Abstract
The analytic performance of a low-cost, research-stage DNA test for the most carcinogenic human papillomavirus (HPV) genotypes (HPV16, HPV18, and HPV45) in aggregate was evaluated among carcinogenic HPV-positive women, which might be used to decide who needs immediate colposcopy in low-resource settings (“triage test”). We found that HPV16/18/45 test agreed well with two DNA tests, a GP5+/6+ genotyping assay (Kappa = 0.77) and a quantitative PCR assay (at a cutpoint of 5000 viral copies) (Kappa = 0.87). DNA sequencing on a subset of 16 HPV16/18/45 positive and 16 HPV16/18/45 negative verified the analytic specificity of the research test. It is concluded that the HPV16/18/45 assay is a promising triage test with a minimum detection of approximately 5,000 viral copies, the clinically relevant threshold.
Keywords: Human papillomavirus (HPV), HPV test, cervical cancer, HPV genotypes
Cervical cancer is one of the most common cancers, and the cause of high mortality in women worldwide (Garcia et al., 2007). Persistent cervical infection with human papillomavirus (HPV) is necessary for cervical carcinogenesis (Schiffman and Castle, 2003). Screening tests based on standardized molecular detection of HPV in cervical cells have been found to be substantially more sensitive and reliable than cytology at detecting high-grade cervical intraepithelial neoplasia (CIN) at an appropriate age (Cox, 2009; IARC, 2005 ). In low-resource settings, where cytology-based programs are not sustainable, low-cost HPV tests may provide a viable, robust cervical cancer screening option.
The careHPV™ Test (QIAGEN Gaithersburg Inc, Gaithersburg, MD, USA), currently under development, is a signal-amplification assay that detects a pool of 14 carcinogenic HPV genotypes in less than three hours, for HPV-based screening in low-resource settings (Qiao et al., 2008). In such settings, different strategies for managing the carcinogenic HPV-positive women may be implemented (Gage and Castle, 2010). For example, some programs may use an immediate screen-and-treat approach (Denny et al., 2010) without diagnostic verification of the presence of cervical pre-cancer by colposcopically directed biopsies, while other programs may require it. Of the latter, the prevalence of carcinogenic HPV DNA may be quite high, 10-20%, which may make colposcopy impractical in low-resource settings that have few trained colposcopists.
Additional triage tests are needed to identify carcinogenic HPV-positive women who are most in need of colposcopic evaluation and/or treatment, thus minimizing the number of women undergoing unnecessary colposcopy while maximizing available resources in low-resource settings.
Distinguishing HPV genotypes that differ in oncogenic potential among carcinogenic HPV positive women might improve clinical management based on patient risk (Berkhof et al., 2006; Bulkmans et al., 2007; Khan et al., 2005). In this report, research data on a newly developed, research-level triage test that works on the careHPV platform and detects the most carcinogenic HPV genotypes, HPV16, HPV18, and HPV45 (Khan et al., 2005; Munoz et al., 2003; Wright et al., 2005) in aggregate among women positive for any carcinogenic HPV genotype are presented.
The research use of the HPV16/18/45 triage test in a convenience sampling of women visiting clinics in Leogane and Blanchard, Port-au-Prince, Haiti, between October 2009 and February 2010 was evaluated. During the study enrollment period, 968 women were recruited to provide physician-collected cervical specimens. The specimens were shipped at ambient temperature to QIAGEN for HPV testing.
The careHPV Test was performed on all 968 specimens, followed by the HPV16/18/45 triage test (“triage test”) on all careHPV-positive and a subset of careHPV-negative specimens. The triage test was performed similarly to the careHPV screening test (Qiao et al., 2008), with the following difference: the triage test includes a pool of complementary RNA probes only to HPV types 16, 18, and 45 out of the 14 different carcinogenic HPV DNA types. Specimen test findings were expressed in relative light units (RLU) and compared with the mean RLU from a threshold or cutoff value (CO), resulting in a RLU/CO ratio. An RLU/CO equal to or greater than 1.0 (equivalent to ~5,000 viral copies) were classified as positive for HPV16, 18, or 45 whereas an RLU/CO less than 1.0 were classified as negative for HPV16, 18, and 45.
