Abstract
Intracisternal A-particle retrotransposons (IAPs) are known, moveable, retrovirus-like elements and are defective in envelope protein synthesis in the mouse genome. Insertion of IAP elements can either interupt or enhance gene function or expression. Using a mouse model called lethal wasting (lew), we recently identified the insertion of an IAP sequence in a gene, 9630033F20Rik, that contains domains involved in glycolysis. The expression pattern of the 9630033F20Rik gene between various normal and diseased tissues was determined by semi-quantitative RT-PCR. The effect of the insertion mutation in 9630033F20Rik on glycolysis in heart, muscle, and brain tissues was further investigated using oligonuleotide microarray analysis. Results indicated that the expression of 9630033F20Rik is ubiquitous and its signal is relatively higher in heart and brain tissues. The insertion caused the deletion of exon 5 and decreased expression of this gene in all the tissues studied in the lew mice. Changes in the expression levels of glycolytic genes mainly occured in muscle tissue, raising a possibility that 9630033F20Rik may function as one of the transcriptional regulators of glycolytic genes in skeletal muscle. However, considering the fact that a single nucleotide mutation in vesicle-associated membrane protein 1 (VAMP1) has been reported as the causal gene for the lew mice, how much of an impact the IAP insertion in the lew mouse phenotype has on glycolytic genes compared to the effect from the VAMP1 mutation responsible for the lew mouse phenotype should be further investigated.
Keywords: Intracisternal type A particle element, insertion, glycolysis, High-Throughput Screening, Phosphoglycerate mutase
Introduction
Endogenous retroviral elements (ERVs) are retroelements that integrate into a chromosome of a germ cell where they can persist as stable integrated proviruses for multiple generations. ERVs are significant genomic mutagens in mice, causing approximately 10% of all reported spontaneous germ line mutations in laboratory strains (Takabatake et al., 2008; Zhang et al., 2008). Recently, their impact on genome function has brought much attention, especially the intracisternal A-particle retrotransposons (IAPs). IAPs are retrovirus-like elements and are defective in envelope protein synthesis and exist without an extracellular stage. The impact of IAPs on genome evolution and their integration sites have been extensively studied recently (Saito et al., 2008, Zhang et al., 2008). In one particular example, Howard et al. (2008) reported on the activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice.
In addition to their own expression and function in the genome, insertion of IAPs in regulatory or normal gene sequences can alter the expression and disrupt the gene's function. Recently, Sun et al. (2008) reported that insertion of an IAP retrotransposon element in the plasma membrane calcium ATPase 2 gene attenuates its expression and produces an ataxic phenotype in joggle mutant mice. Earlier, Zhou et al. (2007) reported that an IAP retrotransposon in the mouse ADAMTS13 gene creates ADAMTS13 variant proteins that are less effective in cleaving von Willebrand factor multimers. Closely relevant to Zhou's short report is the spontaneous muscular dystrophy caused by a retrotransposal insertion in the mouse laminin alpha2 chain gene (Besse et al., 2003).
In this report, we showed presence of an IAP element insertion in the novel gene 9630033F20Rik, which contains domains involved in glycolysis in a mouse model called lethal wasting (lew). A single nucleotide mutation in vesicle-associated membrane protein 1 (VAMP1) has been reported as the causal gene for the lew mice (Nystuen et al., 2007). However, VAMP1 is selectively expressed in the retina and in discrete areas of the brain including the zona incerta and rostral periolivary region and no gross histological abnormalities were observed in those tissues (Nystuen et al., 2007). Our preliminary microarray analysis revealed an inhibition of glycolysis activity, mainly in skeletal muscle. Many muscle wasting related genes were dysregulated in the lew mice, suggesting the possibility that insertion of an IAP sequence in 9630033F20Rik leads to gene expression profile changes. Because 9630033F20Rik has been ruled out as the candidate gene for the lethal wasting disease in lew mice by mapping (Nystuen et al., 2007), the exact impact the IAP insertion has on the lew mouse phenotype is not clear.
