HM95 | Crohn's disease isolate | [36] |
| HM154 | Crohn's disease isolate | [36] |
| HM413 | Crohn's disease isolate | [36] |
| HM419 | Crohn's disease isolate | [36] |
| HM580 | Crohn's disease isolate | [36] |
| HM605 | Crohn's disease isolate | [36] |
| HM615 | Crohn's disease isolate | [36] |
| c1 | Commensal isolate, O81:K16:H- | [23] |
| c2 | Commensal isolate, O6:K39:H- | [23] |
| c3 | Commensal isolate, O77:K96:H18 | [23] |
| c4 | Commensal isolate, O57, O155:K39:H19 | [23] |
| c5 | Commensal isolate, OX184:K-:H10 | [23] |
| c6 | Commensal isolate, O126:K-:H20 | [23] |
| c14 | Commensal isolate, Oru:K18:H19 | [23] |
| c16 | Commensal isolate, O1:K1:H- | [23] |
| c17 | Commensal isolate, O101:K+:H56 | [23] |
| p10A | Ulcerative colitis isolate, inactive eperioperiperiodperiod | [23] |
| p10B | Ulcerative colitis isolate, inactive | [23] |
| p23 | Ulcerative colitis isolate, inactive | [23] |
| p26 | Ulcerative colitis isolate, inactive | [23] |
| p27 | Ulcerative colitis isolate, inactive | [23] |
| p32 | Ulcerative colitis isolate, inactive | [23] |
| p11 | Crohn's disease isolate, inactive | [23] |
| p15 | Crohn's disease isolate, inactive | [23] |
| p31 | Crohn's disease isolate, inactive | [23] |
Figure 1.
Phylogenetic relationship between the isolates. Relationship based on their overall genomic profiles (left side). The Crohn's disease isolates are indicated in green and the ulcerative colitis isolates in pink. CGH data for the three phylogroup identifiers chuA, yjaA and TspE4.C (middle). Phylogenetic group association based on triplex PCR data for the same identifiers (right side).
IBD-specific markers
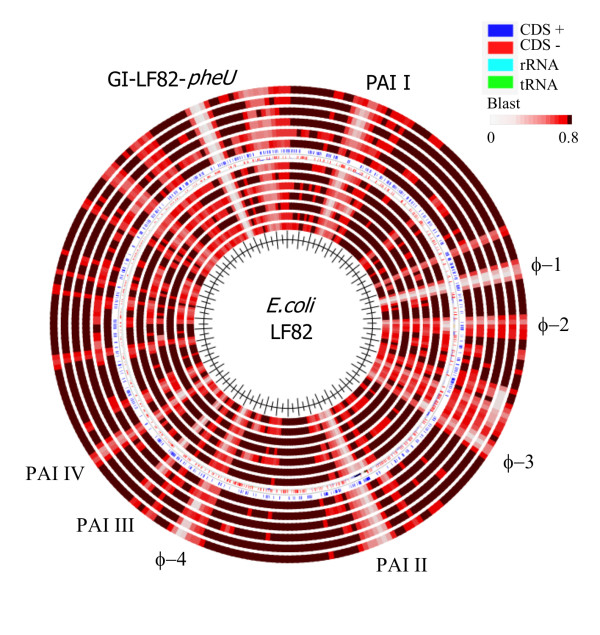
The implication of a specific E. coli pathovar (or pathovars) in the aetiology of IBD has remained elusive, mostly due to the lack of definable IBD marker genes. Recently, the complete genome sequences of two AIEC strains, i.e. LF82, a prototypic CD strain, and CD strain NRG857c, were published [15,16]. This may pave the way for a more systematic search for genetic determinants which can help to classify these pathogens. In order to look for commonalities between LF82 and the IBD isolates used in this study, we compared the inferred genomic sequence of our IBD strains with that of LF82. Initially this led us to examine the prevalence of LF82-associated genomic islands among the IBD isolates. Several of the genomic islands of LF82 appeared to be quite conserved among the IBDs, whereas others were less prevalent, or even LF82-specific (Figure 2). None of the isolates were found to carry to the GI-LF82-pheU island (mainly hypothetical proteins of unknown function) or the φ-1 prophage element, while most of the isolates were predicted to carry the high pathogenicity island (HPI), or PAI-II. The HPI island, which encodes yersiniabactin, is particularly prevalent among ExPEC isolates, and is known to contribute to the persistence of CFT073 in the urinary tract (murine model) [24]. The remaining islands were present in some, but not in all IBD isolates, clearly illustrating the heterogeneity of the population. Although each of these islands may contribute to the pathogenesis or fitness of LF82, most are clearly not descriptive of IBD isolates in general.
Figure 2.
Blast atlas comparison generated by blasting the inferred genomic sequences of the IBD isolates against the prototypic Crohn's disease isolate LF82 using the GeneWiz browser (http://www.cbs.dtu.dk/services/gwBrowser/). Blast lanes from the centre (eight CD isolates followed by five UC isolates): HM95, HM413, HM419, HM580, HM605, HM615, p29, p30, p7, p13, p19A, p19B and p22.
The complete genome sequence of LF82 allowed Miquel et al. to identify 115 LF82-specific genes by comparison with other sequenced E. coli genomes. However, the requirement or contribution of these genes to the pathogenesis of LF82 remains unknown, as does the general prevalence of these genes among IBD isolates. Of the 115 genes, 22 are shared by the other sequenced AIEC strain NRG857c. We analysed the CGH data to examine the prevalence of these genes among our IBD strains. Of the 115 LF82-specific genes, 57 were represented on the CGH microarray. Of these only seven were found present in one or more IBD strains examined, and none were present in all strains. Among the 22 genes, which were shared by NRG857c, only two genes were predicted present (and then only in a single isolate). This clearly reveals that these genes are most likely strain-specific rather than IBD-specific genes. Although some LF82-specific genes that were not present on our microarray might be prevalent among IBD isolates, our results suggest that it may be very difficult to identify IBD-specific genetic markers.