The analytic validity of the triage test by performing DNA sequencing on a subset of the specimens was also evaluated. DNA sequencing was performed on 34 random samples among careHPV positive specimens, with equal numbers of triage positive and negative cases. DNA was purified using a QIAGEN MinElute Media kit and amplified using the PGMY primer mix at QIAGEN (Coutlee et al., 2002). Multiplex sequencing primer mix for bi-directional coverage was conducted at Seqwright. Sequencing resulting in mixed or indeterminate sequences was subjected to sequencing primers in singleplex, and sequences were blasted against the NCBI database.
In addition, two HPV genotyping tests were performed on all women included in this study. Rather than amplifying conserved regions of the HPV genome with consensus PCR primer sets, specimens were tested using a research-use-only HPV type-specific Luminex® (“LMX”) assay based on a well-validated GP5+/6+-based consensus PCR (de Roda Husman et al., 1995) for individual detection of 17 high risk HPV types, including 16, 18, and 45 (Nazarenko et al., 2008). Specimens were also tested using a quantitative PCR (“qPCR”) test (not validated for clinical use) to estimate viral copy number per specimen aliquot. Quantitative PCR was performed using the QIAGEN Rotor-Gene Q real-time PCR Instrument. Type specific primers and probe targeting the HPV E6/E7 region were used. The HPV 16/18/45 multiplex qPCR assay was designed and validated in house. PCR primers and corresponding TaqMan probes were designed for HPV 16, 18, and 45 in the E6/E7 HPV gene region. The dynamic range of each HPV type was determined using a plasmid model. Dynamic range was determined to be at least 6-orders or magnitudes. A correlation coefficient of > 0.99 was achieved for HPV 16, 18, and 45 as measured by qPCR and the input of 10 to 107 genome copies. Each primer/probe pair was tested in Multiplex for specificity against 27 other (non-HPV16/18/45) high-risk and low-risk HPV genotypes at 107 copies/assay of each HPV type plasmid. Primers and probes for each target type were highly specific and did not show cross reactivity at this high target concentration (data not shown). Two cut points for qPCR positivity: detection at any copy number and detection at 5000 or more copies (≥ 5,000 copies), the latter being the threshold used for the FDA-approved digene® HR HPV DNA test (HC2; QIAGEN) was used. The in-house GP5/6+-LMX meet a requirement of 100 copies/assay and the qPCR is more sensitive than qualitative genotyping, with a sensitivity of 10 copies/assay. All screening tests were performed at different laboratories at QIAGEN in a masked and blinded fashion.
To evaluate the analytic performance of the triage test against qPCR and LMX among careHPV-positive women, Kappa values and total and positive percent agreements with 95% binomial confidence intervals (95% CI) were calculated. Paired assay results were tested for statistical differences using an exact McNemar’s χ2 test. The correlation between signal strength/viral load measures of the triage test and qPCR using Spearman rank correlation coefficients among women who tested positive on these two tests and plotted log-transformed levels of triage test results in RLU/CO by log transformed values of qPCR viral copies (sum of viral copy level of HPV 16/18/45) was also evaluated. All statistical analyses were performed using Stata 10.1 (Stata Corp., College Station, TX). Written, informed consent was obtained from all women enrolled in the study and Institutional Review Board approval was provided by the Western Institutional Review Board and Misyon Sante Fanmi Ayisyen, the US and Haitian institutional review boards respectively. Data analysis of the final, anomymized results were deemed exempt from review by the NIH Office of Human Subjects Research.
In this study of 968 women screened by the careHPV Test, the majority (N=848, 88%) tested negative. All 120 careHPV positive women were tested by the HPV16/18/45 triage test; both qPCR and LMX results for 118 women (2 women had neither LMX and qPCR data due to inadequate amount of sample). The concordance between triage test and DNA sequencing among 34 women was 100%, after excluding two samples that failed sequencing.