Materials and Methods
Mice
One pair of heterozygous (lew/+) mice were purchased from the Mouse Mutant Stock Resource colonies at The Jackson Laboratory (TJL). A breeding colony was then established by mating them at the University of Tennessee Health Science Center (UTHSC). Experimental animal procedures and mouse husbandry were performed in accordance with the National Institute of Health's Guide for the Care and Use of Laboratory Animals and approved by the UTHSC Institutional Animal Care and Use Committee.
High-Throughput screening of the lew Locus
Our aim was to identify a gene that is possibly responsible for the lew disease, other than VAMP1. We performed screening using the procedure reported recently (Jiao et al., 2005a, 2005b) around the genomic region of the lew locus, which is located on mouse chromosome 6 between D6Mit55 (114.1mbp) and D6Mit111 (113.8 mbp) (Figure 1A). Liver genomic DNA (gDNA) from +/+, +/lew, and homozygous (lew/lew) mice were extracted for temperature gradient capillary electrophoresis (TGCE) and sequence analysis. Primer pairs flanking the exons of known and predicted genes (including ESTs) within the lew locus were designed with Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Primers were located approximately 50 bp from the 5' or 3' of each exon and, in general, produced 300–400 bp amplicons. A TGCE device (SpectruMedix; State College, PA) was used to analyze amplicons from +/+, +/lew, and lew/lew mouse mixtures. The SpectruMedix system includes a high-throughput capillary electrophoresis instrument capable of analyzing 96 samples every 140 minutes. Heteroduplex analysis was subsequently performed offline using SpectruMedix software. Amplicons from the lew mice were sequenced if they differed from the norm.
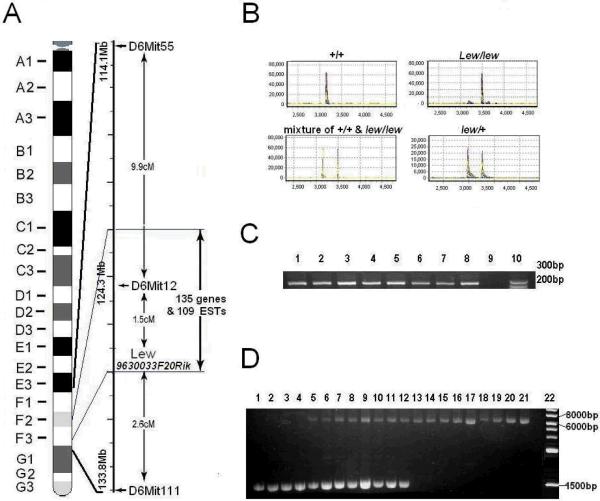
Fig. 1.
Physical localization and identification of IAP insertion of Lew gene. 1A. Location and genetic elements of lew locus (9630033F20Rik) previously mapped on murine chromosome 6; 1B. Sequence analysis of exon 5 amplificant of Lew gene from normal (+/+), heterozygous (lew/+), homozygous (lew/lew) mice and a mixture of +/+ with lew/lew samples. The X-axis represents relative size of the PCR products. Y-axis represents relative strength of signal or the amount of the PCR products. 1C. The PCR amplification products of exon 5 of Lew gene from 8 strains of mice and Lew/Lew mouse. A pair of primers (forward: GACTAGGTCACCGGCACACT, reverse AGGCCTTTACCATCATGTCG) was designed to flank exon 5 of a Lew gene with a size of 252 bps. Samples of lane 1–8 were amplification from C3H/HeJ, C57BL/6J, BALB/cJ, DBA/1J, A/J, PL/J, CAST/Ei/J, SJL/J and lew/lew mice respectively. Lane 10 is DNA marker. ; 1D. Long-range PCR amplification of Lew gene. A pair of primers (forward: GATTAGCTGACAAATGAAATCAAAGA, reverse GTTCTCTGAGGTTCAAGAGACATAAA) was designed to flank exon 5 of a Lew gene with a size of 1511 bps. They were used in long-region PCR to amplify a DNA fragment from mice of+/+, lew/+ and lew/lew genome DNA. Samples of lane 1–4 were amplificants from 4 different normal mice; lanes 5–12 were amplificants from 8 different lew/+ mice and lanes 13–21 were amplification products from 9 different individual lew/lew mice; lane 22 is DNA marker
Total RNA isolation and semi-quantitative RT-PCR
