Abstract
Human telomerase, the reverse transcriptase which extends the life span of a cell by adding telomeric repeats to chromosome ends, is expressed in most cancer cells but not in the majority of normal somatic cells. Inhibition of telomerase therefore holds great promise as anticancer therapy. We have synthesized a novel telomerase inhibitor GRN163L, a lipid—attached phosphoramidate oligonucleotide complementary to template region of the RNA subunit of telomerase. Here, we report that GRN163L is efficiently taken up by human myeloma cells without any need of transfection and is resistant to nucleolytic degradation. The exposure of myeloma cells to GRN163L led to an effective inhibition of telomerase activity, reduction of telomere length and apoptotic cell death after a lag period of 2–3 weeks. Mismatch control oligonucleotides had no effect on growth of myeloma cells. The in vivo efficacy of GRN163L was confirmed in two murine models of human multiple myeloma. In three independent experiments, significant reduction in tumor cell growth and better survival than control mice was observed. Furthermore, GRN163L-induced myeloma cell death could be significantly enhanced by Hsp90 inhibitor 17AAG. These data provide the preclinical rationale for clinical evaluation of GRN163L in myeloma and in combination with 17AAG.
Keywords: telomerase, telomere, myeloma, cancer
Introduction
Telomeres, the specialized nucleoprotein structures, protect chromosomes ends from nucleolytic degradation, end-to-end fusion and unwanted recombination/repair.1 The length of human telomeric DNA in different cell types varies from 500 to 3000 repeats of a conserved TTAGGG sequence.2,3 Normal somatic cells lose 50–100 bp of telomeric DNA with each cell division as DNA replication mechanisms are unable to copy chromosomal DNA proximal to the primase site.4 Progressive telomere shortening in normal replicating somatic cells reaches a critical stage leading to telomere dysfunction and replicative senescence. Short telomeres can also be recognized as damaged DNA and may therefore induce either p53-dependent5 or in the absence of p53, p73-dependent apoptosis.6,7 Therefore, the life span of normal somatic cells is limited to less than ~100 population doublings.
The length of telomeric DNA is maintained by a ribonucleoprotein, human telomerase reverse transcriptase (hTERT), which adds telomeric repeats (TTAGGG) to the ends of human chromosomes.8 Telomerase activity has been detected in the majority of immortal and cancer cells with unlimited proliferative potential.9–11 Although telomerase activity is also present in some normal cells such as hemopoietic stem cells, gastrointestinal epithelial and germ-line cells, most normal somatic tissues do not have detectable telomerase activity.12 In normal cells (lacking telomerase), telomeres shorten with progressive cell division leading to cellular senescence whereas in most (>90%) cancer cells, telomerase is re-activated13–15 and provides unlimited proliferative potential by preventing telomere shortening with each cell division. Consistent with this, the introduction of hTERT gene in human diploid fibroblasts increases their life span in culture.16
Since telomerase activity is present in most cancers but absent or low in majority of normal cells,10,12 telomerase is an attractive therapeutic target. Moreover, the median telomere length in cancer cells is shorter than normal cells,17,18 making them susceptible to crisis and apoptosis earlier than normal cells following telomerase inhibition. A number of telomerase inhibitors have been evaluated in a variety of cancer types.19–23 We have demonstrated that telomerase inhibitors including G-quadruplex interacting agents,18,24,25 peptide nucleic acids and antisense oligonucleotides targeting RNA component of telomerase,26–28 and siRNAs targetinghTERT,29 can induce apoptosis and/or senescence in a variety of human immortal and cancer cells including multiple myeloma cells, in vitro.
In this study, we demonstrate the in vitro and in vivo efficacy of a novel and potent telomerase inhibitor GRN163L. GRN163L is a palmitoyl (C16) lipid—attached N3′–P5′ phosphoramidate oligonucleotide, complementary to the template region of the RNA subunit of telomerase (hTR). Lipid attachment and phosphoramidate chemistry allow efficient uptake of GRN163L by human cells without need for transfection reagent and is resistant to nucleolytic degradation within the cells. GRN163L is the first telomerase inhibitor validated for clinical study, and these data provide preclinical rationale for clinical evaluation of GRN163L in myeloma.
Materials and methods
Telomerase inhibitor
GRN163L, a palmitoyl (C16) lipid-attached N3′–P5′ phosphoramidate oligonucleotide, targeting the template region of RNA subunit of telomerase (hTR) was provided by Geron Corporation (Menlo Park, CA, USA). S7S- and GRN140833-mismatched oligonucleotides were also obtained from Geron Corporation and used as a negative control.
Myeloma cell lines
Human MM cell lines INA6, ARP, OPM1 and MM1S were kindly provided by Dr Renate Burger (University of Erlangen-Nuernberg, Erlangen, Germany), Dr J Epstein (University of Arkansas for Medical Sciences, Little Rock, AR, USA), Dr Edward IB Thompson (University of Texas Medical Branch, Galveston, TX, USA) and Dr Steven Rosen (Northwestern University, Chicago, IL, USA), respectively. ARP, OPM1 and MM1S cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (HyClone, South Logan, UT, USA) whereas INA6, an interleukin 6 (IL-6)-dependent cell line, was cultured in RPMI 1640 medium supplemented with 20% fetal bovine serum (HyClone) and 2.5 ng/ml recombinant human IL-6 (R&D Systems Inc., Minneapolis, MN, USA). All cell lines were maintained in a state of logarithmic growth at 37 °C in humidified air with 5% CO2, as described previously.18,24,25,27 For RNA analysis, cultures were harvested at the same final cell density (5×105 per ml), and processed immediately.