Table 1 shows the detection of HPV 16/18/45 by the triage test, qPCR, and LMX among specimens that tested positive by the careHPV Test. Of these screen-positive specimens, 26% (N=31), 26% (N=30), 38% (N = 44), and 31% (N=37) tested positive for HPV 16/18/45 by the triage test, qPCR (≥ 5,000 copies), qPCR (any copies), and LMX, respectively. Overall, good agreement between all tests was observed, suggesting that the triage test is relatively reliable for HPV 16/18/45 detection. The qPCR (≥ 5000 copies) and LMX results were highly concordant (N=108, 92%). The triage test had the best agreement with qPCR (≥ 5,000 copies), with a Kappa value of 0.87 (95% CI: 0.76-0.97), a percent total agreement of 94.9% (91.0-98.9%), and percent positive agreement of 81.8% (68.9-95.0%). The triage test had the worst agreement with qPCR (any copies), with a Kappa value of 0.69 (95%CI: 0.55-0.83), a percent total agreement of 86.4% (80.3-92.6%), and percent positive agreement of 64.4% (50.5-78.4%). In addition, a low number of false-positives for other HPV genotypes when comparing any of the genotyping tests to the triage test was observed, suggesting a positive attribute of the triage test with potentially appropriate specificity in comparison to the comparison tests. Table 2 shows the test results for the HPV16/18/45 triage test compared to qPCR (≥5,000 copies) and LMX results combined.
Table 1.
Agreement between triage test (HPV 16/18/45) with qPCR and LMX tests among careHPV positive women (N=120)
| HPV16/18/45 Triage Test |
Negative | Positive | Negative | Positive | Total* | Kappa (95% CI) |
% Agreement (95% CI) |
% Positive Agreement (95% CI) |
P- value** |
|---|---|---|---|---|---|---|---|---|---|
| Comparison Tests | Negative | Negative | Positive | Positive | |||||
| qPCR HPV 16/18/45 (≥5000 copies) |
85 | 3 | 3 | 27 | 118 | 0.87 (0.76-0.97) |
94.9% (91.0-98.9) |
81.8% (68.9-95.0) |
1 |
| qPCR HPV 16/18/45 (Any copies) |
73 | 1 | 15 | 29 | 118 | 0.69 (0.55-0.83) |
86.4% (80.3-92.6) |
64.4% (50.5-78.4) |
0.0005 |
| LMX HPV 16/18/45 | 79 | 2 | 9 | 28 | 118 | 0.77 (0.65-0.90) |
90.7% (85.4-95.9) |
71.8% (57.7-85.9) |
0.07 |
qPCR missing for three women, LMX missing for two women.
McNemar’s Exact Test
Table 2.
HPV 16/18/45 triage test results compared to qPCR HPV 16/18/45 and LMX HPV 16/18/45 test results combined among triaged cases (N=118)
| qPCR+/ LMX+ | qPCR+/ LMX− qPCR−/ LMX+ |
qPCR−/ LMX− | ||
|---|---|---|---|---|
| Triage Test Positive (+) | 26 | 3 | 1 | 30 |
|
|
||||
| Triage Test Negative (−) | 3 | 6 | 79 | 88 |
|
|
||||
| 29 | 9 | 80 | 118 | |
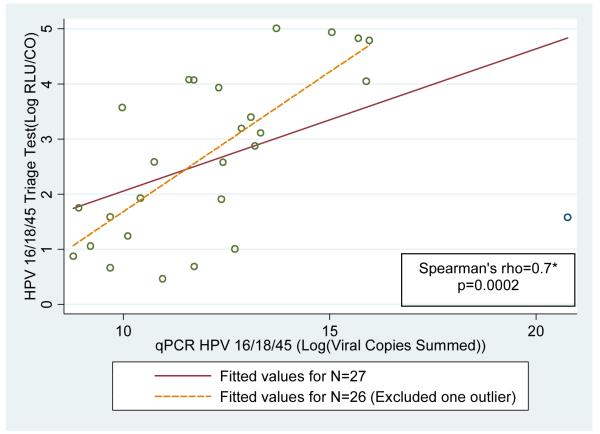
The correlation between signal strength/viral load measures of the triage test and qPCR among women who tested positive on these two tests was evaluated and good correlation (rho= 0.7; p=0.0002 with exclusion of one outlier) was observed. Plots of RLU/CO and log transformed values of qPCR viral copies (sum of viral copy level of HPV 16/18/45) are shown in Figure 1. The data indicate that the correlated aggregate levels of HPV 16/18/45 measured by the triage test and qPCR similarly distinguish positive from negative HPV 16/18/45 women.