Total RNAs were extracted from 12 different tissues, including skeletal muscle, heart, liver, spleen, thymus, lung, intestine, spine, femur, cerebrum, and cerebellum from female normal and lew/lew mice of 13 days of age, with Trizol Reagent (Invitrogen, Carlsbad, CA). The quality of the resultant total RNA was visually determined by observing distinct 28S and 18S ribosomal bands on an agarose gel. The yield and purity of the extracted RNA was quantified by Nano-drop 2000. Samples with a 28S/18S ratio ≥ 1.8 and an A260/A280 ratio of 1.8–2.1 were chosen for this study. Reverse transcription and PCR were conducted using a One-Step RT-PCR kit (Invitrogen). Reactions were performed in a total volume of 50 μl with 8ng/μl of total RNA, dNTP, MgCl2, RNase inhibitor, and primers. First, cDNA synthesis and pre-denaturation were performed in single cycles at 50°C for 40 min and 94°C for 2 min. Next, PCR amplification was performed for 27 cycles: 94°C for 30 sec, 54–58°C for 36 sec, and 72°C for 2 min. The PCR products were analyzed with a 2.0% agarose gel. The quantitative analysis of the PCR products was conducted using Scion Image software (http://rsb.info.nih.gov/nih-image).
DNA sequencing
DNA sequencing was conducted to verify the insertion in the gDNA and cDNA of 9630033F20Rik following our reported procedure (Jiao et al., 2005a). The same primers in the amplification of DNA fragments from either gDNA or mRNA were also used in the sequencing. Sequencing was conducted two times to verify the result for either gDNA or cDNA.
Microarray analysis
To eliminate some individual variation from later analyses and insure more uniform results, individual heart, skeletal muscle, and cerebrum samples from +/+ (n=3) and lew/lew (n=3) mice were pooled into two groups (Lew and WT) prior to total RNA isolation. Microarray assay was conducted following the standard Affymetrix (Santa Clara, CA) protocol. Briefly, 8 μg of total RNA from each group was used for cDNA synthesis by the SuperScript Choice System (Invitrogen), followed by cRNA synthesis using the IVT labeling kit (Affymetrix). Affymetrix's mouse genome 430 2.0 arrays were used for cRNA hybridization overnight (16h). Washing, staining, and image scanning were controlled by GCOS 1.4 software (Affymetrix). Raw data was normalized globally with a target signal of 500. Chips with <3 fold resultant scale factor difference were included for data analysis. The dataset was divided into three subsets: cerebrum, heart, and muscle, each with a pair of WT and Lew sets. Using GCOS 1.4 software, comparison analyses between WT and Lew groups for different tissues were performed based on the Affymetrix change algorithm, in which the Wilcoxon's signed rank test is used to compute p-value of the change call including Increase, Marginal Increase, No Change, Marginal Decrease, or Decrease. In addition, the intensity log ratio of Lew vs. WT was calculated for each probe set based on the Signal Log Ratio Algorithm. To identify differentially expressed genes between different groups, an arbitrary cutoff value of 2.0 fold changes was chosen based on published literature.
Results
Initial gene screening
Our initial screening is for identification of the possible causal gene for the lew disease. First, we screened the last exon of every candidate gene within a targeted region (Figure 1A). Our data indicated that there was no sequence difference in these exons between normal and lew/lew mice (data not shown). Next, we designed a second pair of primers, flanking the first exon of each gene. From these primers we again did not detect any difference between the normal and the lew/lew mice (Data not shown). Finally, we designed our third pair of primers, flanking an exon in the middle of each gene. This time, we noticed that a 252 bp DNA fragment, which includes the exon 5 of a novel gene, named 9630033F20Rik, was not amplified from the lew/lew mice (Figure 1B).