Uptake and time course of GRN163L retention within myeloma cells
Myeloma cells were treated with fluorescein isothiocyanate (FITC)-labeled GRN163L at various concentrations for a period of 6 h. The drug was then removed from the medium, cells were continued to grow in culture and the amount of FITC fluorescence retained per cell was measured using a fluorescence-activated cell flow analyzer (FACScan, Becton-Dickinson, San Jose, CA, USA) at various intervals. The half-life of intracellular FITC label was estimated from median fluorescence values obtained at different time points. To visualize the intracellular localization, treated cells were also examined for FITC fluorescence using a multiphoton fluorescence microscope (Bio-Rad, Hercules, CA, USA). Since FITC label could interfere with fluorescence-based assays and the ability of GRN163L to bind or inhibit telomerase, all subsequent experiments were conducted with nonfluorescent GRN163L.
Assay of telomerase activity
Telomerase activity was assayed using a fluorescence-based TRAPEZE XL telomerase detection kit (Intergen, Purchase, NY, USA). TRAPEZE XL telomerase detection kit provides a refined and fluorometric version of the original Telomeric Repeat Amplification Protocol (TRAP) assay. The kit utilizes ‘fluorescence energy transfer’ primers to generate fluorescently labeled TRAP products and thus allows a highly sensitive and quantitative nonisotopic detection of telomerase activity in vitro. Lysates (1000 cell equivalents) of untreated myeloma cells and those treated with GRN163L were mixed with TRAPEZE XL reaction mix containing Amplifuor primers, and incubated at 30 °C for 30 min. Amplified telomerase products were quantitated with a fluorescence plate reader. Telomerase activity (in TPG units) was calculated by comparing the ratio of telomerase products to an internal standard for each lysate, as described by the manufacturer.
Growth kinetics
Myeloma cells (5×105 per dish) were treated with GRN163L or mismatch control oligonucleotides and the live cell number was determined weekly by trypan-blue exclusion. Aliquots of cells were removed for telomere length, gene expression and apoptosis assays, and the remaining cells replated at the same cell number (5×105 cells per dish) and at the same concentration of the drug or control oligonucleotide. Aliquots for telomere length assay were stored at −150 °C, whereas gene expression profile and apoptosis were processed immediately.
Apoptosis assay
Apoptotic cells were detected using an Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA). GRN163L-treated myeloma cells (1×106 cells per ml) were mixed with annexin V-FITC and incubated in the dark for 15 min at room temperature (RT). A portion of cell suspension (50 μl) was placed onto a glass slide, covered with a cover slip and viewed immediately using a fluorescence microscope equipped with FITC (green) and PI (propidium iodide; red) filters. Two-hundred cells, representing at least five distinct microscopic fields, were analyzed to assess the fraction of FITC- and PI-labeled cells for each sample.
Estimation of telomere length by Telomere-FISH
Cells were treated with KaryoMax (Invitrogen, Carlsbad, CA, USA), hypotonic solution and fixed in glacial acetic acid—ethanol (1:3). For the direct comparison of samples, cell suspensions in the fixative were droped onto a single slide using the eight-well applicator (the plastic top of CultureSlide, Becton Dickinson Labware, Bedford, MA, USA), dropping each sample to the designated well. The air-dried specimens were than processed under virtually identical hybridization conditions, under one coverslip. Q-fluorescent in situ hybridization (FISH) was done using the Cy3-PNA (C3TA2)3 probe (Applied Biosystems/BostonProbes, Bedford, MA, USA). Images were acquired using a ×60/1.40 PlanApo Nikon objective, Nikon Eclipse E6000 microscope equipped with the SD-300-V Optical Head, and Spectral Acquisition v.2.0 software (Applied Spectral Imageing, Vista, CA, USA). Eighty nuclei were analyzed per each sample. The length of a telomere is directly related to its integrated fluorescence intensity value. The quantification of probe signals was done by FISHView v.2.1.1 software (Applied Spectral Imageing) according to the manufacturers’ recommendations.
Gene expression analysis
Total RNA was isolated utilizing an ‘RNeasy’ kit (Qiagen Inc., Valencia, CA, USA) and gene expression profile was evaluated using HG-U133 array (Affymetrix, Santa Clara, CA, USA) representing ~33 000 human genes as described previously.7 GeneChip arrays were scanned on a GeneArray Scanner (Affymetrix Inc., Santa Clara, CA, USA). Array normalization, expression value calculation and clustering analysis were performed using the dChip Analyzer. The Invariant Set Normalization method was used to normalize arrays at probe level to make them comparable, and the model-based method was used for probe selection and to compute expression values.29,30 These expression levels were assigned standard errors based on replicates, which were subsequently used to compute 90% confidence intervals of fold changes in intergroup comparisons. The lower confidence bounds of ‘fold change’ were conservative estimates of the actual changes. Expression of key genes involved in DNA damage sensing, repair, cell-cycle arrest and apoptosis was analyzed.
Animals
Severe combined immunodeficiency (SCID) mice (CB-17), obtained from Taconic (Germantown, NY, USA) were maintained and monitored in our Animal Research Facility. All animal studies were conducted according to protocols approved by the Institutional Animal Care and Use Committee. Animals were killed when their tumors reached 2 cm in diameter or when paralysis or major compromise in their quality of life occurred.
SCID-hu mouse model
Human fetal bone grafts were implanted into SCID mice (SCID-hu mice) as previously described.29–35 Approximately 8 weeks following bone implantation, 5×106 human IL-6-dependent INA6 cells were injected directly into human bone in SCID-hu host. Production of human paraprotein in mouse serum was monitored as an indicator of myeloma cell growth. At least two consecutive measurements of increasing levels of circulating human immunoglobulin signified human MM cell growth.
Human MM xenograft murine model
CB 17 SCID-mice were subcutaneously (s.c.) inoculated in the interscapular area with 2.5×106 OPM1 cells in 100 μl RPMI 1640 medium. Following appearance of palpable tumors, mice were injected intraperitoneally with phosphate-buffered saline (PBS) alone or GRN163L at concentrations described in figure legends. At the time of killing, tumors were excised and cell-cycle profiles of tumor cells derived from control and treated mice were analyzed using PI staining and flow cytometry. Briefly, cells (1×106) were washed with PBS, permeabilized by a 30 min exposure to cold 70% ethanol at 4 °C, washed with PBS, incubated with PI (5 μg/ml) in 500 ml PBS containing 10 μg/ml of RNase for 30 min at RT, and analyzed for DNA content by Cytomics FC 500 Flow Cytometer (Beckam Coulter, Fullerton, CA, USA).