Adhesins and biofilm-related genes
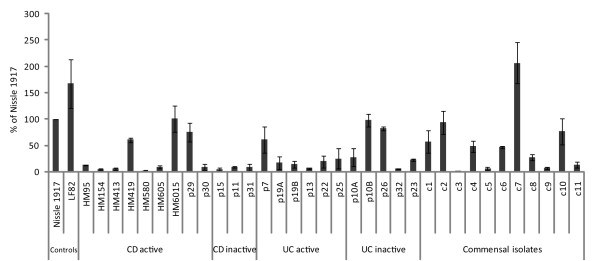
The ability to adhere to and colonize epithelial surfaces is a critical step in the development of many bacterial infections. Although the role of mucosa-associated E. coli in the development or exacerbation of IBD remains controversial, the association of E. coli isolates with the mucosal surface is intimately linked with the disorder(s). In order to determine the general adhesion and virulence potential of the IBD isolates, the genomic profiles were examined for a number of known and putative colonization and virulence factors. Several fimbrial operon genes were found among the isolates, which could contribute to adhesion of these strains (Table 2). Interestingly, the majority of the isolates were predicted to carry P-fimbria-related genes; also, two UC and five CD isolates showed positive haemagglutination (Table 3). Other adhesion-related genes identified among some of the isolates included the sil genes (adhesin for cattle colonization), shlA (autotransporter), a filamentous haemoagglutinin and hek (adhesion determinant) (Additional file 1, Table S1). The isolates were also found to carry a number of biofilm-related genes such as the fim, flu and csg genes encoding type 1 fimbriae, Ag43 (autotransporter protein) and curli, respectively (Table 3). There is mounting evidence that bacterial biofilms plays an important role in intestinal colonization and there is a growing interest in the role of biofilm formation in inflammatory bowel disease [25]. However, there was little if any indication that the IBD isolates displayed any consistently better biofilm-forming capacity in vitro in LB than isolates obtained from healthy control individuals (Figure 3). Several of the isolates also carried a number of genes related to invasion, including ibeA, invA and tia (Additional file 1, Table S1).
Table 2.
Prevalence of fimbrial operons among the IBD isolatesc
| UC (5)a | CD (8)a | MG1655 | LF82 | CFT073b | |
|---|---|---|---|---|---|
| sfm | 4 | 4 | - | - | + |
| ybg | 0 | 2 | + | + | - |
| ycb | 0 | 0 | + | - | - |
| yra | 0 | 3 | + | - | - |
| glt/yhc | 0 | 2 | + | - | - |
| yad | 2 | 1 | - | + | + |
| yag, matB | 5 | 8 | + | + | + |
| sfa/foc operon | 2 | 0 | - | - | + |
| F9, yde | 3 | 4 | - | + | + |
| yeh | 5 | 7 | + | + | + |
| yfc | 4 | 4 | - | + | + |
| pap | 1 | 4 | - | - | + |
| ygi | 2 | 4 | - | + | + |
| auf | 4 | 3 | - | + | + |
| fim operon | 5 | 6 | + | + | + |
| pix | 0 | 0 | - | - | - |
| CS1 | 1 | 2 | - | - | - |
| F17-like fimbriae | 2 | 0 | - | - | - |
| CS12-like fimbriae | 0 | 0 | - | - | - |
| lpf/lpf2 | 0 | 0 | - | + | - |
| bfp | 0 | 0 | - | - | - |
a () total number of isolates included in the analysis
b Same results were obtained by both CGH analysis and genome sequence inspection
c Present/ absent calls are based on the entire fimbrial operons. Some fimbriae may however still be functional even in the absence of some fimbrial genes.
Table 3.
Phenotypic and genotypic characteristics of the IBD strains
| Strains | Source | Group | Pellicle | flu | fima | fimHa | YA | csg | pga | CRb | Motilityc | pap | HAd | hly | Haemolysis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MG1655 | Commensal isolate | A | + | + | BEAICDFGH+ | + | + | CABDEFG+ | ABCD+ | + | + | - | - | - | - |
| Nissle 1917 | Probiotic strain | B2 | + | + | BEAICDFGH+ | + | + | CABDEFG+ | ABCD+ | ++ | + | IA+ | - | ABCD+ | - |
| CFT073 | UPEC strain | B2 | + | + | BEAICDFGH+ | + | + | CABDEFG+ | ABCD+ | + | + | IAHCDJKEFG+ | + | ABCD+ | + |
| p7 | Ulcerative colitis | B2 | + | + | BEAICDFGH+ | + | + | CABDEFG+ | ABCD+ | + | ++ | IHCDJKFG+ | - | ABCD+ | + |
| p13 | Ulcerative colitis | B2 | + | - | BEAICDFGH+ | + | + | CABDEFG+ | ABCD+ | + | ++ | - | - | B+ | - |
| p19A | Ulcerative colitis | B2 | + | + | BEAICDFGH+ | + | - | CABDEFG+ | ABCD+ | ++ | ++ | IAHCDJKEFG+ | + | ABCD+ | + |
| p19B | Ulcerative colitis | D | + | + | BEAICDFGH+ | + | - | CABDEFG+ | ABCD+ | ++ | ++ | IAEFG+ | - | B+ | - |
| p22 | Ulcerative colitis | B2 | + | + | BEAICDFGH+ | + | - | CABDEFG+ | ABCD+ | + | - | IACDJKEFG+ | + | ABCD+ | + |
| p29 | Crohn's disease | A | + | + | BEACDFGH+ | + | - | CABDEG+ | ABCD+ | ++ | ++ | F+ | - | - | - |
| p30 | Crohn's disease | B2 | + | + | BECDFGH+ | + | - | CABDEFG+ | ABCD+ | ++ | ++ | IAHCDJKEFG+ | + | - | - |
| HM95 | Crohn's disease | A | + | - | BEAICDFGH+ | + | + | CABDEFG+ | ABCD+ | +++ | ++ | AF+ | - | B+ | - |
| HM413 | Crohn's disease | B2 | + | + | BEAICDFGH+ | + | + | CABDEFG+ | ABCD+ | +++ | ++ | - | - | B+ | - |
| HM419 | Crohn's disease | A | + | + | BEAICDGH+ | + | - | CABDEG+ | ABCD+ | + | - | IA+f | +f | - | - |
| HM580 | Crohn's disease | D | + | + | BEAICDFGH+ | + | - | CABDEFG+ | -e | - | - | IAHCDJKEFG+ | + | - | - |
| HM605 | Crohn's disease | B2 | + | - | BEACDFGH+ | + | - | CABDEG+ | ABCD+ | ++ | - | IAHCDJKEFG+ | + | - | - |
| HM615 | Crohn's disease | B2 | + | + | BEAICDFGH+ | + | + | CABDEFG+ | ABCD+ | ++ | + | IAHCDJKEFG+ | + | - | - |
Abbreviations: YA: Yeast agglutination, HA: Haemagglutination
aBased on CGH data
bCongo-red binding
cMotility on LB plates, average of four plates. '-', non-motile; '+', average motility; '++', high motility (i.e. covered the whole plate, 80 mm)
dSame results were obtained with and without mannose
eMedium signal intensities were observed for this strain, i.e. that it cannot be excluded that it carries a divergent version of this operon
fThe reason for this discrepancy remains unknown.