After verification of the non-amplification of the exon5-fragment by different PCR conditions and variable concentrations of DNA and primers, we examined all the other exons of 9630033F20Rik located on Chr 6 between 127905895 – 127927250 bp using specific primers for each exon. We did not find any sequence differences between normal and lew/lew mice in these exons.
Detection of an insertion mutation in exon 5 of 9630033F20Rik
To determine whether exon 5 of 9630033F20Rik can be amplified from other mouse strains, we conducted PCR amplification using DNA from multiple normal strains and a lew/lew sample using the same pair of primers that could not amplify exon 5 from 9630033F20Rik. As expected, all strains of normal mice (C3H/HeJ, C57BL/6J, BALB/cJ, DBA/1J, A/J, PL/J, CAST/Ei/J, and SJL/J) had a DNA fragment of normal size, while the lew/lew mouse did not give a DNA fragment (Figure 1C).
To determine the cause of the 252 bp fragment absence, we designed more pairs of primers covering different parts of exon 5 for PCR amplification using DNA samples from lew mice. Our assays showed that the primers with no amplification are located in the genome of lew mice (data not shown). Thus, although the primers exist in the lew mice, the PCR did not produce the expected, or any other sized, DNA fragment
In order to determine whether the non-amplification is due to a large insertion located within exon 5, we conducted PCR amplification again using the same pair of primers that flank exon 5. This time, we used a long range PCR kit that is specifically designed for the amplification of large DNA fragments. As shown in Figure 1D, a much larger DNA fragment was obtained from the lew/lew mice.
Determination of the nature of insertion within the 9630033F20Rik gene
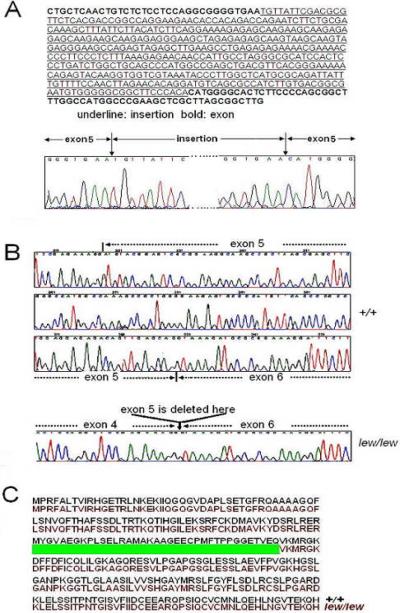
In order to determine the insertion sequences within exon 5 of 9630033F20Rik, we purified the DNA fragment of insertion and conducted DNA sequencing of this fragment. Figure 2A shows partial sequences of inserted DNA sequences. By BLAST search, we found that the insertion sequence belongs to the intracisternal type A particle element (IAP) (ACCESSION: AF532996), which resembles a retrovirus sequence. We also noticed a short sequence of exon5, GGTGAA, was repeated with the insertion of IAP.
Fig. 2.
Consequences of the IAP insertion mutation. 2A. Insertion of partial IAP sequence in the DNA sequence of exon 5 of Lew gene from lew/lew mice. 2B. a stretch of cDNA sequences of Lew gene in +/+ and lew/lew mice. The deleted section of cDNA sequence in lew/lew mice was marked by a dashed arrow. 2C. Predicted amino acid sequences from lew/lew and +/+ mice. Sequences from +/+ and lew/lew mice were in black and red, respectively. The deleted predicted sequence in lew/lew mice was represented by a green bar.
In order to investigate the consequence(s) of the IAP insertion on the cDNA from 9630033F20Rik, we amplified cDNA from 9630033F20Rik by conducting RT-PCR amplification using RNA from normal and lew mice. A pair of primers flanking exon 2 to exon 6 were used in the RT-PCR. The cDNA fragments were purified and sequenced. As shown in Figure 2B, the entire exon 5 of 9630033F20Rik has been left out in the cDNA sequences of lew mice. Figure 2C shows the predicted influences of this mutation on 9630033F20Rik translation. A protein from lew mice is 37 amino acids shorter compared to the normal protein.