Results
Uptake and fate of GRN163L into human myeloma cells in culture
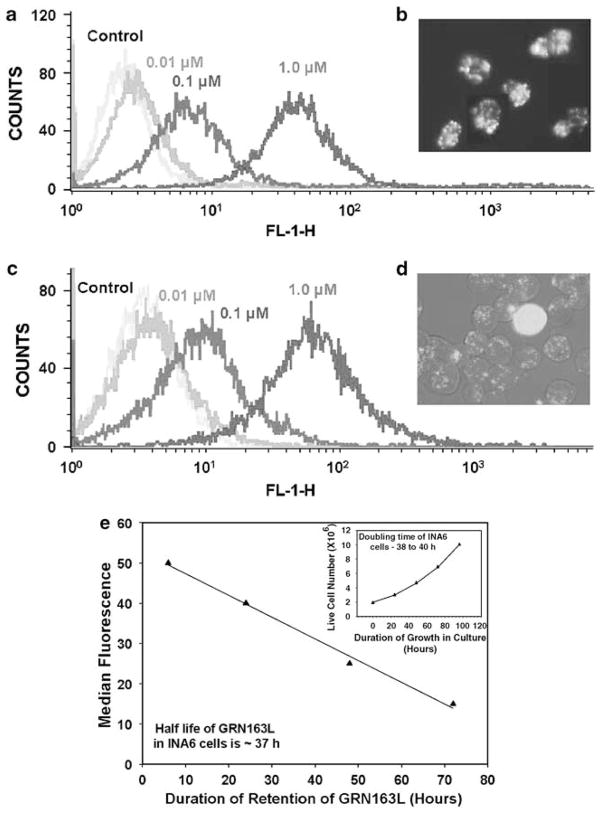
Myeloma cells were treated with FITC-labeled GRN163L at various concentrations for a period of 6 h. The drug was then removed, cells were continued to grow in culture and the amount of fluorescence retained per cell was measured using a fluorescence-activated cell flow analyzer. Intensities of median fluorescence indicate that the drug is efficiently taken up at 0.1 and 1 μM concentrations by both INA6 and ARP myeloma cells after 6 h of treatment (Figures 1a and c), without need for a transfection procedure. To determine the intracellular localization of GRN163L, the cells treated with FITC-labeled GRN163L at 1 μM for 24 h were viewed under a confocal microscope. As seen in Figure 1b (INA-6) and 1D (ARP), the drug was efficiently taken up by both myeloma cell lines and the fluorescence was predominantly nuclear with subnuclear foci visible in many cells. An efficient uptake of GRN163L without any need of transfection reagent was also observed for MM1S cells (not shown). To determine the time course of GRN163L retention in myeloma cells, INA6 cells were treated with FITC-labeled GRN163L (1 μM) for various durations and the amount of the fluorescence retained per cell was measured using a fluorescence-activated cell analyzer. The half-life of GRN163L, estimated from median fluorescence values obtained at different time points in INA6 cells (Figure 1e) is 37 h, not significantly different from the doubling time for the cell life, 38–40 h (Figure 1e inset). These data thus suggest dilution of FITC-GRN163L through cell division, instead of degradation within cell.
Figure 1.
Uptake and half-life of GRN163L in human myeloma cells. (a) Human myeloma cells (INA6) were treated with fluorescein isothiocyanate (FITC)-labeled GRN163L at 0.01, 0.1 and 1.0 μM for 6 h and the intensity of FITC fluorescence was measured using a fluorescence-activated cell sorting (FACS) analyzer. Median fluorescence values shown indicate that the drug is efficiently taken up at 0.1 and 1 μM. (b) Myeloma cells (INA6) were treated with FITC-labeled GRN163L at 1.0 μM for 24 h and viewed under a confocal microscope (Axiovert 200; Zeiss). Uptake of GRN163L can be seen as green fluorescence. (c) Myeloma cells (ARP) were treated with FITC-labeled GRN163L at 1.0 μM for 6 h and the intensities of FITC fluorescence were measured as described above. (d) Myeloma cells (ARP) were treated with FITC-labeled GRN163L at 1.0 μM for 24 h and viewed under a confocal microscope. Uptake of GRN163L can be seen as green fluorescence. (e) Myeloma cells (INA6) were treated with FITC-labeled GRN163L at 1.0 μM for various durations and the amount of FITC fluorescence retained was measured using a fluorescence-activated cell flow analyzer. Decline of median FITC fluorescence overtime is shown. (Inset) Rate of myeloma (INA6) cell growth in culture, indicating a doubling time of 38–40 h.
Inhibition of telomerase activity by GRN163L in human myeloma cells
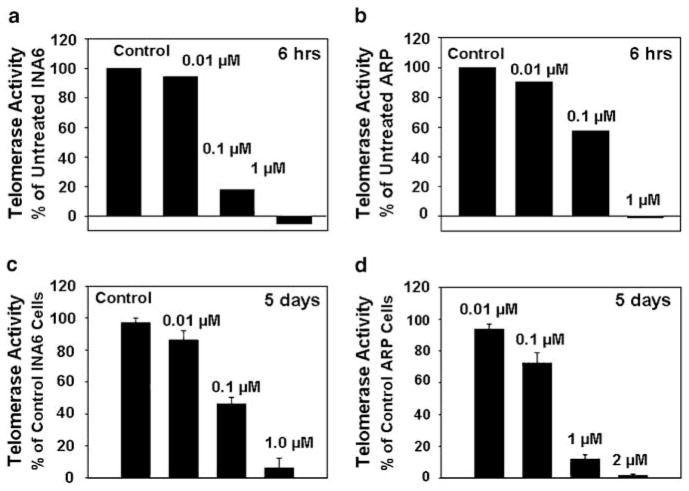
We next evaluated if the uptake of GRN163L was also associated with the inhibition of telomerase activity. We therefore treated myeloma cells with the drug at 0.01, 0.1 and 1.0 μM concentrations for various durations and evaluated for telomerase activity using TRAPEZE XL telomerase detection kit (Intergen). GRN163L at 1 μM led to a complete inhibition of telomerase activity in both INA6 and ARP cells following 6 h of treatment (Figures 2a and b). The inhibition of telomerase activity was not seen at 0.01 μM in either cell line whereas at 0.1 μM the activity was inhibited by >60% in both myeloma cell lines (Figures 2a and b). A complete inhibition of telomerase activity was maintained at day 5, following exposure to 1 μM GRN163L (Figures 2c and d). Inhibition of telomerase activity following treatment with 1 μM GRN163L was also seen for MM1S and OPM1 myeloma cell lines (data not shown). To rule out GRN163L oligonucleotide binding to RNA component of telomerase after lysis of the cells, the cells were washed at least six times after the addition of drug, potentially diluting the residual-free oligonucleotide within cells by >100-fold further upon lysis.