Figure 3.
Comparison of the biofilm-forming capabilities of the IBD isolates and E. coli isolates obtained from IBD patients during inactive periods, and healthy control persons. Biofilm formation was monitored in LB medium using a crystal-violet, microtitre-based biofilm assay. Standard deviations are based on 3 independent experiments each comprising three replicates.
Classical virulence genes
The IBD isolates did not carry any of the classical virulence determinants characteristic of intestinal pathogenic E. coli (Table 4). This correlates well with previous studies, which have also found little evidence of the involvement of specific types of intestinal E. coli pathotypes in the etiology of IBD [26]. However, several of the UC strains were found to carry the hly genes and displayed haemolytic activity (Table 3).
Table 4.
Prevalence of various types of E. coli toxins and effectors among the IBD isolates
| Virulence factor | Pathotype | UC (5)a | CD (8)a | Nissle 1917 | CFT073 |
|---|---|---|---|---|---|
| Shiga toxin I | EHEC | 0 | 0 | - | - |
| Shiga toxin 2 | EHEC | 0 | 0 | - | - |
| EHEC haemolysin (ehx) | EHEC | 0 | 0 | - | - |
| Serine protease, EspP | EHEC | 0 | 0 | - | - |
| Serine protease, EspC | EPEC | 0 | 0 | - | - |
| Urease gene cluster | EHEC | 0 | 0 | - | - |
| Cell-cycle inhibiting factor (cif) | EPEC, EHEC | 0 | 0 | - | - |
| Type III secretion effector, Tir | EPEC, EHEC | 0 | 0 | - | - |
| Type III secretion effector, EspF | EPEC, EHEC | 0 | 0 | - | - |
| Type III secretion effector, EspH | EPEC, EHEC | 0 | 0 | - | - |
| Inhibition of lymphocytes activation (Lif/Efa) | EPEC, EHEC | 0 | 0 | - | - |
| Heat-labile enterotoxin (LT), alpha-subunit (eltA) | ETEC | 0 | 0 | - | - |
| Type III secretion effector, IpaB | EIEC | 0 | 0 | - | - |
| Shigella enterotoxin (ShET2) | EAEC, EIEC | 0 | 0 | - | - |
| Haemolysin (hly) | ExPEC | 2 | 0 | - | + |
| Cytotoxic necrotizing factor 1 (cnf1) | ExPEC, MNEC | 2 | 0 | - | - |
| Serine protease, Sat | ExPEC | 4 | 2 | + | + |
| Haemoglobin-binding protease (tsh) | ExPEC, APEC | 0 | 0 | - | -b |
| Cytolethal distending toxin (cdt) | Several | 0 | 0 | - | - |
Abbreviations: Enterohaemorrhagic E. coli (EHEC), Enteropathogenic E. coli (EPEC), Enterotoxigenic E. coli (ETEC), Enteroinvasive E. coli (EIEC), Enteroaggregative E. coli (EAEC), Extra-intestinal pathogenic E. coli (ExPEC)
a() total number of isolates
bCFT073 carries a haemoglobin protease with homology to this, which is not represented on the chip
Other virulence and fitness factors
In order to look for other potential genetic markers of IBD, we compared the inferred genomic profiles of the isolates with a range of sequenced commensal strains belonging to different phylogenetic groups. Initially we looked for genes that were present in all the IBD isolates but absent in the six commensal strains selected for the analysis (i.e. MG1655 (A), HS (A), IAI1 (B1), SE11 (B1), ED1a (B2) and SE15 (B2)). We also looked for genes common to all isolates within each disease category. No probes could specifically differentiate the UC and the CD isolates from the commensal strains. The UC isolates shared only one gene that was not present among the six commensal strains (i.e. ECIAI39_1981, encoding a hypothetical protein of bacteriophage origin). In CFT073, this gene is located in PAI-CFT073-icdA. Even within each phylogenetic group, only a few genes were conserved. Among the group A strains, only three genes were identified that were not present among the commensal group A strains, including two hypothetical proteins and a putative TonB-like protein. Among group B2, only the ECIAI39_1981 gene was identified. This clearly shows that the IBD isolates represent a heterogeneous population.