The effect of the IAP insertion on the expression of 9630033F20Rik in lew mice
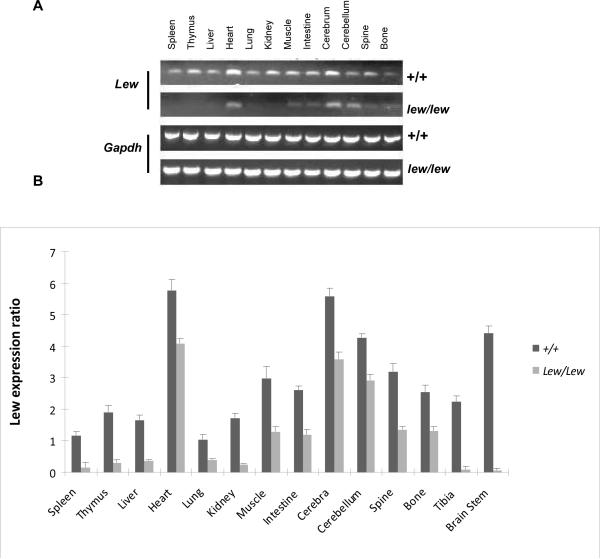
To determine the expression pattern of 9630033F20Rik between different tissues in normal and lew/lew mice, we conducted semi-quantitative RT-PCR using 12 different tissues. We used a pair of primers (forward: GGGCAGTTTCTGAGCAATGT; reverse: TCCAACAGGGAAAACCTCTG) that covered exon 3 to exon 6 with a size of 394 bps for normal mice. The amplification indicated that the 9630033F20Rik expression is ubiquitous and its expression level is relatively higher in heart, cerebrum, and muscle tissues (Figure 3A). Compared with the normal mice, all tissues from lew/lew mice had lower expression levels (Figure 3B).
Fig. 3.
Expression pattern of Lew in different tissues from normal +/+ and disease lew/lew mice by semi-quantitative RT-PCR. A pair of primers (forward:GGGCAGTTTCTGAGCAATGT, reverse: TCCAACAGGGAAAACCTCTG) was designed to cover from exon 3 to exon 6 of Lew gene with a size of 394 bps. Three individual 14 days old +/+ and three same age matched three lew/lew mice RNA samples from 12 different tissues were used for this experiment and a duplicate well was run for each sample. The figure only shows one sample from each type on the gel image. After electrophoresis (3A) and scanning, all PCR product bands were analyzed by using the software Scion Image and relative mRNA expression was estimated by normalization with Gapdh. The final ratios were calculated by dividing the relative values of different tissues by the lowest value for lung of normal mice (3B).
The effect of the insertion mutation on glycolytic gene expression in different tissues of lew mice
By domain analysis, we noticed that 9630033F20Rik contains a stretch of overlay domains involved in glycolysis, including a fructose-2, 6-bisphosphatase (GpmB) domain, and a phosphoglycerate mutase (PGAM) 1 (GpmA) domain. So it is quite likely that 9630033F20Rik is important somehow in glycolysis. To determine the glycolytic gene expression profile in the situation of the 9630033F20Rik mutation, we performed microarray assay to simultaneously measure all possible changes in glycolytic gene expression as well as other related changes (Table 1). Since muscle mass, cardiac function, and mental performance are three major affected aspects in muscular dystrophies (Emery, 2002), we surveyed the expression patterns of all other glycolytic genes in muscle, heart, and cerebrum tissues from lew/lew mice. The results indicated that most of the glycolytic genes, including Pgam2, Eno1, Ldh1, Pfkl, Tpi1, Eno3, Pgam1, Fbp2, Pgk1 and Pkm2, in diseased muscle tissues were down-regulated, while in heart tissues only several glycolytic genes, such as Bpgm, Pfkl, and Pgam1, were down-regulated. In the cerebrum, no extra gene down-regulation was observed in diseased cerebrum tissues. However, a couple of genes were slightly up-regulated in heart or cerebrum tissues. Consistent with this altered glycolytic gene expression, the global change in muscle tissue was remarkably greater than the other two tissues. Interestingly, the largest expression changes were in the two major muscle wasting related genes Fbxo32 and Trim63 (two proteosome ubiquitin transferase E3 enzymes) in heart and skeletal muscle tissues but not in the brain.