Figure 2.
Effect of GRN163L on telomerase activity in MM cells. Myeloma cells were treated with a mismatch control oligonucleotide or GRN163L at various concentrations and telomerase activity was determined by fluorometric detection of telomeric extension products, at various time points. Cell lysate (equivalent of 1000 cells) was mixed with TRAPEZE XL reaction mix containing Amplifluor primers, incubated for 30 min at 30 °C and the products of telomerase activity were quantitated using a fluorescence plate reader. (a) Telomerase activity in INA6 cells treated with GRN163L for 6 h is presented as percentage of activity in control cells; (b) telomerase activity in ARP cells treated with GRN163L for 6 h is presented as percentage of activity in control cells; (c) telomerase activity in INA6 cells treated with GRN163L for 5 days is presented as percentage of activity in control cells; (d) telomerase activity in ARP cells treated with GRN163L for 5 days is presented as percentage of activity in control.
Growth inhibition following treatment of myeloma cells with GRN163L
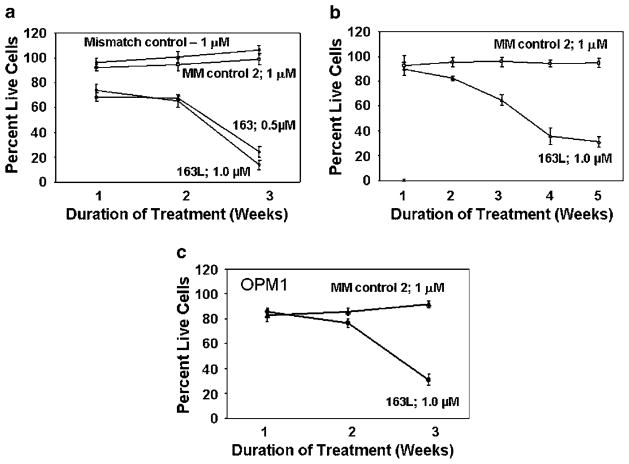
INA6, ARP, MM1S and OPM1 cells were treated with GRN163L or mismatch oligonucleotides (S7S and GRN140833) and the viable cell number determined weekly. For all cell lines tested, the treatment with GRN163L led to a decline in viable cell number to less than 5–10% of the starting cell number over a period of 3–5 weeks. Treatment with the drug at 1 μM for 3 weeks resulted in cell death in 96±4% INA6, 68±4% ARP cells and 70±2.7% OPM1 cells, respectively (Figures 3a–d). Complete cell death was observed following exposure to the drug at 2 μM for all cell lines tested (data not shown). Treatment with mismatch control oligonucleotides (S7S or GRN 140833) had no effect on growth of myeloma cells (Figure 3).
Figure 3.
Effect of GRN163L on myeloma cell survival. MM cells were cultured in regular growth medium containing GRN163L or one of two mismatch control oligonucleotides (S7S or GRN140833) at concentrations shown. Cells were harvested at different time points as indicated and proliferative potential was assessed by trypan-blue exclusion. The growth curves show the mean of three independent experiments with s.e.m. (a) IL-6-dependant INA6 myeloma cells; (b) ARP myeloma cells; (c) OPM1 myeloma cells.
Effect of GRN163L on telomere length
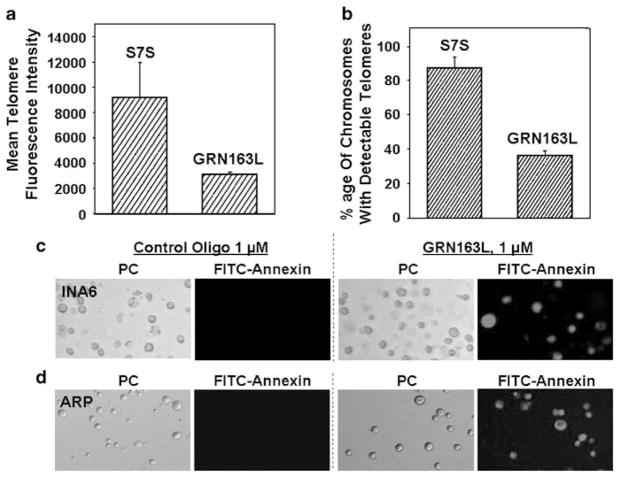
We next evaluated change in telomere length following exposure of myeloma cells to GRN163L. The effect of telomerase inhibition on telomeres of individual chromosomes was examined by Telomere-FISH analysis (Figures 4a and b). Cells treated with GRN163L or control oligonucleotide were fixed and telomeres were hybridized to Cy3-labeled telomeric probe. The impact of drug treatment on telomeres was determined by measuring intensity of telomeric signals and number of detectable telomere in each cell. Mean telomere fluorescence intensity of interphase chromosomes was reduced by 2.4-fold in INA6 myeloma cells treated with GRN163L compared to those treated with control oligonucleotide (Figure 4a). GRN163L treatment was also associated with reduction in number of chromosomes with detectable telomeres. Majority (>90%) of chromosomes in control oligotreated INA6 cells had telomeric signal; however, following GRN163L treatment ≤40% of chromosomes had detectable telomeric signal, indicating complete erosion of telomeric ends on these chromosomes (Figure 4b). A similar reduction in telomere length was also observed with other MM cell lines (data not shown).