Given the heterogeneous nature of the genomic profiles, we proceeded to compare each individual isolate with the commensal strains (Additional file 1, Table S1). Except for the adhesin- and toxin-encoding genes already mentioned, several of the isolates were also predicted to carry lateral flagella. Interestingly, p22 was also found to carry the genes for lateral flagella, a trait which appears to be particular prevalent among group D strains. This may give some indication as to why this strain grouped with the D strains rather than the B2 strains (Figure 1). Several of the isolates were found to carry genes relating to propandiol utilization; however, none of the isolates were predicted to carry all of the pdu operon genes. Interestingly, several of the IBD isolates carried the gipA gene, a Peyer's patch-specific virulence factor, which is also present in LF82. A few isolates also carried vapA, coding for a putative virulence associated protein, as well as a range of ExPEC-related virulence genes (see subsequent section).
UC- and CD-specific genes
The manifestations of UC and CD are distinctly different, as are the pro-inflammatory mediators seen in the two IBD phenotypes. We therefore decided to compare the genomic profiles of the two groups to see whether the E. coli isolates associated with UC and CD would display divergent genotypes. Overall, there were no genes that could entirely differentiate one group from the other, although a few probes were more associated with one group than the other (Additional file 2, Table S2). This clearly highlights the heterogeneity of the populations. It should be noted that isolates were not all obtained from the same origin, which could affect the analysis.
Similarity to UTI isolates
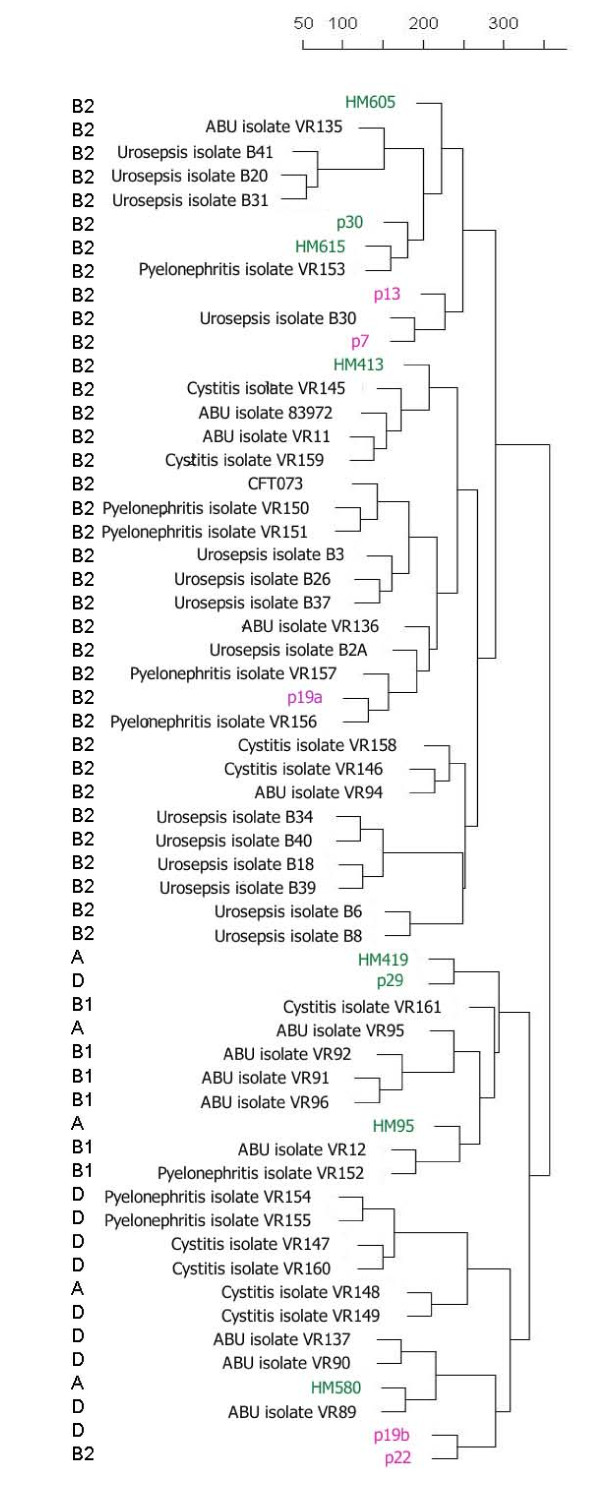
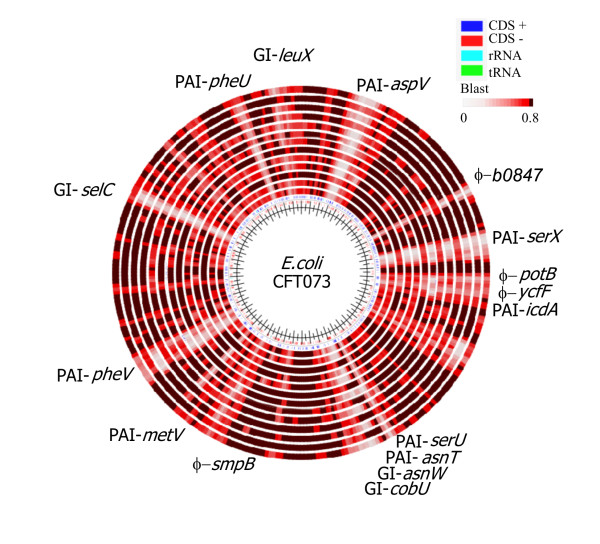
Previous studies have shown that many IBD isolates carry a range of ExPEC-associated virulence determinants. Therefore, the IBD genomic profiles were compared with the CGH data obtained for a range of clinical ExPEC isolates (in this case UTI and urosepsis isolates). Based on their overall genomic profiles, there was no clear delineation between the various disease groups. The phylogenetic analysis grouped the isolates mainly according to their phylogenetic group, and little if any grouping of the IBD isolates was observed (Figure 4). We subsequently examined the prevalence of a range of CFT073-associated islands among the IBD isolates. CFT073 carries a number of well-characterized genomic islands [24], which are all represented on the CGH microarray. Interestingly, several of the IBD isolates were predicted to carry a number of these islands, some of which are not present in the sequenced commensal isolates, such as the CFT073 islands located at aspV, serX, serU, asnW (pks), cobU, metV and leuX (Figure 5). The pks island, encoding the machinery for the synthesis of colibactin (capable of inducing breaks in double-stranded DNA in eukaryotic cells), was present in three of five UC isolates, while only two of eight CD isolates carried this island. A few of the UC isolates were predicted to carry PAI-CFT073-serU, which have previously been shown to encode an immuno-modulatory protein TcpC, which is important for upper urinary tract infections [27,28]. Several of the IBD isolates also carried PAI-CFT073-pheV and PAI-CFT073-pheU, which encode P fimbriae and haemolysin; some strains were also found to express the phenotypes associated with these factors (Table 3). A few CD isolates carried GI-CFT073-selC. In accordance with the presence of a range of UPEC-associated islands, the IBD isolates were also predicted to carry and express a number of ExPEC virulence/fitness genes (Tables 2, 3 and 5). The presence of a number of CFT073-associated PAIs and expression of ExPEC virulence genes among the IBD strains clearly reveals that the IBD strains strongly resemble ExPEC strains.