Table 1.
Effect of Lew mutation on gene expression in lew mice (fold changes)
| Gene category | Gene symbol | Muscle | Heart | Cerebrum |
|---|---|---|---|---|
| Total number of altered genes | 2304 | 1024 | 257 | |
| Glycolysis-associated genes | 9630033F20Rik | −2.3 | −1.3 | −1.9 |
| Eno1 | −2.8 | 0 | 0 | |
| Ldh1 | −2.1 | −1.2 | 0 | |
| LOC433182 | −2 | 0 | 0 | |
| Pgam2 | −2 | 0 | 0 | |
| Pfkl | −1.9 | −1.6 | 0 | |
| Tpi1 | −1.9 | 0 | 0 | |
| Eno3 | −1.7 | 0 | 1.7 | |
| Fbp2 | −1.7 | 1.6 | 0 | |
| Pgam1 | −1.7 | −1.6 | 0 | |
| Hk1 | −1.6 | 0 | 0 | |
| Pfkm | −1.6 | 0 | 0 | |
| Pgm1 | −1.6 | 0 | 0 | |
| Pkm2 | −1.6 | 0 | 1.2 | |
| Bpgm | −1.5 | −3 | 0 | |
| Pgk1 | −1.5 | 0 | 0 | |
| Pgm2 | −1.5 | 1.3 | 0 | |
| Ldh2 | 0 | 1.5 | 1.2 | |
| Muscle wasting-associated genes | Fcmd | −1.9 | 1.3 | 0 |
| Fkrp | −1.4 | 0 | 0 | |
| Sepn1 | −1.6 | −1.6 | 0 | |
| Col6a1 | −1.9 | −1.4 | 0 | |
| Col6a2 | −2.1 | −1.6 | 0 | |
| Col6a3 | −1.9 | −1.5 | 0 | |
| Large | −2.5 | −1.6 | 0 | |
| Fbxo32 | 29.9 | 3.5 | 0 | |
| Trim63 | 4.9 | 2.8 | 0 |
Discussion
Our data indicated that there is an IAP insertion in 9630033F20Rik, which seems to be a novel glycolytic gene. There are two possibilities for the absence of the 252 bp DNA fragment in the lew mice: insertion or deletion mutations. An insertion mutation will cause a larger size PCR product, which is not able to be detected using conditions for normal PCR products. In the event of a deletion mutation, the PCR product should at least include the sequence of one primer from the primer pair. Our data clearly demonstrated that there is a large insertion within exon 5 of 9630033F20Rik. Our data further suggests the possibility that this insertion may lead to the inhibition or reduction of glycolysis. Glycolysis serves two major roles: the conversion of glucose into pyruvate and the generation of carbon skeletons for biosynthesis. It is a 10-step process taking place in the cytosol. PGAMs are involved in step eight of this process; interconversion of 2- and 3-phosphoglycerate. Fructose-2,6-bisphosphatase serves a key regulatory role for phosphofructokinase-1 and fructose-1,6-bisphosphate and thus determines whether glycolysis or gluconeogenesis occurs. The IAP elements are members of retrovirus-like structures consistently observable in a variety of mouse tumor cells. They have been implicated in a number of gene mutations (Gwynn et al., 1998). Recently, a spontaneous mouse model of muscle wasting, named dyPas/dyPas, was identified as a mutant due to an IAP insertion in the Lama2 gene, encoding for the extracellular matrix laminin α2 subunit (Besse e tal., 2003). The glycolytic process is tightly controlled in response to conditions both inside and outside the cell.