Figure 4.
Telomere shortening and apoptosis in MM cells treated with GRN163L. INA6 myeloma cells were treated with mismatch control oligonucleotide (S7S) or GRN163L at 1 μM for 3 weeks and telomeres were examined by Telomere-fluorescent in situ hybridization (FISH) analysis. The impact of drug treatment on individual telomeres was determined by measuring intensity of telomeric signals and number of detectable telomeres. (a) Mean telomere fluorescence intensity on interphase chromosomes in control and GRN163L-treated cells; (b) Percentage of chromosomes with detectable telomeric signal, in control and GRN163L-treated cells. Myeloma cells were treated with control (S7S) oligonucleotide or GRN163L and analyzed for apoptosis using Annexin V-FITC Apoptosis Detection Kit. FITC-labeled apoptotic cells within the same microscopic field were viewed and photographed by phase contrast (PC) or by fluorescence emitted at 518nm (FITC filter). Apoptotic cells appear bright green. (c) INA6 myeloma cells, treated with 1 μM control (S7S) oligonucleotide or GRN163L for 2 weeks; (d) ARP myeloma cells, treated with 1 μM control (S7S) oligonucleotide or GRN163L for 4 weeks.
Apoptotic cell death following GRN163L treatment
INA6 and ARP myeloma cells were treated with 1 μM GRN163L for 2 and 4 weeks, respectively, and apoptotic cells detected using an Annexin V-FITC Apoptosis Detection Kit (BD Biosciences). Following GRN163L treatment, 52% of INA6 cells and 87% of ARP cells stained positive for annexin V (Figure 4b, whereas <1% annexin V-positive cells were detected when INA6 and ARP lines were treated with mismatch oligonucleotide (S7S) (Figures 4c and d). These data indicate that the mechanism of cell death following treatment with GRN163L was predominantly apoptotic.
Gene expression profiles following GRN163L treatment
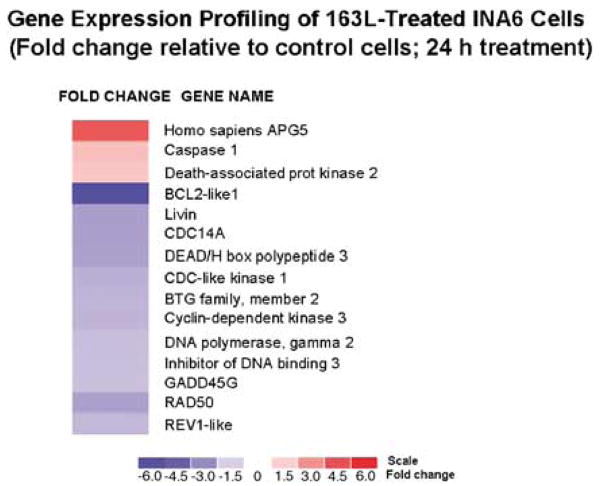
We treated myeloma cells with mismatch control (S7S) or GRN163L oligonucleotides at 1 μM for 24 h and analyzed gene expression profiles using HG-U133 GeneChip arrays (Affymetrix) representing approximately 33 000 genes. The impact of genes implicated in cell proliferation, cell cycle, apoptosis, oncogenesis, tumor suppression, telomere maintenance, and DNA repair and recombination were examined. Of ~5000 genes implicated in these important growth-related activities, the expression of only 15 was changed ≥2-fold relative to control cells (Figure 5). The most noted were homo sapiens APG5 and death-associated protein kinase 2, implicated in the induction of apoptosis.
Figure 5.
Gene expression profile following GRN163L treatment of MM cells. INA6 myeloma cells were treated with 1 μM control oligonucleotide or GRN163L for 24 h and gene expression was evaluated using Human Genome U133 GeneChip arrays (Affymetrix). Fold change in gene expression in GRN163L-treated cells relative to control cells is shown by intensities of red (indicating upregulation) and blue (indicating downregulation) colors. The scale is shown at the bottom.
Effect of combining GRN163L-mediated telomerase inhibition with other agents
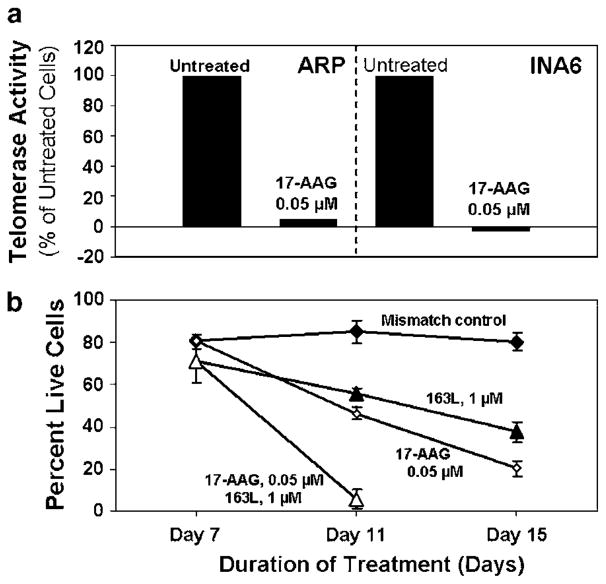
As hsp90 forms an important component of telomerase complex and hsp90 inhibitor has demonstrated activity in MM, we evaluated efficacy of combination of GRN163L with the hsp90 inhibitor 17AAG. As seen in Figure 6a, 17AAG was able to completely inhibit telomerase activity in both INA6 and ARP myeloma cells at 0.05 μM concentration. To evaluate the effect of combination of GRN163L and 17AAG, INA6 myeloma cells were treated with GRN163L at 1 μM for 7 days and 17AAG was then added to the culture at 0.05 μM. Addition of 17AAG to myeloma cells pretreated with GRN163L for 1 week led to complete growth arrest within 4 days, compared to continued growth of cells not pretreated with GRN163L (Figure 6b).