Figure 4.
Phylogenetic relationship between the IBD isolates and a range of UTI and urosepsis isolates. The Crohn's disease isolates are indicated in green and the ulcerative colitis isolates in pink.
Figure 5.
Blast atlas comparison generated by blasting the inferred genomic sequences of the IBD isolates against the prototypic pyelonephritis isolate CFT073 using the GeneWiz browser service. Blast lanes from the middle: HM95, HM413, HM419, HM580, HM605, HM615, p29, p30, p7, p13, p19A, p19B and p22.
Table 5.
Prevalence of ExPEC virulence and fitness genes among the IBD isolates
| Product | CFT073 | p7 | p13 | p19A | p19B | p22 | p29 | p30 | HM95 | HM413 | HM419 | HM580 | HM605 | HM615 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adhesins | ||||||||||||||
| F1C fimbriae | + | +/- | - | + | +/- | + | - | +/- | +/- | +/- | +/- | +/- | +/- | +/- |
| P fimbriae | + | +/- | - | + | +/- | +/- | +/- | + | +/- | - | +/- | + | + | + |
| Pix fimbriae | - | - | +/- | - | - | - | - | - | - | - | - | - | - | - |
| F17 fimbriae | - | + | - | + | - | - | - | - | +/- | - | - | - | - | - |
| CS12 fimbriae | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| yapH | + | + | + | + | - | - | - | - | + | + | - | - | - | - |
| ShlA/HecA/FhaA exoprotein family | + | + | + | + | - | + | - | - | + | + | - | - | - | - |
| Toxins | ||||||||||||||
| RTX family exoprotein | + | + | - | + | - | - | - | - | - | + | - | - | - | - |
| Haemoglobin protease | + | + | + | + | - | + | - | + | - | + | - | - | + | + |
| hly | + | + | +/- | + | +/- | +/- | - | - | +/- | +/- | - | - | - | - |
| sat | + | + | - | + | + | + | + | - | - | - | - | + | - | - |
| cdt | - | - | - | - | - | - | - | - | - | + | - | - | - | - |
| cnf1 | - | + | - | + | - | - | - | - | - | - | - | - | - | - |
| Nutrition | ||||||||||||||
| iro | + | - | + | + | - | + | +/- | +/- | - | + | - | - | + | + |
| pgt | + | +/- | - | - | - | - | +/- | +/- | - | - | - | - | + | + |
| Other | ||||||||||||||
| tcpC | + | - | - | + | - | - | - | - | - | - | - | - | - | - |
| shiA homolog | + | + | - | + | + | + | + | + | + | - | + | + | + | + |
| pntC | - | - | + | - | - | - | - | - | - | - | - | - | - | - |
| ibeA | - | - | + | - | - | - | - | - | - | - | - | - | - | - |
| K1 capsule genes | - | - | - | - | - | - | - | + | - | - | - | - | + | + |
| K2 capsule genes | + | + | - | + | - | - | - | - | - | - | - | - | - | - |
| K15 capsule genes | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| putative FMN-dependent dehydrogenase | - | + | - | + | - | - | - | - | - | - | - | - | - | - |
| PAI-CFT073-asnW (pks) | + | + | - | + | +/- | + | +/- | - | +/- | + | - | - | - | - |
| GimB island | - | - | - | - | - | - | - | + | - | - | - | - | + | + |
+, - and +/- denotes presence, absence and partly present, respectively.
Discussion
The data presented here shows that E. coli IBD isolates represent a heterogeneous population, and that the single most important determinant for the genomic profile of the isolates is their phylogenetic group origin. The data also suggests that it may be difficult to identify IBD-specific, or even CD- or UC-specific, genetic markers. This correlates well with previous reports of Crohn's disease and colon cancer isolates, which also showed that these isolates by no means represent uniform populations [29]. The results may also explain why the pursuit for IBD-specific genetic determinants among E. coli isolates essentially has remained fruitless. However, although no IBD-specific markers could be identified in this study (taking into account 57 of the 115 LF82-specific genes previously identified), it cannot be excluded that the LF82-specific genes that were not represented on the chip, or other E. coli genes missing on the chip, could be IBD-specific. As with many pathogens, virulence is often multifactorial and made up by an assortment of diverse virulence factors. The LF82-specific genes may therefore still prove to be important for the IBD-inducing capacity of that particular strain. Nevertheless, it indicates that it may be difficult if not impossible to find unique IBD genes. Our findings indicate that IBD-induction from E. coli strains is multifactorial and that a range of gene products can trigger the disease. The apparent lack of IBD- and group-specific genetic determinants among the isolates also suggests that the host-microbe interactions and the altered environment of the gut likely play the most important roles in the aetiology of IBD. Due to the predisposition of the patient, the bacterial colonization may invoke an autoimmune reaction, which escalates, leading to the clinical symptoms of IBD.