Three types of enzymes including hexokinase, phosphofructokinase, and pyruvate kinase are generally regarded as the major regulators of glycolysis. Interestingly, our data indicated that the transcripts for all of these three enzymes, such as Hk1, Pfkm and Pkm2, were downregulated in the diseased muscle tissues. Moreover, four major high-abundance distal glycolytic genes, i.e. Pgam2, Eno1, Eno3, and Ldh1, were significantly down-regulated in the affected muscle. 9630033F20Rik contains a fructose-2, 6-bisphosphatase domain, which is involved in both the synthesis and degradation of fructose-2, 6-bisphosphate, a major regulatory molecule that controls glycolysis/gluconeogenesis (Rider et al., 2004). Therefore, the mechanisms for decreased expression of many glycolytic genes may, at least partly, be due to an imbalance in fructose-2, 6-bisphosphate. In addition, other studies have shown that some glycolytic enzymes have additional non-glycolytic functions in transcriptional regulation, apoptosis regulation, and cell motility stimulation (Kim and Dang, 2005). In this regard, it is quite possible that 9630033F20Rik may function through more than one mechanism to alter glycolytic gene expression in mammals.
It is not clear whether the impaired glycolysis expression is relevant to muscle wasting in lew mice because VAMP1 has been regarded as the causal gene for the lew disease. It is noticed that VAMP1 is expressed in brain and eye tissues (Nystuen et al., 2007). One would expect that the downregulation of many genes in muscle as well as in heart tissues may be due to the IAP insertion in 9630033F20Rik. However, VAMP1 mutations in brain and eye tissues may also cause the problem in muscle and heart tissues as well. Muscle phosphoglycerate mutase deficiency has previously been identified as a cause of human metabolic myopathy (Dimauro et al., 1981). A recent report indicates that a severe muscle enolase (Eno3) deficiency causes metabolic myopathy (Comi et al., 2001). Since glycolysis serves dual roles in two basic processes: energy metabolism and biosynthesis, the precise role of impaired glycolysis in the development of muscle wasting needs to be investigated further. A prevailing concept in the understanding of molecular mechanisms of muscle wasting is that Fbxo32 and Trim63 play an important role in most kinds of catabolic condition- and aging- related to muscle wasting (Cao et al., 2005). They are E3 ubiquitin-conjugating enzymes which have been found to be expressed at high levels in many animal models of muscle wasting and human patients with muscular atrophy. A recent microarray study validates this opinion using mouse and rat models of muscle wasting (Lecker et al., 2004). In this study, we observed a remarkable increase of their expression in muscle and heart tissues, suggesting that this extends to the new congenital muscular dystrophy.
In summary, we have identified an IAP insertion in a novel gene which potentially functions in the glycolysis pathway. However, further study is needed to confirm both the function of this gene and the impact of the IAP insertion.
ACKNOWLEDGEMENTS
Support for this work is from Center of Genomics and Bioinformatics (WG) and Center in Connective Tissue Research (WG), at University of Tennessee Health Science Center; Veterans Administration (WG); National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health (R01 AR51190 to WG; AR051440 to HWJ.) and the Muscular Dystrophy Association (grant #3189 to HWJ).