Figure 6.
Effect Hsp90 inhibitor 17AAG on GRN163L-induced MM cell death. (a) INA6 and ARP myeloma cells were treated with 17AAG at 0.05 μM for 24 h and telomerase activity in lysates (1000 cell equivalent) was assessed using TRAPEZE XL Telomerase Detection Kit, as described above; (b) INA6 myeloma cells were treated with 1 μM mismatch control oligonucleotide or GRN163L for 7 days and the cells in each treatment flask were then divided into two aliquots. Cells were then cultured either in the presence of GRN163L or mismatch oligonucleotides alone or with addition of 17AAG at 0.05 μM. Live cell number was determined at different time points by trypan-blue exclusion.
In vivo efficacy of GRN163L
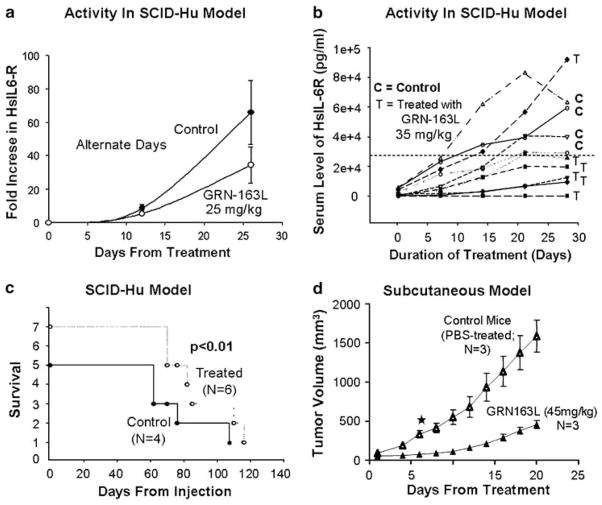
In vivo efficacy of GRN163L was demonstrated in two different murine models (SCID-hu and s.c.) of human multiple myeloma. For SCID-hu mouse model, human fetal bone grafts were implanted into SCID mice and 5×106 INA6 cells were injected directly into human bone implanted in SCID-hu host. Production of human soluble IL-6 receptor was monitored in mouse serum as an indicator of myeloma cell growth. Intraperitoneal administration of GRN163L at 25 mg/kg (Figure 7a) and 35 mg/kg (Figure 7b) on alternate days led to a significant decline in shuIL6-R at both dose levels compared to control group. Additionally, GRN163L-treated mice also had an improved survival compared to control mice (Figure 7c). Efficacy of GRN163L was also tested in s.c. murine model in which SCID mice were s.c. inoculated in the interscapular area with 2.5×106 MM1S myeloma cells. Following appearance of palpable tumors, mice were injected intraperitoneally with PBS alone or GRN163L. Significant reduction in tumor size was observed following 3 weeks treatment with daily intraperitoneal injections of 45 mg/kg GRN163L (Figure 7d).
Figure 7.
In vivoefficacy of GRN163L in multiple myeloma. (a–c) Activity of GRN163L in severe combined immunodeficiency (SCID)-hu mice. Human fetal bone chips were implanted in CB-17 SCID mice and enhanced green fluorescent proteins (EGFP)-transduced IL-6-dependent human myeloma cells were then injected in fetal bone chips. Following detection of human soluble IL-6 receptor in mice serum, the mice were treated with GRN163L. (a) GRN163L (25 mg/kg) was injected intraperitoneally at alternate days and impact on tumor growth was evaluated by measuring human soluble IL-6 receptor in murine serum; (b) GRN163L (35 mg/kg) was injected intraperitoneally at alternate days and impact on tumor growth was evaluated by measuring human soluble IL-6 receptor in mice serum; (c) survival curve of control and GRN163L-treated mice described in panel (b); (d) SCID mice were inoculated subcutaneously in the interscapular area with 5×106 OPM1 myeloma cells and following appearance of tumors, the mice were treated intraperitoneally with phosphate-buffered saline (PBS) alone or GRN163L 35 mg/kg/day.
Discussion
This study shows that (1) GRN163L, an oligonucleotide targeting RNA component of telomerase, is efficiently taken up by human myeloma cells and is not significantly degraded within cells; (2) the uptake of GRN163L leads to inhibition of telomerase activity and induces telomere shortening and apoptosis in multiple myeloma cells; (3) GRN163L-induced growth arrest of myeloma cells is enhanced by Hsp90 inhibitor and (4) in vivo efficacy of this agent is observed in s.c. and SCID-hu murine models of human multiple myeloma.
Although a variety of telomerase inhibitors have been shown to inhibit proliferation of cancer cells,18–25,27–29 there has been a need of agents which are specific enough and suited for in vivo utilization in human clinical trials. Our telomerase inhibitor (GRN163L) is phosphoramidate oligonucleotide complementary to the template region of the RNA subunit of telomerase (hTR) and has a lipid moiety attached, which facilitates uptake in human cells without any need of a transfection reagent or procedure. Using FITC-labeled GRN163L, we have shown that this drug is efficiently taken up by human myeloma cells without any transfection within 6 h of treatment, indicating an efficient uptake. Importantly GRN163L is retained in the cell for a prolonged period of time and its intracellular half-life is similar to the doubling time of recipient cells (37–40 h). These data show that the observed decline in the amount of drug overtime was due to its dilution through cell proliferation, and not due to degradation.