The prevalence of various adhesive structures among the isolates varied considerably. Only relatively few fimbriae-encoding genes were shared by all isolates, even within each disease category. Type I fimbriae has previously been shown to be important for the invasive and adhesive characteristic of the prototypic AIEC strain LF82 [30]. It targets the carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) that is expressed on the apical side of ileal epithelial cells [31]. The high level of ileal colonization that is observed in CD patients may be linked to an abnormal expression of CEACAM6, indicating the presence of predisposing factors [31]. Interestingly, while most of the IBD isolates examined in this study were found to carry the fim genes, far from all were able to agglutinate yeast. It therefore seems likely that some of the isolates achieve adherence be means of other surface structures. Interestingly, several strains were found to carry and express P-fimbriae. While usually associated with UTI strains, P fimbriae can promote intestinal colonization [32,33]. However, whether P fimbriae or any of the other adhesive structures carried by these IBD isolates contributes to adherence in the IBD-inflicted gut, remains to be determined.
In recent years, there has been a growing appreciation of the importance of biofilm formation in the development and exacerbation of a range of chronic infections. This prompted us to evaluate the biofilm formation capacity of our IBD isolates. Previous studies have suggested that AIEC strains may be more efficient biofilm producers than non-AIEC isolates [25]. However, in this study there was no indication that the IBD strains displayed superior biofilm capacities to the non-IBD control strains. There was also no correlation between the active or inactive state of the diseases, and the biofilm production displayed by the isolates. Experimental differences and strains variations (incl. demographic origin) may explain such discrepancy. We have also not focused specifically on AIEC strains, unlike the previous study.
Most of the IBD isolates studied here were found to carry, and express, a number of genetic markers that are associated with ExPEC isolates. ExPEC, which predominantly belong to the phylogenetic groups B2 and D, are generally believed to originate from the gut, where they represent a small fraction of the E. coli flora. It might be conceivable that ExPEC-like strains are overrepresented in the microbiota of IBD patients, possibly due to the altered gut environment caused by the underlying genetic and environmental factors. This correlates well with the finding that the levels of strains belonging to group B2 are increased in IBD patients as compared with control subjects [23]. E. coli strains belonging to group B2, and to some extent group D, are known to carry a number of virulence and fitness genes that are not present in strains belonging to groups A and B1. ExPEC strains are associated with a number of extraintestinal infections like urinary tract infections, meningitis, pneumonia and wound infections. ExPEC strains are also usually regarded as being incapable of intestinal infections [34,35]. Meanwhile, they are good long-term colonisers of the human gut and about 20% of E. coli gut isolates are ExPEC strains. Paradoxically, IBD-associated strains seem to be the only ExPEC-type strains that are associated with intestinal infections (such as CD and UC). Our data suggests that these strains are not genetically distinct from other ExPEC strains.
Conclusions
Collectively, our findings indicate that IBD-associated E.coli represent a heterogeneous population of strains, whose genomic profiles greatly resembles that of ExPEC isolates. Arguably, our findings indicate that IBD-induction from E. coli is multifactorial and that different combinations of gene products may trigger the disease in a complex interplay with host parameters and environmental cues. Nevertheless, it still remains to be determined whether IBD-associated E. coli are directly involved in the pathogenesis of IBD or whether their presence is simply a result of the inflammatory response and the altered microbial environment of the IBD gut.
Methods
Bacterial strains and growth media
The strains used in this study are described in Table 1. HM95, HM154, HM413, HM419, HM580, HM605 and HM615 were kindly provided by Barry J. Campbell [36]. Two well-characterized E.coli strains, Nissle 1917 and MG1655, were included as controls in the phenotypic experiments [37,38]. All cultivations were performed in modified LB medium [39] or ABTG with 0.02% casamino acids. All strains were grown in modified LB medium prior to genomic DNA isolation.
Microarray design and sample preparation
The detailed design of the CGH custom microarray has been described elsewhere [21]. The 31 E. coli genomes used for designing the microarray included the prototypic CD strain LF82 and several ExPEC isolates, such as uropathogenic and avian pathogenic E. coli strains, as well as other pathogenic E. coli strains (e.g. EHEC) and a number of K-12 isolates. The microarray consisted of 134,285 probe sets (of 50-75mers) representing 16,098 E. coli target genes. The genomic DNA was isolated using the "IllustraTM bacterial genomicPrep Spin Kit" (GE Healthcare, 28-9042-58), and the samples were diluted to the recommended concentration. Sample preparation was then carried out using the NimbleGen Arrays User's Guide for CGH analysis. All isolates, except the reference strain CFT073, were run as single samples. The supporting microarray data have been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress) with the accession numbers E-MEXP-3090, E-MEXP-3089 and E-MTAB-212.
CGH data analysis
Data analysis was performed in R (statistical software), using the 'oligo' package for analysis of oligonucleotide arrays at the probe-level (Bioconductor) [40]. The RMA algorithm was used to perform background subtraction, normalisation and expression calculation (output in the log2 scale). Blast atlases were created using the GeneWiz whole genome visualization tool [41]. Hierarchical agglomerative clustering was performed in R, using the 'hclust' clustering method, an euclidean distance measure and the complete genomic profile of each strain (log values for all genes represented by more four or more probes). For interpretation of the CGH data, the following cut-off values (log values) were selected for presence/absence call of the individual probes: 6-8 negative, 8-10 borderline/unknown and 10-12 positive. The cut-off values were selected based on comparison of the CGH data obtained for CFT073 (run in triplicates) and the known genome sequence of this strain.
Phylogenetic group determination
For identification of phylogenetic group associations, a triplex PCR method was employed, using primers targeting two genes (chuA and yjaA) and one anonymous DNA fragment (TspE4.C) [42].