References
- Besse S, Allamand V, Vilquin JT, Li Z, Poirier C, Vignier N, Hori H, Guenet JL, Guicheney P. Spontaneous muscular dystrophy caused by a retrotransposal insertion in the mouse laminin alpha2 chain gene. Neuromuscul Disord. 2003;13:216–22. doi: 10.1016/s0960-8966(02)00278-x. [DOI] [PubMed] [Google Scholar]
- Cao PR, Kim HJ, Lecker SH. Ubiquitin-protein ligases in muscle wasting. Int J Biochem Cell Biol. 2005;37:2088–97. doi: 10.1016/j.biocel.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Comi GP, Fortunato F, Lucchiari S, Bordoni A, Prelle A, Jann S, Keller A, Ciscato P, Galbiati S, Chiveri L, Torrente Y, Scarlato G, Bresolin N. Beta-enolase deficiency, a new metabolic myopathy of distal glycolysis. Ann Neurol. 2001;50:202–7. doi: 10.1002/ana.1095. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Miranda AF, Khan S, Gitlin K, Friedman R. Human muscle phosphoglycerate mutase deficiency: newly discovered metabolic myopathy. Science. 1981;212:1277–9. doi: 10.1126/science.6262916. [DOI] [PubMed] [Google Scholar]
- Emery AE. The muscular dystrophies. Lancet. 2002;359:687–95. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- Glass DJ. Signaling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol. 2003;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- Gwynn B, Lueders K, Sands MS, Birkenmeier EH. Intracisternal A-particle element transposition into the murine beta-glucuronidase gene correlates with loss of enzyme activity: a new model for beta-glucuronidase deficiency in the C3H mouse. Mol Cell Biol. 1998;18:6474–81. doi: 10.1128/mcb.18.11.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008 Jan 10;27(3):404–8. doi: 10.1038/sj.onc.1210631. Epub 2007 Jul 9. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Li X, Beamer WG, Yan J, Tong Y, Goldowitz D, Roe B, Gu W. A deletion causing spontaneous fracture identified from a candidate region of mouse Chromosome 14. Mamm Genome. 2005a;16:20–31. doi: 10.1007/s00335-004-2414-0. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Yan J, Zhao Y, Donahue LR, Beamer WG, Li X, Roe BA, Ledoux MS, Gu W. Carbonic anhydrase-related protein VIII deficiency is associated with a distinctive lifelong gait disorder in waddles mice. Genetics. 2005b;171:1239–46. doi: 10.1534/genetics.105.044487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30:142–50. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. Faseb J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Muntoni F, Voit T. The congenital muscular dystrophies in 2004: a century of exciting progress. Neuromuscul Disord. 2004;14:635–49. doi: 10.1016/j.nmd.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Nystuen AM, Schwendinger JK, Sachs AJ, Yang AW, Haider NB. A null mutation in VAMP1/synaptobrevin is associated with neurological defects and prewean mortality in the lethal-wasting mouse mutant. Neurogenetics. 2007 Jan;8(1):1–10. doi: 10.1007/s10048-006-0068-7. Epub 2006 Nov 11. [DOI] [PubMed] [Google Scholar]
- Rider MH, Bertrand L, Vertommen D, Michels PA, Rousseau GG, Hue L. 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: head-to-head with a bifunctional enzyme that controls glycolysis. Biochem J. 2004;381:561–79. doi: 10.1042/BJ20040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito ES, Keng VW, Takeda J, Horie K. Translation from nonautonomous type IAP retrotransposon is a critical determinant of transposition activity: implication for retrotransposon-mediated genome evolution. Genome Res. 2008 Jun;18(6):859–68. doi: 10.1101/gr.069310.107. Epub 2008 May 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XY, Chen ZY, Hayashi Y, Kanou Y, Takagishi Y, Oda S, Murata Y. Insertion of an intracisternal A particle retrotransposon element in plasma membrane calcium ATPase 2 gene attenuates its expression and produces an ataxic phenotype in joggle mutant mice. Gene. 2008 Mar 31;411(1–2):94–102. doi: 10.1016/j.gene.2008.01.013. Epub 2008 Jan 26. [DOI] [PubMed] [Google Scholar]
- Takabatake T, Ishihara H, Ohmachi Y, Tanaka I, Nakamura MM, Fujikawa K, Hirouchi T, Kakinuma S, Shimada Y, Oghiso Y, Tanaka K. Microarray-based global mapping of integration sites for the retrotransposon, intracisternal A-particle, in the mouse genome. Nucleic Acids Res. 2008 Jun;36(10):e59. doi: 10.1093/nar/gkn235. Epub 2008 May 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 2008 Feb 29;4(2):e1000007. doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Bouhassira EE, Tsai HM. An IAP retrotransposon in the mouse ADAMTS13 gene creates ADAMTS13 variant proteins that are less effective in cleaving von Willebrand factor multimers. Blood. 2007 Aug 1;110(3):886–93. doi: 10.1182/blood-2007-01-070953. Epub 2007 Apr 10. [DOI] [PMC free article] [PubMed] [Google Scholar]