Consistent with an efficient uptake and a significant inhibition of telomerase activity, when myeloma cells were cultured in the presence of GRN163L, live cell number declined over a period of 3–5 weeks to <5% of the initial cell number. Although in some malignancies such as cervical cancer the inhibition of telomerase induces apoptotic cell death within a few days, without inducing significant telomere shortening,30 cell death in most cancer cells occurs after a lag period of 3–5 weeks, probably required for reduction in telomere length below critical limit.18,24–27,29,31 Moreover, we observed a marked reduction in telomere length, leading to complete erosion of telomeres on a subset of chromosomes in myeloma cells following GRN163L treatment. These data along with minimal change in gene expression confirm that GRN163L induces myeloma cell death through specific inhibition of telomerase and that it does not have significant nontelomeric effects on myeloma cells. This is quite consistent with our previous observations data showing that treatment with chemical, antisense and siRNA-based telomerase inhibitors does not affect the proliferation of normal cells.29,31,32
Cancer cells vary in genetic and epigenetic factors, including intracellular levels of nucleases. Therefore, the loss of telomeric DNA following inhibition of telomerase activity may not be predictable and the efficacy of the inhibitor may not necessarily correlate with median telomere length. Consistent with this we and others22,32 have observed that, in certain cancer cell lines including myeloma, a substantially large reduction in telomere length can be achieved with relatively shorter duration of treatment with a telomerase inhibitor. Moreover, telomeres on individual chromosomes may be heterogeneous in size. Therefore, the chromosome/s with the smallest telomeres could be lost faster than those with significantly longer telomeres and loss of one or more critical chromosomes may be sufficient to induce cell death. So overall it is uncertain whether baseline telomere length will affect clinical efficacy of telomerase inhibitor.
Although expression of some apoptosis-related genes such as ‘APG5’ and ‘death-associated protein kinase 2’ was also elevated, no change was observed in the expression of p53 or any of its isoforms including p63 and p73, which are implicated in induction of apoptosis following telomere dysfunction.29 Similarly, the treatment of myeloma cells for a short period of 24 h did not change the expression of any of the main regulatory genes implicated in cell-cycle arrest and apoptosis such as p16, p18, TRAIL, caspases 8 and 10.
We have also demonstrated that telomere shortening and apoptosis following treatment of myeloma cells with GRN163L can be expedited with agents that may inhibit telomerase activity or affect telomeres by distinct mechanisms. Hsp90 is involved in telomerase complex formation and it actually forms a part of the complex. We have previously shown that hsp90 inhibitor inhibits telomerase activity and others have also confirmed that hsp90 inhibition prevents incorporation of RNA component into the telomerase protein structure. Additionally, Hsp90 is a molecular chaperone, required for the attainment of specific conformation, stability and proper functioning of a variety of cellular proteins including telomerase33 as well as several other proteins implicated in DNA damage repair and telomere maintenance through alternate pathways.34,35 The rationale for combining Hsp90 inhibitor with GRN163L was to first initiate the destruction of telomeric DNA in cancer cells by treating with a specific inhibitor (GRN163L) and then expedite the degradation of vulnerable telomeres by giving a brief exposure to an agent, which may significantly expedite the destruction of already eroding telomeres through different mechanisms. We chose Hsp90 inhibitor as potentially it could induce a more complete inhibition of telomerase activity, prevent alternate pathway of telomere maintenance, inhibit the repair of eroding telomeres by DNA repair and recombination proteins, and could possibly affect the stability of telomere stabilizing proteins. Here, we show that 17-AAG in combination with GRN163L, significantly enhanced the cell death. This data provide further rationale to combine GRN163L with other agents able to affect telomere shortening or genomic integrity to significantly expedite tumor cell killing.
Finally, we demonstrate the in vivo efficacy of GRN163L in two murine models of human multiple myeloma. In independent experiments using SCID-hu murine model, as well as SCID mice bearing s.c. tumors derived from human myeloma cells, treatment with GRN163L reduced tumor size as well as improved survival.
These preclinical data showing that GRN163L is a potent and specific telomerase inhibitor, provide the rationale for clinical evaluation of GNR163L in patients with relapsed multiple myeloma.
Acknowledgments
This work was supported in part by NIH-P50-100007 Developmental Research Award to MAS, and Merit Review Award from the Department of Veterans Affairs and NIH—RO1-CA124929 (NCM); NIH-P050-100007 and NIH-PO1-78378 (NCM and KCA). NCM is a Leukemia Society Scholar in Translational Research.
References
- 1.Day JP, Marder BA, Morgan WF. Telomeres and their possible role in chromosome stabilization. Environ Mol Mutagen. 1993;22:245–249. doi: 10.1002/em.2850220411. [DOI] [PubMed] [Google Scholar]
- 2.Allshire RC, Gosden JR, Cross SH, Cranston G, Rout D, Sugawara N, et al. Telomeric repeat from T. thermophila cross hybridizes with human telomeres. Nature. 1988;332:656–659. doi: 10.1038/332656a0. [DOI] [PubMed] [Google Scholar]
- 3.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayflick L. The cellular basis for biological aging. In: Finch CE, Hayflick L, editors. Handbook of the Biology of Aging. Van Nostrand Reinhold Co; NY: 1977. pp. 159–186. [Google Scholar]
- 6.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr, Levrero M, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 7.Stiewe T, Putzer BM. p73 in apoptosis. Apoptosis. 2001;6:447–452. doi: 10.1023/a:1012433522902. [DOI] [PubMed] [Google Scholar]