Figure 1. Correlation between quantification of careHPV16/18/45 (triage test) with qPCR HPV 16/18/45 among triaged women concordant for qPCR/triage results (N=27).
* Note: The solid line represents the fitted value for all 27 women who were positive on both triage and qPCR test. One outlier (triage test: log RLU\CO=1.6; qPCR: log viral copies summed=20.8) was removed; the Spearman’s rho and the dotted line fitted line excludes this outlier.
There were relatively few specimens identified as HPV16/18/45 positive, limiting the ability to exhaustively evaluate the new triage test. A small proportion of women, even high-risk women, will test positive for carcinogenic HPV (Clifford et al., 2005) and only a quarter or third of all carcinogenic HPV infections are represented by HPV16, HPV18, and/or HPV45 (de Sanjose et al., 2007). Accordingly, relatively few specimens are expected to test positive. In our study, 12% of specimens tested positive for carcinogenic HPV by careHPV, and 26% of the careHPV positives also tested positive for HPV16/18/45 by the triage test, leaving only 29 of 968 (3%) positive for HPV16/18/45 by the triage test. Thus, while the new triage test appears to achieve the appropriate analytic sensitivity and specificity, much larger studies will be needed to accumulate significant numbers correlated to clinical endpoints for validation.
In conclusion, a research-level HPV16/18/45 triage test agreed well with two comparison tests for aggregate detection of HPV16, 18, and 45. Importantly, the triage test had a minimum detection of approximately 5,000 viral copies, which is the widely accepted as the clinically relevant threshold established by HC2 test results (Poljak et al., 1999). Excessive analytic sensitivity, fewer than 5,000 viral copies, only increases detection of possibly benign, transient infections that bear little risk of cervical pre-cancer and cancer and are best left undetected (Cuzick et al., 2008). Thus, the triage test might assist in risk-stratifying carcinogenic HPV positive women who are most in need of immediate colposcopic evaluation and/or treatment and might permit less aggressive management of women with other, weaker carcinogenic HPV infections in lower-resource settings where careHPV might be employed. Notably, using this algorithm, only 3% of women would have been referred to colposcopy. Further studies are needed to evaluate if the HPV16/18/45 triage test is the most feasible and efficient low-cost triage test to be used among carcinogenic HPV-positive women in resource-limited regions.
Acknowledgements
The evaluation of the HPV16/18/45 triage test data was conducted for research purposes and was supported in part by the intramural research program of the NIH/NCI. The authors would like to recognize Adam Mallonee of QIAGEN for his contribution to performing the genotyping tests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berkhof J, Bulkmans NW, Bleeker MC, Bulk S, Snijders PJ, Voorhorst FJ, Meijer CJ. Human papillomavirus type-specific 18-month risk of high-grade cervical intraepithelial neoplasia in women with a normal or borderline/mildly dyskaryotic smear. Cancer Epidemiol. Biomarkers Prev. 2006;15:1268–73. doi: 10.1158/1055-9965.EPI-05-0764. [DOI] [PubMed] [Google Scholar]
- Bulkmans NW, Berkhof J, Bulk S, Bleeker MC, van Kemenade FJ, Rozendaal L, Snijders PJ, Meijer CJ. High-risk HPV type-specific clearance rates in cervical screening. Br. J. Cancer. 2007;96:1419–24. doi: 10.1038/sj.bjc.6603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford GM, Gallus S, Herrero R, Munoz N, Snijders PJ, Vaccarella S, Anh PT, Ferreccio C, Hieu NT, Matos E, Molano M, Rajkumar R, Ronco G, de Sanjose S, Shin HR, Sukvirach S, Thomas JO, Tunsakul S, Meijer CJ, Franceschi S. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet. 2005;366:991–8. doi: 10.1016/S0140-6736(05)67069-9. [DOI] [PubMed] [Google Scholar]