Statistical analysis
Statistical analysis was performed using Fischer's exact test when appropriate.
Biofilm formation in microtitre plates
Cells were grown overnight in LB and used to inoculate flat-bottomed, non-treated 48-well plates (Nunc) to an OD600 of 0.05. The microtitre plates were incubated statically at 37°C for 16 hours, and the biofilm stained with 0.1% crystal violet for 15 min. Excess dye was removed by washing with PBS. Crystal violet was then solubilised by the addition of 96% ethanol and A595 was measured. All experiments were repeated at least three times.
Yeast agglutination and haemagglutination tests
The presence of type 1 fimbriae was assayed by the ability of the bacteria to agglutinate yeast cells (Saccharomyces cerevisiae) on glass slides. Ten µl of fresh overnight cultures were mixed with 10 µl 5% yeast cells. The experiment was repeated after the cells had been washed and resuspended in LB containing 50 mM methyl-α-D-mannopyranoside.
The capacity of bacteria to express P fimbriae was assayed by haemagglutination with human type A red blood cells (RBCs). RBCs were washed twice with phosphate-buffered saline (PBS) and 10 µl of 5% RBCs were mixed with a single bacterial colony (freshly grown on LB plates) suspended in PBS on glass slides. Any strain showing positive haemagglutination was tested again after 30 min incubation with 1% D-mannose to rule out type 1 fimbriae and to further support that agglutination was likely mediated by P fimbriae.
Motility on LB plates
One µl of overnight culture was stabbed into LB plates containing 0.3% agar. The distance of migration (the diameter of the ring around the inoculation site) was measured after 16 h of incubation at 37ºC. The assay was performed in duplicates and repeated twice.
Haemolytic activity on blood agar plates
Isolated colonies were spot inoculated and production of haemolysin was detected by determining a zone of lysis under each colony on tryptone soya agar plates with sheep blood (Oxoid Deutschland GmbH) after overnight incubation of the tested strains. The assay was repeated independently twice.
Congo Red-binding assay
The ability to express curli fimbriae was evaluated by streaking each strain on modified LB-agar plates (without NaCl) containing 0.004% Congo Red (CR) and 0.002% Coomassie Brilliant Blue G. CR binding was indicated by the presence of red or pink colonies after incubation overnight at 37°C. The assay was performed in duplicates and repeated independently twice.
Authors' contributions
RMV planned, performed and analysed the CGH microarray data and drafted the manuscript. RMV and VH carried out the phenotypic experiments. RMV, VH and PK conceived and designed the study and wrote the manuscript. AMP provided strains and commented on the manuscript. KAK provided strains. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Tables presenting genes present in the different IBD isolates, but in none of the six commensal strains (MG1655, HS, IAI1, SE11, ED1a and SE15).
Table S2. List of probes that showed significant difference between the disease groups UC and CD.
Contributor Information
Rebecca Munk Vejborg, Email: rvejborg@sund.ku.dk.
Viktoria Hancock, Email: vhan@food.dtu.dk.
Andreas M Petersen, Email: munk@dadlnet.dk.
Karen A Krogfelt, Email: kak@ssi.dk.
Per Klemm, Email: pekl@food.dtu.dk.
Acknowledgements
We thank Birthe Jul Bondo for expert technical assistance. This work was supported by Lundbeckfonden (grant no. R17-A1603).
References
- Loftus EVJ. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- Martin HM, Rhodes JM. Bacteria and inflammatory bowel disease. Curr Opin Infect Dis. 2000;13:503–509. doi: 10.1097/00001432-200010000-00012. [DOI] [PubMed] [Google Scholar]
- Bergman L, Krause U. The incidence of Crohn's disease in central Sweden. Scand J Gastroenterol. 1975;10:725–729. [PubMed] [Google Scholar]
- Loftus EVJ, Sandborn WJ. Epidemiology of inflammatory bowel disease. Gastroenterol Clin North Am. 2002;31:1–20. doi: 10.1016/S0889-8553(01)00002-4. [DOI] [PubMed] [Google Scholar]
- Mayberry JF, Probert CSJ, Jayanthi V, Shivananda S. In: Inflammatory bowel disease. Anagnostides AA, Hodgson HJF, Kirsner JB, editor. London: Chapman and Hall; 1991. Epidemiology of inflammatory disease: a European perspective. [Google Scholar]
- Thornton JR, Emmett PM, Heaton KW. Diet and Crohn's disease: characteristics of the pre-illness diet. Br Med J. 1979;2:762–764. doi: 10.1136/bmj.2.6193.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onderdonk AB, Richardson JA, Hammer RE, Taurog JD. Correlation of cecal microflora of HLA-B27 transgenic rats with inflammatory bowel disease. Infect Immun. 1998;66:6022–6023. doi: 10.1128/iai.66.12.6022-6023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath HC, Ikeda JS, Linde HJ, Scholmerich J, Wilson KH, Sartor RB. Varying cecal bacterial loads influences colitis and gastritis in HLA-B27 transgenic rats. Gastroenterology. 1999;116:310–319. doi: 10.1016/S0016-5085(99)70127-7. [DOI] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN, Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Lepage P, Seksik P, Dore J, Marteau P. Temperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J Clin Microbiol. 2006;44:3172–3177. doi: 10.1128/JCM.02600-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/S0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- Matsuda H, Fujiyama Y, Andoh A, Ushijima T, Kajinami T, Bamba T. Characterization of antibody responses against rectal mucosa-associated bacterial flora in patients with ulcerative colitis. J Gastroenterol Hepatol. 2000;15:61–68. doi: 10.1046/j.1440-1746.2000.02045.x. [DOI] [PubMed] [Google Scholar]
- Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect Immun. 1999;67:4499–4509. doi: 10.1128/iai.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash JH, Villegas A, Kropinski AM, Aguilar-Valenzuela R, Konczy P, Mascarenhas M, Ziebell K, Torres AG, Karmali MA, Coombes BK. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC genomics. 2010;11:667. doi: 10.1186/1471-2164-11-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S, Peyretaillade E, Claret L, de Vallee A, Dossat C, Vacherie B, Zineb EH, Segurens B, Barbe V, Sauvanet P. et al. Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82. PloS one. 2010;5:e12714. doi: 10.1371/journal.pone.0012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruis W, Schutz E, Fric P, Fixa B, Judmaier G, Stolte M. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther. 1997;11:853–858. doi: 10.1046/j.1365-2036.1997.00225.x. [DOI] [PubMed] [Google Scholar]
- Sonnenborn U, Schulze J. Actual clincal studies on the efficacy of a probiotic drug. Microecology and Therapy. 1997;26:211–220. [Google Scholar]
- Rembacken BJ, Snelling AM, Hawkey PM, Chalmers DM, Axon AT. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet. 1999;354:635–639. doi: 10.1016/S0140-6736(98)06343-0. [DOI] [PubMed] [Google Scholar]
- Hancock V, Dahl M, Klemm P. Probiotic Escherichia coli strain Nissle 1917 out-competes intestinal pathogens during biofilm formation. J Med Microbiol. 2010;59:392–399. doi: 10.1099/jmm.0.008672-0. [DOI] [PubMed] [Google Scholar]
- Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P. A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol Genet Genomics. 2010;283:469–484. doi: 10.1007/s00438-010-0532-9. [DOI] [PubMed] [Google Scholar]
- Vejborg RM, Hancock V, Schembri MA, Klemm P. Comparative genomics of urinary tract infectious Escherichia coli. Appl Environ Microbiol. 2011. Accepted, March 9, 2011. [DOI] [PMC free article] [PubMed]
- Petersen AM, Nielsen EM, Litrup E, Brynskov J, Mirsepasi H, Krogfelt KA. A phylogenetic group of Escherichia coli associated with active left-sided inflammatory bowel disease. BMC Microbiol. 2009;9:171. doi: 10.1186/1471-2180-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AL, Henderson TA, Vigil PD, Mobley HL. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J Bacteriol. 2009;191:3469–3481. doi: 10.1128/JB.01717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Medina M, Naves P, Blanco J, Aldeguer X, Blanco J, Blanco M, Ponte C, Soriano F, Darfeuille-Michaud A, Garcia-Gil LJ. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC) BMC Microbiology. 2009;9:202. doi: 10.1186/1471-2180-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, Eric Jezek G, Islas-Islas M, Torres AG. Escherichia coli isolated from a Crohn's disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int J Med Microbiol. 2008;298:397–409. doi: 10.1016/j.ijmm.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, Stappert D, Wantia N, Rodriguez N, Wagner H. et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nature medicine. 2008;14:399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- Schubert S, Norenberg D, Clermont O, Magistro G, Wieser A, Romann E, Hoffmann C, Weinert K, Denamur E. Prevalence and phylogenetic history of the TcpC virulence determinant in Escherichia coli. Int J Med Microbiol. 2010;300:429–434. doi: 10.1016/j.ijmm.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Bronowski C, Smith SL, Yokota K, Corkill JE, Martin HM, Campbell BJ, Rhodes JM, Hart CA, Winstanley C. A subset of mucosa-associated Escherichia coli isolates from patients with colon cancer, but not Crohn's disease, share pathogenicity islands with urinary pathogenic E. coli. Microbiology. 2008;154:571–583. doi: 10.1099/mic.0.2007/013086-0. [DOI] [PubMed] [Google Scholar]
- Boudeau J, Barnich N, Darfeuille-Michaud A. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol Microbiol. 2001;39:1272–1284. doi: 10.1111/j.1365-2958.2001.02315.x. [DOI] [PubMed] [Google Scholar]
- Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel JF, Darfeuille-Michaud A. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlerberth I, Hanson LA, Svanborg C, Svennerholm AM, Nordgren S, Wold AE. Adhesins of Escherichia coli associated with extra-intestinal pathogenicity confer binding to colonic epithelial cells. Microb Pathog. 1995;18:373–385. doi: 10.1006/mpat.1995.0034. [DOI] [PubMed] [Google Scholar]
- Wold AE, Thorssen M, Hull S, Eden CS. Attachment of Escherichia coli via mannose- or Gal alpha 1-4Gal beta-containing receptors to human colonic epithelial cells. Infect Immun. 1988;56:2531–2537. doi: 10.1128/iai.56.10.2531-2537.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Fratamico PM, Gunther NW. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis. 2007;4:134–163. doi: 10.1089/fpd.2007.0087. [DOI] [PubMed] [Google Scholar]
- Johnson JR, Russo TA. Extraintestinal pathogenic Escherichia coli: "the other bad E coli". J Lab Clin Med. 2002;139:155–162. doi: 10.1067/mlc.2002.121550. [DOI] [PubMed] [Google Scholar]
- Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- Bachmann BJ. In: Escherichia coli and Salmonella: cellular and molecular biology. 2. Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editor. Washington, D.C.: American Society for Microbiology; 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12; pp. 2460–2488. [Google Scholar]
- Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol. 2004;186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejborg RM, Bernbom N, Gram L, Klemm P. Anti-adhesive properties of fish tropomyosins. J Appl Microbiol. 2008;105:141–150. doi: 10.1111/j.1365-2672.2007.03718.x. [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen AG, Jensen LJ, Brunak S, Stærfeldt H-H, Ussery DW. A DNA structural atlas for Escherichia coli. J Mol Biol. 2000;299:907–930. doi: 10.1006/jmbi.2000.3787. [DOI] [PubMed] [Google Scholar]
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Tables presenting genes present in the different IBD isolates, but in none of the six commensal strains (MG1655, HS, IAI1, SE11, ED1a and SE15).
Table S2. List of probes that showed significant difference between the disease groups UC and CD.