- 8.Blackburn EH, Greider CW, Henderson E, Lee MS, Shampay J, Shippen-Lentz D. Recognition and elongation of telomeres by telomerase. Genome. 1989;31:553–560. doi: 10.1139/g89-104. [DOI] [PubMed] [Google Scholar]
- 9.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 11.Lord RV, Salonga D, Danenberg KD, Peters JH, DeMeester TR, Park JM, et al. Telomerase reverse transcriptase expression is increased early in the Barrett’s metaplasia, dysplasia, adenocarcinoma sequence. J Gastrointest Surg. 2000;4:135–142. doi: 10.1016/s1091-255x(00)80049-9. [DOI] [PubMed] [Google Scholar]
- 12.Broccoli D, Young JW, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci USA. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 14.Shay JW. Telomerase in human development and cancer. J Cell Physiol. 1997;173:266–270. doi: 10.1002/(SICI)1097-4652(199711)173:2<266::AID-JCP33>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 15.Shay JW, Wright WE. The reactivation of telomerase activity in cancer progression. Trends Genet. 1996;12:129–131. doi: 10.1016/0168-9525(96)30018-8. [DOI] [PubMed] [Google Scholar]
- 16.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 17.Bechter OE, Eisterer W, Pall G, Hilbe W, Kuhr T, Thaler J. Telomere length and telomerase activity predict survival in patients with B cell chronic lymphocytic leukemia. Cancer Res. 1998;58:4918–4922. [PubMed] [Google Scholar]
- 18.Shammas MA, Shmookler Reis RJ, Akiyama M, Koley H, Chauhan D, Hideshima T, et al. Telomerase inhibition and cell growth arrest by G-quadruplex interactive agent in multiple myeloma. Molecular Cancer Therapeutics. 2003;2:825–833. [PubMed] [Google Scholar]
- 19.Fu W, Begley JG, Killen MW, Mattson MP. Anti-apoptotic role of telomerase in pheochromocytoma cells. J Biol Chem. 1999;274:7264–7271. doi: 10.1074/jbc.274.11.7264. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima A, Tauchi T, Sashida G, Sumi M, Abe K, Yamamoto K, et al. Telomerase inhibition enhances apoptosis in human acute leukemia cells: possibility of antitelomerase therapy. Leukemia. 2003;17:560–567. doi: 10.1038/sj.leu.2402825. [DOI] [PubMed] [Google Scholar]
- 21.Sumi M, Tauchi T, Sashida G, Nakajima A, Gotoh A, Shin-Ya K, et al. A G-quadruplex-interactive agent, telomestatin (SOT-095), induces telomere shortening with apoptosis and enhances chemosensitivity in acute myeloid leukemia. Int J Oncol. 2004;24:1481–1487. [PubMed] [Google Scholar]
- 22.Tauchi T, Shin-Ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, et al. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: involvement of ATM-dependent DNA damage response pathways. Oncogene. 2003;22:5338–5347. doi: 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- 23.Seimiya H, Oh-hara T, Suzuki T, Naasani I, Shimazaki T, Tsuchiya K, et al. Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors MST-312, MST-295, and MST-1991. Mol Cancer Ther. 2002;1:657–665. [PubMed] [Google Scholar]
- 24.Shammas MA, Koley H, Beer DG, Li C, Goyal RK, Munshi NC. Growth arrest, apoptosis, and telomere shortening of Barrett’s-associated adenocarcinoma cells by a telomerase inhibitor. Gastroenterology. 2004;126:1337–1346. doi: 10.1053/j.gastro.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 25.Shammas MA, Reis RJ, Li C, Koley H, Hurley LH, Anderson KC, et al. Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clin Cancer Res. 2004;10:770–776. doi: 10.1158/1078-0432.ccr-0793-03. [DOI] [PubMed] [Google Scholar]
- 26.Akiyama M, Hideshima T, Shammas MA, Hayashi T, Hamasaki M, Tai YT, et al. Effects of oligonucleotide N3′→P5′ thio-phosphoramidate (GRN163) targeting telomerase RNA in human multiple myeloma cells. Cancer Res. 2003;63:6187–6194. [PubMed] [Google Scholar]
- 27.Shammas MA, Liu X, Gavory G, Raney KD, Balasubramanian S, Shmookler Reis RJ. Targeting the single-strand G-rich overhang of telomeres with PNA inhibits cell growth and induces apoptosis of human immortal cells. Exp Cell Res. 2004;295:204–214. doi: 10.1016/j.yexcr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Shammas MA, Simmons CG, Corey DR, Reis RJ. Telomerase inhibition by peptide nucleic acids reverses ‘immortality’ of transformed human cells. Oncogene. 1999;18:6191–6200. doi: 10.1038/sj.onc.1203069. [DOI] [PubMed] [Google Scholar]
- 29.Shammas MA, Koley H, Batchu RB, Bertheau RC, Protopopov A, Munshi NC, et al. Telomerase inhibition by siRNA causes senescence and apoptosis in Barrett’s adenocarcinoma cells: mechanism and therapeutic potential. Mol Cancer. 2005;4:24. doi: 10.1186/1476-4598-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yatabe N, Kyo S, Kondo S, Kanaya T, Wang Z, Maida Y, et al. 2-5A antisense therapy directed against human telomerase RNA inhibits telomerase activity and induces apoptosis without telomere impairment in cervical cancer cells. Cancer Gene Ther. 2002;9:624–630. doi: 10.1038/sj.cgt.7700479. [DOI] [PubMed] [Google Scholar]
- 31.Shammas MA, Simmons CG, Corey DR, Shmookler Reis RJ. Telomerase inhibition by peptide nucleic acids reverses ‘immortality’ of transformed human cells. Oncogene. 1999;18:6191–6200. doi: 10.1038/sj.onc.1203069. [DOI] [PubMed] [Google Scholar]
- 32.Shammas MA, Shmookler Reis RJ, Li C, Koley H, Hurley LH, Anderson KC, et al. Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clin Cancer Res. 2004;10:770–776. doi: 10.1158/1078-0432.ccr-0793-03. [DOI] [PubMed] [Google Scholar]
- 33.Workman P. Overview: translating Hsp90 biology into Hsp90 drugs. Curr Cancer Drug Targets. 2003;3:297–300. doi: 10.2174/1568009033481868. [DOI] [PubMed] [Google Scholar]
- 34.Noguchi M, Yu D, Hirayama R, Ninomiya Y, Sekine E, Kubota N, et al. Inhibition of homologous recombination repair in irradiated tumor cells pretreated with Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Biochem Biophys Res Commun. 2006;351:658–663. doi: 10.1016/j.bbrc.2006.10.094. [DOI] [PubMed] [Google Scholar]
- 35.Powers MV, Workman P. Targeting of multiple signalling pathways by heat shock protein 90 molecular chaperone inhibitors. Endocr Relat Cancer. 2006;13 (Suppl 1):S125–S135. doi: 10.1677/erc.1.01324. [DOI] [PubMed] [Google Scholar]