- Coutlee F, Gravitt P, Kornegay J, Hankins C, Richardson H, Lapointe N, Voyer H, Franco E. Use of PGMY primers in L1 consensus PCR improves detection of human papillomavirus DNA in genital samples. J Clin Microbiol. 2002;40:902–7. doi: 10.1128/JCM.40.3.902-907.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JT. History of the use of HPV testing in cervical screening and in the management of abnormal cervical screening results. J. Clin. Virol. 2009;45(Suppl 1):S3–S12. doi: 10.1016/S1386-6532(09)70002-2. [DOI] [PubMed] [Google Scholar]
- Cuzick J, Arbyn M, Sankaranarayanan R, Tsu V, Ronco G, Mayrand MH, Dillner J, Meijer CJ. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine. 2008;26(Suppl 10):K29–41. doi: 10.1016/j.vaccine.2008.06.019. [DOI] [PubMed] [Google Scholar]
- de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 1995;76(Pt 4):1057–62. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- de Sanjose S, Diaz M, Castellsague X, Clifford G, Bruni L, Munoz N, Bosch FX. Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: a meta-analysis. Lancet Infect. Dis. 2007;7:453–9. doi: 10.1016/S1473-3099(07)70158-5. [DOI] [PubMed] [Google Scholar]
- Denny L, Kuhn L, Hu CC, Tsai WY, Wright TC., Jr. Human papillomavirus-based cervical cancer prevention: long-term results of a randomized screening trial. J Natl Cancer Inst. 2010;102:1557–67. doi: 10.1093/jnci/djq342. [DOI] [PubMed] [Google Scholar]
- Gage JC, Castle PE. Preventing cervical cancer globally by acting locally: if not now, when? J Natl Cancer Inst. 2010;102:1524–7. doi: 10.1093/jnci/djq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Jemal A, Ward EM, Center MM, Hao Y, Siegel RI, Thun MJ. Global Cancer Facts & Figures 2007. American Cancer Society; Atlanta, GA: 2007. [Google Scholar]
- IARC . Cervix Cancer Screening. IARC Press; 2005. [Google Scholar]
- Khan MJ, Castle PE, Lorincz AT, Wacholder S, Sherman M, Scott DR, Rush BB, Glass AG, Schiffman M. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J. Natl. Cancer Inst. 2005;97:1072–9. doi: 10.1093/jnci/dji187. [DOI] [PubMed] [Google Scholar]
- Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003;348:518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Nazarenko I, Kobayashi L, Giles J, Fishman C, Chen G, Lorincz A. A novel method of HPV genotyping using Hybrid Capture sample preparation method combined with GP5+/6+ PCR and multiplex detection on Luminex XMAP. J. Virol. Methods. 2008;154:76–81. doi: 10.1016/j.jviromet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Poljak M, Brencic A, Seme K, Vince A, Marin IJ. Comparative evaluation of first- and second-generation digene hybrid capture assays for detection of human papillomaviruses associated with high or intermediate risk for cervical cancer. J. Clin. Microbiol. 1999;37:796–7. doi: 10.1128/jcm.37.3.796-797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao YL, Sellors JW, Eder PS, Bao YP, Lim JM, Zhao FH, Weigl B, Zhang WH, Peck RB, Li L, Chen F, Pan QJ, Lorincz AT. A new HPV-DNA test for cervical-cancer screening in developing regions: a cross-sectional study of clinical accuracy in rural China. Lancet Oncol. 2008;9:929–36. doi: 10.1016/S1470-2045(08)70210-9. [DOI] [PubMed] [Google Scholar]
- Schiffman MH, Castle P. Epidemiologic studies of a necessary causal risk factor: human papillomavirus infection and cervical neoplasia. Journal of the National Cancer Institute. 2003;95:E2. doi: 10.1093/jnci/95.6.e2. [DOI] [PubMed] [Google Scholar]
- Wright JD, Li J, Gerhard DS, Zhang Z, Huettner PC, Powell MA, Gibb RK, Herzog TJ, Mutch DG, Trinkaus KM, Rader JS. Human papillomavirus type and tobacco use as predictors of survival in early stage cervical carcinoma. Gynecol. Oncol. 2005;98:84–91. doi: 10.1016/j.ygyno.2005.03.038. [DOI] [PubMed] [Google Scholar]